Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is an inherited monogenic renal disease characterised by the accumulation of clusters of fluid-filled cysts in the kidneys and is caused by mutations in PKD1 or PKD2 genes. ADPKD genetic diagnosis is complicated by PKD1 pseudogenes located proximal to the original gene with a high degree of homology. The next generation sequencing (NGS) technology including whole exome sequencing (WES) and whole genome sequencing (WGS), is becoming more affordable and its use in the detection of ADPKD mutations for diagnostic and research purposes more widespread. However, how well does NGS technology compare with the Gold standard (Sanger sequencing) in the detection of ADPKD mutations? Is a question that remains to be answered. We have evaluated the efficacy of WES, WGS and targeted enrichment methodologies in detecting ADPKD mutations in the PKD1 and PKD2 genes in patients who were clinically evaluated by ultrasonography and renal function tests. Our results showed that WES detected PKD1 mutations in ADPKD patients with 50% sensitivity, as the reading depth and sequencing quality were low in the duplicated regions of PKD1 (exons 1–32) compared with those of WGS and target enrichment arrays. Our investigation highlights major limitations of WES in ADPKD genetic diagnosis. Enhancing reading depth, quality and sensitivity of WES in the PKD1 duplicated regions (exons 1–32) is crucial for its potential diagnostic or research applications.
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is an inherited renal disease characterised by the accumulation of clusters of fluid-filled cysts in the kidneys, with reported incidence ranging between 1:400 and 1:1000 worldwide1. It is a progressive, monogenic disease that impairs kidney function and eventually causes end-stage renal disease (ESRD). In addition, ADPKD patients may develop extrarenal manifestations, including hepatic and pancreatic cysts, cerebral and aortic aneurysms, cardiac valvular abnormalities and hypertension2.
ADPKD is primarily caused by mutations in 2 genes: PKD1 (MIM 601313) and PKD2 (MIM 173910). Mutations in PKD1, which is located on chromosome 16 (16p13.3), account for approximately 85% of genetically resolved ADPKD cases, whereas mutations in PKD2, located on chromosome 4 (4q21-11), account for the remaining 15%3. PKD1 encodes polycystin-1, which is an integral membrane protein that complexes with polycystin-2, a calcium-permeable cation channel involved in intracellular Ca2+ homoeostasis. The polycystin proteins form a functional complex in kidney tubular primary cilia, which is believed to be involved in adhesion, proliferation and differentiation of tubular epithelial cells4.
ADPKD shows some degree of complexity suggested by the observed heterogeneity at the genic and allelic levels5,6. At the genic level, mutations in PKD1 are associated with a more severe disease phenotype and earlier age of ESRD onset compared with mutations in PKD2 (54.3 years for PKD1 and 74 years for PKD2)7,8. At the allelic level, truncating mutations are associated with earlier mean age of ESRD onset than non-truncating mutations (55.6 and 67.9 years, respectively)9.
Genetic analysis and mutation screening of ADPKD cases is more technically challenging than that of other monogenic diseases. One reason is that both PKD1 and PKD2 are highly variable genes, and molecular analysis reveals no clear hot spots where mutations are likely to occur along the entire length of either gene3,10. Mutations in these genes are often unique to a single family, with recurrent mutations accounting for only 30% of the total detected mutations11–15. The autosomal dominant polycystic kidney disease mutation database (PKDB) established by the Mayo Clinic lists 1243 definite pathogenic PKD1 variants out of the 2055 listed variants and 374 definite pathogenic PKD2 variants out of the 463 listed variants (http://pkdb.mayo.edu). In addition, the existence of pseudogenes for PKD1 results in another level of complexity for ADPKD genetic testing. PKD1 lies in a segmentally duplicated region, such that the first 32 exons are replicated 6 times in pseudogenes located in a region 13–16 Mb proximal to the original PKD1 (16p.13.1), and they share 97.6–97.8% sequence homology with the original PKD1 gene16–18. As these pseudogene regions are less amenable to selection pressure, they tend to have high mutation rates compared with those of the parent gene19. These duplicated regions represent a diagnostic challenge for ADPKD, as conventional sequencing is not effective for specifically targeting the genuine PKD1 regions20. To overcome this issue, novel sequencing strategies have been developed by utilising the rare sequence variations between the pseudogenes and original PKD1 to specifically target exons 1–32 of the genuine PKD1 gene. These techniques utilised long-range PCR (LR-PCR) to specifically generate amplicons from genuine regions and exclude the pseudo-regions, providing a more reliable molecular diagnostic tool for ADPKD11,21. ADPKD genetic diagnosis may complement the current diagnosis protocol, which relies primarily on ultrasonography22, particularly for younger, at-risk individuals as well as those with late onset disease for whom imaging-based diagnosis may be equivocal.
