Abstract
Background
Triple-negative breast cancer (TNBC) remains difficult to be targeted. SET and cancerous inhibitor of protein phosphatase 2A (CIP2A) are intrinsic protein-interacting inhibitors of protein phosphatase 2A (PP2A) and frequently overexpressed in cancers, whereas reactivating PP2A activity has been postulated as an anti-cancer strategy. Here we explored this strategy in TNBC.
Methods
Data from The Cancer Genome Atlas (TCGA) database was analyzed. TNBC cell lines were used for in vitro studies. Cell viability was examined by MTT assay. The apoptotic cells were examined by flow cytometry and Western blot. A SET-PP2A protein-protein interaction antagonist TD19 was used to disrupt signal transduction. In vivo efficacy of TD19 was tested in MDA-MB-468-xenografted animal model.
Findings
TCGA data revealed upregulation of SET and CIP2A and positive correlation of these two gene expressions in TNBC tumors. Ectopic SET or CIP2A increased cell viability, migration, and invasion of TNBC cells. Notably ERK inhibition increased PP2A activity. ERK activation is known crucial for Elk-1 activity, a transcriptional factor regulating CIP2A expression, we hypothesized an oncogenic feedforward loop consisting of pERK/pElk-1/CIP2A/PP2A. This loop was validated by knockdown of PP2A and ectopic expression of Elk-1, showing reciprocal changes in loop members. In addition, ectopic expression of SET increased pAkt, pERK, pElk-1 and CIP2A expressions, suggesting a positive linkage between SET and CIP2A signaling. Moreover, TD19 disrupted this CIP2A-feedforward loop by restoring PP2A activity, demonstrating in vitro and in vivo anti-cancer activity. Mechanistically, TD19 downregulated CIP2A mRNA via inhibiting pERK-mediated Elk-1 nuclear translocation thereby decreased Elk-1 binding to the CIP2A promoter.
Interpretation
These findings suggested that a novel oncogenic CIP2A-feedforward loop contributes to TNBC progression and targeting SET to disrupt this oncogenic CIP2A loop showed therapeutic potential in TNBC.
Research in context.
Evidence before this study
Protein phosphatase 2A (PP2A), a serine/threonine phosphatase, functions as a tumor suppressor that regulates multiple oncogenic pathways such as inactivating pAkt and pERK. SET and CIP2A are intrinsic inhibitors of PP2A and frequently overexpressed in cancers. Restoring PP2A activity has been implicated as a potential anti-cancer strategy.
Added value of this study
We found upregulation of SET and CIP2A and positive correlation of these two gene expressions in triple-negative breast cancer (TNBC) tumors. Notably, ERK inhibition increased PP2A activity, reduced pElk-1 and CIP2A expression. We have identified a feedforward loop consisting of pERK/pElk-1/CIP2A/PP2A and that SET inhibition by a small molecule (TD19) can disrupt this CIP2A-feedforward loop by restoring PP2A activity. Moreover, this SET inhibitor enhanced cisplatin cytotoxicity in association with CIP2A-downregulation in TNBC cells.
Implications of all the available evidence
Our data have disclosed a novel oncogenic CIP2A-feedforward loop that contributes to TNBC progression which can be therapeutically targeted using TD19, a novel SET/PP2A protein-protein interaction inhibitor
Alt-text: Unlabelled Box
1. Introduction
Protein phosphatase 2A (PP2A) functions as a serine/threonine phosphatase that regulate multiple cellular signaling pathways such as inactivating pAkt and pERK through direct dephosphorylation [1]. PP2A has been implicated as an important tumor suppressor and its loss of function has been identified in several solid cancers including breast cancer [2,3]. Accordingly, PP2A controls the cell cycle as well as cell apoptosis [4]. Although loss of PP2A activity is crucial for tumor growth, mutations in PP2A subunits are very rare in breast cancers [5,6]. The trimeric form of PP2A consists of catalytic (PP2Ac), scaffold (PP2AA) and regulatory (PP2AB) subunits. Alterations in the A subunit that impair integration of the C and/or B subunits have only been observed in breast cancers at a low frequency [5], suggesting that other mechanisms can affect PP2A activity. Indeed, some cellular PP2A-interacting proteins, such as SET (I2PP2A, inhibitor 2 of PP2A) and cancerous inhibitor of PP2A (CIP2A), inhibit PP2A activity through direct interaction with PP2A [4].
Both SET and CIP2A have been shown to be up-regulated in a variety of cancers and their expression generally correlates with poor prognosis [[7], [8], [9]]. In breast cancer, SET and CIP2A have been shown frequently overexpressed. Knockdown of SET and CIP2A decreases tumorigenesis [9]. In particular, CIP2A levels were elevated in TNBC compared with non-TNBC and associated with high histological grade and lymph node metastasis [10]. CIP2A has been shown to interact directly with c-MYC and impair its degradation by inhibiting PP2A activity [11]. Previous studies have indicated CIP2A also suppresses PP2A-dependent dephosphorylation of pAkt (Ser473) [[12], [13], [14], [15]], and plays a determinant role in drug-induced apoptosis of several known and investigational anticancer agents, such as bortezomib, tamoxifen, erlotinib derivatives, natural compounds, and small molecules [7,14,[16], [17], [18], [19]], comprehensively reviewed by De et al. [18] and Soofiyani et al. [19]. In addition, CIP2A expression can be controlled by the transcription factor Elk-1 in TNBC cells [14,16].
In contrast, SET inhibits PP2A activity via binding to both N-terminus and C-terminus regions of PP2A [20]. Previous studies have reported that SET activates the transcription factor AP-1, downregulates Akt signaling, inhibits the DNase activity of NM23-H1 tumor-suppressor, or negatively regulates p53 acetylation result in its suppression [[21], [22], [23]]. Given the importance of PP2A inhibition in maintaining the activation c-Myc- and Akt-driven oncogenic survival signals, CIP2A and SET are attractive and potential therapeutic targets for cancer therapy. Collectively, restoring PP2A activity, such as by PP2A-activating drugs (for example a peptide drug OP449, and a sphingolipid analogue FTY720), has been implicated as a potential anti-cancer strategy [[24], [25], [26]].
In the present study, we found SET and CIP2A were upregulated in TNBC patients and the gene expression level of SET was positively correlated with that of CIP2A. SET- or CIP2A- overexpressing TNBC cells demonstrated increased cell viability, migration, and invasion. Moreover, we noted that ERK inhibition increased PP2A activity. Since that PP2A is known for dephosphorylating (inactivating) ERK [1], whereas ERK phosphorylation is also crucial for Elk-1 activation (phosphorylation) and nuclear translocation [[27], [28], [29]], therefore we hypothesized an oncogenic feedforward loop consisting of pERK/pElk-1/CIP2A/PP2A. We validated the loop by knockdown of PP2A and ectopic expression of Elk-1. In addition, ectopic expression of SET increased pAkt, pERK, pElk-1 and CIP2A expressions. We then used TD19, a novel SET/PP2A protein-protein interaction inhibitor that has been reported to induce cancer cell apoptosis by increasing PP2A activity [[30], [31], [32]], to decipher the potential relevance of SET and CIP2A in TNBC. We found that this SET antagonist TD19 exerted anti-tumor effects through inhibiting SET-PP2A interaction and CIP2A/PP2A/pAkt-mediated pathway. We concluded that a novel oncogenic CIP2A-feedforward loop contributes to TNBC progression and targeting SET to disrupt this oncogenic CIP2A loop showed therapeutic potential in TNBC.
2. Materials and methods
2.1. Reagents and antibodies
SET antagonist TD19 (EMQA), a derivative of erlotinib with chemical name of N4-(3-ethynylphenyl)-6,7-dimethoxy-N2-(4-phenoxyphenyl) quinazoline-2,4-diamine), was kindly provided from Dr. Chung-Wai Shiau's lab. The chemical structure and synthesis of TD19 has been disclosed in previous studies [31,33], and its mechanisms as a SET inhibitor has been also reported in prior studies [30,32]. Akt inhibitor (MK-2206) inhibitor and ERK inhibitor (SCH772984) were purchased from Selleckchem (Houston, TX, USA). TD19 at different concentrations were dissolved in dimethyl sulfoxide (DMSO) and added to cells in Dulbecco's Modified Eagle Medium (DMEM) containing 1% FBS. Antibodies used for Western blots of pElk-1 and Elk were obtained from Santa Cruz Biotechnology (San Diego, CA, USA), Other such as CIP2A, PARP, PP2Ac, Akt, pAkt (Ser473), beta-actin, caspase-3, ERK, pERK, pBcl2 (Ser70), Bcl2, pc-Myc (S62), c-Myc, Lamin B, Tubulin, GFP and Myc-tag were purchased from Cell Signaling (Danvers, MA, USA). Cycloheximide (CHX) was purchased from Sigma-Aldrich (St Louis, MO, USA). MTT was purchased from Sigma-Aldrich.
2.2. Cell culture
The MDA-MB-231, MDA-MB-453, MDA-MB-468, HCC1937, Hs 578 T, BT-20, MCF7 and MCF 10A cells were obtained from American Type Culture Collection (Manassas, VA). All cells were cultivated in DMEM (Gibco) with 10% FBS, 2-mM L-glutamine, 0.1-mM non-essential amino acids and 1% penicillin/streptomycin in a 5% CO2 atmosphere at 37 °C.
2.3. Migration and invasion
The migration and invasion assays were performed in 24-well plate for 12 and 20 h respectively. Cells (2 × 104) in 200 microliter of serum free medium were seeded onto upper Cell Culture Insert with 8 μm pores (Greiner Bio One) for migration assay. For invasion assay, the Cell Culture Insert was coated with Matrigel matrix (Corning) and air-dried the Matrigel layer. Rehydrate the dried Matrigel layer for 2 h by adding serum free medium for subsequent assay. Cells (4 × 104) in 200 microliter of serum free medium were seeded onto upper Cell Culture Insert with 8 μm pores. The lower chamber contained 900 microliter of complete medium. The cells migrated or invaded to the Cell Culture Insert membrane which were fixed with methanol for 10 min and stained with 0.005% crystal violet for 1 h. The numbers of migrated or invaded cells were counted under the microscope from 10 random fields.
