Abstract
Background
As part of the tumor microenvironment, the gastric microbiota play vital roles in tumor initiation, progression and metastasis, but stomach microhabitats are not always uniform. We aimed to characterize differences of gastric microbiota in stomach microhabitats associated with gastric cancer (GC) development.
Methods
A cohort of 276 GC patients without preoperative chemotherapy was enrolled retrospectively, and 230 normal, 247 peritumoral and 229 tumoral tissues were obtained for gastric microbiota analysis targeting the 16S rRNA gene by MiSeq sequencing. The microbial diversity and composition, bacterial co-occurrence correlations and predictive functional profiles were compared across different microhabitats.
Findings
GC-specific stomach microhabitats, not GC stages or types, determine the composition and diversity of the gastric microbiota. Most notably, bacterial richness was decreased in peritumoral and tumoral microhabitats, and the correlation network of abundant gastric bacteria was simplified in tumoral microhabitat. Helicobacter pylori (HP), Prevotella copri and Bacteroides uniformis were significantly decreased, whereas Prevotella melaninogenica, Streptococcus anginosus and Propionibacterium acnes were increased in tumoral microhabitat. Higher HP colonisation influenced the overall structure of the gastric microbiota in normal and peritumoral microhabitats. PiCRUSt analysis revealed that genes associated with nucleotide transport and metabolism and amino acid transport and metabolism were significantly enriched in tumoral microbiota, while gastric acid secretion was significantly higher in HP positive group of the tumoral microbiota.
Interpretation
Our present study provided new insights into the roles of gastric microbiota in different stomach microhabitats in gastric carcinogenesis, especially the pathogenesis of HP.
Fund
National Natural Science Foundation of China.
Keywords: Gastric cancer, Gastric microbiota, Helicobacter pylori, Stomach microhabitat, Tumor microenvironment
Research in context.
Evidence before this study
Gastric microbiota were one of the important elements of the tumor microenvironment, which participated in gastric cancer (GC) initiation, progression and metastasis. Previous studies have investigated the characteristics of the gastric microbiota in different gastric diseases, from superficial gastritis, atrophic gastritis, intestinal metaplasia to intestinal-type GC. Recently, several studies found that the gastric mucosal microbiota dysbiosis changed across stages of gastric carcinogenesis and several specific bacterial markers had been identified that could distinguish GC from other gastric diseases. However, most of these studies had always considered the whole stomach as one habitat, the differences of gastric microbiota among different stomach microhabitats in patients with GC were still unclear.
The value of this study
Stomach microhabitats are not always uniform; instead, they vary considerably across sites within the stomach, at the same site over time, and with the health status. The specific changes among different stomach microhabitats might help to reveal the real role of the gastric bacteria on gastric carcinogenesis. With a larger cohort of GC patients without preoperative chemotherapy enrollment, we firstly demonstrated that GC-specific stomach microhabitats determine the composition and diversity of the gastric microbiota. The most important bacterial carcinogen, Helicobacter pylori (HP), were significantly decreased in tumoral microhabitat, while the higher relative abundance of HP influenced the overall structure of the gastric microbiota in normal and peritumoral microhabitats. Inconsistent with previous studies, we found that bacteria from different stomach microhabitats showed different correlation network and functions.
Implications of all the available evidence
We report novel findings on the relationship between gastric microbiota and GC. Stomach microhabitats determine the overall structure and composition of the gastric microbiota, which provide new insights into the roles of gastric microbiota in different stomach microhabitats on gastric carcinogenesis, especially the pathogenesis of HP.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer (GC) is the leading cause of cancer-related mortality worldwide. According to the latest cancer statistics in China, 679,000 newly diagnosed GC cases and 498,000 GC related deaths occurred in 2015, indicating that GC is the second most common cancer after lung cancer [1]. Although the overall incidence and mortality have steadily declined in the past several decades [2], 80–90% of GC patients are diagnosed with advanced-stage disease, with a 5-year survival rate of <30% [3]. Due to its high mortality and significant regional disparity in its distribution, GC remains an important public health burden in China.
GC is a multifactorial disease, and alterations of the tumor microenvironment are required for GC initiation, progression and metastasis. As part of the tumor microenvironment, gastric microbiota has attracted increasing attention, as it can affect cancer growth and spread in many ways. However, gastric microbiota has been relatively understudied compared to the gut microbiota. Due to acidic conditions and other antimicrobial factors, the human stomach is thought to be exclusively inhabited by Helicobacter pylori (HP) and viewed as an inhospitable environment for microorganisms. However, recent advances in sequencing technologies have provided a broader picture of the gastric microbiota [[4], [5], [6], [7], [8], [9], [10], [11]]. The tremendous complexity of the gastric microbiota shows significant differences with the microbiota described in the mouth and oesophagus, indicating that the stomach may be home to a distinct microbial ecosystem. The high number and immense diversity of bacteria within the stomach can influence metabolism, tissue development, inflammation, and immunity [12,13], and gastric dysbiosis has been linked to various pathological conditions, including GC [14]. Aviles-Jimenez et al. demonstrated a gradual shift in the gastric microbiota profiles from non-atrophic gastritis (NAG) to intestinal metaplasia (IM) to intestinal-type GC [8], and Tseng et al. reported changes in the gastric microbiota before and after surgical treatment (subtotal gastrectomy) of GC [15]. Recently, Coker et al. demonstrated that the microbial composition changes with GC progression and they identified the differences in bacterial interactions across the different stages of gastric carcinogenesis [16].
The various habitats throughout the body contribute to the diversity and composition of the microbiota and are in a dynamic state of change according to their distinct atmospheric and nutritional compositions that provide a setting for symbiotic interactions among the various microbes within that ecosystem and the host. However, the gastric microbiota shows remarkable heterogeneity at different sites within the stomach and at the same site at different physiological and pathophysiological states [15]. Distinct pH, oxygen, nutrients, ions, and chemicals may vary considerably in tumor tissues and adjacent tumor-free tissues, which can be considered as different microhabitats that preferentially support the colonisation of certain bacterial strains. Previous studies have found that tumor-inciting microbes, such as HP, are gradually eliminated by changes in the tumor microenvironment. These changes result in an environment that is no longer hospitable to the microbe [17,18]. As different bacterial species preferentially inhabit tumor microenvironments, changes in a particular microbial species may modify the gastric microbiota towards a more carcinogenic bacterial community. To establish the role of the gastric microbiota in GC, it is important to determine the differences in the colonisation of gastric microbiota in tumors compared to tumor-free tissues.
In the present study, a large number of GC subjects without preoperative chemotherapy was enrolled retrospectively, and mucosal tissues were collected from different stomach microhabitats. We aimed to assess the diversity and composition of the gastric microbiota across tumoral and peritumoral microhabitats, and compare them to those in the same subjects with normal gastric mucosal morphology. We emphasized the major shifts in the gastric tumor microbiota relative to that of matched, normal gastric tissue from the same individual, which allow us to survey microbial communities in stomach microhabitats with an intrinsic control for the effects of environment and host genetics. This study will provide a better understanding of the microbial transition in tumors and tumor-free tissues, and of the association between bacterial colonisation and GC development. Ultimately, this will help to identify novel microbiome-related diagnostic tools and therapeutic interventions.
2. Materials and methods
2.1. Patients
In total, 276 GC patients without preoperative chemotherapy were enrolled from March 2009 to August 2013 from the First Affiliated Hospital, School of Medicine, Zhejiang University (Zhejiang, China). Stomach tissues were obtained from patients with primary GC who accepted gastrectomy. Finally, 229 tumoral tissues, 247 peritumoral tissues and 230 normal tissues were selected for microbiota analysis, which were based on DNA amount and quality appropriate for 16S rRNA gene amplification and sequence analysis. The tumor and tumor-free tissues were collected and confirmed by pathological diagnosis. >99% of the GC was moderately/poorly differentiated. According to Lauren, GC can be divided into adenocarcinomas of diffuse and intestinal types [19]. 96 intestinal-type patients, 56 diffuse-type patients and 124 mixed-type patients were included. Patients were determined to be HP positive (HP+) by positive result in rapid urease test or histopathology. The tumor and tumor-free (2-5 cm adjacent to the cancer tissue, Peritumor; >5 cm adjacent to the cancer tissue, Normal) tissues were collected, which were confirmed by pathological diagnosis. The clinical and pathological staging were based on the 7th edition American Joint Committee on Cancer (AJCC) cancer staging manual of GC TNM Staging [20]. Tumor stage was categorized as follows: stage I, invades mucosa or submucosa; stage II, invades mucularis mucosa; stage III, invades adventitia; stage IV, invades adjacent structures. 142 cases of early-stage GC (AJCC pathologic stage I and II) and 134 cases of late-stage GC (AJCC pathologic stage III and IV) were included [21]. The following criteria were used to exclude subjects: body mass index (BMI = weight in kilograms divided by the height in meters squared) > 30; use of antibiotics, probiotics, prebiotics, or synbiotics in the previous month; preoperative chemotherapy, radiotherapy, or other biological treatment before gastrectomy. Basic demographic data, clinical data and information about possible confounders of microbiota analysis were collected at the time of inclusion for all GC patients (Table 1). All research was approved by the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (China). Informed written consent was obtained from each of the patients before enrollment.
