Abstract
Estrogen/ERα signaling is critical for breast cancer progression and therapeutic treatments. Thus, identifying new regulators of this pathway will help to develop new therapeutics to overcome chemotherapy resistance of the breast cancer cells. Here, we report Ajuba directly interacts with ERα to potentiate ERα target gene expression, and biologically Ajuba promotes breast cancer cell growth and contributes to tamoxifen resistance of these cells. Ajuba constitutively binds the DBD and AF2 regions of ERα, and these interactions can be markedly enhanced by estrogen treatment. Mechanistically, Ajuba recruits DBC1 and CBP/p300 and forms a ternary complex to co-activate ERα transcriptional activity and concomitantly enhances ERα acetylation. Moreover, components of this complex can be found at endogenous promoters containing functional ERα responsive elements. Taken together, these data demonstrate that Ajuba functions as a novel co-activator of ERα and that Ajuba/DBC1/CBP/p300 ternary complex may be a new target for developing therapeutics to treat breast cancer.
INTRODUCTION
Breast cancer is the most prevalent malignancy and the second leading cause of cancer-related death in women worldwide. Extensive investigations have established the critical role of estrogen signaling in breast cancer development and therapeutic treatments (1). Accordingly, a large number of chemical drugs targeting estrogen signaling have been developed, such as tamoxifen, anastrazole and letrozole. In most cases, tamoxifen antagonizes estrogen mediated transcriptional activation and finally inhibits cell growth. Although the clinical application of tamoxifen has really brought encouraging outcomes, most patients unexceptionally relapse sooner or later due to the existence of tamoxifen-resistant cancer cells.
Estrogen exerts its biological effects by functioning as the native ligand of estrogen receptors (ERs) including ERα and ERβ. ERα possesses typical nuclear receptor structure: AF1 domain, DNA-binding domain (DBD) and Ligand-binding domain (LBD) from N-terminus to C-terminus. In addition to binding estrogen, LBD also contains AF2 domain and mediates ERα dimerization. Through AF1 or AF2 domain, ERα recruits various cofactors by binding to NR-boxes (L-X-X-L-L) or CORNR-boxes (L/I-X-X-I/V-L) resided in these cofactors to either activate or repress its target gene expression. Generally, the recruitment of cofactors by AF2 is estrogen-dependent, while the recruitment of cofactors by AF1 is estrogen-independent. In addition, many cofactors also bind to ERα independent the NR or CORNR motif (2). The DBD domain mediates ERα interaction with estrogen response element (ERE). In addition, various modifications can occur in these domains which have great influence on the ERα activity (3,4). For instance, EGF-activated MAPK can phosphorylate ERα at ser118, resulted in ERα binding to DNA in the absence of Estrogen (5–7). CBP/p300 also acetylates ERα at K266/268 and K302/303 to enhance its DNA binding activity and transcriptional activity (8,9).
DBC1 (Deleted in Breast Cancer-1), also known as CCAR2 or p30DBC, was initially discovered as a gene deleted in human chromosome 8p21 in breast cancer (10). However, subsequent studies found that DBC1 is over-expressed in many tumor types including breast, gastric and colorectal cancer, and the expression level of DBC1 is well correlated with poor prognosis (11–16). Recently, DBC1 was demonstrated as an inhibitor of SIRT1 and PARP1, indicating the critical role of DBC1 in cell survival and aging (17,18). Additionally, several studies have shown that DBC1 can participate in transcriptional regulation (19). DBC1 interacts with ERβ, LXRα and Rev-erbα and functions as a transcriptional co-repressor (20–22), while DBC1 acts as a co-activator of RARα and AR to promote their transcriptional activities (23,24). In breast cancer cells, DBC1 enhances the transcriptional activities of ERα and PEA3, suggesting a positive role of DBC1 in breast cancer progression (25,26).
Ajuba is characterized as three tandem LIM domains at the C-terminus and belongs to the Ajuba/Zyxin subfamily of LIM proteins, which includes Ajuba, Zyxin, LIMD1, LPP, WTIP and TRIP6. Ajuba contains both nuclear export sequence (NES) and nuclear localization sequence (NLS) and thus can shuttle between cytoplasm and nucleus. In cytoplasm, Ajuba directly interacts with Rac, α-catenin and F-actin to maintain cell-cell adhesion and regulate cytoskeleton assembly (27–29). Ajuba also interacts with many signaling transducers to modulate the signal propagation from cytoplasm to nucleus such as JAK/STAT (30), Hippo (31–35), WNT/β-catenin (36). In nucleus, Ajuba can regulate gene expression by associating with transcriptional factors. Ajuba recruits 14-3-3 and PRMT5 to Snail and mediates the inhibition of E-cadherin expression (37,38).
We previously identified that Ajuba contains three functional nuclear-binding motifs (NR-boxes or CORNR-boxes) mediating the interaction of Ajuba with RARα to negatively regulate its transcriptional activity (39). Conversely, we further found that Ajuba directly interacts with PPARγ and LXRα, but independent of these conserved NR-boxes, and recruits CBP/p300 to enhance their transcriptional activities respectively (40,41). The versatile binding activities and differential functions between Ajuba and PPARγ, LXRα and RARα suggest that Ajuba may broadly interact with nuclear hormone receptors via multiple mechanisms. PPARγ, LXRα and RARα all belong to the type II NR subfamily, it is interesting and important to examine the binding behavior between Ajuba and the type I subfamily of nuclear receptors, such as ERα. In this study, we report that Ajuba directly interacts with ERα independent of its NR-boxes, or the motif for PPARγ binding, and their interaction can be enhanced by estrogen stimulation; mechanistically, Ajuba recruits CBP/p300 and DBC1 to increase the acetylation of ERα and potentiates ERα target gene expression; and high level of Ajuba promotes breast cancer cell growth and contributes to the tamoxifen resistance.
MATERIALS AND METHODS
Plasmids and siRNA
The plasmids of Myc-Ajuba, Myc-Wtip, Myc-Limd1, Myc-Lpp, Myc-Zyxin, Myc-P300 and HA-CBP have been previously described (41). The Flag-tagged or HA-tagged ERα and DBC1 were cloned into pcDNA-3.1-vector. The serial truncations of ERα and GST-tagged ERα were subcloned from plasmid of pcDNA3.1-Flag-ERα. The 3xERE-luciferase reporter plasmid was kindly gifted by Dr Jianhua Wang (Shanghai Tumor Hospital, Fudan University). The TFF1-promoter was cloned into a pGL3.0-basic vector. The sequence of siRNA-DBC1 has been described previously and was commercially synthesized (18).
Cell culture, transfection, retroviral infection and luciferase reporter assay
The breast cancer cell lines MCF7 and T47D, were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 2 mM l-glutamine, and penicillin (50 U/ml)/streptomycin (50 μg/ml) at 37°C under 5% CO2 in a humidified chamber. The detailed methods of transfection and infection were previously described (41). The luciferase reporter assay was performed in 293T cells and the cells were cultured in phenol-red free media with 5% charcoal-stripped FBS for at least 1 day before transfection. The mixture of transfection was prepared in phenol-red free medium without FBS. The transfected cells were cultured in phenol-red free media with 5% charcoal-stripped FBS for 24 h and then were treated with 20 nM E2 (β-estradiol, E8875, Sigma) or ethanol for 12 h, and the detailed luciferase reporter assay has been previously described (39).
Co-immunoprecipitation(co-IP) assay, GST-pulldown, western blottings and antibodies
The co-IP assay and western blotting were described previously (41). Transfected 293T cells or T47D cells were cultured for at least 2 days in phenol-red free media with 5% charcoal-stripped FBS before the 12 h-treatment with E2 in the co-IP assays. GST-tagged ERα or DBC1and His tagged Ajuba were expressed in BL21 respectively, and purified by Glutathione Sepharose beads (17-0756-01, GE Healthcare) or Ni-beads (17-5318-06, GE Healthcare). For the in vitro binding assays, the purified proteins of GST-ERα or DBC1 were mixed with His-Ajuba and E2 (100nM) was added into the mixture for 12 hours.