2.4. Western blot
Cells treated with drugs at the indicated concentrations for different periods of time were collected and prepared for Western blot, and lysates were analyzed. The protocol of Western blot analysis was described as previously report [34].
2.5. Apoptosis analysis
Drugs-induced cell apoptosis were measured by sub-G1 analyzing of Propidium iodide (PI)-stained cells using flow cytometry and Western blot analyzing cleavage-PARP and caspase-3. For flow cytometric analyzing, cells were collected and washed with phosphate buffered saline (PBS), furthered by fixation of 100% ethanol at −20 °C for 24 h. After fixation, cells were removed from ethanol and washed with PBS. Cells were re-suspended in PBS containing 4 μg/milliliter PI and analyzed by flow cytometry (BD Bioscience).
2.6. Cell viability assay
Cells were seeded to 96-well plates for 24 h and treated with indicated concentration of TD19 and cisplatin for 72 h. The treated cells were added 0.5 mg/milliliter MTT to each well and incubated for 2 h at 37 °C. The violet precipitates were subsequently dissolved in 100 microliter of DMSO. The absorbance at 570 nm was measured on an UQuant reader.
2.7. PP2A activity
The phosphatase activity of PP2A were assessed using PP2A immunoprecipitation phosphatase assay kit (Millipore, Billerica, MA, USA) as previously describe [34]. Briefly, the whole-cell extracts or tumor homogenates were prepared in 20 mM imidazole HCl, 2 mM EDTA, 2 mM EGTA with proteases inhibitor (10 μg/milliliter each of aprotinin, leupeptin, pepstatin, 1 mM benzamidine, and 1 mM PMSF). And then, pNPP Ser/Thr Assay Buffer with anti-PP2Ac antibody and Protein A agarose slurry was added and incubated for 24 h at 4 °C. Protein phosphatase activity was determined by measuring the generation of free phosphate by adding 750 μM phosphopeptide substrate. After 10 min of incubation at 30 °C, the malachite green was added and determined by measuring optical density at 650 nm.
2.8. Transfection
Cells were transfected with Myc-tagged CIP2A (gene name KIAA1524), Elk-1-GFP, Myc-DDK-tagged-Human SET or pCMV-6 expression plasmids separately. Briefly, cells were seeded into 6-cm dish for 24 h and transfected with indicated plasmid using Lipofectamine 2000 (Thermo Fisher Scientific, CM, USA) following to manufacturer's instructions. After 48 h, cells were collected for Western blotting and sub-G1 analyzing. For small interfering RNA Gene knockdown, MDA-MB-468 cells were seeding 24 h and transfected with PP2Ac Smart-pool small interfering RNAs (siRNA) or control siRNA (final concentration of 50 nM) using DharmaFECT 1 Transfection Reagent (T-2001-01) according to manufacturer's instructions. After transfection 48 h, the transfected cells were treated with TD19 or DMSO for 48 h, cell were collected and assayed for Western blot and apoptosis analysis by flow cytometry. SiRNAs including the control (D-001810-10) and PP2Ac (L-003598-01) were purchased from Dharmacon (Chicago, IL, USA).
2.9. The construction for the CIP2A promoter into luciferase reporter plasmids
Briefly, reporter plasmids contain various lengths of CIP2A promoter from nucleotide −2000/−1, −1000/−1, −400/−1, −300/−1, −150/−1, −110/−1, and − 62/−1 were amplified by PCR from the genomic DNA of cells and cloned into the reporter Firefly vector (pGL4.17, Promega, Madison, WI, USA). The nucleotide sequence of the clones was verified by sequencing.
2.10. Luciferase assay
The promoter activity was determined by using the dual-luciferase reporter assay kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Briefly, MDA-MB-468 cells were co-transfected with the different length of CIP2A promoter luciferase reporter constructs and pGL4.74-renilla (Renilla luciferase reporter) as an indicator for normalization of transfection efficiency. After 24 h of post-transfection, cells were treated with TD19 (3 μM) or DMSO for 24 h. Cells were lysed and analyzed for dual-luciferase assay. The reporter assay was repeated three times in parallel for statistical analysis.
2.11. Nuclear cytoplasmic fractionation
Briefly, MDA-MB-468 cells were seeding for 24 h and treated with TD19 or DMSO for 24 h. Cells then collected and nuclear and cytoplasmic extracts were prepared using Nuclear and Cytoplasmic Extraction reagents (Thermo Fisher, CM, USA) according to the manufacturer's instructions. The cell fraction extracts were validated and quantified by Western blotting for markers that specifically express in each of fractions.
2.12. Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was performed using Pierce Agarose ChIP Kit (Thermo Fisher Scientific, CM, USA) according to the manufacturer's instruction. Briefly, MDA-MB-468 cells were treated with TD19 or DMSO for 24 h. After drug treatment, cells were washed with PBS and subsequently added 1% formaldehyde for fixing cross-linking between chromatin (DNA) and proteins at room temperature for 10 min. Cells were furthered lysed for DNA digestion by Micrococcal Nuclease at 37 °C for 15 min; Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific) were added in cell lysis step to avoid protein degradation. Lysates were purified by centrifugation at 12,500 ×g for 5 min at 4 °C. Immunoprecipitation was assayed by adding Elk-1, or rabbit IgG antibodies as negative control. The immunocomplex was precipitated by incubation with 25 microliter of protein A/G sepharose beads for 1 h at 4 °C. The protein/DNA complex was eluted using 200 microliter of elution buffer from the beads. Cross-linking of protein–DNA was disrupted by adding 8 microliter of 5 M NaCl at 95 °C for 15 min. The DNA was purified using spin columns and used in the PCR reaction for amplification of the CIP2A promoter region (−139 to −16 nt). Anti-RNA polymerase II antibody and GAPDH primers were provided by the manufacturer as a positive control for chromatin immunoprecipitation assay.
2.13. Combination index calculation
Cells were treated with TD19 (0.6–2.4 μM) and cisplatin (1.2–4.8 μM). The combination index (CI) was determined using the Chou and Talalay method and the software package CompuSyn (Biosoft, Ferguson, MO, USA). A CI value of <1 was defined as synergism.
2.14. Reverse transcription-PCR
MDA-MB-468 cells were treated with TD19 or DMSO for 24 h, total RNA was extracted from cells using TRIzol Reagent (Invitrogen, San Diego, CA, USA) and RT-PCR was performed following to the manufacturer's instructions (Thermo Fisher Scientific, CM, USA). RT-PCR products were validated using specific primers for human CIP2A (forward primer, 5′-TGGCAAGATTGACCTGGGATTTGGA-3′; reverse primer, 5′-AGGAGTAATCAAACGTGGGTCCTGA-3′), and using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene as an internal control (forward primer, 5′-CGACCACTTTGTCAAGCTCA-3′; reverse primer, 5′-AGGGGTCTACATGGCAACTG-3′).
2.15. Xenograft tumor growth
All animal experiment was conducted under an approved by Institutional Animal Care and Use Committee of Taipei Veterans General Hospital. Female BALB/c nu/nu mice (5–7 weeks of age) were purchased from the National Laboratory Animal Center (Taipei, Taiwan) and maintained in an SPF-environment. Each mouse was subcutaneously injected in the dorsal flank with 5 × 106 MDA-MB-468 cells suspended in 100 microliter of a 1:1 mixture of PBS and Matrigel (BD Biosciences, Bedford, MA, USA) under isoflurane anesthesia. The volume of tumor was measured using the standard formula width2 × length × 0.52. When tumors reached around 200 mm3, mice were orally administered of TD19 (10 mg/kg body weight) or control PBS as vehicle three times a week for around 6 weeks. All the mice were sacrificed on day 43, and the xenografted tumors were harvested and assayed for subsequent experiments.
2.16. Immunohistochemical staining
Immunohistochemical staining procedure has been described [13]. Briefly, primary antibodies against CIP2A (ab84547, Abcam, UK), pAkt (pAkt1/2/3 Thr 308, Santa Cruz, sc-16,646-R), and SET (A302-262A, Bethyl Laboratories) were used at 1:100, 1:200, and 1:500 dilution for overnight incubation, respectively. The slides were then counterstained with hematoxylin stain solution. Rabbit IgG was used as a control for antibody specificity. To detect the apoptosis, tumor tissue samples were stained the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end labeling (TUNEL) method with S7100 ApopTag® peroxidase in situ Apoptosis Detection Kit (Merck Millipore Corporation, Darmstadt, Germany), according to the manufacturer's instructions. Staining signals were detected using the EnVisionTM system (Dako, USA).
2.17. Determining histology score (H-score)
IHC stainings were assessed semiquantitatively by a H-score, ranging from 0 to 300, as defined by multiplying the percentage of positive-stained carcinoma cells (from 0 to 100) by the staining intensity (from 0 to 3) [35]. The intensity of staining was scored as negative (score 0), weak (score 1), moderate (score 2), and strong staining (score 3). The determination of H-score was performed by a pathologist (Dr. PY Chu) specialized in breast pathology independently with blinding of the clinical information of tumor specimen. The third quartile H-score of SET (score of 180) was selected for defining low versus high expressing tumors and clinical-pathological correlation.
