Abstract
Hippo pathway is involved in tumorigenesis, and its regulation in cytosol has been extensively studied, but its regulatory mechanisms in the nuclear are not clear. In the current study, using a FBS-inducing model following serum starvation, we identified KDM3A, a demethylase of histone H3K9me1/2, as a positive regulator for hippo target genes. KDM3A promotes gene expression through two mechanisms, one is to upregulate YAP1 expression, and the other is to facilitate H3K27ac on the enhancers of hippo target genes. H3K27ac upregulation is more relevant with gene activation, but not H3K4me3; and KDM3A depletion caused H3K9me2 upregulation mainly on TEAD1-binding enhancers rather than gene bodies, further resulting in H3K27ac decrease, less TEAD1 binding on enhancers and impaired transcription. Moreover, KDM3A is associated with p300 and required for p300 recruitment to enhancers. KDM3A deficiency delayed cancer cell growth and migration, which was rescued by YAP1 expression. KDM3A expression is correlated with YAP1 and hippo target genes in colorectal cancer patient tissues, and may serve as a potential prognosis mark. Taken together, our study reveals novel mechanisms for hippo signaling and enhancer activation, which is critical for tumorigenesis of colorectal cancer.
INTRODUCTION
Hippo signaling pathway is firstly discovered in drosophila and highly conserved in human beings (1–3). Its proper activation is important for cell fate decision, organ size control and regeneration (3). Its dysregulation has been connected with tumorigenesis and inflammation (1,3,4). In mammals, the activation of hippo signaling pathway involves a phosphorylation cascade consisting macrophage stimulating 1/2 (MST1/2), large tumor suppressor kinase 1/2 (LATS1/2) and transcription activator Yes associated protein 1 (YAP1)/tafazzin (TAZ). Phosphorylation of YAP1 restrains the protein in the cytoplasm for degradation. When hippo pathway is silent, dephosphorylated YAP1 is translocated into nuclear, interacts with TEA domain transcription factor 1–4 (TEAD1–4) and subsequently activates the transcription of target genes (1–3,5,6), which can be inhibited by VGLL4 (7–9). TEAD family proteins are key transcription factors in hippo signaling. Their binding to chromatin is considered to remain constant no matter whether the pathway is activated or not (10,11). Though the regulation of hippo pathway in cytosol has been extensively studied, the regulation of TEADs-dependent transcription in the nuclear still remains elusive. It is still not clear how TEAD1 is recruited to chromatin and whether chromatin environment is involved.
Upon receiving upstream signals, the activation of signaling pathways often results in the activation of transcription factors, which bind enhancers on chromatin and activate transcription. Histone modifications are one of the major parts of epigenetic regulators, and transcriptional enhancers are marked by histone modifications (12–14). H3K4me1 is enriched on enhancers and lysine methyltransferase 2C/D (KMT2C/D, also known as MLL3/4) are the key enzymes in mammalian cells (15–17). H3K27ac is an important mark for active enhancer, catalyzed by E1A binding protein p300 (EP300) and CREB binding protein (CREBBP/CBP) (18). The combination of H3K4me1 and H3K27ac has now been widely used to identify distal enhancers across the genome (19–21). The latest studies demonstrated that enhancers exist not only close to transcription start sites but also at distal regions, and some of them are even several hundred kilo-base away (14,22). Interestingly, a transcription factor often binds to thousands of enhancers but only regulates the expression of hundreds of genes, suggesting multiple enhancers are responsible for one gene. However, we still do not know much how the activity of enhancers are regulated and how the enhancer-gene network works.
H3K9me2 is a transcription repressive mark on chromatin, mainly catalyzed by histone methyltransferases EHMT2/G9a and EHMT1/GLP (23,24). Unlike the heterochromatin mark H3K9me3, H3K9me2 is mostly localized on euchromatin (24,25). H3K9me2 is one of histone modifications firstly identified, and it inhibits transcription through chromatin compaction and crosstalk with DNA methylation (24). H3K9me2 is dynamic regulated by multiple histone demethylase, including lysine demethylase 3A/B, 4A-D (KDM3A/B, KDM4A-D) and others (26). Many of these proteins have been shown related with tumorigenesis (26,27). For example, KDM3A is over expressed in colorectal and breast cancers, and responsible for H3K9me2 removal on oncogenes (25,28,29). KDM4A was reported to regulate site-specific copy gain and DNA re-replication, and promote cellular transformation by inhibiting p53 signaling (30,31). All these suggest the methylation of H3K9 is tightly related with cancer, but the underlying mechanisms still require further investigation.
In the current study, we identified KDM3A as a key regulator critical for hippo signaling and revealed novel mechanisms for recruitment of TEAD1 to target enhancers. KDM3A regulates the expression of YAP1, and more importantly, KDM3A removes H3K9me2 on enhancers and is required for p300 recruitment, which is critical for enhancer activation, TEAD1 recruitment, and transcription of hippo target genes.
MATERIALS AND METHODS
Reagents and cell lines
Antibodies against KDM3A (Abcam ab91252), β-Actin (Abclonal AC006), hippo signaling antibody sampler kit (CST 8579, including LATS1, MST1, MST2, SAV1, pYAP S127, YAP, MOB1, pMOB1 Thr35), H3K9me2 (Abcam ab1220), TEAD1 (CST 12292), p300 (Santa Cruz sc-585x), H3K4me1 (CST 5326), H3K27ac (Abcam ab4729), H3K27me3 (Millipore 07-449), Pol II (Abcam ab817), H3K4me3(Millipore 04-745), were purchased from indicated commercial sources. Protein G-Sepharose beads were purchased from GE Healthcare. PCR primers were custom synthesized by BGI and siRNAs were by GenePharma (Supplementary Table S3). Bix01294, UNC0631were purchased from Selleck. Cell lines were purchased from Cell Bank of Chinese Academy and cultured under recommended conditions.
Transfection of plasmids and siRNAs
siRNAs were carried out using Lipo2000 according to the manufacturer's instructions. The siRNA sequences used are shown in Table 1. Cell Culture and building of Stable Cell lines—Human colorectal cancer cells HCT116 were grown in DMEM with 10% fetal bovine serum, 0.1% penicillin and streptomycin. To establish KDM3A knockdown, KDM3A knockdown with YAP overexpression, and YAP overexpression stable cell lines, HCT116 cells were infected with retroviral containing the indicated plasmids. KDM3A sgRNA was designed and CRISPR-CAS9 technique was used to establish KDM3A knockout cell line.
Immunoprecipitation
The cells were harvested and lysed in NP40 Lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 0.5% NP40) or high salt lysis buffer (20 mM HEPES pH 7.4, 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% triton X-100, 1 mM DTT) with proteinase inhibitors. The supernatant was then incubated with protein G beads (GE Healthcare) and desired antibody at 4°C for 4 h. The beads were spin down and washed for three times with lysis buffer. The final drop of wash buffer was vacuumed out and SDS loading buffer was added to the beads followed by western blot. All immunoblotting images are representative of at least three independent experiments. Data shown in the bar figures are the average values from three independent experiments with the SDs. P-values were calculated using a two-tailed t test.
RNA interference, reverse transcription and quantitative PCR
The indicated cells were transfected with siRNA and were scraped down and collected by centrifugation. Total RNA was extracted with RNA extraction kit (Aidlab) according to manufacturer's manual. Approximately 1 μg of total RNA was used for reverse transcription with a first strand cDNA synthesis kit (Toyobo). The amount of mRNA was assayed by quantitative PCR. β-Actin was used to normalize the amount of each sample. Assays were repeated at least three times. Data shown were average values ± SD of three representative experiments.