The antibodies used for western blottings, co-IP and ChIP assays were: anti-Myc (13-2500, Invitrogen), anti-Flag (F3165, F7425, Sigma), anti-HA (MMS-101P, Covance), anti-acetyl-H3 (06-559, sc-369, Millipore), anti-acetyl-lysine (9441s, Cell signaling and 05-515, Millipore), anti-DBC1 (A300-432A or 434A, Bethyl Laboratories), anti-β-actin (60008-1-lg, Proteintech). The antibodies of ERα (sc8002 and sc-543), CBP (A-22), anti-p300 (N-15), normal IgG (sc-2025 and sc-2027) were purchased from Santa Cruz. The rabbit Ajuba antibody was described previously (47).
RNA-extraction, reverse transcription and qPCR
T47D or MCF7 cells were treated with 20nM E2 or ethanol for 12 h before RNA extraction and qRT-PCR. To test the effect of tamoxifen on ERα target genes expression, MCF7 and T47D cells cultured in normal media were directly treated with tamoxifen for 24 h before RNA extraction and qRT-PCR. The procedure of RNA extraction and qRT-PCR was previously described (41). The primers (5′-3′) used were listed below:
β-actin: F-GGACTTCGAGCAAGA GATGG; R-AGCACTGTGTTGGCGTACAG.
TFF1: F- GTGCAAATAAGGGCTGCTGT; R-GCAGATCCCTGCAGAAGTGT.
Greb1: F-CAAAGAATAACCTGTTGGCCCTGC; R-CTGCGTTTAGTGAGGGGT-GAT. SGK3: F-ACAATATCCTTCACAAACCCCTAA; R-CCTCCAATACACTGG -CATTCACT.
Chromatin immunoprecipitation assay (ChIP)
T47D cells were treated with 100 nM E2 or ethanol for 1 h before being cross-linked by using formaldehyde. The detailed method for ChIP assay has been previously described (41). The primers (5′-3′) used in qPCR were listed below:
TFF1: F-CTAGACGGAATGGGCTTCATG and R-GAGTCTCCTCCAACCTGACC-TTA; Greb1: F-TTGAGCAAAAGCCACAAAGTAG and R-GCTGCGGCAATCA -GAAGTAT; SGK3: F-CTTTGTTCCTGATCTGTTCGACAC and R-ATGTTTCCA -AGAGCAAATAGCAGT.
Cell growth and viability assays
The T47D cells and derivatives were seeded into 96-well (5000–10000 cells per well) and cultured in phenol-red free media containing 5% charcoal-stripped FBS with 20 nM E2 or ethanol. The cell growth was detected at indicated day by applying Cell Counting Kit-8 (Dojindo Laboratories) and referred to as the absorption at OD450. In the tamoxifen toxic assay, T47D or MCF7 derived cells were seeded into 96-well and cultured in normal media for 1 day, and then continued to be cultured in tamoxifen or ethanol containing media for 6 days. The cell viability was detected by using Cell Counting Kit-8, and the values applied in the final graph were normalized to normal control cells.
Statistical analysis
Data shown as mean ± SD were analyzed by independent Student's t-test.
RESULTS
Ajuba interacts with ERα independent of the conserved NR boxes
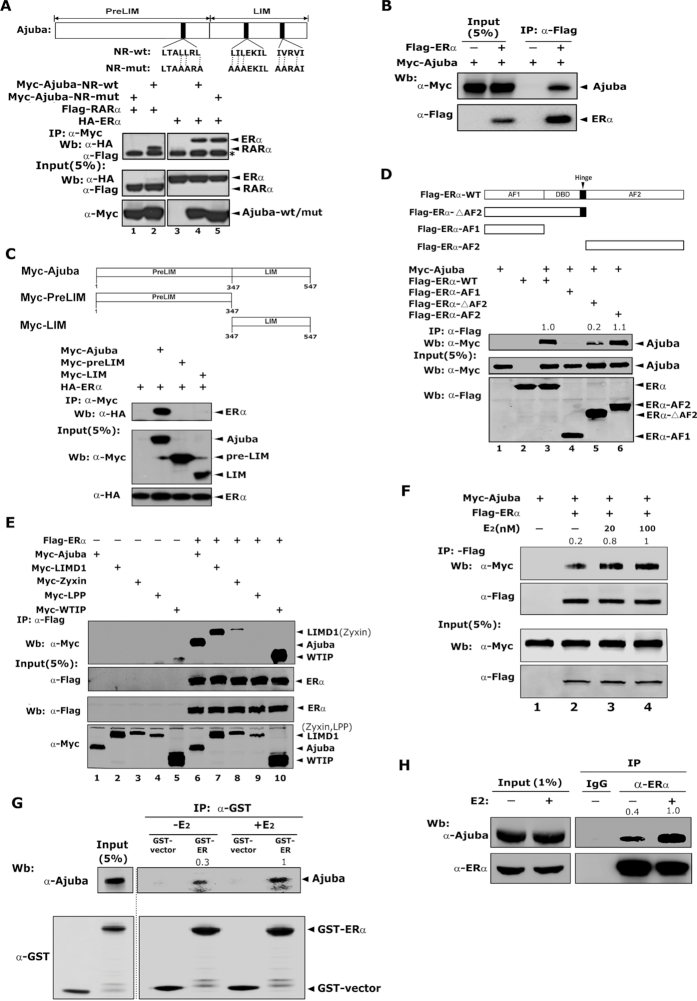
To examine the interaction between Ajuba and ERα, we transiently transfected Myc-Ajuba, Myc-Ajuba-NR-mut and HA-ERα or HA-RARα into 293T cells, co-IP assays were performed with antibody specifically against HA epitope and the co-eluted proteins were detected by western blotting assays using antibody against the Myc epitope. Consistent to prior report, RARα readily co-immunoprecipitated wild type Ajuba, but failed to co-immunoprecipitate Ajuba NR mutant (Figure 1A, lanes 1 and 2). ERα co-immunoprecipitated both wild type Ajuba and Ajuba-NR-mutant with similar affinity, suggesting that Ajuba interacts with ERα via a different mechanism other the conserved NR boxes (Figure 1A, lanes 4 and 5). In addition, the association between Ajuba and ERα was further confirmed by using Flag-ERα to pulldown Myc-Ajuba (Figure 1B). To determine the regions of Ajuba mediating its interaction with ERα, we constructed two truncation mutants encoding the LIM or preLIM regions of Ajuba respectively and co-IP assays were performed accordingly. Neither preLIM nor LIM mutants immunoprecipitated ERα (Figure 1C).
Figure 1.
Ajuba interacts with ERα independent of the conserved NR boxes. (A) The three conserved NR-boxes in Ajuba were shown on the upper panel. Plasmids were transiently transfected into 293T cells and the co-IP assay was carried out with Myc antibody. * IgG-heavy chain. (B) Flag-ERα and Myc-Ajuba plasmids were transfected into 293T cells and co-IP assay was performed by using Flag-M2-beads. (C) The preLIM and LIM regions of Ajuba were illustrated on the upper panel. The full length or truncations of Ajuba were transiently co-transfected into 293T cells together with Flag-ERα plasmid. The co-IP assay was performed by using Flag antibody. (D) The functional domains of ERα were shown on the upper panel. The interaction between full-length or truncations of ERα with Ajuba in 293T cells was detected by co-IP assay and western blotting. The relative amount of co-eluted Myc-Ajuba was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled. (E) The plasmids encoding Ajuba, LIMD1, Zyxin, Wtip and Lpp were respectively co-transfected with Flag-ERα plasmid into 293T cells and co-IP assay was performed. (F) The plasmids encoding Flag-ERα and Myc-Ajuba were transfected into 293T cells, and the resulting cells were cultured in phenol-red free media containing 5% charcoal stripped FBS for 2 days and then treated with E2 (20 or 100 nM) or ethanol for 12 h. The co-IP assay was performed by using Flag-M2 beads. The relative amount of immunoprecipitated Myc-Ajuba was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled. (G) GST-ERα and His-Ajuba proteins were expressed in E. coli BL21, and GST-pulldown assay was performed in the presence of E2 (100 nM) or ethanol. The relative amount of pulled-down His-Ajuba was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled. (H) T47D cells treated with 100 nM E2 or ethanol for 12 h were harvested and co-IP assay was performed by using ERα antibody or IgG control. The relative amount of immunoprecipitated Ajuba was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled.