2.18. TCGA data analysis
Patient tumor subtypes and gene expression data for breast cancers was accessed from TCGA public data portal (https://tcga-data.nci.nih.gov/tcga/). Level 3 released gene level expression data for expression data for RNAseq were downloaded. The data processing and quality control were conducted by Broad Institute's TCGA workgroup. The RNAseq gene expression level 3 data contains RNAseq by Expectation-Maximization (RSEM) [36]. RSEM values of CIP2A (gene name KIAA1524) and SET gene expression were retrieved and analyzed according to clinical factors. For survival analysis from the TCGA dataset of TNBC cases, RSEM z-scored values using z-score transformation [37] were retrieved and a cut-off value of 2 was defined for higher versus lower CIP2A expression.
2.19. Statistical analysis
Data are expressed as mean ± SD or SE. Statistical comparisons were based on nonparametric tests, and statistical significance was defined as a P value <0.05 by Student's t-test. For survival analysis, DFS curves of patients were generated by the Kaplan-Meier method and compared by Log-rank test. All statistical analyses were performed using SPSS for Windows software, version 12.0 (SPSS, Chicago, IL, USA).
3. Results
3.1. SET is upregulated in patients with TNBC and relates to poor survival
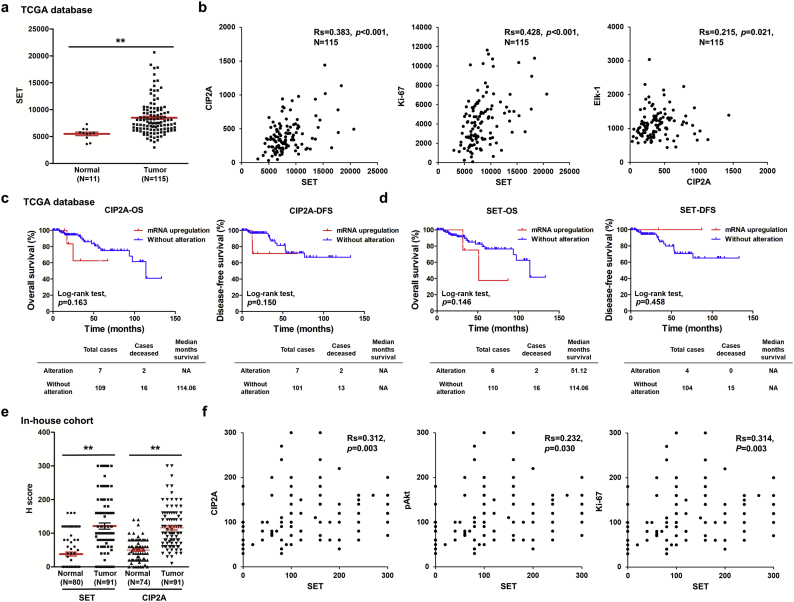
To examine the clinical significance of two cellular inhibitors of PP2A, SET and CIP2A, the data from The Cancer Genome Atlas (TCGA) database was analyzed. We found patients with TNBC showed increased CIP2A level in TCGA database previously [14]. The transcript level of SET was higher in TNBC tissues than normal tissues (Fig. 1a) and positive correlated with CIP2A and Ki-67 (Fig. 1b, left and middle). It has been reported that Elk-1 upregulated CIP2A expression by interacting with the proximal CIP2A promoter [38], we found the expression level of CIP2A slightly positive correlated with Elk-1 level (Fig. 1b, right). CIP2A gene alterations, including copy number variation and mRNA dysregulation, were linked to poor disease-free survival (DFS) but not overall survival (OS). SET gene alterations showed a trend toward poor OS (Supplementary Fig. 1a). Upregulation of mRNA is the highest alteration frequency of CIP2A and SET gene (Supplementary Fig. 1b). Moreover, samples with CIP2A or SET gene amplification or mRNA upregulation also harbored higher corresponding transcripts level compared with those without (Supplementary Fig. 1c). However, CIP2A or SET mRNA upregulations were not significantly associated with lower OS and DFS in TNBC patients (Fig. 1c). Further, we collected samples from 91 TNBC patients and performed IHC staining. The mean SET H-score was 121.2 (0−300) for TNBC tumor tissues compared with 38.0 (0–160) for normal tissues. The mean CIP2A H-score was 115.8 (10−300) for TNBC tumor tissues compared with 49.8 (0–140) for normal tissues. IHC stainings were assessed semiquantitatively by a H-score. The H-score of SET and CIP2A in TNBC tumor tissues were higher than normal tissues (Fig. 1e). The expression level of SET was positively correlated with CIP2A, pAkt and Ki-67 levels (Fig. 1f). To analyze the clinical significances of SET expression in TNBC, we divided patients into high and low expression SET subgroups based on the third quartile of H score of SET and analyzed for relationship of clinical variables. SET expressions did not correlate with age, stage, grade, tumor necrosis, lymphovascular invasion and CIP2A while associated with pAkt and Ki-67 (Supplementary Table 1). The in-house cohort exhibited high levels of SET and CIP2A show a trend toward lower DFS in TNBC (Supplementary Fig. 1d). These data indicated SET was upregulated in TNBC tissues and correlated with CIP2A and Ki-67.
Fig. 1.
SET and CIP2A were up-regulated in the TNBC tumor tissues and correlated with CIP2A and Ki-67. (a-b) Level 3 data of mRNA expressions from TNBC samples and normal tissue samples were downloaded from the TCGA and Broad GDAC Firehose data portal. The mRNA RSEM of all samples were selected and analyzed for comparing abundances by GraphPad Prism 5 software. The transcript levels of SET, CIP2A, Ki-67 and Elk-1 were measured by RNA sequencing in TCGA data. The transcript levels of SET in TNBC tumor samples and normal tissue samples were measured by RNA sequencing in TCGA data (a). The correlation of SET, CIP2A, Ki-67 and Elk-1 were analyzed. Rs, Spearman's rank correlation coefficient (b). (c-d) Overall survival (OS) and disease-free survival (DFS) curves were plotted for TNBC cases with or without alteration of SET and CIP2A expressions. (e-f) The tissue sections were quantitatively scored according to the percentage of positive cells and staining intensity. SET and CIP2A expression scores in TNBC tissue specimens and normal tissues (e). The relative protein levels of CIP2A, pAkt and Ki-67 in TNBC tumor samples were positively correlated with those of SET levels (f). Rs, Spearman's rank correlation coefficient. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.2. SET and CIP2A contribute to aggressiveness of TNBC cells
Data showed most of TNBC cell lines harbored higher levels of CIP2A and SET than epithelial cell MCF 10A and luminal-type MCF7 cells (Supplementary Fig. 2a). To evaluate the role of CIP2A and SET in TNBC progression, TNBC cell lines were transiently transfected with CIP2A-, SET-expressing or control plasmids and evidenced by Western blot analysis. Interestingly, the level of CIP2A also elevated in SET-expressing cells (Fig. 2a). Results showed that CIP2A and SET enhanced cell viability, migration and invasion abilities of MDA-MB-231 and MDA-MB-468 cells (Fig. 2b-d) while knockdown of CIP2A and SET suppressed these effects (Supplementary Fig. 2b-e). These data indicated CIP2A and SET could act as oncogenes of TNBC in vitro.
Fig. 2.
SET and CIP2A enhance cell viability and abilities of migration and invasion in TNBC cells. (a-d) MDA-MB-231 and MDA-MB-468 cells were transiently transfected with empty vector, CIP2A- or SET-expression plasmids for 48 h and evidenced by Western blot (a, left). The results were quantified (a, right). The transfected cells were assayed by MTT (b) migration (c) and invasion assays (d). Data are representative of three independent experiments. Columns, mean (N = 3); bars, SD. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.3. ERK inhibition suppresses CIP2A/PP2A axis
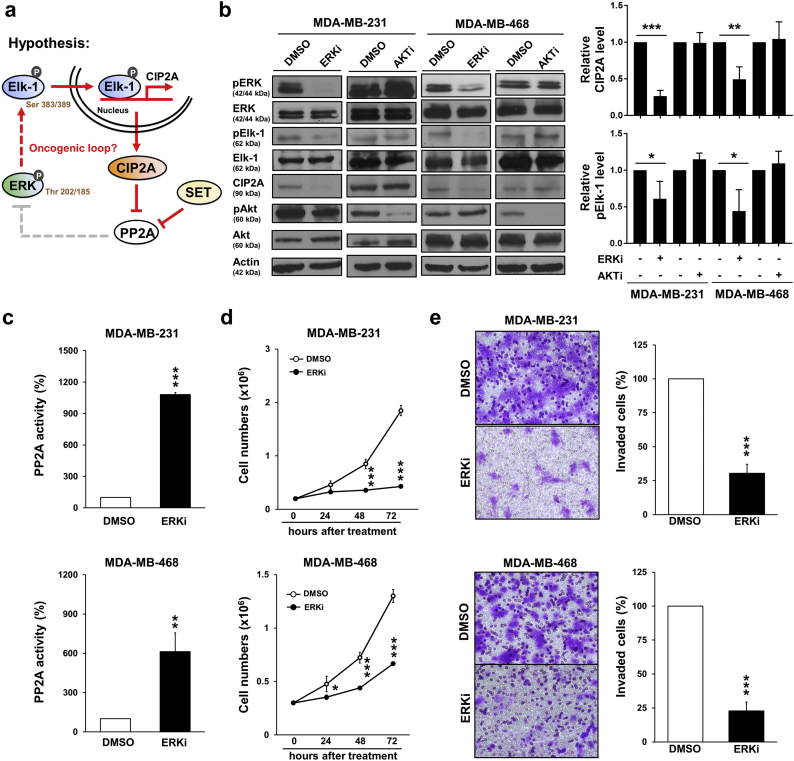
PP2A is known for dephosphorylating ERK [1], and ERK phosphorylation is crucial for Elk-1 activation [28]. We would like to explore whether ERK inhibition disrupted Elk-1/CIP2A/PP2A axis (Fig. 3a). ERK inhibition but not AKT inhibition reduced Elk-1 phosphorylation and CIP2A expression (Fig. 3b). Intriguingly, ERK inactivation significantly increased PP2A activity (Fig. 3c). Moreover, ERK inhibitor suppressed cell proliferation (Fig. 3d) which might partly result from cell apoptosis and cell cycle arrest (Supplementary Fig. 3a-b). The invasion ability of TNBC cells were also reduced by ERK inactivation (Fig. 3e). Taken together, these results hinted the presence of ERK/Elk-1/CIP2A/PP2A oncogenic loop.