Since the completion of the human genome project, the demand for faster and more economical sequencing methods has led to the development of next generation sequencing (NGS) technologies as alternative/complementary methods to traditional testing. The massively parallel sequencing platforms of NGS facilitate faster sequencing with higher DNA throughput at lower cost than traditional Sanger sequencing23,24. This encouraged wider utilisation of technologies such as whole genome sequencing (WGS) and whole exome sequencing (WES) in medical diagnosis and research. While the human exome includes <2% of the whole genome, approximately 85% of disease-causing mutations and functional variants are located within these coding regions25,26. This makes WES more cost-effective than WGS in the identification of rare causes of genetic diseases as well as predisposing variants in known diseases and disorders25. ADPKD is one of the diseases that could benefit from NGS in the clinical settings. Moreover, this monogenic condition is considerably associated with intrafamilial phenotypic variability of the disease progression rate and the extrarenal manifestations, despite sharing the same mutation5. Such observations within ADPKD families suggest the involvement of heritable modifier genes5,6. WES and WGS may help in uncovering the modifier genes responsible for the significant phenotypic variability observed, which may aid in better understanding of the disease pathology. However, the reliability and efficiency of NGS-based strategies as diagnostic tools for ADPKD are yet to be examined and evaluated, specifically for the complex PKD1 gene in terms of its large size, the absence of mutation hot spots and the existence of the PKD1 duplicated regions—the pseudogenes16,18. Numerous studies have demonstrated that the high level of sequence similarity between the pseudogene sequences and their parent genes can obscure the detection of the pathogenic mutations, as the unintended detection of pseudo-mutations is possible21,27. Such false-positive results could affect the specificity and reliability of the analysis; therefore, it is essential to evaluate the clinical utility of NGS strategies for the diagnosis of ADPKD.
In the current study, we evaluated the efficiency of WES, WGS and targeted enrichment for sequencing PKD1 and PKD2 to detect ADPKD mutations in patients who have been clinically evaluated by ultrasonography, renal function analysis and long-range PCR (LR-PCR). Our results highlight major limitations of WES in the detection of PKD1 mutations in ADPKD patients.
Results
Clinical evaluation revealed typical ADPKD symptoms
In the current study, total 51 individuals from 6 typical ADPKD families were enrolled. Twenty-six individuals had already been diagnosed with ADPKD according to the unified criteria for ultrasonographic diagnosis of ADPKD (Supplementary Fig. 1)16. The other 25 individuals were at risk, and their disease status was unknown when they consented to participate in the study. Following ultrasonographic analysis and RFTs, 7 individuals were diagnosed with ADPKD, whereas the remaining 18 individuals were healthy. All results were confirmed by genetic testing using Sanger sequencing of PKD1 and PKD2.
Detection of PKD1 mutations using LR-PCR and Sanger sequencing
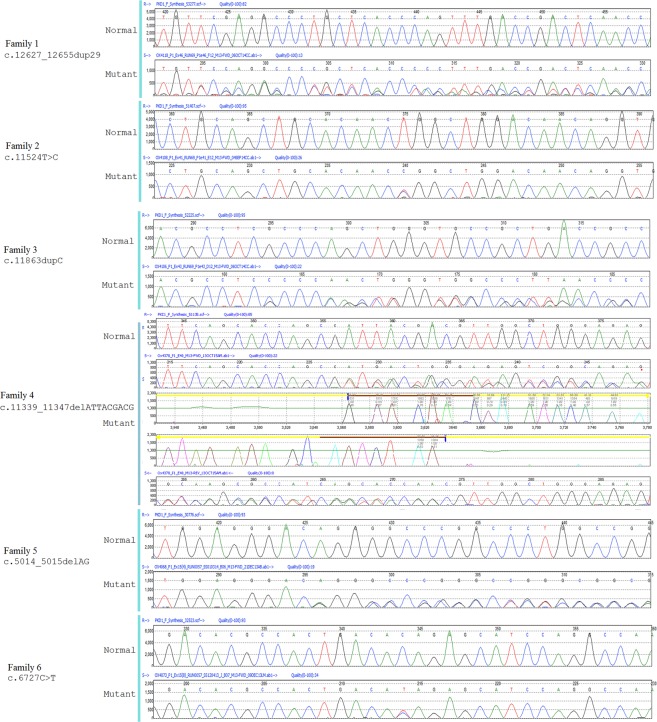