To determine the regions of ERα mediating the interaction with Ajuba, plasmids encoding serial ERα truncations of AF1, AF2 and the deletion of AF2 region (ΔAF2) were constructed respectively and were co-expressed along with Myc-Ajuba in 293T cells. The co-IP assays demonstrated that AF2 domain alone and the full length of ERα showed similar binding affinity to Ajuba (Figure 1D, lanes 3 and 6), but AF1 region failed to bind Ajuba (Figure 1D, lane 4). ΔAF2 displayed a weaker interaction with Ajuba (Figure 1D, lane 5). These observations indicate that AF2 is the major region mediating the interaction with Ajuba and the DBD-hinge region contains a weak binding activity for Ajuba.
To examine the binding activity of ERα to other members of Ajuba/Zyxin family, we co-expressed ERα with Ajuba, Limd1, Wtip, Zyxin or Lpp in 293T cells, and the co-IP assays were performed. ERα showed similar binding activity with Ajuba, Limd1 and Wtip (Figure 1E, lanes 6, 7 and 10), and weakly interacted with Zyxin (Figure 1E, lane 8). No interaction was observed between ERα and Lpp (Figure 1E, lane 9). These data indicate that ERα selectively interacts with members of the Ajuba/Zyxin family.
Estrogen enhances the interaction between Ajuba and ERα
To determine if the interaction between ERα and Ajuba is affected by estrogen stimulation, 293T cells transfected with plasmids of Flag-ERα and Myc-Ajuba were cultured in phenol-red free media containing 5% charcoal stripped FBS for 2 days and then treated with E2 at doses of 20nM or 100 nM or ethanol for 12 h and the resulting cells were prepared for co-IP assays. Estrogen treatment markedly increased the binding between ERα and Ajuba in a dose dependent manner (Figure 1F, lanes 3 and 4). To further confirm this observation, we expressed GST-ERα and His-Ajuba in E. coli BL21, and in vitro GST-pulldown assays were performed in the presence of estrogen or ethanol. Consistently, Ajuba interacted with ERα without E2, but the E2 treatment markedly enhanced their interaction (Figure 1G). To further examine the interaction between ERα and Ajuba at the endogenous level, we treated T47D cells as that in 293T cells aforementioned, and the resulting cells were prepared for co-IP assays with antibody specifically against ERα. Similarly, ERα co-eluted Ajuba without E2, and E2 significantly increased their binding at endogenous level (Figure 1H). Taken together, these observations demonstrate that ERα directly and constitutively binds Ajuba and estrogen can markedly increase their interaction.
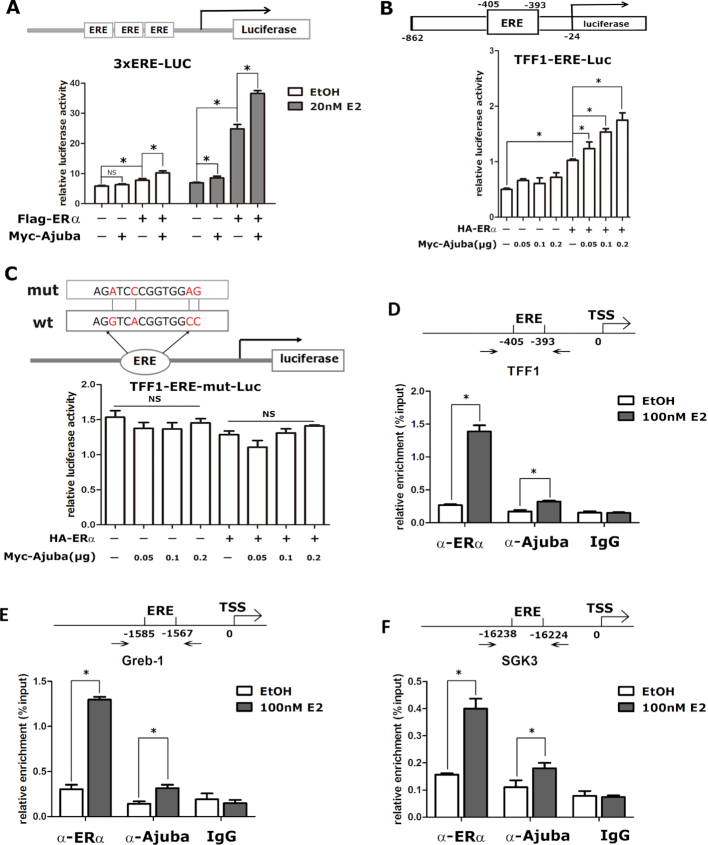
Ajuba enhances the transcriptional activity of ERα
To examine the effect of Ajuba on ERα transcriptional activity, a luciferase reporter containing three tandem EREs (Figure 2A, upper), together with Ajuba and/or ERα plasmids, was transiently co-expressed in 293T cells. In the absence of E2, Ajuba alone showed no induction on the reporter, and ERα alone or in combination with Ajuba slightly increased the luciferase activity. In the presence of E2, Ajuba slightly increased the reporter activity, while ERα strikingly activated the reporter activity which was further significantly enhanced by its combination with Ajuba (Figure 2A, lower).
Figure 2.
Ajuba enhances ERα transcriptional activity. (A) The pGL3.0-ERE-Luc reporter contains three tandem ERE elements and were transiently transfected into 293T cells. The resulting cells were treated with 20nM E2 or ethanol for 12hrs and were prepared for the luciferase reporter assay. The luciferase activity was normalized to the value of β-gal activity (three repeats, * means P < 0.05). (B and C) Wild-typed (B) or mutated (C) TFF1 promoter driven luciferase reporter plasmids were co-transfected with ERα and/or Ajuba plasmids into 293T cells. The transfected cells were treated with 20 nM E2 before being harvested for luciferase reporter assay and the value was normalized to β−gal (Three repeats, * means P < 0.05, NS means no significance). (D–F) The location of ERE in the promoter or enhancer of ERα target genes was shown in the upper panel. The T47D cells were treated with 100 nM E2 or ethanol for 1 h and prepared for the ChIP assay. The relative enrichment of Ajuba and ERα on the promoter or enhancer of TFF1 (D), Greb1 (E) and SGK3 (F) was detected by qPCR and normalized to input (three repeats, * means P < 0.05).
TFF1 is a classic target gene of ERα and is widely used as a reporter to evaluate the activity of ERα. Thus, we further examined the effect of Ajuba and ERα on TFF1 promoter luciferase reporter (Figure 2B, upper). Similar to that of the ERE reporter, in the presence of E2, ERα alone evidently increased the TFF1 promoter activity and the addition of Ajuba further elevated the transcriptional activity of ERα in a dose dependent manner (Figure 2B, lower). Conversely, the effect of ERα and Ajuba was abolished when the ERE in the TFF1 promoter was inactively mutated (Figure 2C). To determine if Ajuba can be recruited to the endogenous promoter of ERα target genes, we carried out the ChIP assays in T47D cells and found E2 treatment stimulated both ERα and Ajuba to bind the ERE-region within the promoter or enhancer region of selected ERα target genes TFF1, Greb1 and SGK3 (Figure 2D–F). Collectively, these data demonstrate that Ajuba functions as a co-activator of ERα to enhances its target gene transcription.