Fig. 3.
Inhibition of ERK increases PP2A activity as well as the abilities of cell growth and invasion. (a) Working hypothesis of CIP2A oncogenic loop. (b) MDA-MB-231 and MDA-MB-468 cells were treated with DMSO, SCH772984 (ERK inhibitor, 0.1 μM) or MK-2206 (AKT inhibitor, 10 μM) for 48 h. The cell lysates were then analyzed by Western blot (left). The results were quantified (right). (c) MDA-MB-231 and MDA-MB-468 cells were treated with DMSO or SCH772984 (ERK inhibitor, 0.1 μM) for 48 h and then assayed for PP2A activity. (d) MDA-MB-231 and MDA-MB-468 cells were treated with DMSO or SCH772984 (ERK inhibitor, 0.1 μM). The cells were counted by trypan blue exclusion method at 24, 48 and 72 h. (e) MDA-MB-231 and MDA-MB-468 cells were treated with DMSO or SCH772984 (ERK inhibitor, 0.1 μM) for invasion transwell assay (20 h). Data are representative of three independent experiments. Columns, mean (N = 3); bars, SD. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
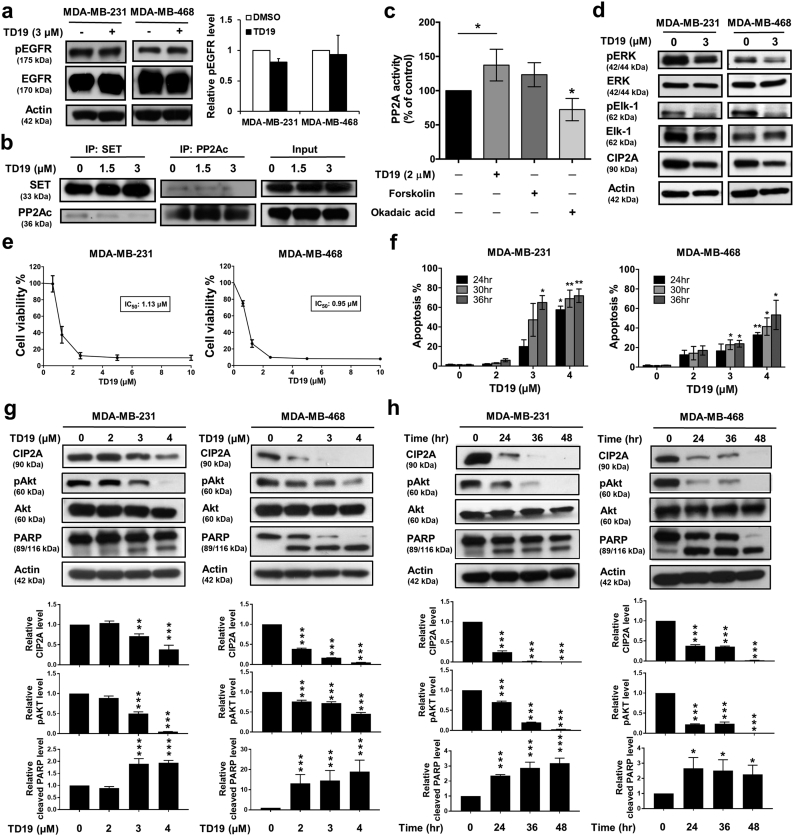
3.4. SET inhibitor triggers cell apoptosis through CIP2A/PP2A/pAkt signaling
To analyze the effects of disruption of SET and PP2A protein-protein interaction, TNBC cells were treated with TD19 for further analysis. We found erlotinib derivative TD19 did not decrease EGFR phosphorylation (Fig. 4a and Supplementary Fig. 4a). TD19 could interrupt the association between SET and PP2Ac (Fig. 4b). We further examined the effect of TD19 in PP2A regulation and found the phosphatase activity of PP2A was elevated in TD19-treated cells. Forskolin, a PP2A activator, showed a trend toward increase in PP2A activity; however, the negative control okadaic acid significantly decreased PP2A activity (Fig. 4c). We also found TD19 decreased ERK and Elk-1 phosphorylation and CIP2A expression in dose-dependent manner (Fig. 4d and Supplementary Fig. 4b). Additionally, TD19 inhibited cell viability and elevated the apoptotic cells (Fig. 4e-f and Supplementary Fig. 4c-d). TD19 downregulated CIP2A and pAkt as well as induced PARP cleavage in a dose-dependent and time-dependent manner (Fig. 4g-h and Supplementary Fig. 4e-f). We further examined the other PP2A downstream targets, which involved in apoptotic signaling, in TD19-treated TNBC cells. The phosphorylation of Bcl2 was decreased by TD19 whereas the phosphorylation of c-Myc was not (Supplementary Fig. 4g), suggesting TD19-elicited PP2A activation might induce cell apoptosis also through Bcl2 other than Akt signaling.
Fig. 4.
TD19 triggers cell apoptosis through CIP2A/PP2A/pAkt signaling.
(a) TD19 exhibited limited effect on EGFR phosphorylation in MDA-MB-231 and MDA-MB-468 cells. (b) TD19 disrupted the interaction between SET and PP2Ac. (c) Analysis of PP2A activity in TD19-treated MDA-MB-468 cells. Cells were incubated with DMSO, TD19 (3 μM), forskolin (40 μM, as a positive control) or okadaic acid (20 nM, as a negative control) for 24 h. Cell lysates were assayed for PP2A activity. (d) Cells were incubated with DMSO or TD19 (3 μM) for 24 h. The cell lysates were analyzed by Western blot. (e) Cells were treated with TD19 at the indicated doses for 48 h and cell viability was assessed by MTT assay. (f) Cells were treated with TD19 at the indicated doses for 24, 30, and 36 h. Apoptotic cells were determined by flow cytometry (sub-G1 analysis of PI- stained cells). (g) MDA-MB-231 and MDA-MB-468 cells were treated with TD19 at the indicated doses for 24 h and further analyzed by Western blot (up). The results were quantified (down). (h) MDA-MB-231 and MDA-MB-468 cells were treated TD19 with 4 μM and 3 μM respectively for indicated time. Whole-cell extracts were prepared and assayed by Western blot (up). The results were quantified (down). Points, mean (N = 3); columns, mean (N = 3); bars, SD. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
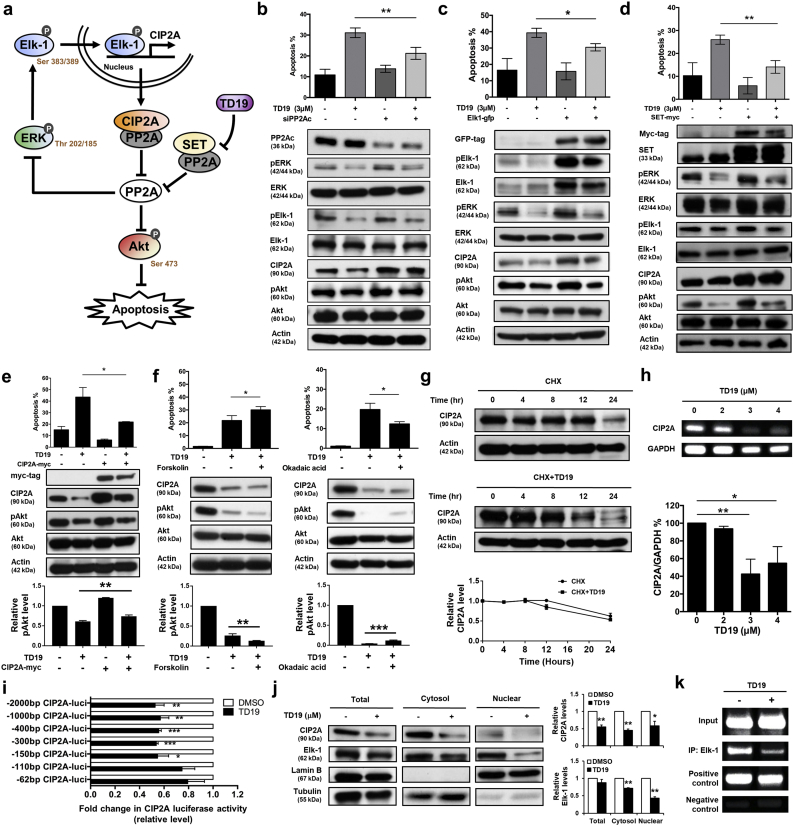
3.5. Targeting SET disrupts oncogenic CIP2A feedforward loop
ERK activates Elk-1 resulting in Elk-1 transcriptional regulation of CIP2A expression which leads to a reduction in PP2A activity. PP2A inactivated Akt resulting in cell apoptosis. Our data revealed that TD19 led to ERK and Elk-1 inactivation, CIP2A reduction as well as PP2A activity elevation. We hypothesized the presence of ERK/Elk-1/CIP2A/PP2A oncogenic loop (Fig. 5a). To validate this hypothesis, we analyzed the molecular events within TD19 treatment. TD19 suppressed pERK, pElk-1, CIP2A and pAkt as well as induced cell apoptosis which were restored by PP2Ac knockdown, Elk-1 and SET expression in MDA-MB-468 cells (Fig. 5b-d and Supplementary Fig. 5a-c). SET knockdown downregulated pERK, pElk-1, CIP2A and pAkt while did not increase apoptotic cells. Furthermore, TD19 could still induce apoptosis in the background of SET knockdown, albeit slightly reduced apoptosis (Supplementary Fig. 5d). These data indicated that TD19, at least in part, exerted its anti-TNBC activity through disrupting SET and CIP2A/PP2A/ERK/Elk-1 loop and thus downstream pAkt. However, other unidentified mechanisms may also contribute to the TD19-induced apoptosis. Cell apoptosis was significantly increased in TNBC cells with TD19 treatment which reduced by CIP2A restoration (Fig. 5e). TD19-induced cell apoptosis was further enhanced by PP2A agonist forskolin whereas repressed by PP2A inhibitor okadaic acid treatment (Fig. 5f). Furthermore, to decipher the molecular mechanism of TD19-suppressed CIP2A expression, the cycloheximide (CHX) chase analysis was conducted in MDA-MB-468 cells. The stability of CIP2A protein did not alter by TD19 treatment (Fig. 5g). However, both of the transcript and the transcriptional activity of CIP2A promoter were significantly decreased by TD19 (Fig. 5h-i). TD19 reduced cytosol and nuclear Elk-1 and the binding between Elk-1 protein and CIP2A promoter (Fig. 5j-k). Mcl-1 has been reported regulated by Elk-1 [39]. We also found the transcriptional activity of Mcl-1 promoter and the binding of Elk-1 to Mcl-1 promoter were reduced by TD19 (Supplementary Fig. 5e-f). These data demonstrated the existence of pERK/pElk-1/CIP2A/PP2A oncogenic loop and SET inhibitor disrupted this feedforward loop.