Table 1.
Summary of the study subjects' characteristics.⁎
| Characteristics | Patients (n = 276) |
|---|---|
| Age (means ± SD) | 61.11 ± 11.82 |
| Gender(Female/Male) | 81/195 |
| Weight(Kg, means ± SD) | 61.24 ± 10.19 |
| Height(cm, means ± SD) | 164.8 ± 7.1 |
| BMI(means ± SD) | 22.46 ± 3.32 |
| Complications, no | |
| Hypertension | 74 |
| Diabetes mellitus | 17 |
| Tumor localization, no | |
| Proximal stomach | 35 |
| Body/Fundus | 109 |
| Antrum | 132 |
| Tumor differentiation, no | |
| High differentiated | 2 |
| Moderately/poor differentiated | 274 |
| Lauren typing, no | |
| Intestinal type | 96 |
| Diffuse type | 56 |
| Mixed type | 124 |
| Tumor stage, no | |
| I (Ia, Ib) | 57 |
| II (IIa, IIb) | 85 |
| III (IIIa, IIIb, IIIc) | 121 |
| IV | 13 |
| HP infection, no | |
| Positive | 193 |
| Negative | 83 |
| Antibiotics use, no | 0 |
| PPI use, no | 276 |
| Pre-operative chemotherapy, no | 0 |
| Sample collection | |
| Normal, no | 230 |
| Tumor stage (I/II/III/IV, no) | 44/68/108/10 |
| HP infection (positive/negative, no) | 166/64 |
| Peritumor, no | 247 |
| Tumor stage (I/II/III/IV, no) | 46/72/116/13 |
| HP infection (positive/negative, no) | 182/65 |
| Tumor, no | 229 |
| Tumor stage (I/II/III/IV, no) | 48/68/102/11 |
| HP infection (positive/negative, no) | 152/77 |
BMI: Body mass index; HP, Helicobacter pylori; no, number; PPI, Proton pump inhibitors; SD, standard deviation.
2.2. Samples collection, DNA isolation, amplicon library construction and sequencing
After gastrectomy, tissues were rinsed with sterile water, flash frozen in liquid nitrogen, and characterized by staff pathologists. Bacterial genomic DNA was extracted from tissue samples by using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions with minor modifications [22]. Amplicon libraries were constructed with Illumina sequencing-compatible and barcode-indexed bacterial PCR primers 319F/806R, which target the V3-V4 regions of 16S rRNA gene [23]. All PCR reactions were performed with KAPA HiFi HotStart ReadyMix using the manufacturer's protocol (KAPA Biosystems) and approximately 50 ng of extracted DNA per reaction. Thermocycling conditions were set at 95 °C for 1 min, 55 °C for 1 min, then 72 °C for 1 min for 30 cycles, followed by a final extension at 72 °C for 5 min. All PCR reactions were performed in 50 μl triplicates and combined after PCR. The amplicon library was prepared using a TruSeq™ DNA sample preparation kit (Illumina Inc., San Diego, CA, USA). Prior to sequencing, the PCR products were extracted with the MiniElute® Gel Extraction Kit (QIAGEN) and quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Electron Corporation) and Qubit 2.0 Fluorometer (Invitrogen). The purified amplicons were then pooled in equimolar concentrations and the final concentration of the library was determined by Qubit (Invitrogen). Negative DNA extraction controls (lysis buffer and kit reagents only) were amplified and sequenced as contamination controls. Sequencing was performed on a MiSeq instrument (Illumina) using a 300 × 2 V3 kit together with PhiX Control V3 (Illumina).
2.3. Bioinformatic analysis
The 16S rRNA gene sequence data set generated from the MiSeq run were first merged and demultiplexed into per samples using the QIIME version 1.9.0 with default parameters [24]. Chimera sequences were detected and removed using the USEARCH software based on the UCHIME algorithm [25]. Open-reference operational taxonomic unit (OTU) pick was then performed with USEARCH V7 referenced against Greengenes database version 13.8 at 97% sequence similarity [26,27]. OTUs with a number of sequences < 0.005% of the total number of sequences were discarded as recommended [28]. The result was an OTU table, which was used for downstream analysis.
For taxonomic assignment, the most abundant sequences were chosen as the representative sequences of corresponding OTUs. Taxonomic assignment of individual datasets were classified against the Greengenes database version 13.8 using both RDP classifier and UCLUST version 1.2.22 implemented in QIIME [27,29]. Any sequences that were identified as members of Eukarya, Archaea, Mitochondria, Chloroplasts and Cyanobacteria lineages, were removed. Alpha diversity was calculated with QIIME software with Python scripts base on the sequence similarity at 97% level, including index of observed species, abundance-based coverage estimator (ACE), Chao1 estimator, Shannon, Simpson, Evenness and PD whole tree. Sequence coverage was assessed in mothur by rarefaction curves and Good's coverage [30,31]. Beta diversity was measured by unweighted and weighted UniFrac distance calculated with 10 times of subsampling by QIIME. These distances were visualized by principal coordinate analysis (PCoA) [32]. Hierarchical clustering was performed using Spearman's rank correlation coefficient as a distance measure and a customized script developed in R version 3.5.1. The output file was further analyzed using Statistical Analysis of Metagenomic Profiles software package (STAMP) version 2.1.3 [33].
For the predictive functional analyses, PiCRUSt software package version 1.0.0 was used to identify predicted gene families and associated pathways from inferred metagenomes of taxa of interest identified from the compositional analyses, which was based on the fact that phylogeny and function are closely linked [34]. Predicted functional genes were categorized into Clusters of Orthologous Groups (COG) and into Kyoto Encyclopedia of Genes and Genome (KEGG) orthology (KO), and compared across patient groups using STAMP. Pathways and enzymes were assigned using KEGG database options built into the pipeline. The pathways that were nonprokaryotic, had fewer than 2 sequences in each cohort, or had a difference in mean proportions < 0.1% were excluded from analysis. The characterization of microorganismal features differentiating the gastric microbiota was performed using the linear discriminant analysis (LDA) effect size (LEfSe) method (http://huttenhower.sph.harvard.edu/lefse/), which emphasizes both statistical significance and biological relevance [35]. With a normalized relative abundance matrix, LEfSe uses the Kruskal-Wallis rank sum test to detect features with significantly different abundances between assigned taxa and performs LDA to estimate the effect size of each feature. A significant alpha at 0.05 and an effect size threshold of 2 were used for all biomarkers discussed in this study.
Correlation analysis was performed using sparse compositional correlation (SparCC) algorithm on the complete OTU table collapsed to the genus level, which was introduced by Friedman and Alm and was known for its robustness to the compositional effects that are influenced by the diversity and sparsity of correlation in human microbiome data sets [36]. SparCC was employed to represent co-abundance and co-exclusion networks between OTUs. For SparCC, 1000 bootstrap replicates were used to calculate significance values, and considered correlation coefficients greater or <0.2 and −0.2 respectively and p-values < .05. This set of iterative procedures were applied separately to normal, peritumor and tumor data sets to infer the basis correlation values within and/or between paired sampling sites. Visualization of the network was achieved using Cytoscape version 3.4.0.
2.4. Statistical analysis
For continuous variables, independent t-test, White's nonparametric t-test, and Mann-Whitney U test were applied. For categorical variables between groups, Pearson chi-square or Fisher's exact test was used, depending on assumption validity. For taxon among subgroups, ANOVA test was applied (Tukey-Kramer was used in Post-hoc test, Effect size was Eta-squared). For correlation analyses, Spearman's rank correlation test was used. False-discovery rate (FDR) was calculated according to Benjamini-Hochberg, FDR-corrected p values were denoted as QFDR and was used when performing all untargeted screening analyses of different taxa. Statistical analysis was performed using the SPSS V19.0 (SPSS Inc., Chicago, IL) and STAMP V2.1.3 [33]. GraphPad Prism version 6.0 (San Diego, CA) was used for preparation of graphs. All tests of significance were two sided, and p < .05 or corrected p < .05 was considered statistically significant.
2.5. Accession number
The sequence data from this study were deposited in the GenBank Sequence Read Archive with the accession number SRP128749.