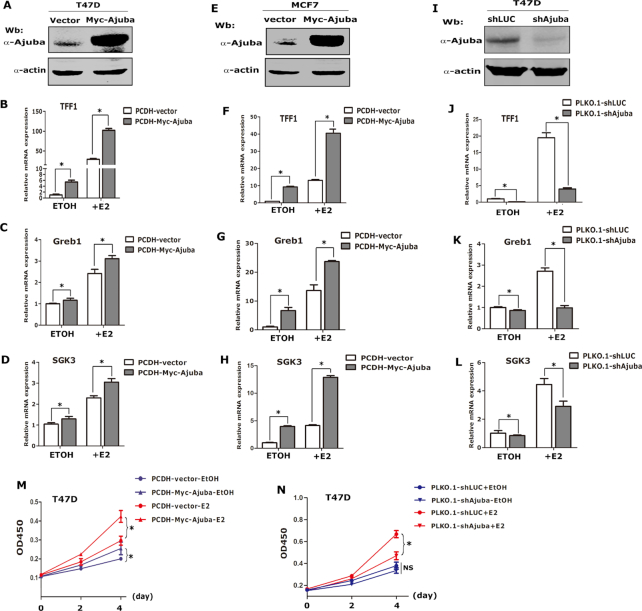
Ectopic expression of Ajuba stimulates endogenous ERα target gene expression and promotes breast cancer cell growth
To explore the biological function of the interaction between Ajuba and ERα, we stably expressed Ajuba in T47D and MCF7 cells respectively and the expression of TFF1, Greb1 and SGK3 was detected by qRT-PCR assays. In the absence of E2, overexpression of Ajuba moderately induced the expression of these target genes, while in the presence of E2, Ajuba strongly induced expression of these target genes (Figure 3A–H). Moreover, depletion of Ajuba using specific targeting shRNAs significantly dampened the expression of these target genes at basal level or in the presence of E2 in T47D cells (Figure 3I–L).
Figure 3.
Ajuba enhances ERα target gene expression and promotes breast cancer cell growth. (A) Ajuba was stably expressed in T47D cells and western blot assay showed the expression level of Ajuba. (B–D) The T47D-Ajuba and T47D-vector cells were cultured in phenol-red free media containing 5% charcoal stripped FBS for 2 days and then treated with 20 nM E2 or ethanol for 12 h and the relative mRNA expression of TFF1 (B), Greb1 (C) and SGK3 (D) was detected by qRT-PCR and normalized by the expression of β-actin (three repeats, * means P < 0.05). (E) Ajuba was stably expressed in MCF7 cells. (F–H) The expression of TFF1 (F), Greb1 (G) and SGK3 (H) in MCF7-Ajuba and vector-control cells was detected by qRT-PCR (three repeats, * means P < 0.05). (I) Ajuba was stably knocked-down in T47D cells (shLuc stands for sh-luciferase). (J–L) The expression of TFF1 (J), Greb1 (K) and SGK3 (L) in T47D-shAjuba and sh-Luc control cells was detected by qRT-PCR (three repeats, * means P < 0.05). (M–N) CCK8 assays were carried out to examine the cell growth in Ajuba overexpressed or knocked-down T47D cells respectively (three repeats, * means P < 0.05).
To examine whether Ajuba can affect the cell growth induced by estrogen signaling, the CCK8 assays were performed to evaluate the cell growth rate in T47D cells stably expressing Ajuba. Without E2, Ajuba slightly increased cell growth, while drastically increased the cell growth in the presence of E2 (Figure 3M). Conversely, the estrogen stimulated cell growth was evidently reduced when Ajuba was depleted in T47D cells (Figure 3N). Collectively, these data indicate that Ajuba is an important inducer of ERα target genes and promotes cell growth in breast cancer cells.
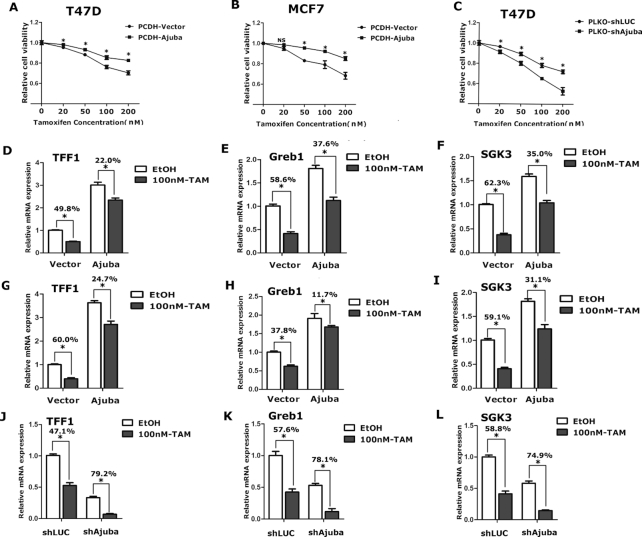
Ajuba antagonizes the Tamoxifen-toxic effect on breast cancer cells.
Tamoxifen (TAM) is the most often used antagonist of ERα in chemotherapy for breast cancer treatment. Through competitively binding to ERα, TAM significantly inhibits estrogen induced target genes expression and inhibits cell growth. We speculated Ajuba might override TAM effect and contributes to TAM-resistance of breast cancer cells. We performed CCK8 assays in breast cancer cells stably expressing Ajuba or shRNA specifically targeting Ajuba with/without TAM. Indeed, TAM efficiently inhibited cell growth in a dose dependent manner (Figure 4A–C). Overexpression of Ajuba evidently dampened the inhibition of TAM on T47D cells (Figure 4A). Similar results were also observed in MCF7 cells (Figure 4B). Conversely, depletion of Ajuba sensitized T47D cells to TAM treatment (Figure 4C).
Figure 4.
Ajuba antagonizes the Tamoxifen-toxic effect in breast cancer cells. (A–C) Ajuba-overexpressed or knocked-down T47D and MCF7 cells were treated with TAM or ethanol for 6 days and cell viability was examined by CCK8 assay. The results were normalized to the normal control cells (ethanol-treated groups) (three repeats, * means P < 0.05, NS means no significance). (D–F) T47D-Ajuba and vector control cells were treated with TAM (100 nM) or ethanol for 4 days, the relative mRNA expression of ERα target genes was detected by qRT-PCR. The results were normalized by ethanol-control group and the inhibitory percent of TAM was labeled (three repeats, * means P < 0.05). (G–I) The effect of TAM (100 nM) on the expression of ERα target genes in MCF7-Ajuba and vector control cells were detected by qRT-PCR. The results were normalized by ethanol-control group and the inhibitory percent of TAM was labled (three repeats, * means P < 0.05). (J–L) Knocking-down Ajuba in T47D cells magnified inhibitory effect of TAM (100 nM) on the expression of ERα target genes. The results were normalized by ethanol-control group and the inhibitory percent of TAM was labeled (three repeats, * means P < 0.05).
Moreover, we examined the effect of Ajuba on TAM-mediated ERα target gene expression in T47D and MCF7 cells respectively. Consistent to prior report, TAM evidently decreased the expression of TFF1, Greb1 and SGK3, which was still significant under the over-expression of Ajuba (Figure 4D–I). However, the inhibitory percent of TAM was significantly weakened by Ajuba over-expression in T47D cells (Figure 4D–F), and MCF7 cells (Figure 4G–I). Conversely, knocking-down of Ajuba expression intensified the inhibitive effect of TAM on the expression of these target genes in T47D cells (Figure 4J–L). Taken together, these data demonstrate that Ajuba can antagonize the toxic effect of TAM and may contribute to the TAM resistance in breast cancer cells.
Ajuba directly interacts with DBC1 and enhances DBC1 and ERα interaction
To explore the molecular mechanism by which Ajuba activates ERα transcriptional activity, we performed co-IP assay followed by Mass Spectrometry (MS) analysis as described (37), and found that DBC1 was a potential Ajuba interacting protein. DBC1 has been shown as a co-activator of ERα (25), we speculated that Ajuba and DBC1 might cooperatively activate ERα transactivity. To verify the association between Ajuba and DBC1, we co-expressed DBC1 and Ajuba in 293T cells and the co-IP assays showed that Ajuba and DBC1 could readily pulldown each other (Figure 5A and B), and their interaction was further confirmed by endogenous co-IP assay in T47D cells (Figure 5C).