Tissue microarray and IHC
The tissue microarray slide were purchased from Shanghai Outdo Biotech. The slide contained 12 colorectal cancer samples, 3 normal colon tissues and 1 adjacent prostate cancer tissue. The slide was de-paraffinized, rehydrated and subjected to heat-mediated antigen retrieval. For immunohistochemistry (IHC) analysis, the sections were incubated with 3% H2O2 for 15 min at room temperature to quench endogenous peroxidase activity. After incubating in normal goat serum for 1 h, sections were treated with primary antibody at 4°C overnight. IHC analysis of tumor samples was performed using primary antibodies against YAP (dilution 1:50; CST). The sections were then washed three times in PBS and treated for 30 min with biotinylated goat-anti-rabbit IgG secondary antibodies. After washing three times in PBS, sections were incubated with streptavidin-conjugated HRP. After washing three times in PBS for 5 min each, specific detection was developed with 303-diaminobenzidine (DAB-2031). Images were taken by using an Olympus camera and matched software.
Cell viability assay
Cells were seeded on the 24-well plate about 10 000 cells per well, incubated at 37°C for 72 h. Then 100 μl MTT (5 μg/μl) was added into every well, incubated for 4 h at 37°C. After that, 400 μl lysate buffer (50% DMF + 30% SDS, pH 4.7) was added into each well. Signals were collected by Microplate System. Data shown in the bar figures are the average values of three independent experiments with the SDs. P values were calculated using a two-tailed t test.
Real time cell analysis (RTCA) of cell proliferation and migration
Cell proliferation and migration were analyzed with RTCA assay as described before (32). Cells were cultured at 6000 per well in CIM-Plate wells coated with (invasion) or without (proliferation) matrigel. The cell index signals were read by xCELLigence RTCA DP Analyzer (ACEA Bioscience). Invasion and migration are monitored continuously over a 48-hour period. Each experiment was repeated three times and results were presented as mean ± SD.
ChIP assay
ChIP assay was performed as previously described (33). Briefly, ∼1 × 107 cells were fixed with 1% formaldehyde and quenched by glycine. The cells were washed three times with PBS and then harvested in ChIP lysis buffer (50 mM Tris–HCl, pH7.6, 1 mM CaCl2, 0.2% Triton X-100). DNA was digested to 150–300 bp by MNase (SIGMA) before extensive centrifugation. Four volume of ChIP dilution buffer (20 mM Tris–HCl, pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS) was added to the supernatant. The resulted lysate was then incubated with protein G beads and antibodies at 4°C overnight. The beads were washed five times and DNA was eluted by Chip elution buffer (0.1 M NaHCO3, 1% SDS, 20 μg/ml proteinase K). The elution was incubated at 65°C overnight and DNA was extracted with DNA purification kit (TIANGEN). The purified DNA was assayed by quantitative PCR with Biorad MyIQ. Assays were repeated at least three times. Data shown were average values ± SD of representative experiments. The sequences of primers are in Supplementary Table S1. The enrichment of histone modifications was normalized with input. Histone H3 ChIP was also performed and the results were also showed after normalized with H3, which are included in the file of Supplementary data.
Cell cycle analysis with flow cytometry
Cells were harvested after digestion with 0.05% Trypsin–EDTA. The cells were then washed twice with PBS and fixed in ice-cold 70% ethanol overnight. Fixed cells were washed twice with PBS and stained in PBS containing propidium iodide (PI, 50 μg/ml) and RNase (100 μg/ml) for 30 min at 37°C. Cell cycle analysis was performed on an Epics XL-MCL flow cytometer (Beckman Coulter) with System II (version 3.0) software (Beckman Coulter). Additional analysis of cell cycle distribution was determined using FlowJo software.
Xenograft experiments of cancer cells
The 5-week-old male BALB/C nude mice were purchased from Beijing HFK Bioscience Co. Ltd. A colorectal cancer mouse model was established by injecting subcutaneously the animals with 1 × 106 HCT116 cells in the flank region. Tumor volumes were measured twice a week using calipers. Tumor volumes were derived as V = 0.5 × length × width2. After three weeks of injection, the tumors were harvested and weighed. All animal xenograft experiments were performed following the university laboratory animal guidelines and were approved by the Animal Experimentations Ethics Committee of Wuhan University.
Pipeline of RNA-seq analysis
RNA-seq library was performed by using Illumina TruSeq library construction kit. Using 5 μg total RNA as initiation, and then prepared according to the manufacturer's instruction. mRNA-seq libraries were sequenced using HiSeq2000 for 100 bp paired-end sequencing. Quality control of mRNA-seq data was performed using Fatsqc and low quality bases were trimmed. All RNA-seq data were mapped to the human genome (hg19) by TopHat (version 2.1.1) and allow maximum two mismatch. The gene expression level was calculated by Cufflinks with default parameters and gene ontology analysis was performed using DAVID (https://david.ncifcrf.gov) (34,35).
Pipeline of ChIP-Seq analysis
ChIP-seq was performed as described before (33). Library was prepared using the sequencing library preparation kit from Vazyme (ND604) according to the manufacturer's protocol. DNA was prepared for end repair and ‘A’ tailing, adaptor ligation, and library amplification. ChIP-Seq libraries were sequenced on HiSeq 2500 for 150 bp paired end sequencing.
Quality control of ChIP-Seq data was performed using Fastqc. Low quality bases and adaptor contamination were deleted. Raw reads were aligned to the human genome (hg19) with Bowtie2 (version 2.1.0), and only uniquely mapped reads retained. The redundant reads were removed using SAMtools. MACS2 (version 2.1.1) was used to call peaks. The genomic location of the peaks and their distance to the TSS of annotated genes were using the annotatePeaks.pl of HOMER. (http://homer.ucsd.edu/homer/index.html,v4.9,2-20-2017).
The enhancers were identified as the published protocol (18). Briefly, after mapping H3K27ac ChIP-seq reads to genome, significant H3K27ac peaks were defined by MACS2 (P-value cut-off 1E–9). Distal peaks (1.5 kb away from gene TSS) were stitched together with one another if their distance was shorter than 12.5 kb. All these individual and stitched distal H3K27ac peaks were identified as enhancer.
The distance between TEAD1 peaks and the transcription start sites (TSSs) of annotated genes were using the annotatePeaks.pl of HOMER. The presence of TEAD1 peaks and the genomic locations of TEAD1 were the criteria used to define enhancers: TEAD1 peaks located away from TSS (±2 kb) were defined as distal enhancers. The read counts were normalized by computing the numbers of reads per kilo base of bin per million of reads sequenced (RPKM).The ChIP-seq signals at TSS (±10 kb) or enhancer center (±2 kb or ± 1 kb) were computed by generated the RPKM values in each 100-bp bin among the defined region.
Survival analysis
The survival analysis based on gene expression via OncoLnc (http://www.oncolnc.org). OncoLnc link TCGA survival data to mRNA expression levels. The datasets of colorectal cancer are based on TCGA-COAD (Colon Adenocarcinoma).
RESULTS
Identification of KDM3A as a positive regulator for hippo target genes
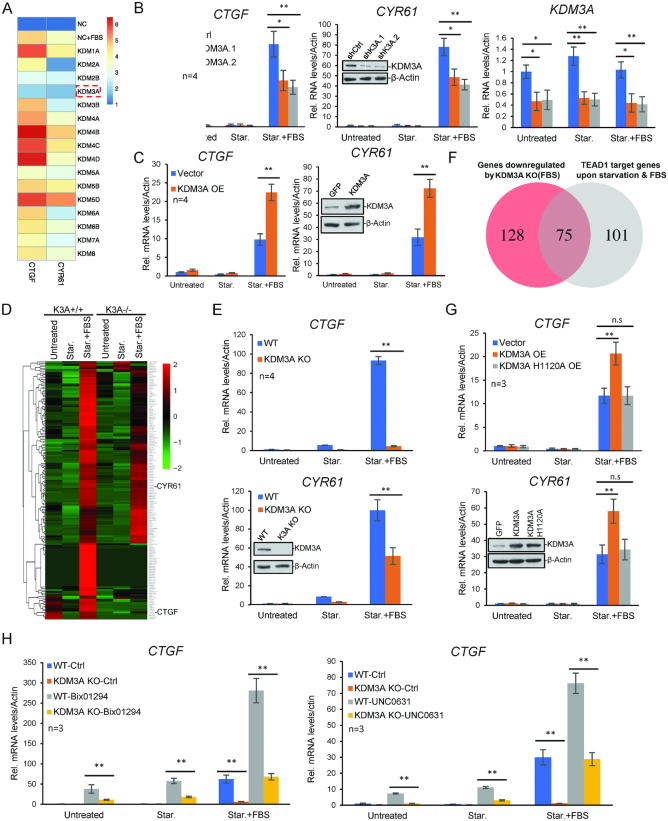
To investigate the role of protein methylation in hippo signaling pathway, we utilized a siRNA library targeting all the lysine methyltransferases and demethylases, and screened for regulators of the FBS-induced expression of two hippo target genes, connective tissue growth factor (CTGF) and cysteine rich angiogenic inducer 61 (CYR61) (Figure 1A). Serum was withdrawn from cultured HCT116 for 24 h and FBS was added to activate hippo target genes. One hour later, the expression of CTGF and CYR61 was measured with RT-PCR. We identified a number of candidates (Figure 1A), and found that KDM3A knockdown has the strongest effect on repressing the expression of the two hippo genes. Further considering KDM3A has been identified as a potential oncogene (25), we chose it for further mechanistic study.