Genetic analysis of the enrolled subjects revealed 6 PKD1 mutations that segregated with the disease in the 6 ADPKD families included in the current study (Fig. 1 and Table 1). Four of the 6 mutations found are novel PKD1 mutations. In family 1, we detected a novel duplication in exon 46; a stretch of 29 bases from position 12627 to 12655 were duplicated, causing a frameshift after position 12655 (c.12627_12655dup; Phe4219Cysfs). In family 2, we detected a novel missense mutation in exon 41 caused by substitution of thymidine to cytosine at position 11524 that results in tryptophan substitution with arginine at position 3842 of the protein (c11524T > C; p.Trp3842Arg). The mutation showed a score of 1 using Polyphen-2 and 0 using SIFT, which suggests pathological significance. In family 3, we detected a novel PKD1 mutation in exon 41 caused by duplication of the cytosine at position 11863, resulting in a frameshift at position 3955 that leads to a premature stop codon because of the substituted amino acid at position 6 (c.11863dupC, p.Gln3955Profs*6). As this mutation is predicted to cause a frameshift, we define it as pathogenic. In family 4, a novel deletion of 9 bases was detected in exon 40 of PKD1 (c.11339_11347del9), resulting in the deletion of 3 amino acids from the protein (p.Asp3780_D3782del). In family 5, we detected a two-base deletion mutation in exon 15 (c.5014_5015delAG). This deletion caused a frameshift from position 1672, leading to a premature stop codon at position 98 after the substitution (p.Arg1672fs98*). This particular mutation is the most frequent mutation detected in ADPKD families9,15. In family 6, a known nonsense mutation was detected in exon 15. This truncating mutation resulted from a single base change (c.6727 C > T), causing a premature stop codon p.Gln2243*10,28.
Figure 1.
PKD1 mutations detected using targeted sequencing. A total of 6 mutations were found, 4 of which are novel.
Table 1.
ADPKD mutations detected by LR-PCR and Sanger’s sequencing.
| Family | Exon | Variant DNA | Variant Protein | Mutation Type | Reference |
|---|---|---|---|---|---|
| 1 | 46 | c.12627_12655dup29 | p.Phe4219fs | Duplication/Frameshift | Novel |
| 2 | 41 | c.11524 T > C | p.Trp3842Arg | Missense | Novel |
| 3 | 43 | c.11863dupC | p.Gln3955fs*6 | Duplication/Frameshift | Novel |
| 4 | 40 | c.11339_11347delATTACGACG | p.Asp3780_Asp3782del | Deletion/Framshift | Novel |
| 5 | 15† | c.5014_5015delAG | p.Arg1672Glyfs*98 | Deletion/Frameshift | [15] |
| 6 | 15 | c.6727 C > T | p.Gln2243* | Nonsense | [28] |
†Mutation could not be detected by WES.
Identification of PKD1 and PKD2 variants using WES
WES of the samples from the 26 ADPKD patients (known to have ADPKD before study enrolment) identified 27 variants in PKD1 (including 6 novel variants) and 5 variants in PKD2. Of the 27 PKD1 variants, 12 were synonymous changes, 12 were non-synonymous changes and the remaining 3 were insertions/deletions. For PKD2 variants, only 1 synonymous and 4 non-synonymous changes were identified (Table 2). Five of the 6 ADPKD mutations identified using Sanger sequencing were identified by WES, whereas the remaining mutations (c.5014_5015delAG, found in family 5) could not be detected in the 8 patients included in WES analysis from this family (Tables 1 and 2). In addition, the mutation found in family 6 (16:2158441-SNV, c.6727 C > T) was detected only in one patient out of 6 included in the WES analysis from this family. While 4 of the 5 ADPKD mutations detected using WES were frameshift insertions and deletions, the remaining reported mutation was a missense mutation (16:2141795-SNV, c.11524 T > C; p.Trp3842Arg), and its pathological impact was evaluated and confirmed using the pathological prediction and conservation scores (Supplementary Tables 1,2).
Table 2.
PKD1 and PKD2 variants detected by WES. ADPKD mutations highlighted in bold.