Figure 5.
Ajuba recruits DBC1 to enhance ERα transcriptional activity. (A, B) plasmids encoding DBC1 and Ajuba were transfected into 293T cells and co-IP assay was performed by using Myc antibody (A) or Flag-M2-beads (B). (C) The endogenous interaction between DBC1 and Ajuba was detected in T47D cells by co-IP assay. (D) GST-DBC1 and His-Ajuba was respectively expressed in E. coli BL21, and in vitro binding assay was performed. (E) The plasmids of full-length and truncations of Ajuba protein were co-transfected into 293T cells with DBC1 plasmids and co-IP assay was carried out. (F) Increasing amount of plasmids encoding Ajuba were co-expressed along with DBC1 and ERα in 293T cells and co-IP assay showed that Ajuba enhanced the interaction between DBC1 and ERα (the relative amount of immunoprecipitated HA-DBC1 was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled). (G) TFF1 promoter driven luciferase reporter assay was performed in 293T cells and the transfected cells were treated with 20 nM E2 before detecting luciferase activity and the value was normalized to β-gal (three repeats, * means P < 0.05). (H) T47D-Ajuba and vector control cells were treated with 100 nM E2 for 1 h. ChIP assay was performed by using DBC1 antibody and the result was normalized by input (three repeats, * means P < 0.05). (I) MCF7-Ajuba and vector control cells were transfected with siRNA of DBC1 for 48 h, the expression of TFF1 was detected by qPCR and the expression of DBC1 and ERα were analyzed by western blotting (three repeats, * means P < 0.05).
To test if their interaction is direct, we expressed DBC1 and Ajuba protein in E. coli respectively, and in vitro binding assays showed GST fused DBC1 could readily pulldown His-tagged Ajuba protein (Figure 5D). To determine which domain of Ajuba mediates the interaction with DBC1, we co-transfected the full-length and its two truncation mutants of Ajuba with DBC1 plasmids into 293T cells, and the co-IP assays showed that both the preLIM region and full-length of Ajuba could interact with DBC1, but not the LIM region (Figure 5E). Collectively, these data demonstrate that Ajuba can directly interact with DBC1 protein through its preLIM region.
To determine if Ajuba, DBC1 and ERα can form a ternary complex, we co-expressed HA-DBC1, Myc-Ajuba and Flag-ERα proteins in 293T cells and performed co-IP assays with Anti-Flag M2 beads. Indeed, ERα co-immunoprecipitated with DBC1 as previously reported (Figure 5F, lane2). Notably, Ajuba significantly enhanced the interaction pf DBC1 and ERα in a dose dependent manner (Figure 5F, lanes 3–5), indicating that Ajuba, DBC1 and ERα can co-exist in one ternary complex and Ajuba can enhance the association between DBC1 and ERα.
Ajuba and DBC1 cooperatively enhance ERα transactivity
To examine the effect of Ajuba and DBC1 on ERα transactivity, we first performed luciferase reporter assay using the TFF1-Luc in 293T cells. Both co-expression of ERα with Ajuba or DBC1 alone moderately increased ERα transcriptional activity, while the combination of DBC1 and Ajuba maximally stimulated ERα transactivity (Figure 5G). To examine if Ajuba affects DBC1 binding to the endogenous ERα target promoters, we performed ChIP assays using an antibody specifically against DBC1 in T47D cells stably expressing Ajuba or vector control. The co-eluted DNA fragments were examined by qPCR using primer set flanking the ERE-containing region of the TFF1 promoter. Indeed, DBC1 could bind ERE-containing region of the TFF1 promoter, and the binding activity was further enhanced by Ajuba over-expression (Figure 5H).
To further examine the effect of DBC1 and Ajuba on ERα target gene expression, we transiently knocked-down DBC1 expression by siRNA in MCF7 cells stably expressing Ajuba or Vector control and total RNAs were extracted and were subjected for qRT-PCR analyses. Consistent to prior report (25), knocking-down DBC1 resulted in slightly decreased expression of TFF1 in MCF7-vector cells. Remarkably, the expression of TFF1 in MCF7-Ajuba cells was significantly dampened by knocking-down DBC1 (Figure 5I). Taken together, these data demonstrate that Ajuba can enhance the binding activity of DBC1 to its target promoter, and in turn DBC1 may be required for Ajuba to maximally induce ERα target genes.
Ajuba promotes ERα transcriptional activity by recruiting CBP/p300 and increasing the acetylation of ERα
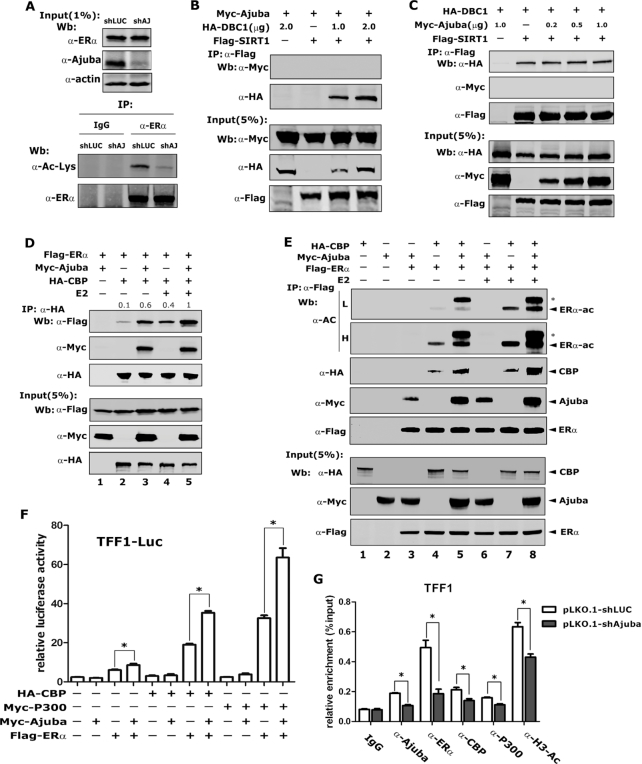
DBC1 was previously shown to promote the acetylation of ERα by inhibiting the activity of deacetylase SIRT1. Thus, we speculated that Ajuba might affect the acetylation of ERα via associating with DBC1. Total ERα was immunoprecipitated using an ERα specific antibody and a pan-Ac-Lys antibody was used to determine the acetylation level of ERα. Remarkably, the acetylation of ERα was readily detected in T47D-vector cells, but was evidently decreased in T47D-shAjuba cells, indicating that Ajuba is required to maintain the acetylation of ERα (Figure 6A).
Figure 6.
Ajuba promotes ERα transcriptional activity by recruiting CBP/p300 and increasing the acetylation of ERα. (A) The acetylation of ERα in T47D-shLuc and shAjuba cells was detected by western blotting after the enrichment of the endogenous ERα by using ERα antibody. (B) co-IP assay in transfected 293T cells showed that Ajuba did not bind SIRT1. (C) co-IP assay was performed in transfected 293T cells and the interaction between DBC1and SIRT1 was analyzed. (D) Transfected 293T cells were treated with 100 nM E2 or ethanol for 12 h and co-IP assay showed that E2 enhanced the interaction between Ajuba, CBP and ERα (the relative amount of immunoprecipitated Flag-ERα was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled). (E) Ajuba and E2 could increase ERα acetylation. Plasmids were transfected into 293T cells and the proteins were enriched by Flag antibody, and the acetylation of ERα was detected by using acetyl-Lysine antibody (* means the band of acetylated Ajuba, L means low exposure and H means high exposure). (F) Transfected 293T cells were treated with 20 nM E2 and TFF1-promoter driven luciferase reporter assay was performed and the value was normalized to β-gal (three repeats, * means P < 0.05). (G) shAjuba-T47D and shLuc- T47D cells were treated with 100 nM E2 for 1 h and ChIP assay was carried out by using indicated antibodies. The result was normalized by input and 5–10% as input (three repeats, * means P < 0.05).