Fig. 5.
Targeting SET disrupts oncogenic CIP2A feedforward loop in TNBC cells.
(a) Illustration of CIP2A/PP2A/pERK/pElk-1 feedforward loop. (b-d) MDA-MB-468 cells were transfected with siRNA against PP2Ac (b), Elk-1- (c) or SET-expressing plasmids (d) for 24 h. Then the cells were treated with TD19 (3 μM) or DMSO. Cells were assayed for sub-G1 analysis (up). Whole-cell lysates were further analyzed by Western blot (down). (e) Ectopic expression of myc-tagged CIP2A reduces the apoptotic effect of TD19 in MDA-MB-468 cells (up). Whole-cell extracts were analyzed by Western blot (middle). The results were quantified (down). (f) Co-treatment of PP2A agonist forskolin enhanced TD19-induced apoptosis. MDA-MB-468 cells were co-treated with DMSO (control) or TD19 (3 μM) and forskolin (40 nM) for 24 h (left). Pretreatment of PP2A inhibitor okadaic acid protected cells from TD19-induced apoptosis. MDA-MB-468 cells were pretreated with or without okadaic acid (20 nM) for 1 h; then washed and treated with DMSO (control) or TD19 (3 μM) for 24 h (right). Cells were assayed for sub-G1 analysis (up). Cell lysates were analyzed by Western blot (middle). The results were quantified (down). (g) Limited effect of TD19 on CIP2A protein degradation. MDA-MB-468 cells were treated with 100 μg/milliliter pan-translation inhibitor cycloheximide (CHX) with or without TD19 (2 μM) for the indicated period of time, then the stability of CIP2A protein in whole-cell lysates was assessed by Western blot (up). The results were quantified (down). (h) TD19 inhibited CIP2A transcription. MDA-MB-468 cells were treated with TD19 at 0, 2, 3 and 4 μM for 24 h; then RNA was isolated for RT-PCR analysis. (i) Promoters with different lengths of deletion were constructed. Followed by transfecting with the mutant clone or the wide-type promoter for 24 h, MDA-MB-468 cells were subsequently exposed to TD19 (3 μM) or DMSO for 24 h. Cell lysates were then assayed for dual luciferase activity. (j) TD19 decreased Elk-1 translocation from the cytosol to the nucleus. Nuclear and cytoplasmic extracts were prepared from MDA-MB-468 cells treated with TD19 (3 μM) or DMSO for 24 h. Cell lysates were Western blotted for CIP2A and Elk-1. Lamin B and Tubulin were used as a loading control (up). The results were quantified (down). (k) TD19 disturbed binding of Elk-1 to the CIP2A promoter region. Chromatin immunoprecipitation assays of the CIP2A promoter were performed. Soluble chromatin was immunoprecipitated with Elk-1 or IgG (negative control, NC) antibodies. Immunoprecipitates were subjected to PCR with primer pairs specific to the CIP2A promoter (−139 to −16 nt). Anti-RNA polymerase II antibody and GAPDH primers were used as a positive control for the assay technique and reagent integrity. Data are representative of three independent experiments. Columns, mean (N = 3); bars, SD. Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
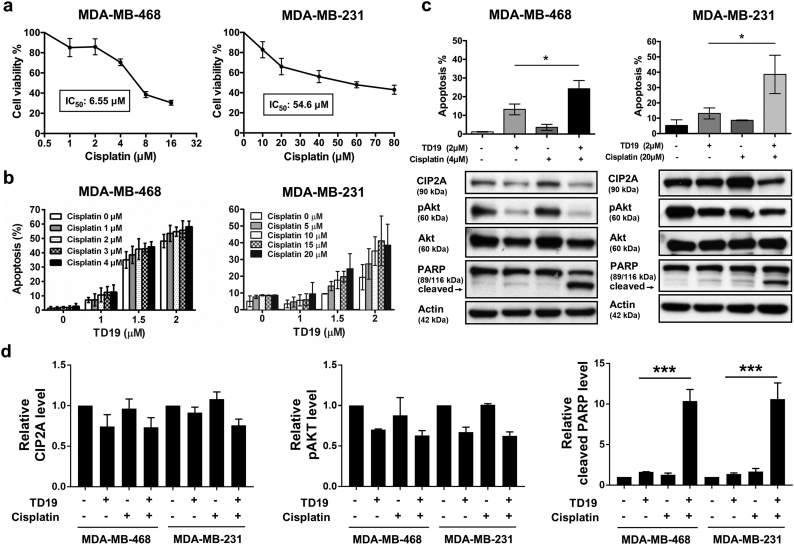
3.6. Combination of SET inhibitor and cisplatin enhances apoptotic effects
It has been reported that combination of chemotherapy and SET inhibitor exhibited a synergistic effect in decreasing cell viability and clonogenic activity [40]. Several clinical studies reported that platinum-based agents use as first-line treatment of patients with TNBC resulted in longer progression-free survival, but studies indicated that eventually becomes drug resistance [41]. To test the other therapeutic options, we tested the in vitro effect of TD19 plus cisplatin. MDA-MB-468 cells were sensitive to cisplatin than MDA-MB-231 cells (Fig. 6a). TD19 plus cisplatin enhanced cell apoptosis in TNBC cells (Fig. 6b). Combination of TD19 and cisplatin induced PARP cleavage whereas did not alter TD19-reduced phosphorylation of Akt, ERK and Elk-1 (Fig. 6c-d and Supplementary Fig. 6).
Fig. 6.
Combination of TD19 and cisplatin enhances apoptotic effects in association with CIP2A downregulation.
(a) Dose-escalation effects of cisplatin on cell viability in MDA-MB-468 and MDA-MB-231 cells. After 48-h treatment, cell viability was assessed by MTT assay. (b) MDA-MB-468 and MDA-MB-231 cells were treated with TD19 and cisplatin at indicated concentration for 48 h. The apoptotic cells were analyzed by flow cytometry. (c) Apoptotic cells and the molecular events were determined by flow cytometry and Western blot. Data are representative of three independent experiments. (d) The quantitative data of Fig. 6C was shown. Points, mean (N = 3); columns, mean (N = 3); bars, SD; Student's t-test, *, P < 0.05; **, P < 0.01; ***, P < 0.001.
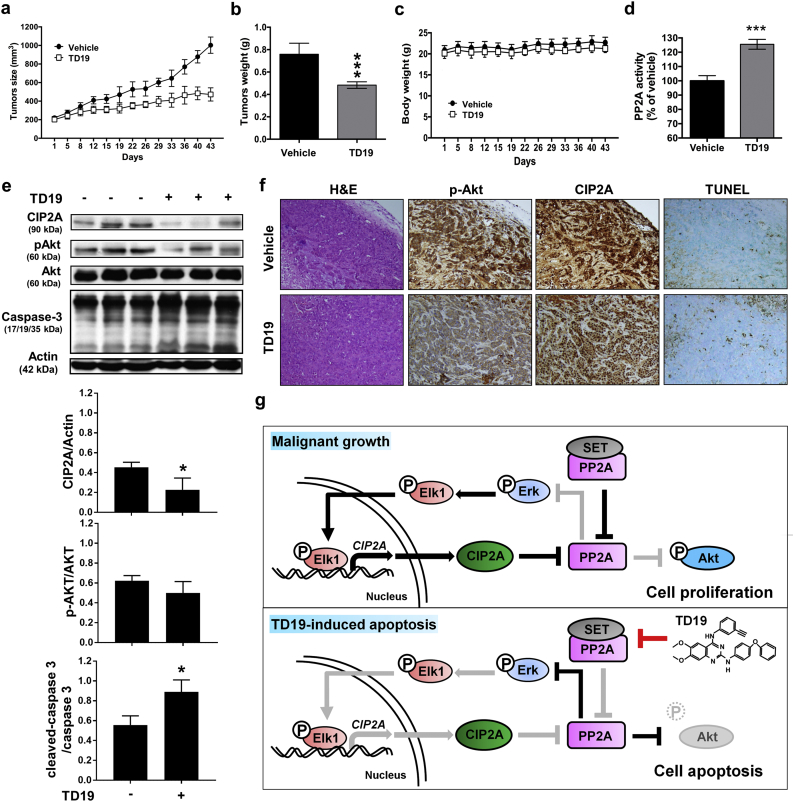
3.7. Targeting SET decreases tumor growth in vivo
To evaluate the capacity of SET inhibitor in TNBC treatment in vivo, the MDA-MB-468 xenograft was performed. TD19 treatment significantly reduced tumor size and tumor weight without body weight alteration (Fig. 7a-c). The phosphatase activity of PP2A in TD19-treated tumor was elevated (Fig. 7d). The levels of CIP2A and pAkt were suppressed accompanied by cleaved Caspase-3 and TUNEL-positive cells elevation (Fig. 7e-f and Supplementary Fig. 7). These data exhibited the therapeutic effect of SET inhibitor in TNBC treatment in vivo.