Figure 1.
KDM3A regulates the expression of hippo target genes. (A) siRNAs targeting histone methylases and demethylases were transfected into HCT116. Forty eight hours after transfection, serum was withdrawn, and 24 h later, FBS was added into the medium. One hour later, the expression of CTGF and CYR61 was assayed with quantitative RT-PCR. The color represents the logarithmic value of FBS-induced gene expression folds. (B) KDM3A was knocked down with two different siRNAs in HCT116 and the expression of CTGF and CYR61 was determined by qRT-PCR. Untreated represents cells continuously cultured in the standard DMEM medium, Star. for cell with serum starvation, and Star. + FBS for cells with FBS addition after 24 h starvation. (C) KDM3A was exogenous expressed in HCT116 and the expression of CTGF and CYR61 was determined by qRT-PCR. (D) KDM3A knockout cell line was generated with CRISPR/Cas9 technique. The expression of CTGF and CYR61 was determined by qRT-PCR. (E) The heat map shows the expression (FPKM) of all FBS-induced DEGs with or without KDM3A. (F) Venn diagram to show the overlapped genes between KDM3A-regulated FBS-responsive genes and TEAD1 direct targeting genes. (G)KDM3A wild type or H1120A mutant was transfected in HCT116 and the expression of CTGF and CYR61 was determined by qRT-PCR. (H) KDM3A WT or KO cells were treated with Bix01294 (10 μM) or UNC0631 (10 μM) for 24 h. The expression of CTGF was determined by qRT-PCR. Transcript levels were determined relative to actin mRNA levels and normalized relative to control cells. * means P-value ≤ 0.05, ** for P-value ≤ 0.01. All the results represent the means (±SD) of at least three independent experiments.
Firstly, we verified the effect of KDM3A on the activation of CTGF and CYR61. We used two independent siRNAs to knock down KDM3A in HCT116, and induced hippo target genes with FBS after serum starvation. The results confirmed that KDM3A deficiency attenuates the activation of CTGF and CYR61 by FBS (Figure 1B). When over-expressing KDM3A in HCT116, CTGF activation was significantly promoted (Figure 1C). We also generate a KDM3A knockout cell line with CRISPR/CAS9. CTGF and CYR61 activation in the KDM3A−/− cell line was also greatly impaired (Figure 1D). We further confirmed the result by RNA-Seq. We analyzed all the FBS-activated genes in wild type cells and found the genes of hippo pathway was significantly enriched. Consistent with previous results, the activation of a large portion of them was greatly reduced in KDM3A−/− cells, including CTGF and CYR61 (Figure 1E). We also performed TEAD1 ChIP-Seq analysis and found totally 75 TEAD1 direct targeting genes were overlapped with the FBS-responsive genes whose RNA levels were down-regulated after KDM3A knockdown (Figure 1F). We further knocked down KDM3A in HeLa and several colon cancer cell lines, including DLD1, SW480 and RKO. The results show that KDM3A also regulates CTGF and CYR61 expression in these cells (Supplementary Figure S1). All these indicated that KDM3A is required for the activation of hippo target genes.
The enzyme activity of KDM3A is required for its function in hippo signaling
To study whether the enzyme activity of KDM3A is required, we generated a demethylase-dead mutant replacing histidine at 1120 residue by alanine (36). When exogenous expressed in HCT116, in comparison with wild type KDM3A, the catalytic dead mutant had no effect on the activation of hippo target genes, suggesting its demethylase activity is required (Figure 1G). Since KDM3A is a specific demethylase for histone H3K9me1/2 and no chemical inhibitor has been reported, we utilized two inhibitors for H3K9me2 methyltransferase G9a, BIX-01294 and UNC0631 (37,38), to inhibit H3K9me2 on the chromatin. The treatment with the two inhibitors both significantly enhanced the activation of CTGF and CYR61 (Figure 1H). These suggest KDM3A promotes the expression of Hippo target genes probably through removal of H3K9me2.
YAP1 expression targeted by KDM3A
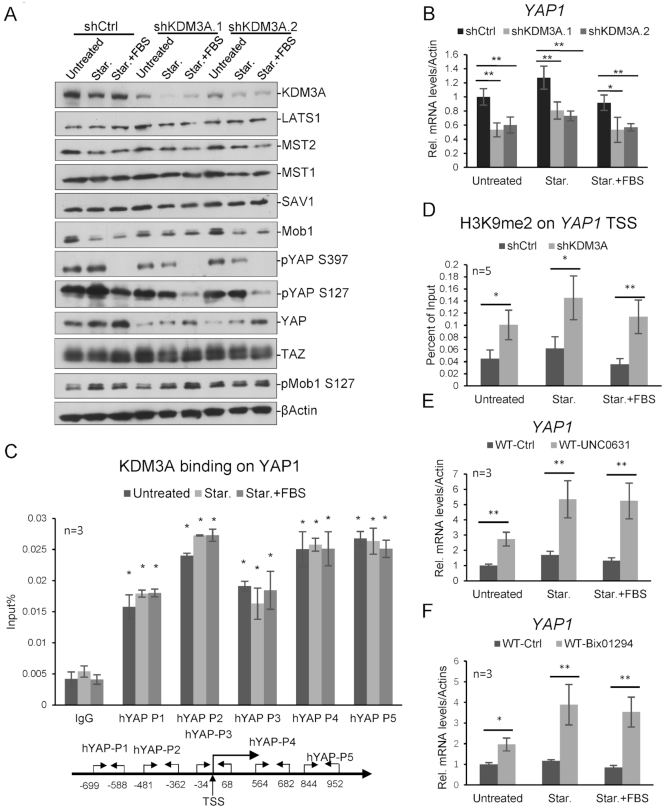
To investigate the underlying mechanisms for KDM3A-regulating hippo pathway, we knocked down KDM3A transiently with two different shRNAs and performed western blotting, one shRNA targeting luciferase as control. We found that KDM3A deficiency caused disappearance of YAP1 protein but not other tested key proteins in hippo pathway, suggesting KDM3A may regulate YAP1 expression (Figure 2A). We further found that YAP1 mRNA level in the above samples decreased dramatically after KDM3A knockdown (Figure 2B). When we knocked down KDM3A in other cell lines, similar results were observed (Supplementary Figure S2).
Figure 2.
KDM3A regulates YAP1 expression. (A) HCT116 WT or KDM3A knockdown cell lines were starved for 24 h and then treated with FBS for 1 h. Cell lysates were immunoblotted with indicated antibodies. Two different shRNA constructs were used. (B) Cells in (A) were harvested and YAP1 mRNA level were determined relative to β-actin and control cells. (C) KDM3A ChIP assay shows KDM3A binding on YAP1 relative to Input. The primer sets for qPCR are indicated at the bottom. (D) H3K9me2 ChIP analysis in HCT116 WT or shKDM3A.1 stable cell line shows the enrichment of H3K9me2 on YAP1, relative to IgG and control, with primer set p3. (E, F) Cells were treated with two H3K9me2 inhibitors, UNC0631 (E) and Bix01294 (F), respectively. YAP1 expression were determined with quantitative RT-PCR. * means P-value ≤ 0.05, ** for P-value ≤ 0.01. The results in all experiments represent the means (±SD) of at least three independent experiments.