To examine if Ajuba interacts with SIRT1 or forms a complex with SIRT1 and DBC1, we co-expressed Myc-Ajuba, Flag-SIRT1 and HA-DBC1 into 293T cells, and co-IP assays were performed. Consistent to previous report, strong interaction between SIRT1 and DBC1 was observed, but there is no interaction between SIRT1 and Ajuba regardless of co-expression of DBC1 (Figure 6B). In addition, the interaction between DBC1 and SIRT1 was not affected by the expression of Ajuba (Figure 6C). These results suggest Ajuba does not participate in the DBC1/SIRT1 complex.
CBP/p300 can acetylate ERα to increase its DNA-binding and transcriptional activities (8,9). To examine the hypothesis that Ajuba may help to recruit CBP/p300 to ERα, we carried out a co-IP assays in transfected 293T cells and found that the association between ERα and CBP was weak in the absence of E2 and evidently enhanced by the E2 treatment (Figure 6D, lanes 2 and 4). Notably, the association between ERα and CBP was apparently enhanced by Ajuba with or without E2 (Figure 6D, lanes 3 and 5), and concomitantly Ajuba significantly increased the acetylation level of ERα (Figure 6E). Notably, the band above the acetylated ERα was acetylated Ajuba (Figure 6E Lanes 5, 8). The luciferase report assays showed that Ajuba or CBP/p300 could increase the transcriptional activity of ERα, and co-transfection of Ajuba and CBP or p300 maximally enhanced the transcriptional activity of ERα (Figure 6F). We next performed ChIP assays in T47D cells and the results showed that the knocking-down of Ajuba strikingly impaired the binding of CBP and p300 to the promoter of TFF1 and accompanied by the decreased acetylation of H3 (Figure 6G). In addition, the ERα binding also significantly decreased when Ajuba was knocked down (Figure 6G). Collectively, these data indicate that Ajuba enhances the acetylation and the DNA binding activity of ERα.
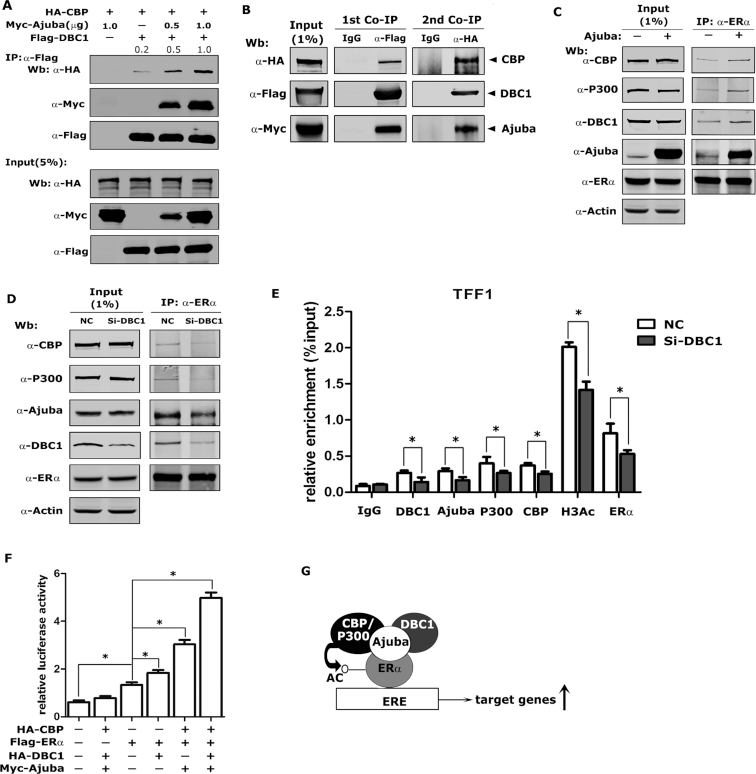
Ajuba, DBC1 and CBP/p300 form a ternary complex to activate ERα target gene transcription
To examine if Ajuba, DBC1 and CBP can form a complex, we co-expressed Flag-DBC1, HA-CBP and Myc-Ajuba at doses of 0.5 and 1.0 μg plasmids in 293T cells and co-IP assays were performed with Flag antibody (M2 beads). In the absence of Ajuba expression, DBC1 and CBP showed weak interaction, while co-expression of Ajuba could markedly enhance their interaction in dose dependent manner (Figure 7A). We further confirmed these observations by performing two-step co-IP assays: the co-IP assays were first performed with Flag antibody followed by using HA antibody (Figure 7B).
Figure 7.
Ajuba, DBC1 and CBP/p300 forms a ternary complex to enhance ERα target gene transcription. (A) Plasmids were transfected into 293T cells and the Co-IP assay was performed by using Flag-M2 beads (the relative amount of immunoprecipitated HA-CBP was semi-quantified by grayscale analysis and the mean values of the three repeats were labeled). (B) Myc-Ajuba, HA-CBP and Flag-DBC1 plasmids were co-transfected into 293T cells and sequential co-IP assays were performed. The first round co-IP assay was carried out by using Flag antibody or normal IgG and then the 3XFlag-peptides eluted product was subjected to the second round co-IP assay by using HA antibody or normal IgG. (C) The endogenous co-IP assay was performed in T47D-PCDH-Vector or T47D-PCDH-Myc-Ajuba cells by using ERα antibody. (D) siRNA-DBC1 was transfected into T47D cells for 48 h, and the co-IP assay was performed by using ERα antibody. (E) The siRNA-NC or siRNA-DBC1 was transfected into T47D cells and the ChIP assay was performed after the E2-treatment and the results were normalized to input and 5–10% as input (three repeats, * means P < 0.05). (F) TFF1-luciferase reporter assays were performed in 293T cells. Transfected cells were treated with 20 nM E2 for 12 h before harvesting and the value was normalized to β-gal (three repeats, * means P < 0.05). (G) The model of Ajuba regulating ERα transcriptional activity. Ajuba forms a complex with DBC1 and CBP to enhance the acetylation and transactivity of ERα.
Next, we performed endogenous co-IP assays using antibody specifically against ERα in T47D cells stably expressing Myc-Ajuba or vector to further examine the interaction between Ajuba, DBC1, CBP/p300 and ERα. Ajuba, DBC1, CBP/p300 were co-eluted with ERα from whole cell lysates prepared from T47D-vector cells, while over-expression of Ajuba evidently enhanced the association between ERα and DBC1 and CBP/P300 (Figure 7C). In contrast, knocking-down DBC1 in T47D cells resulted in decreased interaction of ERα with Ajuba and CBP/P300 (Figure 7D). In addition, ChIP assays showed that knocking-down DBC1 significantly decreased the binding of CBP/p300, Ajuba and ERα to the promoter of TFF1 and accompanied by the decreased acetylation level of H3 (Figure 7E). To examine the synergistic role of the ternary complex in regulating the transcriptional activity of ERα, we performed a luciferase reporter assay in 293T cells and found that both DBC1 and Ajuba/CBP could significantly activate ERα transactivity and the co-expression of the three proteins could maximally activate the transcriptional activity of ERα (Figure 7F). Taken together, these results indicate Ajuba recruits DBC1 and CBP/p300 and forms a ternary complex to enhance ERα target genes transcription (Figure 7G).
DISCUSSION
In this paper, we report that Ajuba functions as an atypical co-activator of ERα by recruiting CBP/p300 and DBC1 to increase the acetylation of ERα and enhances ERα target gene transcription, and biologically promotes breast cancer growth and attributes to tamoxifen-resistance of breast cancer cells.
Ajuba is structurally constituted by three tandem LIM motifs in the C-terminus and proline/glycine rich preLIM domain in the N-terminus. The LIM motifs are double zinc fingers and function as protein-protein interaction modules, while the preLIM region is predicted to be non-structured, which confers Ajuba more flexibility and uncertainty for protein bindings. Additionally, Ajuba contains both NLS and NES signals and can shuttle between cytoplasm and nucleus. These properties make Ajuba a versatile scaffold and can participate in assembly of variety of protein complexes, which is well represented by the interaction between Ajuba and nuclear hormone receptors (41–43).