Fig. 7.
Targeting SET decreases tumor growth of MDA-MB-468 xenograft nude mice.
(a-d) The MDA-MB-468 xenograft bearing mice were administered orally with TD19 at a dose of 10 mg/kg or vehicle (PBS) three times weekly. The tumor size (a), tumor weight (b), body weight (c) and PP2A activity (d) were measured. (e) Western blot analysis of the expression level of CIP2A, pAkt, Akt, Caspase-3 and actin in MDA-MB-468 xenografts treated with control or TD19 (up). The results were quantified (down). (f) H&E stain and IHC staining for CIP2A and pAkt as well as TUNEL staining for apoptosis detection in the MDA-MB-468 xenografts treated with control or TD19. (g) Working model of TD19-induced apoptosis in TNBC. Power fields 100×. Points, mean (N = 6); columns, mean (N = 3); bars, SD. Student's t-test, ***, P < 0.001.
4. Discussion
In this study, we found SET and CIP2A contributed to the aggressive nature of TNBC. We discovered that a positive feedforward loop of CIP2A composed of CIP2A, PP2A, pERK and pElk-1 contributed to CIP2A expression. Furthermore, this CIP2A feedforward loop can be disrupted by a SET protein-protein interaction inhibitor, TD19. SET inhibitor TD19 induced cell apoptosis via ERK/Elk-1/CIP2A axis which elicited PP2A activity and inactivation of Akt (Fig. 7g).
SET protein, first characterized in acute undifferentiated leukemia in 1994 [42,43], is later found to be an intrinsic cellular inhibitor of PP2A [20,44]. SET has been known as an oncoprotein that is overexpressed in various cancers, including acute myeloid leukemia [45], lymphoid malignancies [46], head and neck cancers [47], NSCLC [48], and hepatocellular carcinoma [32]. Retrospectively, CIP2A level is elevated in TNBC and serves as prognostic marker of TNBC [10,49]. Here, we showed the clinical significance of SET in TNBC. Our results exhibited both of SET and CIP2A were upregulated in TNBC tumor tissues compared to normal tissues and linked with poor overall survival (Fig. 1). Overexpression of SET or CIP2A promoted cell viability, migration and invasion of TNBC cells (Fig. 2). Although SET may promote cancer aggressiveness though other interacting partners, such as inhibition of the tumor suppressor NM23-H1 [22] and p53 [23], or forming fusion proteins [50,51], the interaction of SET with PP2A has drawn a major attention in the development of anti-cancer strategies [25,52,53]. Recently, Hung has nicely reviewed the biological role, clinical relevance and potential therapeutic implication of SET protein in cancers [25].
Notably, currently there have been several available drugs/compounds aiming to interfere with the interaction of SET and PP2A, such as a sphingosine analogue fingolimod (FTY720) [54], an apolipoprotein E [55], peptide mimetic OP449 (also named COG449) [40] and TD19 [30,32]. FTY720 is an approved drug for multiple sclerosis, a devastating neurological disease characterized by chronic neuronal inflammatory demyelination of the central nervous system. The mechanism of action claimed for multiple sclerosis therapy is that FTY720 acts as a modulator of the sphingosine-1-phosphate receptor (S1P-R) on the surface of lymphocytes, prevents egress of lymphocytes from secondary lymphoid organs, and thus ameliorates CNS infiltration of these lymphocytes [56]. Although FTY720 has been demonstrated as a SET/PP2A protein-protein interaction inhibitor, this “off-target” mechanism of action remains in preclinical model and FTY720 has not yet been tested in clinical trials as an anti-cancer drug. In contrast, OP449 was designated as a SET/PP2A protein-protein interaction inhibitor [40] and its anti-cancer efficacy has been tested in several preclinical cancer models [46,57], including breast cancer [9].
ERK inhibitor suppressed CIP2A and induced PP2A activity in TNBC cells (Fig. 3). High protein level of ERK in patients with TNBC is associated with short overall survival [58]. Mounting evidence mentions that targeting Raf/MEK/ERK pathway is a potential therapeutic strategy for TNBC [59]. Antagonizing inhibitors of PP2A is considered as a therapeutic strategy of TNBC. TD19 disrupted the interaction between SET and PP2A, enhanced PP2A activity and inactivated ERK and Elk-1. In addition, TD19 dephosphorylated Akt as well as induced cell apoptosis (Fig. 4). Recently, a phase II trial demonstrates that adding an Akt inhibitor ipatasertib to paclitaxel enhances efficacy in patients with metastatic TNBC; patients receiving ipatasertib plus paclitaxel has longer progression-free survival than patients who received paclitaxel alone [60]. These findings indicate the need for further investigations of the efficacy of Akt-inhibiting agents in TNBC [61]. TD19 elevated PP2A activity which dephosphorylating ERK and Akt. Therefore, that TD19 indirectly inactivated ERK and Akt may also impair TNBC progression.
Despite that it is not our aim to compare the efficacies of these PP2A-activity modulators (TD19, ERK inhibitor, and forskolin), we used these agents with different mechanisms to demonstrate the existence of a novel CIP2A/PP2A/pERK/pElk-1 feedforward loop, and that interfering SET could perturb this loop. In terms of mechanisms of action, forskolin has been known as a cAMP inducer and reduces phosphorylation of PP2Ac at Tyr307 on the C-terminal tail of PP2A C-subunit, thereby activating PP2A [62,63]. The finding that ERK inhibitor (SCH772984) increased PP2A activity is novel and has led us to discover the CIP2A/PP2A/pERK/Elk-1 feedforward loop. In contrast, TD19, of which chemical structure was derived from erlotinib with distinct different drug effect, has been shown increasing PP2A activity by interfering SET/PP2A protein-protein interaction [30,32]. Interestingly, forskolin has been shown induce apoptosis though its cAMP-induction, ERK inhibitor (SCH772984) can induce apoptosis through ERK inhibition [64]. In contrast, TD19 induced apoptosis, at least in part, through disrupting SET and CIP2A/PP2A/pERK/pElk-1 loop and thus downstream pAkt. Collectively, the magnitude of PP2A activity increment in vitro might not necessarily well-correlated with apoptosis induction.
Our data suggested the presence of pERK/pElk-1/CIP2A/PP2A oncogenic loop, and TD19 could interfere with this regulation (Fig. 5). The potential of inhibitors which targeting both of CIP2A and SET simultaneously is valuable evaluated for cancer treatment [65,66]. Previous studies also mention that the existence of a positive CIP2A/c-Myc/E2F1 positive feedback loop in chronic myeloid leukemia. SET has been reported inhibiting c-Myc degradation and stabilizing E2F1 through PP2A inhibition. Therefore, SET might participate in the regulation of CIP2A/c-Myc/E2F1 axis [67].
Several studies revealed that SET/PP2A signaling participated in the regulation of drug resistance. SET-suppressed NDRG1 mediates cisplatin resistance in human lung cancer cells [68]. Knockdown of SET promotes cisplatin sensitivity in head and neck squamous cell carcinoma [69]. PP2A inhibition determines poor outcome and doxorubicin resistance in early breast cancer and its activation shows promising therapeutic effects [70]. Our data showed that combination of TD19 and cisplatin exhibited a synergistic effect in TNBC cells (Fig. 6). Similarly, TD19 also enhances the chemosensitivity of NSCLC cells to paclitaxel [30]. Our finding that TD19 enhanced cisplatin cytotoxicity (Fig. 6) has particularly clinical relevance. Recently a large randomized phase III trial ‘the Triple Negative Breast Cancer Trial’ compared first-line treatment of either carboplatin or docetaxel in 376 patients with metastatic TNBC, and found that in subjects with germline-BRCA mutation, carboplatin had double the overall response rate of docetaxel (68% versus 33%, respectively; biomarker, treatment interaction P = .01) [71]. A retrospective study of Chinese population showed that platinum-based chemotherapy, in particular cisplatin-based regimens using at the first line may improve ORR and progression-free survival (PFS), but not overall survival. These clinical results suggest the clinical benefit of platinums (including cisplatin) in metastatic TNBC but also indicate a room for improving therapeutic benefit of platinums. Both MDA-MB-231 and MDA-MB-468 cells have been reported to harbor BRCA allelic loss but without BRCA mutations [72]. According to Lehmann [73], MDA-MB-468 belongs to basal-like subtype (by Lehmann's classification) and more sensitive to cisplatin, whereas MDA-MB-231 is mesenchymal like subtype, and less sensitive to cisplatin (comparing to MDA-MB-468). In line with Lehmann, our data showed that either in cisplatin-sensitive MDA-MB-468 or less sensitive MDA-MB-231 cells, targeting SET to disrupt CIP2A/PP2A/pAkt signaling is a feasible strategy to enhance cisplatin cytotoxicity, suggesting the possibility to test such combination strategy in future studies.
Our current study exerts some limitation, first the detail mechanisms of action of the SET/PP2Ac interaction inhibitor TD19 remains to be fully illustrated, such as whether TD19 influences the binding of other PP2A interactors. Moreover, our data show that TD19 induces apoptosis partly via SET and CIP2A/PP2A/pERK/pElk-1 axis, other unidentified pathways may also contribute to the anti-cancer activity of TD19. There are still unsolved questions and future studies are needed to fully decipher the relevant networks in SET, CIP2A and PP2A.