3. Results
3.1. Altered gastric mucosal microbiota in GC microhabitats
In the present study, the possible confounders of microbiota analyses such as the gender and age of GC patients in each GC stomach microhabitat, were not significantly different (p > .05). To investigate the gastric microbiota in different stomach microhabitats, we obtained 39,188,435 high-quality reads with an average of 55,507 reads per sample for the microbiota analysis (Table 2). Good's estimator of coverage was nearly 100%, indicating that the identified reads represented the majority of bacterial sequences present in the stomach. Diversity indices, such as Shannon, Simpson and Heip evenness, were significantly decreased in peritumoral microhabitats (Fig. 1a–c), while richness indices, such as ACE, observed species and phylogenetic diversity (PD) whole tree, were also decreased in peritumoral and tumoral microhabitats (Fig. 1d–f). In normal microhabitats, higher species richness and more low-abundance OTUs were observed (Fig. 1g–h), with more than twice the number of unique OTUs obtained (Fig. 1i). Due to significant inter-individual variation, principal coordinate analysis (PCoA) could not separate the three microhabitats into different clusters (Fig. S1).
Table 2.
Comparison of phylotype coverage and diversity estimation of the 16S rRNA gene libraries at 97% similarity.
| Group | No. of reads | No. of OTUsa | Good's (%) | ACE | Chao 1 | PD whole tree | Shannon | Simpson | Heip |
|---|---|---|---|---|---|---|---|---|---|
| Normal | 12,109,166 | 9908 | 99.995% | 10,145 | 10,312 | 417.81 | 7.4155 | 0.9176 | 0.017152 |
| Peritumor | 14,701,752 | 7647 | 99.994% | 7911 | 8184 | 324.24 | 6.6757 | 0.8920 | 0.013567 |
| Tumor | 12,377,517 | 6837 | 99.996% | 6964 | 7132 | 299.67 | 7.7465 | 0.9660 | 0.031440 |
The operational taxonomic units (OTUs) were defined at the 97% similarity level.
Fig. 1.
The diversity and richness of the gastric microbiota in different stomach microhabitats. The diversity indices, such as Shannon (a), Simpson (b) and Heip evenness (c), and the richness indices, such as ACE (d), observed species (e) and PD whole tree (f), were used to evaluate the overall structure of the gastric microbiota in the three stomach microhabitats. Data are presented as mean ± standard deviation. Unpaired t-tests (two-tailed) were used to analyse variation among the three stomach microhabitats. Rarefaction curves were used to estimate the richness (at a 97% level of similarity) of the gastric microbiota among the three groups (g). The vertical axis shows the number of OTUs expected after sampling the number of tags or sequences shown on the horizontal axis. Rank abundance curves of bacterial OTUs derived from the three groups, which indicated that the majority of the OTUs were present at low abundance in the gastric microbiota samples with greater sequencing depth. (h). The Venn diagram illustrates the overlap of OTUs in the gastric microbiota among the three microhabitats (i).
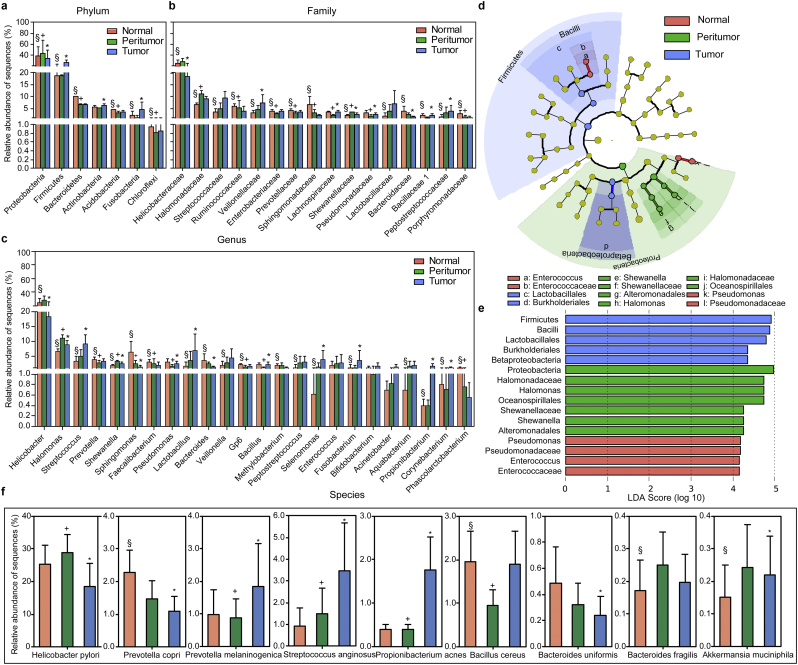
Generally, the gastric microbiota was dominated by Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, Acidobacteria, and Fusobacteria in descending order (Fig. 2a), which is distinct from the dominant phyla identified in the gut microbiota [37,38]. The Proteobacteria/Firmicutes ratio was significantly increased in peritumoral microhabitats (6.41 ± 10.63 in normal microhabitat Vs. 11.09 ± 17.40 in peritumoral microhabitat, p = .000; 11.09 ± 17.40 in peritumoral microhabitat Vs. 4.66 ± 11.67 in tumoral microhabitat, p = .000; 6.41 ± 10.63 in normal microhabitat Vs. 4.66 ± 11.67 in tumoral microhabitat, p = .177; Mann-Whitney U test). The top 16 families and 24 genera of the gastric microbiota are shown in Fig. 2b and c. Notably, the abundant genera, such as Helicobacter, Halomonas and Shewanella, were enriched in the peritumoral microhabitat, while Streptococcus, Selenomonas, Fusobacterium, Propionibacterium, and Corynebacterium were enriched in the tumoral microhabitat. A heatmap depicting the most abundant genera identified in the gastric microbiota demonstrated correlations between the stomach microhabitats and the abundance of selected genera (Fig. S2). Discriminant analyses using LEfSe showed that 16 bacterial phylotypes were significantly different among the three microhabitats (Fig. 2d–e). More differential bacterial phylotypes were also identified in the stomach microhabitats (Fig. S3). We also observed that the species of HP, Prevotella copri, Prevotella melaninogenica, Streptococcus anginosus, Propionibacterium acnes, Bacillus cereus, Bacteroides uniformis, Bacteroides fragilis and Akkermansia muciniphila were significantly different across the three groups (Fig. 2f). In contrast to previous reports, our deep sequencing data indicated that HP, P. copri and B. uniformis were significantly decreased, while P. melaninogenica, S. anginosus and P. acnes were increased in the tumoral microhabitat [39].
Fig. 2.
Different bacterial taxa among the three stomach microhabitats. Comparisons of the relative abundance of dominant bacterial taxa at the level of bacterial phylum (a), family (b) and genus (c). LEfSe identifies the taxa with the greatest differences in abundance among the three stomach microhabitats (d). Only the taxa meeting a significant LDA threshold value of >2 are shown (e). Nine differentially abundant bacterial species were also identified among the three microhabitats (p < .05, f; Mann-Whitney U tests). Data are presented as mean ± standard deviation. Mann-Whitney U tests were used to analyse variation among the three stomach microhabitats. §, p < .05 between normal and peritumoral tissues; +, p < .05 between peritumoral and tumoral tissues; *, p < .05 between normal and tumoral tissues.
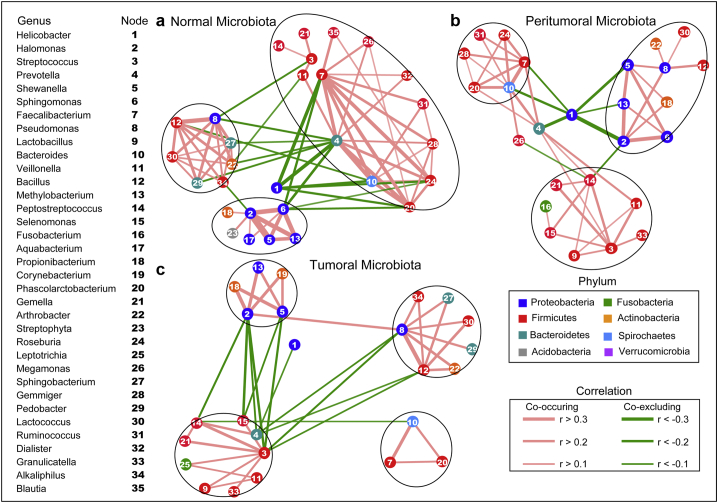
The overall structure of the gastric microbiota is the result of dynamic interactions between community members. A SparCC algorithm with FDR adjustments was employed to generate correlation-based microbial interaction networks based on the relative abundance of OTUs across the three microhabitats (Fig. 3). The correlation networks formed different bacterial clusters in the three groups, with a more complex network of interactions in normal microhabitats than that in peritumoral and tumoral microhabitats, especially within or between the predominant Proteobacteria and Firmicutes phyla. The most dominant member, Helicobacter, was negatively correlated with Prevotella, Bacteroides, Faecalibacterium, Phascolarctobacterium and Roseburia. Those genera showed mainly positive correlations within the same bacterial clusters. The genera of Halomonas, Shewanella, Methylobacterium and Sphingomonas demonstrated strong positive correlations in normal microbiota. However, most of these correlations were no longer significant in peritumoral and tumoral microbiota, especially the interactions between Helicobacter and other genera in the two stomach microhabitats. One new, strong negative correlation was formed between Helicobacter and Halomonas in the peritumoral microbiota. We also observed that other correlations between different genera were weakened or even lost in the tumoral microbiota.