We previously demonstrated that Ajuba broadly interacts with multiple nuclear hormone receptors via different binding motifs. Ajuba was first shown to bind RARα via the three conserved nuclear-binding motifs and negatively regulates RARα transcriptional activity. Interestingly, their interaction could be disrupted upon ligand (all trans-Retinoid Acid) binding to RARα (39). Conversely, we also discovered Ajuba functioning as a co-activator of PPARγ and LXRα by recruiting CBP/p300. Although Ajuba enhances transcriptional activities of both PPARγ and LXRα, the interaction manners between Ajuba and PPARγ or LXRα are different: Ajuba interacts with PPARγ through its preLIM region, and their interaction is ligand independent (41); while Ajuba interacts with LXRα through its LIM domain in a ligand dependent manner (40). These observations suggest that Ajuba is an atypical co-regulator of nuclear hormone receptors. This claim is further supported by the binding manner between Ajuba and ERα: although the full-length of Ajuba can interact with ERα, neither the preLIM nor the LIM regions alone binds ERα; moreover, the interaction between Ajuba and ERα does not require ligand, but can be markedly enhanced by the ligand binding to ERα. This may be attributed to both DBD and AF2 regions of ERα can interact with Ajuba: the interaction of Ajuba with DBD is independent of ligand, while the interaction with AF2 domain of is dependent on ligand binding. To fully understand the binding behavior of Ajuba and the nuclear hormone receptors, the detailed binding motifs should be identified and we certainly have interests to explore further.
DBC1 has been reported as an important co-activator of ERα, but the underlying mechanism remains elusive (25). Acetylation of ERα at two sites K266/268 and K302/303 can significantly enhance its transcriptional activity. The acetylation of K302/303 results in increased transcriptional activity and hormone sensitivity of ERα (9), and the K266/268 is located in the DBD domain and its acetylation results in increased DNA-binding activity and transcriptional activity (8). However, those acetylation of ERα can be eliminated by SIRT1, while DBC1, by forming a complex with SIRT1 and ERα, can overcome the effect of SIRT1 to maintain the acetylation level of ERα (25). Here, we found that DBC1 can enhance the acetylation of ERα via a different mechanism. DBC1 forms a complex with Ajuba, CBP/p300 and ERα and can enhance interaction of ERα with Ajuba and CBP/p300, which results in increased acetylation of ERα and its transcriptional activity. These observations extend the role of DBC1 in the regulation of ERα transcriptional activity. Interestingly, DBC1 can directly interact with Ajuba and SIRT1 respectively, but these three proteins do not co-exist in one complex, indicating that the two complexes Ajuba/DBC1, SIRT1/DBC1 are mutually exclusively existed in the cells, and exert different functions. Notably, we also observed that Ajuba and DBC1 affect the DNA-binding activity of ERα on endogenous promoters evidenced by ChIP assays. These effects may be contributed to their effect on ERα acetylation, and/or on the DNA binding activity of ERα.
Many drugs targeting ERα signaling have been developed and widely used as the first line chemotherapeutics in ER positive breast cancer treatment, including tamoxifen, raloxifene and exemestane (44,45). However, drug-resistance always emerges and limits their therapeutic effect. To a large extent, drug resistance is attributed to the ligand independent function of ERα. Many mechanisms have been reported to mediate the estrogen-independent function of ERα, including phosphorylation of Ser118 of ERα (5,46). Here, we found Ajuba can interact with ERα in the absence of estrogen, which may explain the Ajuba effect on ERα target genes expression and cell proliferation in the absence of estrogen. In breast cancer lines, decreasing Ajuba expression level sensitizes the toxic effect of tamoxifen, suggesting Ajuba may be a target for override the tamoxifen resistance.
In summary, our results demonstrate that Ajuba can enhance acetylation of ERα and potentiate ERα transcriptional activity; biologically, elevated expression of Ajuba promotes breast cancer cells growth and results in tamoxifen-resistance; mechanistically, Ajuba recruits DBC1 and CBP/p300 to ERα to enhance its transcriptional and DNA binding activities, as well as histone acetylation. These observations suggest that Ajuba/DBC1/CBP/p300 ternary complex may be a new target for therapeutics to breast cancer treatment.
ACKNOWLEDGEMENTS
We thanks Professor Cheng Luo (Shanghai Institute of Materia Medica, Chinese Academy of Sciences) for critical discussion.
FUNDING
National Science Foundation of China [81872131, 31671415, 81471000]; Shanghai Committee of Science and Technology [15410724200]. Funding for open access charge: National Science Foundation of China.
Conflict of interest
The authors declare no conflict of interest.
REFERENCES
- 1. Koren S., Bentires-Alj M.. Breast tumor Heterogeneity: Source of fitness, hurdle for therapy. Mol. Cell. 2015; 60:537–546. [DOI] [PubMed] [Google Scholar]
- 2. Manavathi B., Samanthapudi V.S.K., Gajulapalli V.N.R.. Estrogen receptor coregulators and pioneer factors: the orchestrators of mammary gland cell fate and development. Front. Cell Dev. Biol. 2014; 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zwart W., de Leeuw R., Rondaij M., Neefjes J., Mancini M.A., Michalides R.. The hinge region of the human estrogen receptor determines functional synergy between AF-1 and AF-2 in the quantitative response to estradiol and tamoxifen. J. Cell Sci. 2010; 123:1253–1261. [DOI] [PubMed] [Google Scholar]
- 4. Benecke A., Chambon P., Gronemeyer H.. Synergy between estrogen receptor α activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Rep. 2000; 1:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anbalagan M., Rowan B.G.. Estrogen receptor alpha phosphorylation and its functional impact in human breast cancer. Mol. Cell. Endocrinol. 2015; 418:264–272. [DOI] [PubMed] [Google Scholar]
- 6. Kato S., Endoh H., Masuhiro Y., Kitamoto T., Uchiyama S., Sasaki H., Masushige S., Gotoh Y., Nishida E., Kawashima H. et al.. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995; 270:1491–1494. [DOI] [PubMed] [Google Scholar]
- 7. Bunone G., Briand P.A., Miksicek R.J., Picard D.. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996; 15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 8. Kim M.Y., Woo E.M., Chong Y.T., Homenko D.R., Kraus W.L.. Acetylation of estrogen receptor alpha by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol. Endocrinol. 2006; 20:1479–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang C., Fu M., Angeletti R.H., Siconolfi-Baez L., Reutens A.T., Albanese C., Lisanti M.P., Katzenellenbogen B.S., Kato S., Hopp T. et al.. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J. Biol. Chem. 2001; 276:18375–18383. [DOI] [PubMed] [Google Scholar]
- 10. Hamaguchi M., Meth J.L., von Klitzing C., Wei W., Esposito D., Rodgers L., Walsh T., Welcsh P., King M.C., Wigler M.H.. DBC2, a candidate for a tumor suppressor gene involved in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:13647–13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim J.E., Chen J., Lou Z.. p30 DBC is a potential regulator of tumorigenesis. Cell Cycle. 2009; 8:2932–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sung J.Y., Kim R., Kim J.E., Lee J.. Balance between SIRT1 and DBC1 expression is lost in breast cancer. Cancer Sci. 2010; 101:1738–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee H., Kim K.R., Noh S.J., Park H.S., Kwon K.S., Park B.H., Jung S.H., Youn H.J., Lee B.K., Chung M.J. et al.. Expression of DBC1 and SIRT1 is associated with poor prognosis for breast carcinoma. Hum. Pathol. 2011; 42:204–213. [DOI] [PubMed] [Google Scholar]