To study whether KDM3A directly regulates YAP1 transcription, we performed KDM3A ChIP assay and measured its binding to YAP1 promoter. Five pairs of primers for YAP1 promoter were designed and the results indicated that KDM3A binds to these regions (Figure 2C). ChIP assay with H3K9me2 also indicated H3K9me2 on YAP1 significantly increased after KDM3A knockdown (Figure 2D). H3K9me2 inhibitors, BIX-01294 and UNC0631, were also applied to HCT116, respectively. Inhibition of H3K9me2 significantly increased YAP1 expression in the cell (Figure 2E and F). All these indicate that KDM3A regulates YAP1 transcription through removal of H3K9me2.
Regulation of hippo target genes by KDM3A independent of YAP1
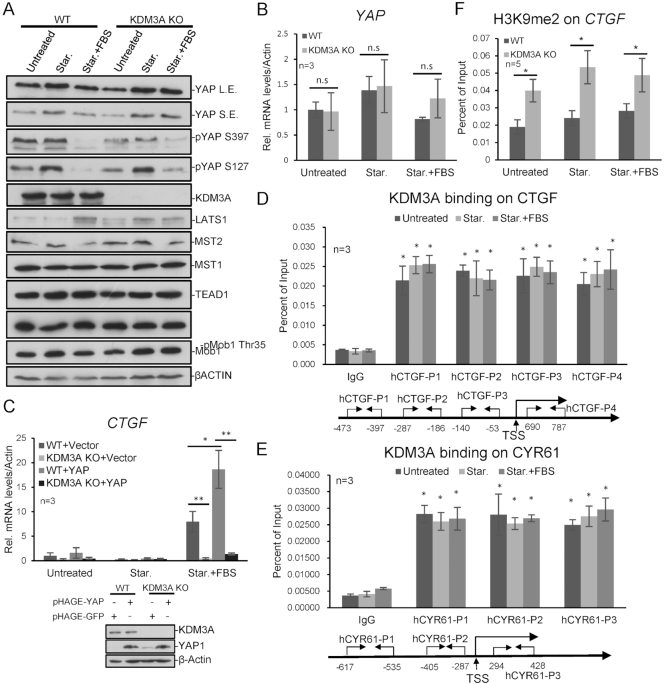
The above data suggest that KDM3A promotes the activation of hippo target genes through modulating YAP1 transcription. Surprisingly, when we analyzed the level of hippo pathway proteins in KDM3A−/− cell line, we found a clone that KDM3A knockout did not down-regulate YAP1 level (Figures 1E, 3A and B). We examined other KDM3A knockout clones and observed similar results (data not shown). We deduce that YAP1 expression may be critical for cell survival, and during cell line selection, cells altered the chromatin environment and kept YAP1 expression in the absence of KDM3A for survival.
Figure 3.
KDM3A directly regulates the expression of Hippo signaling downstream genes. (A) KDM3A WT or KO cell lines were starved for 24 h and then treated with FBS for 1 h. Cell lysates were immunoblotted with indicated antibodies. (B) YAP1 mRNA level was determined in KDM3A WT and KO cell lines with quantitative RT-PCR. (C) YAP1 was exogenous expressed in KDM3A WT and KO cell lines. KDM3A depletion represses CTGF expression induced by YAP1. (D, E) KDM3A ChIP analysis in HCT116 shows KDM3A binding on hippo target genes CTGF (D) and CYR61 (E). The primer sets for qPCR are indicated at the bottom. (F) H3K9me2 ChIP analysis in KDM3A WT or KO cell line shows the enrichment of H3K9me2 on CTGF, relative to IgG, with primer set p3. * means P-value ≤ 0.05, ** for P-value ≤ 0.01. The results in all experiments represent the means (±SD) of at least three independent experiments.
Interestingly, in the above KDM3A−/− cells, FBS-induced CTGF and CYR61 expression was still greatly attenuated (Figure 1C, E and 3C). While exogenous expressing YAP1 in the cells, KDM3A depletion also impaired YAP1-induced CTGF expression (Figure 3C). YAP1 exogenous expression activated the transcription regardless the endogenous YAP1 level, which should overwhelm YAP1 down-regulation caused by KDM3A deficiency, however, KDM3A deficiency still greatly inhibited CTGF expression, suggesting KDM3A maybe also functions downstream of YAP1. We guessed that KDM3A might directly regulate hippo target genes and performed KDM3A ChIP assay to verify it. As expected, KDM3A directly binds to CTGF and CYR61 on chromatin (Figure 3D and E), and consistently, H3K9me2 on CTGF and CYR61 significantly increased in both KDM3A−/− and knockdown cells, but not on GAPDH gene (Figure 3FSupplementary Figure S3).
H3K27ac is critical for regulation of hippo target genes
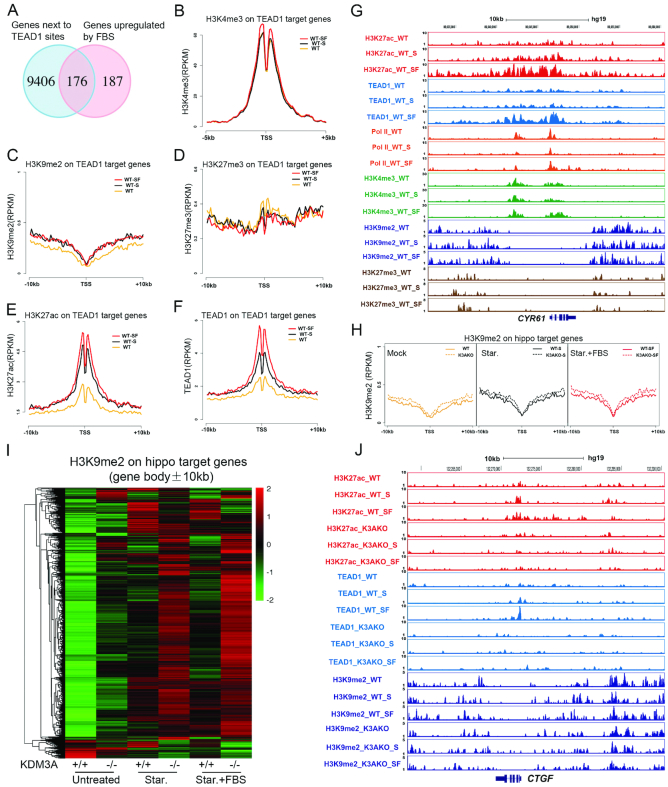
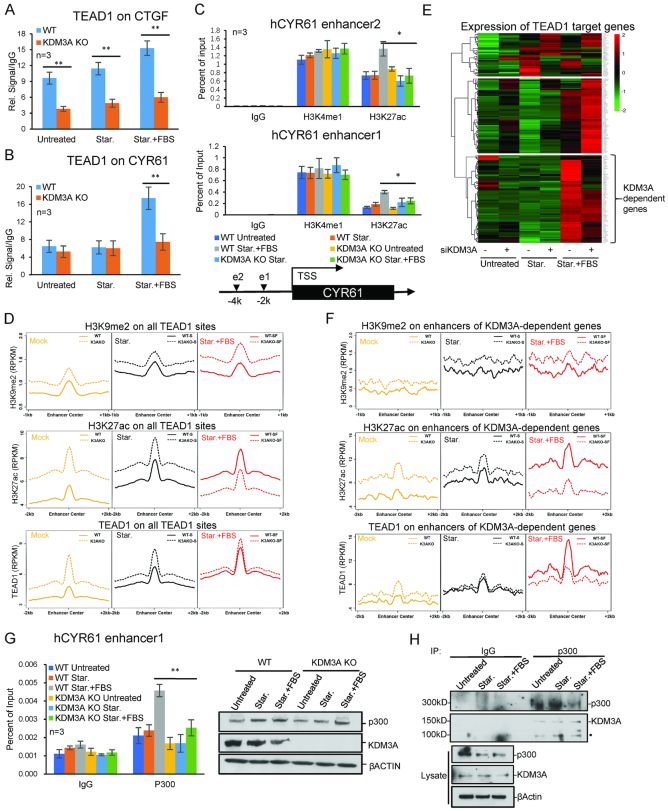
To further investigate the epigenetic regulation of hippo target genes, we performed ChIP-Seq with TEAD1 and several histone modifications. Totally, we identified 9582 TEAD1 binding sites across the whole genome, and their closest genes were considered as potential target genes (Figure 4A). 363 up-regulated genes induced by FBS (≥2-fold) were identified in the previous RNA-Seq study (Figure 4A), and 176 overlapped genes were identified as hippo target genes upon FBS treatment after starvation in HCT116 (Figure 4A and Supplementary Table S1). Then the enrichment of histone modifications on these 176 genes were calculated. H3K4me3 at transcription start sites (TSS) is a hallmark of transcription activation (39). Interestingly, the average H3K4me3 level on the TSS of hippo target genes only increase a very tiny level after FBS treatment (Figure 4B). Two repressive marks, H3K9me2 and H3K27me3 did not change obviously either (Figure 4C and D). Only H3K27ac increased significantly after starvation and further increased upon FBS addition (Figure 4E). The binding of TEAD1 around TSS showed a similar pattern as H3K27ac (Figure 4F). It seems that H3K27ac is more relevant with gene activation than other assayed histone modifications.