Fig. 3.
Correlation strengths of the abundant gastric microbiota in different stomach microhabitats. Correlation network of the abundant gastric microbiota in normal microhabitat (a), peritumoral microhabitats (b) and tumoral microhabitats (c). The correlation coefficients were calculated with the Sparse Correlations for Compositional data algorithm (SparCC). Cytoscape version 3.4.0 was used for network construction. Red and green lines represent positive and negative correlations, respectively. The correlation network became simpler. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Microhabitat-specific gastric microbiota across different GC stages and types
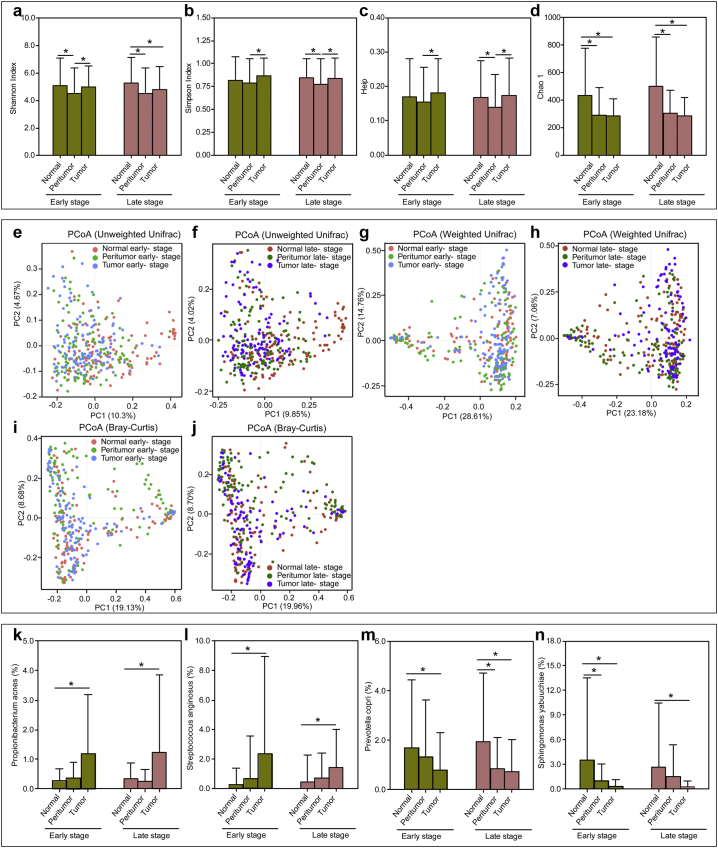
To identify whether the gastric microbiota was different based on GC stages, we examined the taxonomic differences between early-stage and late-stage GC in stomach microhabitats. We enrolled 142 early-stage and 134 late-stage patients, including 112/118, 118/129, and 116/113 for normal, peritumoral, and tumoral tissues, respectively. Interestingly, the bacterial diversity was not significantly different between two stages in the same microhabitat (Fig. 4a–d). Decreased diversity in peritumoral microbiota and decreased richness in peritumoral and tumoral microhabitats were observed in both stages. In addition, PCoA could not distinguish stage-related changes in these groups based on the unweighted UniFrac distance, weighted UniFrac distance and Bray-Curtis distance (Fig. 4e–j). Altered profiles of the gastric microbiota showed similar trends across different stages (Fig. S4a–c). We also found that P. acnes, S. anginosus, P. copri and Sphingomonas yabuuchiae were different among the three microhabitats in the same GC stage (Fig. 4k–n). LEfSe showed subtle clade differences among three early-stage and late-stage GC microhabitats (Figs. S5 and S6).
Fig. 4.
Overall structure of the early-stage and late-stage gastric microbiota in the three stomach microhabitats. The α-diversity indices such as Shannon (a), Simpson (b), Chao 1 (c) and Heip evenness (d) were estimated by QIIME. Data are presented as mean ± standard deviation. Unpaired t-tests (two-tailed) were used to analyse variation among the three microhabitats in the same GC stage. Plots of principal coordinate analysis (PCoA) of the gastric microbiota in the early-stage and late-stage based on the unweighted UniFrac distance (e and f), weighted UniFrac distance (g and h) and Bray-Curtis distance (i and j). Propionibacterium acnes (k), Streptococcus anginosus (l), Prevotella copri (m) and Sphingomonas yabuuchiae (n) were different among the three microhabitats in the same GC stage. Data are presented as mean ± standard deviation. Mann-Whitney U tests were used to analyse variation among the three stomach microhabitats. *p < .05.
Overall, 96 intestinal-type patients, 56 diffuse-type patients and 124 mixed-type patients were included. Shannon was clearly lower in the peritumoral microhabitat, and Chao 1, ACE, PD whole tree and observed species were also significantly decreased in peritumoral and tumoral microhabitats, when compared between the same GC types (Fig. S7). In stomach microhabitats, several non-dominant bacterial phylotypes were different between intestinal and diffuse GC types (Fig. S8). However, there was no significant difference in the composition of the gastric microbiota between intestinal- and diffuse-GC types in the same stomach microhabitat. In combination with GC stages, these data suggested that the specific stomach microhabitats affect the diversity and composition of the gastric microbiota, regardless of GC type.
3.3. HP infection (HPI) and gastric mucosal dysbiosis
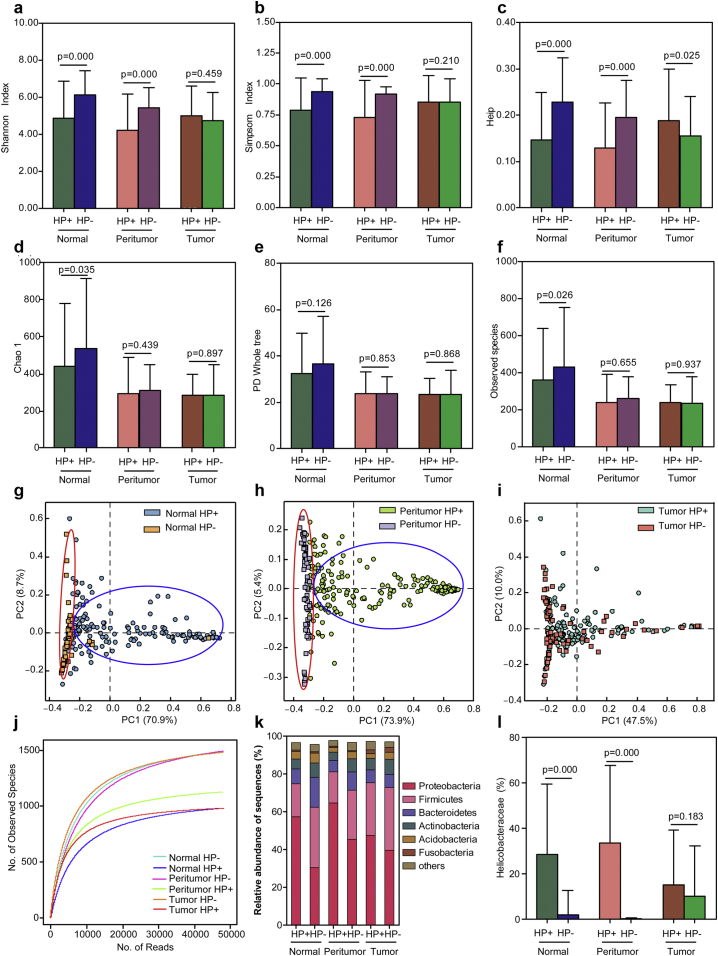
To investigate whether HP colonisation influences the overall structure and composition of the gastric microbiota in specific microhabitats, we examined changes in the gastric microbiota in patients with histopathological HP+ and H. pylori negative (HP−). Approximately 70% of GC patients were diagnosed as histopathological HP+. Except for tumoral tissues, our results found a tendency towards a reduction in bacterial diversity (lower in the HP+ group; higher in the HP− group; Fig. 5a–c), while the observed species and Chao1, but not PD whole tree, were significantly higher in the normal HP− group (Fig. 5d–f) than those in both normal and peritumoral tissues. Interestingly, PCoA divided the gastric microbiota into different bacterial clusters between HP+ and HP− groups in normal and peritumoral microbiota (Fig. 5g–h), but not in tumoral microbiota (Fig. 5i). Combined with the higher HP colonisation found in these microhabitats, our results indicated that HPI may be one of the major determinants for the bacterial diversity of the gastric microbiota. The rarefaction curves showed that the richness of the gastric microbiota in HP− groups was significantly higher than that in HP+ groups (Fig. 5j). The dominant phyla of the gastric microbiota are shown in Fig. 5k, with a higher Proteobacteria/Firmicutes ratio in HP+ groups of normal and peritumoral tissues compared to matched HP− groups. Despite these samples being histopathologically HP−, the Helicobacteraceae family could also be detected by sequencing, especially in the tumoral microbiota (Fig. 5l).