In summary, we discover a novel oncogenic CIP2A-feedforward loop in TNBC and targeting SET to disrupt oncogenic CIP2A feedforward loop shows therapeutic potential in TNBC.
Acknowledgments
Acknowledgements
The laboratory works were completed using facilities from Medical Science & Technology Building of Taipei Veterans General Hospital. Research was supported by Biobank, Taipei Veterans General Hospital. The authors would like to thank Dr. Kuen-Feng Chen, a co-founder of TD19, for his indispensable contributions on CIP2A/PP2A/pAkt in previous studies.
Funding sources
This research was supported by grants from the Taiwan Clinical Oncology Research Foundation; the Yen Tjing Ling Medical Foundation (CI-107-10); the Ministry of Science and Technology, Taiwan (MOST 104-2628-B-075-001-MY3, 105-2314-B-075-044-MY3); Yang-Ming Branch of Taipei City Hospital (10601-62-020, 10701-62-030); Taipei Veterans General Hospital (V106C-101, V107C-025, V107C-016, V106D27-002-MY2-2); from Taipei Veterans General Hospital and National Taiwan-University Hospital, and from the Ministry of Health and Welfare, Executive Yuan, Taiwan (MOHW107-TDU-B-212-112015 for the Center of Excellence for Cancer Research at Taipei Veterans General Hospital).
Conflicts of interest
The authors declare that they have no competing interests.
Authors' contributions
LMT and CYL were responsible for coordination and manuscript editing as well as acting as corresponding authors. CYL, TTH and JLC drafted the manuscript. TTH, KYL and YTC conducted in vitro experiments. TTH and WLW conducted in vivo experiments. PYC conducted histopathological experiments. TTH, KYL, CTH, WLW, MSD and CWS helped in data interpretation and statistical analysis. CYL and LMT supervised the study. All authors read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.032.
Appendix A. Supplementary data
Supplementary material
References
- 1.Mumby M. PP2A: unveiling a reluctant tumor suppressor. Cell. 2007;130(1):21–24. doi: 10.1016/j.cell.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 2.Eichhorn P.J., Creyghton M.P., Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795(1):1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Curtis C., Shah S.P., Chin S.F. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seshacharyulu P., Pandey P., Datta K., Batra S.K. Phosphatase: PP2A structural importance, regulation and its aberrant expression in cancer. Cancer Lett. 2013;335(1):9–18. doi: 10.1016/j.canlet.2013.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sablina A.A., Hahn W.C. SV40 small T antigen and PP2A phosphatase in cell transformation. Cancer Metastasis Rev. 2008;27(2):137–146. doi: 10.1007/s10555-008-9116-0. [DOI] [PubMed] [Google Scholar]
- 6.Calin G.A., di Iasio M.G., Caprini E. Low frequency of alterations of the alpha (PPP2R1A) and beta (PPP2R1B) isoforms of the subunit A of the serine-threonine phosphatase 2A in human neoplasms. Oncogene. 2000;19(9):1191–1195. doi: 10.1038/sj.onc.1203389. [DOI] [PubMed] [Google Scholar]
- 7.Tseng L.M., Liu C.Y., Chang K.C., Chu P.Y., Shiau C.W., Chen K.F. CIP2A is a target of bortezomib in human triple negative breast cancer cells. Breast Cancer Res. 2012;14(2):R68. doi: 10.1186/bcr3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu P., Xu X.L., Huang Q., Zhang Z.H., Zhang Y.B. CIP2A with survivin protein expressions in human non-small-cell lung cancer correlates with prognosis. Med Oncol. 2012;29(3):1643–1647. doi: 10.1007/s12032-011-0053-3. [DOI] [PubMed] [Google Scholar]
- 9.Janghorban M., Farrell A.S., Allen-Petersen B.L. Targeting c-MYC by antagonizing PP2A inhibitors in breast cancer. Proc Natl Acad Sci U S A. 2014;111(25):9157–9162. doi: 10.1073/pnas.1317630111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H., Qiu H., Song Y. Cip2a promotes cell cycle progression in triple-negative breast cancer cells by regulating the expression and nuclear export of p27Kip1. Oncogene. 2017;36(14):1952–1964. doi: 10.1038/onc.2016.355. [DOI] [PubMed] [Google Scholar]
- 11.Junttila M.R., Puustinen P., Niemela M. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130(1):51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 12.Liu C.Y., Huang T.T., Huang C.T. EGFR-independent Elk1/CIP2A signalling mediates apoptotic effect of an erlotinib derivative TD52 in triple-negative breast cancer cells. Eur J Cancer. 2017;72:112–123. doi: 10.1016/j.ejca.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Liu C.Y., Hsieh F.S., Chu P.Y. Carfilzomib induces leukaemia cell apoptosis via inhibiting ELK1/KIAA1524 (Elk-1/CIP2A) and activating PP2A not related to proteasome inhibition. Br J Haematol. 2017;177(5):726–740. doi: 10.1111/bjh.14620. [DOI] [PubMed] [Google Scholar]
- 14.Liu C.Y., Hu M.H., Hsu C.J. Lapatinib inhibits CIP2A/PP2A/p-Akt signaling and induces apoptosis in triple negative breast cancer cells. Oncotarget. 2016;7(8):9135–9149. doi: 10.18632/oncotarget.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu H.C., Hung M.H., Chen Y.L. Erlotinib derivative inhibits hepatocellular carcinoma by targeting CIP2A to reactivate protein phosphatase 2A. Cell Death Dis. 2014;5 doi: 10.1038/cddis.2014.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C.Y., Hung M.H., Wang D.S. Tamoxifen induces apoptosis through cancerous inhibitor of protein phosphatase 2A-dependent phospho-Akt inactivation in estrogen receptor-negative human breast cancer cells. Breast Cancer Res. 2014;16(5):431. doi: 10.1186/s13058-014-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen K.F., Liu C.Y., Lin Y.C. CIP2A mediates effects of bortezomib on phospho-Akt and apoptosis in hepatocellular carcinoma cells. Oncogene. 2010;29(47):6257–6266. doi: 10.1038/onc.2010.357. [DOI] [PubMed] [Google Scholar]
- 18.De P., Carlson J., Leyland-Jones B., Dey N. Oncogenic nexus of cancerous inhibitor of protein phosphatase 2A (CIP2A): an oncoprotein with many hands. Oncotarget. 2014;5(13):4581–4602. doi: 10.18632/oncotarget.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soofiyani S.R., Hejazi M.S., Baradaran B. The role of CIP2A in cancer: a review and update. Biomed Pharmacother. 2017;96:626–633. doi: 10.1016/j.biopha.2017.08.146. [DOI] [PubMed] [Google Scholar]
- 20.Li M., Guo H., Damuni Z. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry. 1995;34(6):1988–1996. doi: 10.1021/bi00006a020. [DOI] [PubMed] [Google Scholar]
- 21.Al-Murrani S.W., Woodgett J.R., Damuni Z. Expression of I2PP2A, an inhibitor of protein phosphatase 2A, induces c-Jun and AP-1 activity. Biochem J. 1999;341(Pt 2):293–298. [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Z., Beresford P.J., Oh D.Y., Zhang D., Lieberman J. Tumor suppressor NM23-H1 is a granzyme A-activated DNase during CTL-mediated apoptosis, and the nucleosome assembly protein SET is its inhibitor. Cell. 2003;112(5):659–672. doi: 10.1016/s0092-8674(03)00150-8. [DOI] [PubMed] [Google Scholar]
- 23.Wang D., Kon N., Lasso G. Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature. 2016;538(7623):118–122. doi: 10.1038/nature19759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal A., MacKenzie R.J., Pippa R. Antagonism of SET using OP449 enhances the efficacy of tyrosine kinase inhibitors and overcomes drug resistance in myeloid leukemia. Clin Cancer Res. 2014;20(8):2092–2103. doi: 10.1158/1078-0432.CCR-13-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hung M.H., Chen K.F. Reprogramming the oncogenic response: SET protein as a potential therapeutic target in cancer. Expert Opin Ther Targets. 2017;21(7):685–694. doi: 10.1080/14728222.2017.1336226. [DOI] [PubMed] [Google Scholar]
- 26.Cristobal I., Madoz-Gurpide J., Manso R., Gonzalez-Alonso P., Rojo F., Garcia-Foncillas J. Potential anti-tumor effects of FTY720 associated with PP2A activation: a brief review. Curr Med Res Opin. 2016;32(6):1137–1141. doi: 10.1185/03007995.2016.1162774. [DOI] [PubMed] [Google Scholar]
- 27.Lavaur J., Bernard F., Trifilieff P. A TAT-DEF-Elk-1 peptide regulates the cytonuclear trafficking of Elk-1 and controls cytoskeleton dynamics. J Neurosci. 2007;27(52):14448–14458. doi: 10.1523/JNEUROSCI.2279-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruzalegui F.H., Cano E., Treisman R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene. 1999;18(56):7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- 29.Yang S.H., Shore P., Willingham N., Lakey J.H., Sharrocks A.D. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999;18(20):5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hung M.H., Wang C.Y., Chen Y.L. SET antagonist enhances the chemosensitivity of non-small cell lung cancer cells by reactivating protein phosphatase 2A. Oncotarget. 2016;7(1):638–655. doi: 10.18632/oncotarget.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao T.T., Wang C.Y., Lai C.C. TD-19, an erlotinib derivative, induces epidermal growth factor receptor wild-type nonsmall-cell lung cancer apoptosis through CIP2A-mediated pathway. J Pharmacol Exp Ther. 2014;351(2):352–358. doi: 10.1124/jpet.114.215418. [DOI] [PubMed] [Google Scholar]
- 32.Hung M.H., Chen Y.L., Chu P.Y. Upregulation of the oncoprotein SET determines poor clinical outcomes in hepatocellular carcinoma and shows therapeutic potential. Oncogene. 2016;35(37):4891–4902. doi: 10.1038/onc.2016.21. [DOI] [PubMed] [Google Scholar]