Figure 4.
Dynamic change of histone modifications on Hippo target genes. (A) Venn diagram shows the overlapped genes between the TEAD1 related genes and FBS upregulated DEGs (≥2-fold) in HCT116. (B–F) The average signals of H3K4me3 (B), H3K9me2 (C), H3K27me3 (D), H3K27ac (E) and TEAD1 (F) enrichment on TEAD1 target genes. (G) The UCSC browser view shows H3K27ac, TEAD1, POL2, H3K4me3, H3K9me2 and H3K27me3 enrichment around CYR61. (H) Heat maps show the enrichment of H3K9me2 (RPKM) on hippo target genes (gene body ±10 kb) KDM3A WT and KO cells. (I) The average H3K9me2 levels around the TSS of TEAD1 target genes (TSS ± 10 kb) in KDM3A WT and KO cell lines. (J) The UCSC browser view shows H3K27ac, TEAD1 and H3K9me2 enrichment around CTGF. The sequencing data were obtained from two biological replicates.
RNA Pol II accumulates on the promoters of hippo target genes before FBS treatment
Further analysis indicated that under normal and starvation conditions, RNA Pol II seemed to accumulate on the promoters of hippo target genes, and then released after FBS addition (Figure 4G, Supplementary Figure S4A and B), suggesting RNA Pol II is perhaps paused on these promoters before FBS treatment. Beside these, TEAD1 and active histone modifications are present on the genes in the normal and starvation conditions (Figure 4G, Supplementary Figure S4A and B). Our results are consistent with the previous report (10), and suggest that the transcription machines, including RNA Pol II, TEAD1 and histone modifications, are pre-assembled on hippo target genes.
Repression of gene expression by activated TEAD1
From the above analysis, we found a group of genes repressed by FBS (Supplementary Figure S4C–F). Among these, several genes related with cell cycle arrest or apoptosis were identified, such as CDKN1B and BBC3/PUMA (Supplementary Figure S4E). Since serum starvation arrests cell cycle and FBS re-initiates it, it is reasonable that FBS represses the expression of cell cycle inhibitor and apoptotic genes. Interestingly, we found that some of them are TEAD1 direct targets, such as BBC3/PUMA (Supplementary Figure S4F). BBC3/PUMA is a well-known apoptotic gene downstream of p53. It will be interesting to understand the underlying mechanism of transcription repression by TEAD1.
Increase of H3K9me2 on hippo target genes with KDM3A deficiency
Then we studied the role of KDM3A-dependent H3K9me2 for hippo target genes. In the KDM3A−/− cells, H3K9me2 level increase on the identified hippo target genes, at a relative small level; as comparison, on the whole genome, KDM3A depletion did not cause significant H3K9me2 increase (Figure 4H, Supplementary Figure S5A–D). We noticed that H3K9me2 level on the gene bodies of hippo target genes is lower than the surrounding chromatin (Figure 4G, Supplementary Figure S4A and B). We deduced that the surrounding chromatin might also be important and added up all the H3K9me2 signals on gene bodies, upstream and downstream 10 kb regions. In the normal condition, H3K9me2 on these genes was at very low level, and after serum starvation and FBS induction, H3K9me2 slightly increased on a portion of genes (Figure 4H and I). While KDM3A was knocked out, H3K9me2 increased on some genes; and under starvation and FBS conditions, KDM3A depletion caused H3K9me2 increase on most of the hippo target genes (Figure 4I, Supplementary Figure S4E). These results indicate that KDM3A is critical and specific for removal of H3K9me2 on hippo target genes during activation.
KDM3A promotes TEAD1 binding to target genes
From TEAD1 ChIP-Seq data, we found that TEAD1 binds the sites very close to TSS of both CTGF and CYR61. Then we analyzed the enrichment of H3K27ac and TEAD1 around their TSS after KDM3A removal. H3K27ac level was largely impaired on CTGF with KDM3A depletion (Figure 4J); and TEAD1 almost completely disappeared under all three conditions (Figures 4J and 5A). For CYR61, at the normal and starvation conditions, KDM3A depletion did not affect H3K27ac and TEAD1 obviously; but upon FBS activation, the bound H3K27ac and TEAD1 were much less in KDM3A−/− cells (Figure 5B, Supplementary Figure S5A–C). GAPDH was used as a negative control here (Supplementary Figure S5A). These suggest that KDM3A is critical for TEAD1 recruitment and H3K27ac level on hippo target genes.
Figure 5.
KDM3A regulates H3K27ac and TEAD1 binding on enhancers. (A, B) ChIP analysis shows TEAD1 enrichment in KDM3A WT and KO cells lines. KDM3A depletion attenuates TEAD1 recruitment to CTGF under all three conditions (A), while on CYR61 only during FBS treatment (B). (C) ChIP analysis shows H3K4me1 and H3K27ac enrichment on CYR61 enhancers in KDM3A WT and KO cells lines. The locations of two enhancers are shown at the bottom. (D) The average H3K9me2, H3K27ac and TEAD1 enrichment on all TEAD1 target sites. (E) Heat map shows the expression (FPKM) and clustering of TEAD1 target genes. (F) The average H3K9me2, H3K27ac and TEAD1 enrichment on enhancers of KDM3A-dependent genes. (G) ChIP assays to show p300 enrichment on CYR61 enhancer under starvation and FBS treatment. (H) Co-immunoprecipitation of p300 and KDM3A in HCT116 cells under starvation and FBS treatment. • labelled non-specific bands. * means P-value ≤ 0.05, ** for P-value ≤ 0.01. The results in all histograms represent the means (±SD) of at least three independent experiments. The sequencing data were obtained from two biological replicates.
KDM3A is required for enhancer activation of hippo target genes
Since H3K9me2 is relative low on the gene body (Figure 4G and Supplementary Figure S4A) and critical for TEAD1 recruitment, we deduced KDM3A might regulate enhancer activity. We analyzed the TEAD1 and H3K27ac ChIP-seq data and predicted the potential enhancers for CTGF and CYR61. The results of ChIP-PCR showed that KDM3A depletion caused down-regulation of TEAD1 on the identified enhancers of CYR61 and CTGF (Figure 5A and B). Based on TEAD1 and H3K27ac ChIP-Seq data, we also identified distal enhancers for CYR61 and CTGF. H3K4me1 and H3K27ac are two hallmarks for transcription enhancers; interestingly, KDM3A depletion only impaired the level of H3K27ac but not H3K4me1 (Figure 5C, Supplementary Figure S6A–D). If we knocked down YAP1 or TEAD1, the modifications on enhancers did not change significantly (Supplementary Figure S7A–D).