Fig. 5.
Altered gastric microbiota in the three stomach microhabitats influenced by Helicobacter pylori infection (HPI). HPI influenced the diversity indices of gastric microbiota in different stomach microhabitats such as Shannon (a), Simpson (b), Heip evenness (c), and the richness indices such as Chao 1 (d), PD whole tree (e) and observed species (f). Data are presented as mean ± standard deviation. Unpaired t-tests (two-tailed) were used to analyse variation between HP+ and HP− groups in the three stomach microhabitats. Plots of principal coordinate analysis (PCoA) of the gastric microbiota influenced by HPI in normal microhabitats (g), peritumoral microhabitats (h) and tumoral microhabitats (i) based on the unweighted UniFrac metric. Rarefaction analysis of the gastric microbiota from HP+ and HP− groups in three stomach microhabitats (j). Taxonomic differences between the gastric microbiota of HP+ and HP− groups in the three microhabitats at the phylum level (k). The different relative abundance of HP in histopathological HP+ and HP− groups in the three microhabitats (l). Data are presented as mean ± standard deviation. Mann-Whitney U tests were used to analyse variation between the HP+ and HP− groups in the three stomach microhabitats.
To identify specific changes of the gastric microbiota that correlated with HPI in each GC microhabitat, we compared the composition of the gastric microbiota between HP+ and HP− samples using LEfSe and STAMP. Consistent with the previous diversity analyses, normal and peritumoral microbiota exhibited similar changing patterns (Figs. S9a and S10a), whereas more altered bacterial phylotypes were found in normal microbiota after HPI. However, the altered phylotypes in tumoral microbiota were mostly non-dominant microorganisms, except Helicobacter (Fig. S11a). The community composition was further demonstrated by the significant differences between the HP+ and HP− groups in each stomach microhabitat using STAMP (Figs. S9b, S10b and S11b). At the species level, P. copri, B. cereus and B. uniformis were enriched in the normal HP− group, while P. copri, Bacteroides plebeius, Akkermansia muciniphila, B. uniformis and B. fragilis were enriched in the peritumoral HP− group. However, only P. acnes and two non-dominant species, such as Acinetobacter schindleri and B. stercoris, were enriched in the tumoral HP− group. In addition, these oscillating genera within each tumoral microhabitat are shown in the heatmaps, with a gradual increase of Shannon in both the HP+ and HP− groups (Figs. S12a–b, S13a–b, and S14a–b). Notably, our data demonstrate that HP is negatively correlated with Shannon regardless of the stomach microhabitat, which confirms the critical roles of HP in bacterial diversity of the gastric microbiota (Figs. S12c, S13c and S14c). ROC analysis was performed to assess the values of gastric microbiota profiling as a diagnostic tool to discriminate between HP+ and HP−. Without HP included, other differential genera provided an area under the curve (AUC) in the ROC analysis of 0.796 in normal microbiota, 0.693 in peritumoral microbiota, and 0.700 in tumoral microbiota to distinguish HPI (Figs. S12d, S13d, S14d). Our present data suggest that HPI contributes to gastric dysbiosis in stomach microhabitats.
3.4. Inferred functional changes in GC-associated gastric mucosal microbiota
The functional content of the gastric microbiota was predicted by PiCRUSt based on closed-reference OTU picking. In our present study, 19 Clusters of Orthologous Groups (COG) functional categories were tested, which identified 6 differentially abundant COGs with a QFDR < 0.05 between normal and peritumoral microbiota, 8 between normal and tumoral microbiota, and 10 between peritumoral and tumoral microbiota (Fig. 6a). Eleven COG categories, including carbohydrate transport and metabolism, coenzyme transport and metabolism, nucleotide transport and metabolism, and amino acid transport and metabolism, exhibited the most significant differences among the three microhabitats (QFDR < 0.05, Fig. 6b; Benjamini–Hochberg FDR method). Among these differential COGs, nucleotide transport and metabolism, amino acid transport and metabolism and inorganic ion transport and metabolism were significantly enriched in the tumoral microbiota. In addition, we also compared 64 Kyoto Encyclopedia of Genes and Genome (KEGG) pathways at level 2. At an FDR of 0.05, we identified more differentially abundant pathways between tumor microbiota and others (Fig. S15). Consistent with the significant alterations of HP-associated gastric microbiota, the KEGG pathways were changed between the HP+ and HP− groups in normal and peritumoral tissues (Fig. S16). Specifically, gastric acid secretion was significantly higher in the HP+ group of the tumoral microbiota. Together, these functional changes in the gastric microbiota may contribute to GC initiation and progression.
Fig. 6.
PiCRUSt-based gastric microbiome study in the three different stomach microhabitats. The different bacterial functions in the three stomach microhabitats were evaluated between each other based on two-sided Welch's t-test (a). Comparisons among the three stomach microhabitats for each COG functional category shown by percentage (b). The Benjamini-Hochberg method was used for multiple testing correction based on the false discovery rate (FDR) by STAMP.
4. Discussion
The GC tumor microenvironment is colonised with site-specific gastric microbiota that foster symbiotic interactions, which has been considered as an important factor in GC initiation and progression. The normal microenvironment acts as a barrier to tumorigenesis, while the tumor microenvironment can induce and promote cancer. Currently, considerable progress has been made towards understanding the altered composition and diversity of the gastric microbiota in gastric diseases, which have always considered the whole stomach as one habitat [4,5,[7], [8], [9], [10], [11],15,16]. Tseng et al. found that GC tissue and neighbouring normal tissue had similar gastric microbiota in early-stage GC patients [15], while Li et al. reported that there was little difference in the gastric microbiota between antrum and body biopsy specimens [40]. However, the stomach microhabitats are not uniform; rather, they vary considerably across sites within the stomach, at the same site over time, and with health status. The different nutritional compositions within the stomach microhabitats contribute to altered diversity and composition of the gastric microbiota. It is more difficult to obtain healthy stomach tissues as controls; therefore, our present, large-scale trial study screened those confirmed GC tumoral, and peritumoral tissue, and neighbouring normal tissue from the same GC patient to investigate discrepancies in the gastric microbiota. In contrast to previous studies, our self-control study found alterations of diversity and composition of the gastric microbiota in tumor and tumor-free microhabitats, which affect both bacteria-bacteria interactions as well as bacteria-host interactions that occur in stomach microhabitats during GC development. Previous studies on colorectal cancer have suggested that different bacterial species preferentially inhabit the tumor sites but not tumor-free sites [41,42]. The altered gastric microbiota influences inflammation and immunity both locally at the mucosal level and systemically. Our previous study demonstrated that patients with GC had increased regulatory T cells (Tregs) in peripheral blood and carcinoma tissue [43]. Disorders of the intimate interactions between the gastric microbiota in the tumor microenvironment and immune system may contribute to GC development by eliciting tumor-promoting immune responses.
To our knowledge, this is the first large-scale trial to explore specific changes of the gastric microbiota in GC stomach microhabitats using high-throughput sequencing techniques. Previous studies have found changes in the gastric microbiota across stages of gastric carcinogenesis in patients with NAG, IM and GC [8,16,44]. A Chinese pilot study reported that gastric microbiota features are associated with cancer risk factors and clinical outcomes in patients with gastric cardia cancer [5]. Consistent with the previous studies, we also found altered diversity and composition of the gastric microbiota in stomach microhabitats. Specifically, the bacterial richness showed a marked decreasing trend from normal to peritumoral to tumoral tissues, demonstrating that the altered microenvironment of the tumoral sites is not suitable for specific bacterial colonisation. The GC stomach microhabitat, but not GC stage, type, or cell differentiation, determined the overall structure of the gastric microbiota. Interestingly, our present study also found that the correlation network in the tumoral microhabitat became much simpler.