- 14. Zhang Y., Gu Y., Sha S., Kong X., Zhu H., Xu B., Li Y., Wu K.. DBC1 is over-expressed and associated with poor prognosis in colorectal cancer. Int. J. Clin. Oncol. 2014; 19:106–112. [DOI] [PubMed] [Google Scholar]
- 15. Cha E.J., Noh S.J., Kwon K.S., Kim C.Y., Park B.-H., Park H.S., Lee H., Chung M.J., Kang M.J., Lee D.G. et al.. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin. Cancer Res. 2009; 15:4453–4459. [DOI] [PubMed] [Google Scholar]
- 16. Radvanyi L., Singh-Sandhu D., Gallichan S., Lovitt C., Pedyczak A., Mallo G., Gish K., Kwok K., Hanna W., Zubovits J. et al.. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J., Bonkowski M.S., Moniot S., Zhang D., Hubbard B.P., Ling A.J.Y., Rajman L.A., Qin B., Lou Z., Gorbunova V. et al.. A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science. 2017; 355:1312–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim J.-E., Chen J., Lou Z.. DBC1 is a negative regulator of SIRT1. Nature. 2008; 451:583–586. [DOI] [PubMed] [Google Scholar]
- 19. Joshi P., Quach O.L., Giguere S.S., Cristea I.M.. A functional proteomics perspective of DBC1 as a regulator of transcription. J. Proteomics Bioinform. 2013; Suppl 2:002. [PMC free article] [PubMed] [Google Scholar]
- 20. Sakurabashi A., Wada-Hiraike O., Hirano M., Fu H., Isono W., Fukuda T., Morita Y., Tanikawa M., Miyamoto Y., Oda K. et al.. CCAR2 negatively regulates nuclear receptor LXRalpha by competing with SIRT1 deacetylase. J. Steroid Biochem. Mol. Biol. 2015; 149:80–88. [DOI] [PubMed] [Google Scholar]
- 21. Chini C.C., Escande C., Nin V., Chini E.N.. DBC1 (Deleted in Breast Cancer 1) modulates the stability and function of the nuclear receptor Rev-erbα. Biochem. J. 2013; 451:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Koyama S., Wada-Hiraike O., Nakagawa S., Tanikawa M., Hiraike H., Miyamoto Y., Sone K., Oda K., Fukuhara H., Nakagawa K. et al.. Repression of estrogen receptor beta function by putative tumor suppressor DBC1. Biochem. Biophys. Res. Commun. 2010; 392:357–362. [DOI] [PubMed] [Google Scholar]
- 23. Garapaty S., Xu C.F., Trojer P., Mahajan M.A., Neubert T.A., Samuels H.H.. Identification and characterization of a novel nuclear protein complex involved in nuclear hormone receptor-mediated gene regulation. J. Biol. Chem. 2009; 284:7542–7552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fu J., Jiang J., Li J., Wang S., Shi G., Feng Q., White E., Qin J., Wong J.. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J. Biol. Chem. 2009; 284:6832–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ji Yu E., Kim S.-H., Heo K., Ou C.-Y., Stallcup M.R., Kim J.H.. Reciprocal roles of DBC1 and SIRT1 in regulating estrogen receptor α activity and co-activator synergy. Nucleic Acids Res. 2011; 39:6932–6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim H.J., Kim S.H., Yu E.J., Seo W.Y., Kim J.H.. A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer. Oncogene. 2014; 34:4500–4508. [DOI] [PubMed] [Google Scholar]
- 27. Marie H., Pratt S.J., Betson M., Epple H., Kittler J.T., Meek L., Moss S.J., Troyanovsky S., Attwell D., Longmore G.D. et al.. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J. Biol. Chem. 2003; 278:1220–1228. [DOI] [PubMed] [Google Scholar]
- 28. Nola S., Daigaku R., Smolarczyk K., Carstens M., Martin-Martin B., Longmore G., Bailly M., Braga V.M.. Ajuba is required for Rac activation and maintenance of E-cadherin adhesion. J. Cell Biol. 2011; 195:855–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pratt S.J., Epple H., Ward M., Feng Y., Braga V.M., Longmore G.D.. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 2005; 168:813–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jia H., Song L., Cong Q., Wang J., Xu H., Chu Y., Li Q., Zhang Y., Zou X., Zhang C. et al.. The LIM protein AJUBA promotes colorectal cancer cell survival through suppression of JAK1/STAT1/IFIT2 network. Oncogene. 2016; 36:2655–2666. [DOI] [PubMed] [Google Scholar]
- 31. Das Thakur M., Feng Y., Jagannathan R., Seppa M.J., Skeath J.B., Longmore G.D.. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr. Biol. 2010; 20:657–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jagannathan R., Schimizzi G.V., Zhang K., Loza A.J., Yabuta N., Nojima H., Longmore G.D.. AJUBA LIM proteins limit hippo activity in proliferating cells by sequestering the hippo core kinase complex in the cytosol. Mol. Cell Biol. 2016; 36:2526–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rauskolb C., Sun S., Sun G., Pan Y., Irvine K.D.. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014; 158:143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reddy B.V., Irvine K.D.. Regulation of Hippo signaling by EGFR-MAPK signaling through Ajuba family proteins. Dev. Cell. 2013; 24:459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tanaka I., Osada H., Fujii M., Fukatsu A., Hida T., Horio Y., Kondo Y., Sato A., Hasegawa Y., Tsujimura T. et al.. LIM-domain protein AJUBA suppresses malignant mesothelioma cell proliferation via Hippo signaling cascade. Oncogene. 2015; 34:73–83. [DOI] [PubMed] [Google Scholar]
- 36. Haraguchi K., Ohsugi M., Abe Y., Semba K., Akiyama T., Yamamoto T.. Ajuba negatively regulates the Wnt signaling pathway by promoting GSK-3beta-mediated phosphorylation of beta-catenin. Oncogene. 2008; 27:274–284. [DOI] [PubMed] [Google Scholar]
- 37. Hou Z., Peng H., Ayyanathan K., Yan K.P., Langer E.M., Longmore G.D., Rauscher F.J. 3rd. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol. Cell Biol. 2008; 28:3198–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hou Z., Peng H., White D.E., Wang P., Lieberman P.M., Halazonetis T., Rauscher F.J.. 14-3-3 binding sites in the Snail protein are essential for Snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010; 70:4385–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hou Z., Peng H., White D.E., Negorev D.G., Maul G.G., Feng Y., Longmore G.D., Waxman S., Zelent A., Rauscher F.J.. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fan H., Dong W., Li Q., Zou X., Zhang Y., Wang J., Li S., Liu W., Dong Y., Sun H. et al.. Ajuba preferentially binds LXRalpha/RXRgamma heterodimer to enhance LXR target gene expression in liver cells. Mol. Endocrinol. 2015; 29:1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Q., Peng H., Fan H., Zou X., Liu Q., Zhang Y., Xu H., Chu Y., Wang C., Ayyanathan K. et al.. The LIM protein Ajuba promotes adipogenesis by enhancing PPARgamma and p300/CBP interaction. Cell Death Differ. 2016; 23:158–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kanungo J., Pratt S.J., Marie H., Longmore G.D.. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 2000; 11:3299–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schimizzi G.V., Longmore G.D.. Ajuba proteins. Curr. Biol. 2015; 25:R445–R446. [DOI] [PubMed] [Google Scholar]
- 44. Osborne C.K., Schiff R.. Mechanisms of endocrine resistance in breast cancer. Annu. Rev. Med. 2011; 62:233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turner N.C., Neven P., Loibl S., Andre F.. Advancesin the treatment of advanced oestrogen-receptor-positive breast cancer. Lancet. 2016; 389:2403–2414. [DOI] [PubMed] [Google Scholar]
- 46. Le Romancer M., Poulard C., Cohen P., Sentis S., Renoir J.M., Corbo L.. Cracking the estrogen receptor's posttranslational code in breast tumors. Endocrine reviews. 2011; 32:597–622. [DOI] [PubMed] [Google Scholar]
- 47. Hou Z., Peng H., White D.E., Negorev D.G., Maul G.G., Feng Y., Longmore G.D., Waxman S., Zelent A., Rauscher F.J. 3rd. LIM protein Ajuba functions as a nuclear receptor corepressor and negatively regulates retinoic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]