We then utilized the ChIP-Seq data and studied the effect of KDM3A on the activation of all TEAD1-binding enhancers. After KDM3A removal, the up-regulation of H3K9me2 on TEAD1-binding enhancers is much higher than that on hippo target genes (Figures 4I and 5D, Supplementary Figure S7G). However, the change of TEAD1 and H3K27ac on hippo target genes did not follow the similar trend as H3K9me2. We deduced it is maybe due to the selective regulation of enhancers by KDM3A, and we then clustered hippo target genes and identified a group of KDM3A-dependent genes, which showed impaired FBS-activation with KDM3A deficiency (Figure 5E). We then identified and analyzed their related enhancers. The results clearly showed that during FBS activation, the increased levels of H3K27ac and TEAD1 on their enhancers both decreased in KDM3A−/− cells (Figure 5F).
KDM3A is associated with p300 and required for its recruitment
p300 is the enzyme catalyzing H3K27ac on enhancers. To assess the relationship between KDM3A and p300, ChIP assays were performed and indicated that KDM3A depletion caused less p300 recruited to CTGF and CYR61 enhancers, but the global p300 level was not affected (Figure 5G, Supplementary Figure S5F). Co-immunoprecipitation assay indicates p300 interacts with KDM3A in the above cells (Figure 5H). These data suggest that KDM3A regulates the acetylation of enhancers not only through histone modifications, but also via interaction with p300 and regulating its recruitment.
KDM3A and YAP1 promote tumor formation of colon cancer cells
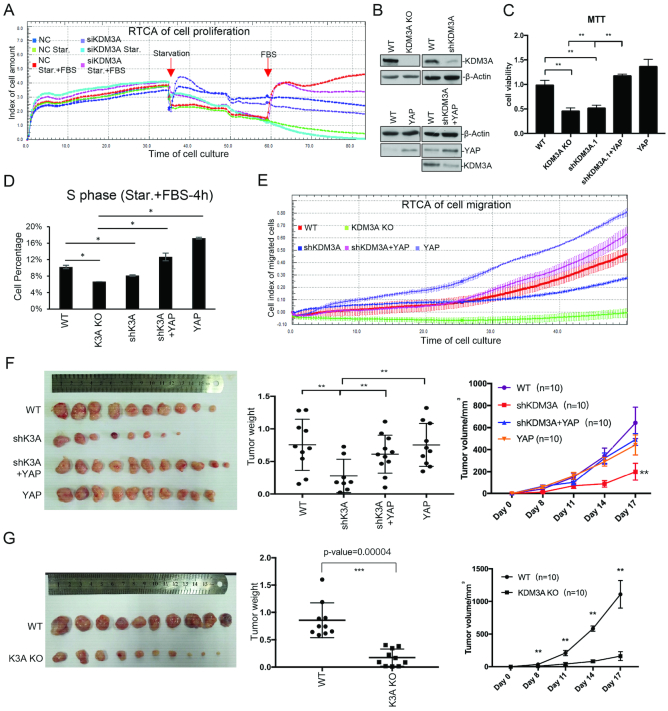
Since proper regulation of hippo pathway is critical for cell growth, development and tumorigenesis, we investigated the function of KDM3A and YAP1 in regulating the proliferation and cell cycle of colon cancer cells. We knocked down KDM3A with siRNA in HCT116 and studied cell proliferation via Real time cell analysis (RTCA). The curves represent the trends of cell growth. Both control and KDM3A knockdown cells grew with similar trends during normal culture and serum starvation. When FBS was added back to the medium and cells start growing again, the KDM3A knockdown cells obviously grew slower than wild type (Figure 6A). We then generated KDM3A stable knockdown cell lines through infection with viral-packaged shRNA, and subsequently stably expressed YAP1 in them (Figure 6B). MTT assay showed that KDM3A knockout or knockdown slowed down cell proliferation, while YAP1 expression rescued it (Figure 6C). Serum starvation can arrest cell cycle and FBS addition causes cell cycle re-entry. We labelled cells with PI and performed flow cytometry. We found that KDM3A knockdown or knockout decreased the percentage of S phase cells after FBS addition, while YAP1 expression increased it (Figure 6D, Supplementary Figure S8A). Cell migration assay determined by RTCA instrument showed that KDM3A knockdown or knockout also caused slower migration, which was rescued by YAP1 expression (Figure 6E).
Figure 6.
KDM3A promotes colon cancer cell proliferation and migration through hippo pathway. (A) KDM3A was knocked down HCT116 via siRNA and cells were seeded at a density of 4000 cells/well in xCELLigence plates. Cell index (cell proliferation) was monitored with RTCA as the manufacturer's protocol. Cells were serum starved 36 h after plating and FBS was added 24 h later. The impedance signals were recorded every 15 min over a period of up to 80 h. (B) Western analysis of wild type, shKDM3A, KDM3A KO, YAP1 overexpression and shKDM3A with YAP1 overexpression stable cell lines. (C) MTT assay to show cell proliferation of the cell lines in (B). (D) The above cell lines were starved and treated with FBS. BrdU and PI were used to label cells before cell percentage was assayed with flow cytometry. The percentage of S phase was shown. (E) Cell migration of the above cell lines was assayed with RTCA according to the manufacturer's protocol. (F, G) The above cell lines were injected into nude mice (n = 10). The image of tumors (left), tumor weight (middle) and tumor growth curve (right) were shown. (G) The KDM3A knockout and control cell lines were injected into nude mice (n = 10). The image of tumors (left), tumor weight (middle) and tumor growth curve (right) were shown. * means P-value ≤ 0.05, ** for P-value ≤ 0.01 and *** for P-value ≤ 0.001.
To evaluate the role of KDM3A-regulated hippo pathway in tumorigenesis, we injected the above cells into nude mice. The results suggested KDM3A knockdown or knockout in HCT116 significantly decreased the weight and volume of tumors, while YAP1 expression in KDM3A knockdown cells rescued the phenotype (Figure 6F and G).
KDM3A regulates hippo pathway in colorectal cancer patients
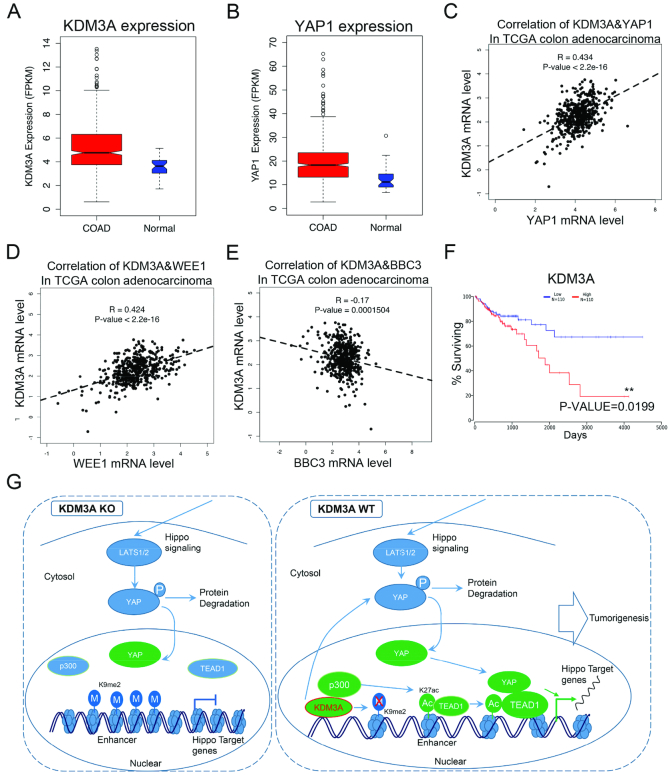
To further study the importance of KDM3A-regulated hippo pathway in colorectal cancer, we utilized TCGA data and found that both KDM3A and YAP1 are highly expressed in colon cancer tissues compared with the adjacent tissues (Figure 7A and B). The co-expression analysis showed that KDM3A mRNA level is nicely correlated with YAP1 (P value < 2.2e–16), which is consistent with our previous results (Figure 7C). WEE1, IFRD1, RND3, PARD6B, ELSMAN1 and UGDH-AS2 are hippo target genes (Supplementary Table S1). We found that in colon cancer tissues, their mRNA levels are correlated with KDM3A (Figure 7D, Supplementary Figure S8B–F). BBC3 is an apoptotic gene repressed after TEAD1 activation (Supplementary Figure S5 and Supplementary Table S2). The expression of KDM3A and BBC3 is negatively correlated (Figure 7E). These suggest KDM3A regulates the expression of hippo target genes in colorectal cancer tissues. Interestingly, the survival rates of KDM3A high-expressed and low-expressed patients showed significant difference, but not YAP1, CTGF or CYR61 (Figure 7F, Supplementary Figure S8G–I), suggesting KDM3A may serve as a potential prognosis marker for colon cancer.