As a class I carcinogen of GC, HP in the tumoral sites is classically believed to contribute to GC tumorigenesis, and mass eradication of HPI significantly decreases the risk of developing cancer in infected individuals without pre-malignant lesions [[45], [46], [47]], reinforcing the theory that HP influences early stages in gastric carcinogenesis. HP is the strongest known risk factor for both diffuse-type and intestinal-type GC, and it uses various mechanisms to dampen host immune responses and persist in the stomach [48]. In fact, HP acts by a “hit and run” mechanism for GC and is no longer present in the intratumoral microhabitat at the time when GC is identified [17]. Our present findings confirmed that HP was significantly decreased in the tumoral microhabitat. The altered tumoral microhabitat with loss of specialized glandular tissue and decreased acid secretion might lead to the decrease of HP [5]. We also demonstrated that the presence or absence of HP led to significantly different population structures in normal and peritumoral microhabitats [49], correlating with the relative abundance of HP. A previous study found that persistent HPI of the gastric mucosa influences gastric inflammatory gene expression resulting in AG, a condition associated with a reduced capacity for gastric acid secretion and an increased risk of GC [49]. The high prevalence of HPI with reduced gastric acid secretion in peritumoral and normal microhabitats might allow for the survival and proliferation of other microbes, such as Halomonas, Prevotella and Streptococcus, which are normally killed by the acidic environment, resulting in the initiation of gastric carcinogenesis [50]. As a carcinogenic pathogen, HP might actively participate in GC by changing the gastric mucosal immunity, especially the imbalance of Treg/Th17. However, HP colonisation in the stomach alone is not sufficient to induce gastric carcinogenesis. Lofgren et al. demonstrated that HP-induced GC is promoted by the presence of a complex microbiota, as HP mono-associated mice developed fewer tumors than their specific pathogen-free counterparts in a hypergastrinaemic transgenic mouse model [51]. This may be explained by increased conversion of dietary nitrates, such as N-nitrosamines and N-nitrosamides, that might be attributed to HPI-associated gastric microbiota alterations, which would ultimately promote GC development [52,53]. Compared with the tumoral microhabitat, the inferred function of the gastric microbiota by PiCRUSt changed significantly in normal and peritumoral microhabitats with or without HP colonisation, and this may contribute to the initiation of gastric carcinogenesis. In contrast to the roles of HP in the promotion of gastric carcinogenesis, HP colonisation in the stomach has been suggested to protect against oesophageal adenocarcinoma. This is due to down-modulation of gastric acid secretion [54], which emphasizes the organ-specific effects of bacterially driven carcinogenesis.
Interestingly, P. melaninogenica, S. anginosus and P. acnes were enriched in tumoral microhabitat and P. copri and B. uniformis decreased significantly, whereas B. fragilis and A. muciniphila showed a similar changing pattern between peritumoral and tumoral tissues. P. acnes, a classic skin bacterium, has recently been identified as a member of the gastric microbiota, which is a microhabitat-preferred bacterium that is found only in mucosal specimens but not in the gastric fluid [4,55,56]. In this study, the tumoral microhabitat was characterized by overabundance of Propionibacterium acnes, which is an important participant in GC tumorigenesis. P. acnes and its products, mainly short-chain fatty acids, trigger a possible corpus-dominant lymphocytic gastritis [57]. Tumoral microhabitat-enriched oral-originated S. anginosus, which was significantly decreased in the normal and peritumoral microhabitats, has been found to be associated with GC, and has significant centralities in the GC ecological network [16]. In combination with other biomarkers, such as Peptostreptococcus stomatis, Parvimonas micra, Slackia exigua and Dialister pneumosintes, S. anginosus distinguished GC from AG with an AUC of 0.81 [16]. Andersson et al. also demonstrated that Streptococcus was the most dominant genus in the stomach in the absence of HPI [58]. P. melaninogenica, an oral and respiratory pathogen, was also observed to be increased in the tumoral microhabitat. Dong et al. found that P. melaninogenica was the dominant species of the genus Prevotella in the stomach microbiota, making up an average of 9.17% for the NAG group and 6.95% for the chronic AG group [59]. However, unlike S. anginosus and P. melaninogenica, P. copri was significantly decreased in peritumoral and tumoral microhabitats. Hollister et al. reported that P. copri was detected in stool specimens but rarely observed in samples from other body sites [60], while our present study showed that P. copri was one of the dominant species in the gastric mucosal microbiota. Using 454 pyrosequencing, Scher et al. demonstrated that the presence of P. copri in fecal microbiota was strongly correlated with disease in new-onset untreated rheumatoid arthritis patients [61]. However, the role of P. melaninogenica and P. copri in the tumor microenvironment in GC pathogenesis requires further investigation. In addition, B. cereus, a food-borne pathogen that causes diarrhoeal disease in human, was found to be significantly decreased in the peritumoral microhabitat. A previous study showed that spores or vegetative B. cereus cells can survive the pH barrier and pepsin of the stomach and reach to the small intestine where they produce toxins in sufficient amounts [62]. As an opportunistic human pathogen, B. fragilis enterotoxin may have the potential to contribute to oncogenic transformation in the colon [63]. As a common member of the colonic microbiota, mucin-degrading A. muciniphila could increase the number of intestinal tumors, the thickness of the intestinal mucus layer, and the density of mucin-producing goblet cells, which are prime candidates for microbiota-borne modulation of intestinal tumorigenesis [64,65]. Consistent with previous studies, both B. fragilis and A. muciniphila were enriched in peritumoral and tumoral tissues, and might participate in the process of gastric carcinogenesis. Further mechanistic studies are required to explore the roles and mechanisms of these differential bacterial species among different GC tumoral environments in GC development. Furthermore, evaluation of the interactions between GC microbiota and the gastric immune system, and specific microbial functions in cancer microenvironments might prove valuable.
In summary, we completed a large-scale analysis of GC tumors using high-throughput sequencing techniques and found that the diversity and composition of the gastric microbiota were significantly altered in different stomach microhabitats. In contrast to previous studies that considered the stomach as a whole, our study observed that the stomach microhabitats determined the overall structure and composition of the gastric microbiota, regardless of different GC stage and type. Interestingly, the carcinogenic pathogen H. pylori was significantly decreased in tumoral sites, which simplified the network of bacterial interactions in the gastric microbiota. The altered composition of the gastric microbiota in the three stomach microhabitats may be associated with its role in gastric carcinogenesis. However, the role of the different bacteria of the gastric microbiota in specific microhabitats remains unclear. Further studies are required to determine the gastric bacteria in tumoral microhabitats and host interactions in the process of gastric carcinogenesis.
Acknowledgments
Acknowledgements
The authors thank all of the participants who recruited patients in this study. We also thank Prof. Emad El-Omar and Dr. Fatima EL-Assaad for the valuable evaluation.
Funding sources
This present work was funded by the grants of Science and Technology Planning Project of Zhejiang Province (2015C33174), Zhejiang Province Key Science and Technology Innovation Team (2013TD13), Major Project of Science and Technology Department of Zhejiang Province (2014C03040-1), Project of Zhejiang Provincial Education Department (Y201326985) and the National Natural Science Foundation of China (81771724, 31700800, 81790633, 81273254, 31870839).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors' contributions
XSL and ZXL conceived and designed the experiments. XSL, XL, FJ, YM, YWC, FPL, CXY, LJL and ZXL performed the experiments. XSL, LS, and ZXL analyzed the data. XSL, LS and ZXL wrote the paper and edited the manuscript. The final manuscript was read and approved by all authors.
Conflicts of interest
The authors declare no conflicts of interest.
Consent for publication
We have obtained consents to publish this paper from all the participants of this study.
Ethics approval and consent to participate
All research conformed to the Helsinki Declaration and was approved by the Ethics Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University (China) and were implemented in accordance with the approved guidelines. Informed written consent was obtained from each of the patients before enrollment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.034.
Appendix A. Supplementary data
Supplementary material
Fig. S1 3D principal coordinate analysis (PCoA) plots of individual gastric mucosal microbiota based on unweighted (a) and weighted (b) UniFrac distance in normal microhabitat (n = 230), peritumoral microhabitat (n = 247) and tumoral microhabitat (n = 229) of gastric cancer patients. Each symbol represents a sample.
Fig. S2 Heatmap of the 76 key genera in gastric microbiota, which indicates the genus-level of gastric bacteria alters in normal microhabitat (n = 230), peritumoral microhabitat (n = 247) and tumoral microhabitat (n = 229) of gastric cancer patients. The colour of the spots in the panel represents the relative abundance (normalized and log10-transformed) of the genus in each sample. The relative abundance of the bacteria in each genus is indicated by a gradient of colour from red (low abundance) to blue (high abundance). The genera were organized by Spearman's correlation analysis based on their relative abundances. The taxonomic classifications of the genus are shown on the right. The corresponding Shannon index in each sample is shown under the heatmap.
Fig. S3 Taxonomic differences of the gastric mucosal microbiota among three stomach microhabitats. Related cladogram showing bacterial taxa significantly enriched in each group using the LDA model results for the bacterial hierarchy (upper panel) and principal coordinate analysis (PCoA) for gastric microbiota at three microhabitats (lower panel).
Fig. S4 Tumor stages of gastric cancer related changes of gastric mucosal microbiota. Cladogram represents the features of gastric mucosal microbiota among early-stage and late-stage of gastric cancer using the LDA model results for the bacterial hierarchy (a). Comparisons of the relative abundance at the level of bacterial phylum and genus in early stage (b) and late-stage (c) among the three stomach microhabitats. The data are presented as the mean ± standard deviation. Mann-Whitney U tests were used to analyse variation among three stomach microhabitats.⁎, p < .05; #, p < .01.
Fig. S5 Taxonomic differences of the gastric mucosal microbiota in early-stage of GC among three stomach microhabitats using LEfSe.