- 33.Chen K.F., Pao K.C., Su J.C. Development of erlotinib derivatives as CIP2A-ablating agents independent of EGFR activity. Bioorg Med Chem. 2012;20(20):6144–6153. doi: 10.1016/j.bmc.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 34.Liu C.Y., Su J.C., Huang T.T. Sorafenib analogue SC-60 induces apoptosis through the SHP-1/STAT3 pathway and enhances docetaxel cytotoxicity in triple-negative breast cancer cells. Mol Oncol. 2017;11(3):266–279. doi: 10.1002/1878-0261.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammed R.A., Green A., El-Shikh S., Paish E.C., Ellis I.O., Martin S.G. Prognostic significance of vascular endothelial cell growth factors -A, -C and -D in breast cancer and their relationship with angio- and lymphangiogenesis. Br J Cancer. 2007;96(7):1092–1100. doi: 10.1038/sj.bjc.6603678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou C., Wu Y.L., Chen G. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 37.Cheadle C., Vawter M.P., Freed W.J., Becker K.G. Analysis of microarray data using Z score transformation. J Mol Diagn. 2003;5(2):73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu C.Y., Huang T.T., Chu P.Y. The tyrosine kinase inhibitor nintedanib activates SHP-1 and induces apoptosis in triple-negative breast cancer cells. Exp Mol Med. 2017;49(8) doi: 10.1038/emm.2017.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend K.J., Zhou P., Qian L. Regulation of MCL1 through a serum response factor/Elk-1-mediated mechanism links expression of a viability-promoting member of the BCL2 family to the induction of hematopoietic cell differentiation. J Biol Chem. 1999;274(3):1801–1813. doi: 10.1074/jbc.274.3.1801. [DOI] [PubMed] [Google Scholar]
- 40.Neviani P., Perrotti D. SETting OP449 into the PP2A-activating drug family. Clin Cancer Res. 2014;20(8):2026–2028. doi: 10.1158/1078-0432.CCR-14-0166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouwman P., Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res. 2014;20(3):540–547. doi: 10.1158/1078-0432.CCR-13-0225. [DOI] [PubMed] [Google Scholar]
- 42.Adachi Y., Pavlakis G.N., Copeland T.D. Identification of in vivo phosphorylation sites of SET, a nuclear phosphoprotein encoded by the translocation breakpoint in acute undifferentiated leukemia. FEBS Lett. 1994;340(3):231–235. doi: 10.1016/0014-5793(94)80144-4. [DOI] [PubMed] [Google Scholar]
- 43.Adachi Y., Pavlakis G.N., Copeland T.D. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J Biol Chem. 1994;269(3):2258–2262. [PubMed] [Google Scholar]
- 44.Li M., Makkinje A., Damuni Z. The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem. 1996;271(19):11059–11062. doi: 10.1074/jbc.271.19.11059. [DOI] [PubMed] [Google Scholar]
- 45.Cristobal I., Garcia-Orti L., Cirauqui C. Overexpression of SET is a recurrent event associated with poor outcome and contributes to protein phosphatase 2A inhibition in acute myeloid leukemia. Haematologica. 2012;97(4):543–550. doi: 10.3324/haematol.2011.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christensen D.J., Chen Y., Oddo J. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood. 2011;118(15):4150–4158. doi: 10.1182/blood-2011-04-351072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leopoldino A.M., Squarize C.H., Garcia C.B. SET protein accumulates in HNSCC and contributes to cell survival: antioxidant defense, Akt phosphorylation and AVOs acidification. Oral Oncol. 2012;48(11):1106–1113. doi: 10.1016/j.oraloncology.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 48.Liu H., Gu Y., Wang H. Overexpression of PP2A inhibitor SET oncoprotein is associated with tumor progression and poor prognosis in human non-small cell lung cancer. Oncotarget. 2015;6(17):14913–14925. doi: 10.18632/oncotarget.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cristobal I., Zazo S., Torrejon B. CIP2A confirms its prognostic value in triple-negative breast cancer. Oncogene. 2017;36(23):3357–3358. doi: 10.1038/onc.2016.474. [DOI] [PubMed] [Google Scholar]
- 50.Chae H., Lim J., Kim M. Phenotypic and genetic characterization of adult T-cell acute lymphoblastic leukemia with del(9)(q34);SET-NUP214 rearrangement. Ann Hematol. 2012;91(2):193–201. doi: 10.1007/s00277-011-1289-x. [DOI] [PubMed] [Google Scholar]
- 51.von Lindern M., van Baal S., Wiegant J., Raap A., Hagemeijer A., Grosveld G. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol. 1992;12(8):3346–3355. doi: 10.1128/mcb.12.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Switzer C.H., Cheng R.Y., Vitek T.M., Christensen D.J., Wink D.A., Vitek M.P. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30(22):2504–2513. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cristobal I., Rincon R., Manso R. Deregulation of the PP2A inhibitor SET shows promising therapeutic implications and determines poor clinical outcome in patients with metastatic colorectal cancer. Clin Cancer Res. 2015;21(2):347–356. doi: 10.1158/1078-0432.CCR-14-0724. [DOI] [PubMed] [Google Scholar]
- 54.Saddoughi S.A., Gencer S., Peterson Y.K. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol Med. 2013;5(1):105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christensen D.J., Ohkubo N., Oddo J. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J Immunol. 2011;186(4):2535–2542. doi: 10.4049/jimmunol.1002847. [DOI] [PubMed] [Google Scholar]
- 56.Sharma S., Mathur A.G., Pradhan S., Singh D.B., Gupta S. Fingolimod (FTY720): first approved oral therapy for multiple sclerosis. J Pharmacol Pharmacother. 2011;2(1):49–51. doi: 10.4103/0976-500X.77118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Farrell A.S., Allen-Petersen B., Daniel C.J. Targeting inhibitors of the tumor suppressor PP2A for the treatment of pancreatic cancer. Mol Cancer Res. 2014;12(6):924–939. doi: 10.1158/1541-7786.MCR-13-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartholomeusz C., Gonzalez-Angulo A.M., Liu P. High ERK protein expression levels correlate with shorter survival in triple-negative breast cancer patients. Oncologist. 2012;17(6):766–774. doi: 10.1634/theoncologist.2011-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giltnane J.M., Balko J.M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med. 2014;17(95):275–283. [PubMed] [Google Scholar]
- 60.Kim S.B., Dent R., Im S.A. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2017;18(10):1360–1372. doi: 10.1016/S1470-2045(17)30450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaloge S., Deforceville L. Targeting PI3K/AKT pathway in triple-negative breast cancer. Lancet Oncol. 2017;18(10):1293–1294. doi: 10.1016/S1470-2045(17)30514-4. [DOI] [PubMed] [Google Scholar]
- 62.Cristobal I., Rincon R., Manso R. Hyperphosphorylation of PP2A in colorectal cancer and the potential therapeutic value showed by its forskolin-induced dephosphorylation and activation. Biochim Biophys Acta. 2014;1842(9):1823–1829. doi: 10.1016/j.bbadis.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 63.Feschenko M.S., Stevenson E., Nairn A.C., Sweadner K.J. A novel cAMP-stimulated pathway in protein phosphatase 2A activation. J Pharmacol Exp Ther. 2002;302(1):111–118. doi: 10.1124/jpet.302.1.111. [DOI] [PubMed] [Google Scholar]
- 64.Cagnol S., Chambard J.C. ERK and cell death: mechanisms of ERK-induced cell death--apoptosis, autophagy and senescence. FEBS J. 2010;277(1):2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 65.Cristobal I., Manso R., Gonzalez-Alonso P., Rojo F., Garcia-Foncillas J. Should we also evaluate SET together with CIP2A for the treatment with second-generation tyrosine kinase inhibitors in chronic myeloid leukemia? Leukemia. 2015;29(10):2117. doi: 10.1038/leu.2015.201. [DOI] [PubMed] [Google Scholar]
- 66.Costa R.L.B., Han H.S., Gradishar W.J. Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review. Breast Cancer Res Treat. 2018;169(3):397–406. doi: 10.1007/s10549-018-4697-y. [DOI] [PubMed] [Google Scholar]
- 67.Lucas C.M., Harris R.J., Holcroft A.K. Second generation tyrosine kinase inhibitors prevent disease progression in high-risk (high CIP2A) chronic myeloid leukaemia patients. Leukemia. 2015;29(7):1514–1523. doi: 10.1038/leu.2015.71. [DOI] [PubMed] [Google Scholar]
- 68.Liu H., Gu Y., Yin J. SET-mediated NDRG1 inhibition is involved in acquisition of epithelial-to-mesenchymal transition phenotype and cisplatin resistance in human lung cancer cell. Cell Signal. 2014;26(12):2710–2720. doi: 10.1016/j.cellsig.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 69.Sobral L.M., Sousa L.O., Coletta R.D. Stable SET knockdown in head and neck squamous cell carcinoma promotes cell invasion and the mesenchymal-like phenotype in vitro, as well as necrosis, cisplatin sensitivity and lymph node metastasis in xenograft tumor models. Mol Cancer. 2014;13:32. doi: 10.1186/1476-4598-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rincon R., Cristobal I., Zazo S. PP2A inhibition determines poor outcome and doxorubicin resistance in early breast cancer and its activation shows promising therapeutic effects. Oncotarget. 2015;6(6):4299–4314. doi: 10.18632/oncotarget.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tutt A., Tovey H., Cheang M.C.U. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med. 2018;24(5):628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elstrodt F., Hollestelle A., Nagel J.H. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66(1):41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- 73.Lehmann B.D., Bauer J.A., Chen X. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material