Figure 7.
KDM3A promotes colon cancer through hippo pathway. (A, B) The data sets of colorectal cancer were downloaded from TCGA database. Boxplots show the expression (FPKM) of KDM3A (A) and YAP1 (B) in cancer and normal tissues. (C–E) The correlation of KDM3A with YAP1(C) and hippo target genes, WEE1 (D) and BBC3 (E), in colon adenocarcinoma. (F) The patient survival curves with high- or low-expressed KDM3A in colon adenocarcinoma. (G) A sketch to illustrate the role of KDM3A in hippo pathway and enhancer activation. All the above cancer-related data were downloaded from TCGA.
DISCUSSION
TEAD family members and YAP1 are key proteins for the activation of hippo target genes. TEADs keep binding to chromatin and the nuclear translocation of YAP1 is the key step for TEAD1 activation and gene expression. However, the molecular events on chromatin controlling the target gene expression are not clear. In the current study, we identified KDM3A as a critical regulator for the activation of hippo target genes. KDM3A functions through two mechanisms. One is to control the transcription of YAP1, and the other is to promote enhancer activation and help TEAD1 to bind its enhancers (Figure 7G).
Upon starvation and FBS treatment, hippo target genes are turned on and off very rapidly, and the chromatin environment surrounding these genes has well prepared for it. Under normal and starvation conditions, when hippo pathway is active, the transcription machinery is pre-assembled, and TEAD1 transcription factor, RNA Pol II and the activation mark, H3K4me3, are highly enriched on the genes, but transcription is poised. When FBS is added and hippo pathway is inhibited, YAP1 translocates into nuclear and transcription of hippo target genes is activated. Then more H3K27ac and TEAD1 presents on enhancers and promoters, but H3K4me3 does not increase obviously. KDM3A is critical for transcription activation. Its deficiency causes increase of H3K9me2 on enhancers of KDM3A-dependent genes, such as CYR61, and the subsequent reduction of H3K27ac, as well as p300 and TEAD1 recruitment. We have performed immunoprecipitation assay to check whether KDM3A interacts with TEAD1 or YAP1. We repeated the experiment for many times, but did not get positive results (data not shown), suggesting KDM3A recruitment to chromatin is not dependent on TEAD1, YAP1 or upstream hippo signaling. In fact, our data showed that KDM3A interacts with p300 even in normal tissue culture condition. The result is consistent with the observations that TEAD1 and KDM3A always bind to chromatin regardless hippo pathway is activated or not. Our data together support a model that KDM3A and p300 sit on enhancers regardless of hippo pathway activation, which is required for TEAD1 binding to chromatin. When hippo pathway is repressed and YAP1 translocates into nuclear, brings more amount of p300, leads to H3K27ac elevation and activates expression of target genes.
KDM3A was reported to promote tumorigenesis through regulating the expression of oncogenes in other cancer types, such as MYC. MYC is also a hippo target genes and our discovery fits the previous studies (25,29). It is possible that KDM3A regulates MYC also through hippo pathway. One of the questions not solved is how KDM3A selectively regulates its target genes in cancer cells. KDM3A depletion does not affect the global change of H3K9me2, but only on a group of genes (40,41). In mammalian cells, multiple H3K9 methylases and demethylases exist and each of them may play different functions. In the future, it needs to be determined how these enzymes are recruited to different genes and selectively regulate H3K9 methylation dynamics on chromatin.
H3K9me2 is one of the first-identified repressive histone modifications. It inhibits transcription mainly through compacting chromatin and crosstalk with other repressive marks (24). Our study indicates that KDM3A deficiency caused increase of H3K9me2, decrease of both H3K27ac and TEAD1 on enhancers and genes. Nevertheless, H3K9me2 is relatively lower on gene bodies, and higher on enhancers of hippo target genes. KDM3A probably regulates the expression of hippo target genes mainly through enhancer activation. H3K4me1 and H3K27ac are hallmarks for active enhancers, while H3K4me1 for enhancer priming (13,14). KDM3A deficiency caused the decrease of H3K27ac but not H3K4me1, suggesting KDM3A and H3K9me2 are important for enhancer activation during FBS induction, but not enhancer priming. Interestingly, in the absence of KDM3A, the average levels of H3K27ac and TEAD1 increased on enhancers under normal and starvation conditions (Figure 5D and F), suggesting at least some enhancers gained more H3K27ac and TEAD1 during transcription poising status along with H3K9me2 increase. It is possible that the cells increased the level of positive marks to balance H3K9me2 inhibitory mark, in order to maintain the balance among pre-assembled transcription machine. However, not all the enhancers gained these, for example, H3K27ac and TEAD1 on CTGF significantly lost along with KDM3A knockout in all three conditions.
YAP1 is critical for proper intestinal regeneration and stem cell proliferation (42). Meanwhile, it may become an oncogene for colorectal cancer (43). We found that the regulation of hippo target genes by KDM3A is critical for tumorigenesis of colorectal cancer. KDM3A deficiency impairs tumor formation while YAP1 expression rescues it. Both KDM3A and YAP1 are highly expressed in colorectal cancer. KDM3A expression is correlated with YAP1, WEE1, IFRD1, RND3, PARD6B, ELMSAN1 and UGDH-AS1, but negatively correlated with BBC3. Among these, WEE1 is a regulator of cell cycle; IFRD1, RND3 and PARD6B are involved in cancer; and BBC3 is an apoptotic gene (44–48). These suggest that KDM3A probably promotes colon cancer through activating their expression. KDM3A is also a potential prognosis marker for colorectal cancer, but not YAP1, CTGF or CYR61. The previous study reported that YAP1 is critical for the function of intestinal epithelial stem cell in mice. We found that YAP1 is highly expressed in the crypt cells of normal human intestinal tissues (Supplementary Figure S7F). The high level of YAP1 in crypt cells is probably the reason that YAP1 and hippo target genes are not good prognosis marks.
Our study showed the relationship between H3K9me2 and the expression of hippo target genes. Not only KDM3A regulates H3K9me2 on their enhancers, treatment of H3K9me2 inhibitors could further activated their expression. These drugs will then be useful to the future studies about hippo pathway. Moreover, KDM3A may serve as a novel drug target in colorectal cancer.
To sum up, in this study we demonstrate the importance of the dynamic H3K9me2 in regulating the activation of hippo target genes. Removal of H3K9me2 by KDM3A is critical for YAP1 expression, enhancer activation and TEAD1 recruitment of hippo target genes. These are critical mechanisms for tumorigenesis of colorectal cancer.
DATA AVAILABILITY
All the NGS sequencing data can be accessed from GEO database (GSE108922).
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: W.H.Y., T.S.B., X.Q. and Z.Q.Y. performed the experiments; L.Q.Y. and L.Q.L. did bioinformatics analysis; Y.M. contributes in the IHC analysis; Z.L. and Y.M. provided critical ideas and comments for the study; W.M. and L.L.Y. provided funding and directed the project; W.M. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2016YFA0502100]; National Natural Science Foundation of China to Min Wu [31470771 and 31771503]; Lian-Yun Li [31670874 and 31521091]; Science and Technology Department of Hubei Province, China [CXZD2017000188]. Funding for open access charge: Ministry of Science and Technology of China [2016YFA0502100].
Conflict of interest statement. None declared.