Fig. S6 Taxonomic differences of the gastric mucosal microbiota in late-stage of GC among three stomach microhabitats using LEfSe.
Fig. S7 Comparison of the diversity and richness of gastric mucosal microbiota between intestinal-type and diffuse-type of gastric cancer in each stomach microhabitat. The diversity indices of gastric microbiota such as Shannon (a), Simpson (b), and the richness indices such as ACE (c), Chao 1 (d), PD whole tree (e) and observed species (f) were used to evaluated the overall structure of gastric microbiota between two types of gastric cancer in each microhabitat. The data are presented as the mean ± standard deviation. Unpaired t-tests (two-tailed) were used to analyse variation between intestinal- and diffuse-type of GC in the three stomach microhabitats.
Fig. S8 Taxonomic differences of the gastric mucosal microbiota between intestinal-type and diffuse-type of gastric cancer. Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between intestinal- and diffuse-type of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR).
Fig. S9 Taxonomic differences of the gastric mucosal microbiota between histopathological H. pylori positive and negative (HP+ and HP−) groups of gastric cancer in normal microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to H. pylori infection (HPI) in normal microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S10 Taxonomic differences of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in peritumoral microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to HPI in peritumoral microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S11 Taxonomic differences of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in tumoral microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to HPI in tumoral microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S12 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in normal microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the normal microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Phascolarctobacterium, Roseburia, Streptococcus, Halomonas, Blautia, Shewanella, Ruminococcus and Dialister that were used to predict the presence of HPI in normal microhabitat (d).
Fig. S13 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in peritumoral microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the peritumoral microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Phascolarctobacterium, Veillonella, Megamonas, Gemmiger, Halomonas, Blautia and Shewanella that were used to predict the presence of HPI in peritumoral microhabitat (d).
Fig. S14 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in tumoral microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the tumoral microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Ruminococcus, Bacteroides, Carnobacterium, Paracoccus, Novosphingobium and Akkermansia that were used to predict the presence of HPI in tumoral microhabitat (d).
Fig. S15 Differentially abundant KEGG pathways were identified between tumor microbiota and others at an FDR of 0.05. The different KEGG pathways at level 2 were identified between normal and peritumoral microbiota (a), normal and tumoral microbiota (b), peritumoral and tumoral microbiota (c). Benjamini-Hochberg method was used for multiple testing correction based on false discovery rate (FDR) by STAMP.
Fig. S16 Differentially abundant KEGG pathways were identified between between histopathological HP+ and HP− groups in each tumoral microhabitat at an FDR of 0.05. The different KEGG pathways at level 2 were identified between histopathological HP+ and HP− groups in normal microhabitats (a), peritumoral microhabitat (b), and tumoral microhabitat (c). Benjamini-Hochberg method was used for multiple testing correction based on false discovery rate (FDR) by STAMP.
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Strong V.E., Wu A.W., Selby L.V., Gonen M., Hsu M., Song K.Y. Differences in gastric cancer survival between the US and China. J Surg Oncol. 2015;112:31–37. doi: 10.1002/jso.23940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bik E.M., Eckburg P.B., Gill S.R., Nelson K.E., Purdom E.A., Francois F. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu G., Hu N., Wang L., Wang C., Han X.Y., Humphry M. Gastric microbiota features associated with cancer risk factors and clinical outcomes: a pilot study in gastric cardia cancer patients from Shanxi, China. Int J Cancer. 2017;141:45–51. doi: 10.1002/ijc.30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin M.E., Bhatnagar S., George M.D., Paster B.J., Canfield D.R., Eisen J.A. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eun C.S., Kim B.K., Han D.S., Kim S.Y., Kim K.M., Choi B.Y. Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter. 2014;19:407–416. doi: 10.1111/hel.12145. [DOI] [PubMed] [Google Scholar]
- 8.Aviles-Jimenez F., Vazquez-Jimenez F., Medrano-Guzman R., Mantilla A., Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep UK. 2014;4:4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amir I., Konikoff F.M., Oppenheim M., Gophna U., Half E.E. Gastric microbiota is altered in oesophagitis and Barrett's oesophagus and further modified by proton pump inhibitors. Environ Microbiol. 2014;16:2905–2914. doi: 10.1111/1462-2920.12285. [DOI] [PubMed] [Google Scholar]
- 10.Yang I., Woltemate S., Piazuelo M.B., Bravo L.E., Yepez M.C., Romero-Gallo J. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep UK. 2016;6 doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira R.M., Pereira-Marques J., Pinto-Ribeiro I., Costa J.L., Carneiro F., Machado J.C. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee Y.K., Mazmanian S.K. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iida N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng C.H., Lin J.T., Ho H.J., Lai Z.L., Wang C.B., Tang S.L. Gastric microbiota and predicted gene functions are altered after subtotal gastrectomy in patients with gastric cancer. Sci Rep UK. 2016;6:20701. doi: 10.1038/srep20701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coker O.O., Dai Z., Nie Y., Zhao G., Cao L., Nakatsu G. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut. 2018;67:1024. doi: 10.1136/gutjnl-2017-314281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ota H., Katsuyama T., Nakajima S., El-Zimaity H., Kim J.G., Graham D.Y. Intestinal metaplasia with adherent Helicobacter pylori: a hybrid epithelium with both gastric and intestinal features. Hum Pathol. 1998;29:846–850. doi: 10.1016/s0046-8177(98)90455-5. [DOI] [PubMed] [Google Scholar]
- 18.Sears C.L., Garrett W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 20.Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 21.Datta J., McMillan M.T., Ruffolo L., Lowenfeld L., Mamtani R., Plastaras J.P. Multimodality therapy improves survival in resected early stage gastric cancer in the United States. Ann Surg Oncol. 2016;23:2936–2945. doi: 10.1245/s10434-016-5224-1. [DOI] [PubMed] [Google Scholar]
- 22.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7 doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald D., Price M.N., Goodrich J., Nawrocki E.P., Desantis T.Z., Probst A. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 28.Navas-Molina J.A., Peralta-Sanchez J.M., Gonzalez A., McMurdie P.J., Vazquez-Baeza Y., Xu Z.J. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good I.J. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- 31.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone C., Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parks D.H., Tyson G.W., Hugenholtz P., Beiko R.G. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langille M.G., Zaneveld J., Caporaso J.G., McDonald D., Knights D., Reyes J.A. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ling Z., Liu X., Luo Y., Yuan L., Nelson K.E., Wang Y. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genomics. 2013;14:390. doi: 10.1186/1471-2164-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 39.Seo I., Jha B.K., Suh S.I., Suh M.H., Baek W.K. Microbial profile of the stomach: comparison between normal mucosa and cancer tissue in the same patient. J Bacteriol Virol. 2014;44:162–169. [Google Scholar]
- 40.Li X.X., Wong G.L.H., To KF, Wong V.W.S., Lai L.H., Chow D.K.L. Bacterial microbiota profiling in gastritis without helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Z.G., Guo B.M., Gao R.Y., Zhu Q.C., Qin H.L. Microbiota disbiosis is associated with colorectal cancer. Front Microbiol. 2015;6:20. doi: 10.3389/fmicb.2015.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jobin C. Colorectal cancer CRC-all about microbial products and barrier function? Nat Rev Gastroenterol Hepatol. 2012;9:694–696. doi: 10.1038/nrgastro.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang X.M., Liu X.S., Lin X.K., Yu H., Sun J.Y., Liu X.K. Role of plasmacytoid dendritic cells and inducible costimulator-positive regulatory T cells in the immunosuppression microenvironment of gastric cancer. Cancer Sci. 2014;105:150–158. doi: 10.1111/cas.12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li T.H., Qin Y., Sham P.C., Lau K.S., Chu K.M., Leung W.K. Alterations in gastric microbiota after H. Pylori eradication and in different histological stages of gastric carcinogenesis. Sci Rep UK. 2017;7:44935. doi: 10.1038/srep44935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan K.F., Zhang L., Gerhard M., Ma J.L., Liu W.D., Ulm K. A large randomised controlled intervention trial to prevent gastric cancer by eradication of Helicobacter pylori in Linqu County, China: baseline results and factors affecting the eradication. Gut. 2016;65:9–18. doi: 10.1136/gutjnl-2015-309197. [DOI] [PubMed] [Google Scholar]
- 46.Wong B.C., Lam S.K., Wong W.M., Chen J.S., Zheng T.T., Feng R.E. Helicobacter pylori eradication to prevent gastric cancer in a high-risk region of China: a randomized controlled trial. JAMA. 2004;291:187–194. doi: 10.1001/jama.291.2.187. [DOI] [PubMed] [Google Scholar]
- 47.Lee Y.C., Chen T.H., Chiu H.M., Shun C.T., Chiang H., Liu T.Y. The benefit of mass eradication of Helicobacter pylori infection: a community-based study of gastric cancer prevention. Gut. 2013;62:676–682. doi: 10.1136/gutjnl-2012-302240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cerf-Bensussan N., Gaboriau-Routhiau V. The immune system and the gut microbiota: friends or foes? Nat Rev Immunol. 2010;10:735–744. doi: 10.1038/nri2850. [DOI] [PubMed] [Google Scholar]