REFERENCES
- 1. Zanconato F., Cordenonsi M., Piccolo S.. YAP/TAZ at the roots of cancer. Cancer Cell. 2016; 29:783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meng Z., Moroishi T., Guan K.L.. Mechanisms of Hippo pathway regulation. Genes Dev. 2016; 30:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu F.X., Zhao B., Guan K.L.. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015; 163:811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Q., Meng F., Chen S., Plouffe S.W., Wu S., Liu S., Li X., Zhou R., Wang J., Zhao B. et al.. Hippo signalling governs cytosolic nucleic acid sensing through YAP/TAZ-mediated TBK1 blockade. Nat. Cell Biol. 2017; 19:362–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansen C.G., Moroishi T., Guan K.L.. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 2015; 25:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu A.M., Wong K.F., Jiang X., Qiao Y., Luk J.M.. Regulators of mammalian Hippo pathway in cancer. Biochim. Biophys. Acta. 2012; 1826:357–364. [DOI] [PubMed] [Google Scholar]
- 7. Jiao S., Li C., Hao Q., Miao H., Zhang L., Li L., Zhou Z.. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat. Commun. 2017; 8:14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang W., Gao Y., Li P., Shi Z., Guo T., Li F., Han X., Feng Y., Zheng C., Wang Z. et al.. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014; 24:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koontz L.M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S., Pan D.. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell. 2013; 25:388–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M. et al.. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008; 22:1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cebola I., Rodriguez-Segui S.A., Cho C.H., Bessa J., Rovira M., Luengo M., Chhatriwala M., Berry A., Ponsa-Cobas J., Maestro M.A. et al.. TEAD and YAP regulate the enhancer network of human embryonic pancreatic progenitors. Nat. Cell Biol. 2015; 17:615–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calo E., Wysocka J.. Modification of enhancer chromatin: what, how, and why. Mol. Cell. 2013; 49:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nizovtseva E.V., Todolli S., Olson W.K., Studitsky V.M.. Towards quantitative analysis of gene regulation by enhancers. Epigenomics. 2017; 9:1219–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 15. Wang C., Lee J.E., Lai B., Macfarlan T.S., Xu S., Zhuang L., Liu C., Peng W., Ge K.. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11871–11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu D., Gao X., Morgan M.A., Herz H.M., Smith E.R., Shilatifard A.. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol. Cell Biol. 2013; 33:4745–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herz H.M., Mohan M., Garruss A.S., Liang K., Takahashi Y.H., Mickey K., Voets O., Verrijzer C.P., Shilatifard A.. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012; 26:2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan J., Jiang Y.Y., Mayakonda A., Huang M., Ding L.W., Lin H., Yu F., Lu Y., Loh T.K.S., Chow M. et al.. Super-Enhancers promote transcriptional dysregulation in nasopharyngeal carcinoma. Cancer Res. 2017; 77:6614–6626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roe J.S., Hwang C.I., Somerville T.D.D., Milazzo J.P., Lee E.J., Da Silva B., Maiorino L., Tiriac H., Young C.M., Miyabayashi K. et al.. Enhancer reprogramming promotes pancreatic cancer metastasis. Cell. 2017; 170:875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M., Park T.J., Deaville R., Erichsen J.T., Jasinska A.J. et al.. Enhancer evolution across 20 mammalian species. Cell. 2015; 160:554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pott S., Lieb J.D.. What are super-enhancers. Nat. Genet. 2015; 47:8–12. [DOI] [PubMed] [Google Scholar]
- 23. Suganuma T., Workman J.L.. Crosstalk among histone modifications. Cell. 2008; 135:604–607. [DOI] [PubMed] [Google Scholar]
- 24. Shinkai Y., Tachibana M.. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011; 25:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao Q.Y., Lei P.J., Zhang X., Zheng J.Y., Wang H.Y., Zhao J., Li Y.M., Ye M., Li L., Wei G. et al.. Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model. Clin. Epigenet. 2016; 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Swami M. Epigenetics: Demethylation links cell fate and cancer. Nat. Rev. Genet. 2010; 11:749. [DOI] [PubMed] [Google Scholar]
- 27. He Y., Korboukh I., Jin J., Huang J.. Targeting protein lysine methylation and demethylation in cancers. Acta Biochim. Biophys. Sin. (Shanghai). 2012; 44:70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krieg A.J., Rankin E.B., Chan D., Razorenova O., Fernandez S., Giaccia A.J.. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol. Cell Biol. 2010; 30:344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wade M.A., Jones D., Wilson L., Stockley J., Coffey K., Robson C.N., Gaughan L.. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2015; 43:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johmura Y., Sun J., Kitagawa K., Nakanishi K., Kuno T., Naiki-Ito A., Sawada Y., Miyamoto T., Okabe A., Aburatani H. et al.. SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat. Commun. 2016; 7:10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Black J.C., Manning A.L., Van Rechem C., Kim J., Ladd B., Cho J., Pineda C.M., Murphy N., Daniels D.L., Montagna C. et al.. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013; 154:541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ju L.G., Zhu Y., Long Q.Y., Li X.J., Lin X., Tang S.B., Yin L., Xiao Y., Wang X.H., Li L. et al.. SPOP suppresses prostate cancer through regulation of CYCLIN E1 stability. Cell Death Differ. 2018; doi:10.1038/s41418-018-0198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhu K., Lei P.J., Ju L.G., Wang X., Huang K., Yang B., Shao C., Zhu Y., Wei G., Fu X.D. et al.. SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res. 2017; 45:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 35. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng M.B., Zhang Y., Cao C.Y., Zhang W.L., Zhang Y., Shen Y.F.. Specific phosphorylation of histone demethylase KDM3A determines target gene expression in response to heat shock. PLoS Biol. 2014; 12:e1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan J.D., Lei P.J., Zheng J.Y., Wang X., Li S., Liu H., He Y.L., Wang Z.N., Wei G., Zhang X. et al.. The selective activation of p53 target genes regulated by SMYD2 in BIX-01294 induced autophagy-related cell death. PLoS One. 2015; 10:e0116782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu F., Barsyte-Lovejoy D., Allali-Hassani A., He Y., Herold J.M., Chen X., Yates C.M., Frye S.V., Brown P.J., Huang J. et al.. Optimization of cellular activity of G9a inhibitors 7-aminoalkoxy-quinazolines. J. Med. Chem. 2011; 54:6139–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006; 75:243–269. [DOI] [PubMed] [Google Scholar]
- 40. Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y.. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007; 450:119–123. [DOI] [PubMed] [Google Scholar]
- 41. Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y.. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006; 125:483–495. [DOI] [PubMed] [Google Scholar]
- 42. Staley B.K., Irvine K.D.. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol. 2010; 20:1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang Z., Liu P., Zhou X., Wang T., Feng X., Sun Y.P., Xiong Y., Yuan H.X., Guan K.L.. Endothelin promotes colorectal tumorigenesis by activating YAP/TAZ. Cancer Res. 2017; 77:2413–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mills C.C., Kolb E.A., Sampson V.B.. Recent advances of Cell-Cycle inhibitor therapies for pediatric cancer. Cancer Res. 2017; 77:6489–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lewis M.A., Sharabash N., Miao Z.F., Lyons L.N., Piccirillo J., Kallogjeri D., Schootman M., Mutch M., Yan Y., Levin M.S. et al.. Increased IFRD1 expression in human colon cancers predicts reduced patient survival. Dig. Dis. Sci. 2017; 62:3460–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paysan L., Piquet L., Saltel F., Moreau V.. Rnd3 in Cancer: A review of the evidence for tumor promoter or suppressor. Mol. Cancer Res. 2016; 14:1033–1044. [DOI] [PubMed] [Google Scholar]
- 47. Cunliffe H.E., Jiang Y., Fornace K.M., Yang F., Meltzer P.S.. PAR6B is required for tight junction formation and activated PKCzeta localization in breast cancer. Am. J. Cancer Res. 2012; 2:478–491. [PMC free article] [PubMed] [Google Scholar]
- 48. Yu J., Zhang L.. PUMA, a potent killer with or without p53. Oncogene. 2008; 27(Suppl. 1):S71–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the NGS sequencing data can be accessed from GEO database (GSE108922).