- 49.Kienesberger S., Cox L.M., Livanos A., Zhang X.S., Chung J., Perez-Perez G.I. Gastric Helicobacter pylori infection affects local and distant microbial populations and host responses. Cell Rep. 2016;14:1395–1407. doi: 10.1016/j.celrep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engstrand L., Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27:39–45. doi: 10.1016/j.bpg.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 51.Lofgren J.L., Whary M.T., Ge Z., Muthupalani S., Taylor N.S., Mobley M. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011;140:210–220. doi: 10.1053/j.gastro.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lertpiriyapong K., Whary M.T., Muthupalani S., Lofgren J.L., Gamazon E.R., Feng Y. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the Helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grosse Y., Baan R., Straif K., Secretan B., El Ghissassi F., Cogliano V. Carcinogenicity of nitrate, nitrite, and cyanobacterial peptide toxins. Lancet Oncol. 2006;7:628–629. doi: 10.1016/s1470-2045(06)70789-6. [DOI] [PubMed] [Google Scholar]
- 54.Peek R.M., Jr., Blaser M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 55.von Rosenvinge E.C., Song Y., White J.R., Maddox C., Blanchard T., Fricke W.F. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J. 2013;7:1354–1366. doi: 10.1038/ismej.2013.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maldonado-Contreras A., Goldfarb K.C., Godoy-Vitorino F., Karaoz U., Contreras M., Blaser M.J. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montalban-Arques A., Wurm P., Trajanoski S., Schauer S., Kienesberger S., Halwachs B. Propionibacterium acnes overabundance and natural killer group 2 member D system activation in corpus-dominant lymphocytic gastritis. J Pathol. 2016;240:425–436. doi: 10.1002/path.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersson A.F., Lindberg M., Jakobsson H., Backhed F., Nyren P., Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong T., Feng Q., Liu F., Chang L.K., Zhou X., Han M. Alteration of stomach microbiota compositions in the progression of gastritis induces nitric oxide in gastric cell. Exp Ther Med. 2017;13:2793–2800. doi: 10.3892/etm.2017.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hollister E.B., Gao C., Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146:1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scher J.U., Sczesnak A., Longman R.S., Segata N., Ubeda C., Bielski C. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2 doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berthold-Pluta A., Pluta A., Garbowska M. The effect of selected factors on the survival of Bacillus cereus in the human gastrointestinal tract. Microb Pathog. 2015;82:7–14. doi: 10.1016/j.micpath.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 63.Toprak N.U., Yagci A., Gulluoglu B.M., Akin M.L., Demirkalem P., Celenk T. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 64.Dingemanse C., Belzer C., van Hijum S.A., Gunthel M., Salvatori D., den Dunnen J.T. Akkermansia muciniphila and Helicobacter typhlonius modulate intestinal tumor development in mice. Carcinogenesis. 2015;36:1388–1396. doi: 10.1093/carcin/bgv120. [DOI] [PubMed] [Google Scholar]
- 65.Sanapareddy N., Legge R.M., Jovov B., McCoy A., Burcal L., Araujo-Perez F. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. ISME J. 2012;6:1858–1868. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Fig. S1 3D principal coordinate analysis (PCoA) plots of individual gastric mucosal microbiota based on unweighted (a) and weighted (b) UniFrac distance in normal microhabitat (n = 230), peritumoral microhabitat (n = 247) and tumoral microhabitat (n = 229) of gastric cancer patients. Each symbol represents a sample.
Fig. S2 Heatmap of the 76 key genera in gastric microbiota, which indicates the genus-level of gastric bacteria alters in normal microhabitat (n = 230), peritumoral microhabitat (n = 247) and tumoral microhabitat (n = 229) of gastric cancer patients. The colour of the spots in the panel represents the relative abundance (normalized and log10-transformed) of the genus in each sample. The relative abundance of the bacteria in each genus is indicated by a gradient of colour from red (low abundance) to blue (high abundance). The genera were organized by Spearman's correlation analysis based on their relative abundances. The taxonomic classifications of the genus are shown on the right. The corresponding Shannon index in each sample is shown under the heatmap.
Fig. S3 Taxonomic differences of the gastric mucosal microbiota among three stomach microhabitats. Related cladogram showing bacterial taxa significantly enriched in each group using the LDA model results for the bacterial hierarchy (upper panel) and principal coordinate analysis (PCoA) for gastric microbiota at three microhabitats (lower panel).
Fig. S4 Tumor stages of gastric cancer related changes of gastric mucosal microbiota. Cladogram represents the features of gastric mucosal microbiota among early-stage and late-stage of gastric cancer using the LDA model results for the bacterial hierarchy (a). Comparisons of the relative abundance at the level of bacterial phylum and genus in early stage (b) and late-stage (c) among the three stomach microhabitats. The data are presented as the mean ± standard deviation. Mann-Whitney U tests were used to analyse variation among three stomach microhabitats.⁎, p < .05; #, p < .01.
Fig. S5 Taxonomic differences of the gastric mucosal microbiota in early-stage of GC among three stomach microhabitats using LEfSe.
Fig. S6 Taxonomic differences of the gastric mucosal microbiota in late-stage of GC among three stomach microhabitats using LEfSe.
Fig. S7 Comparison of the diversity and richness of gastric mucosal microbiota between intestinal-type and diffuse-type of gastric cancer in each stomach microhabitat. The diversity indices of gastric microbiota such as Shannon (a), Simpson (b), and the richness indices such as ACE (c), Chao 1 (d), PD whole tree (e) and observed species (f) were used to evaluated the overall structure of gastric microbiota between two types of gastric cancer in each microhabitat. The data are presented as the mean ± standard deviation. Unpaired t-tests (two-tailed) were used to analyse variation between intestinal- and diffuse-type of GC in the three stomach microhabitats.
Fig. S8 Taxonomic differences of the gastric mucosal microbiota between intestinal-type and diffuse-type of gastric cancer. Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between intestinal- and diffuse-type of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR).
Fig. S9 Taxonomic differences of the gastric mucosal microbiota between histopathological H. pylori positive and negative (HP+ and HP−) groups of gastric cancer in normal microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to H. pylori infection (HPI) in normal microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S10 Taxonomic differences of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in peritumoral microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to HPI in peritumoral microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S11 Taxonomic differences of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in tumoral microhabitat. LEfSe identified the features of gastric microbiota that are discriminative with respect to HPI in tumoral microhabitat using the LDA model results for the bacterial hierarchy, while LDA coupled with effect size measurements identified the most differentially abundant taxa between the two groups (a). Comparisons of the relative abundance of gastric mucosal microbiota at the level of bacterial phylum, family and genus between HP+ and HP− groups of gastric cancer using STAMP with Benjamini-Hochberg correction (QFDR, b).
Fig. S12 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in normal microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the normal microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Phascolarctobacterium, Roseburia, Streptococcus, Halomonas, Blautia, Shewanella, Ruminococcus and Dialister that were used to predict the presence of HPI in normal microhabitat (d).
Fig. S13 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in peritumoral microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the peritumoral microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Phascolarctobacterium, Veillonella, Megamonas, Gemmiger, Halomonas, Blautia and Shewanella that were used to predict the presence of HPI in peritumoral microhabitat (d).
Fig. S14 Comparison of the gastric mucosal microbiota between histopathological HP+ and HP− groups of gastric cancer in tumoral microhabitat. Heatmap indicating the different abundant genera between HP+ and HP− groups of the tumoral microhabitat (a), and the corresponding Shannon index shows from low to high in each group (b). Correlation between Shannon index and the relative abundance of HP (c). Receiver operating characteristic (ROC) curves for differential genera such as Prevotella, Faecalibacterium, Bifidobacterium, Ruminococcus, Bacteroides, Carnobacterium, Paracoccus, Novosphingobium and Akkermansia that were used to predict the presence of HPI in tumoral microhabitat (d).
Fig. S15 Differentially abundant KEGG pathways were identified between tumor microbiota and others at an FDR of 0.05. The different KEGG pathways at level 2 were identified between normal and peritumoral microbiota (a), normal and tumoral microbiota (b), peritumoral and tumoral microbiota (c). Benjamini-Hochberg method was used for multiple testing correction based on false discovery rate (FDR) by STAMP.
Fig. S16 Differentially abundant KEGG pathways were identified between between histopathological HP+ and HP− groups in each tumoral microhabitat at an FDR of 0.05. The different KEGG pathways at level 2 were identified between histopathological HP+ and HP− groups in normal microhabitats (a), peritumoral microhabitat (b), and tumoral microhabitat (c). Benjamini-Hochberg method was used for multiple testing correction based on false discovery rate (FDR) by STAMP.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.