| WES Variant | Exon | Classification | cDNA Change | Protein Change | rs ID |
|---|---|---|---|---|---|
| PKD1 (NM_001009944.2) | |||||
| 16:2140009-Ins | 46 | Frameshift Ins | c.12627_12655dup29 | p.Glu4219fs | Novel |
| 16:2140010-MIX | 46 | Synonymous | c.12630 T > C | p.= | rs7203729 |
| 16:2140294-SNV | 45 | Nonsyn SNV | c.12436 G > A | p.Val4146Ile | rs148478410 |
| 16:2140321-SNV | 45 | Synonymous | c.12409 C > T | p.= | rs79899502 |
| 16:2140454-SNV | 45 | Synonymous | c.12276 A > G | p.= | rs3087632 |
| 16:2140554-SNV | 45 | Nonsyn SNV | c.12176 C > T | p.Ala4059Val | rs3209986 |
| 16:2140680-SNV | 44 | Nonsyn SNV | c.12133 A > G | p.Ile4045Val | rs10960 |
| 16:2141025-Ins | 43 | Frameshift Ins | c.11863_11864dupC | p.Gln3955fs | Novel |
| 16:2141028-SNV | 43 | Nonsyn SNV | c.11860 G > C | p.Ala3954Pro | Novel |
| 16:2141795-SNV | 41 | Nonsyn SNV | c.11524 T > C | p.Trp3842Arg | Novel |
| 16:2142112-Del | 40 | Del | c.11339_11347delATTACGACG | p.Asp3780_Asp3782del | Novel |
| 16:2142113-SNV | 40 | Synonymous | c.11346 C > T | p.= | rs145955373 |
| 16:2144176-SNV | 35 | Nonsyn SNV | c.10535 C > T | p.Ala3512Val | rs34197769 |
| 16:2144182-SNV | 35 | Nonsyn SNV | c.10529 C > T | p.Thr3510Met | rs45478794 |
| 16:2147399-SNV | 33 | Synonymous | c.10326 G > A | p.= | rs141138826 |
| 16:2152387-SNV | 25 | Nonsyn SNV | c.9196 T > C | p.Phe3066Leu | rs77028972 |
| 16:2152388-SNV | 25 | Synonymous | c.9195 G > C | p.= | rs78003543; rs9935834 |
| 16:2156447-SNV | 18 | Synonymous | c.7441 C > T | p.= | rs2003782 |
| 16:2158441-SNV† | 15 | Stopgain | c.6727 C > T | p.Gln2243* | 28 |
| 16:2158570-SNV | 15 | Nonsyn SNV | c.6598 C > T | p.Arg2200Cys | rs140869992 |
| 16:2159405-SNV | 15 | Synonymous | c.5763 G > A | p.= | rs2575313 |
| 16:2159996-SNV | 15 | Synonymous | c.5172 C > T | p.= | rs9935526 |
| 16:2160503-SNV | 15 | Synonymous | c.4665 A > C | p.= | rs71385734 |
| 16:2162887-SNV | 13 | Synonymous | c.3063 T > C | p.= | rs2369068 |
| 16:2164294-SNV | 11 | Synonymous | c.2730 C > T | p.= | rs35965348 |
| 16:2164808-SNV | 11 | Nonsyn SNV | c.2216 G > A | p.Arg739Gln | rs40433 |
| 16:2165470-SNV | 10 | Nonsyn SNV | c.2006G > T | p.Cys669Phe | Novel |
| PKD2 (NM_000297.3) | |||||
| 4:88928968-SNV | 1 | Nonsyn SNV | c.83 G > C | p.Arg28Pro | rs1805044 |
| 4:88929305-SNV | 1 | Synonymous | c.420 G > A | p.= | rs2728118 |
| 4:88929453-SNV | 1 | Nonsyn SNV | c.568 G > A | p.Ala190Thr | rs117078377 |
| 4:88967919-SNV | 6 | Nonsyn SNV | c.1445 T > G | p.Phe482Cys | rs75762896 |
| 4:88989089-SNV | 13 | Nonsyn SNV | c.2398 A > C | p.Met800Leu | rs2234917 |
†WES detected the mutation in 1 patient from a total of 9 ADPKD patients from the same family. Sanger’s sequencing confirmed that all the 9 family patients are carrying the mutation.
Highlighted variants in bold are the ADPKD mutations detected by WES.
Overall, the sensitivity of WES for detecting PKD1 mutations throughout the length of the gene, which reflects the ability of the test to correctly identify the true disease-causing mutations, was 50%; however, for mutations in exons 1–32, it was only 7.14% and for exons 33–46, it was 100%. As there were no false-positive results recorded, the specificity of WES for detection of PKD1 mutations was 100% for the entire length of the gene. As all the ADPKD mutations in enrolled patients were found in PKD1, sensitivity and specificity for PKD2 could not be calculated (Table 3).
Table 3.
Sensitivity and Specificity of WES for detection of PKD1 mutations.
| Sensitivity | Specificity | |
|---|---|---|
| PKD1 exons 1–32 | 7.14% | 100% |
| PKD1 exons 33–46 | 100% | 100% |
| Entire PKD1 | 50% | 100% |
WES of exons 1–32 of PKD1 showed low reading depth, genotype quality and quality
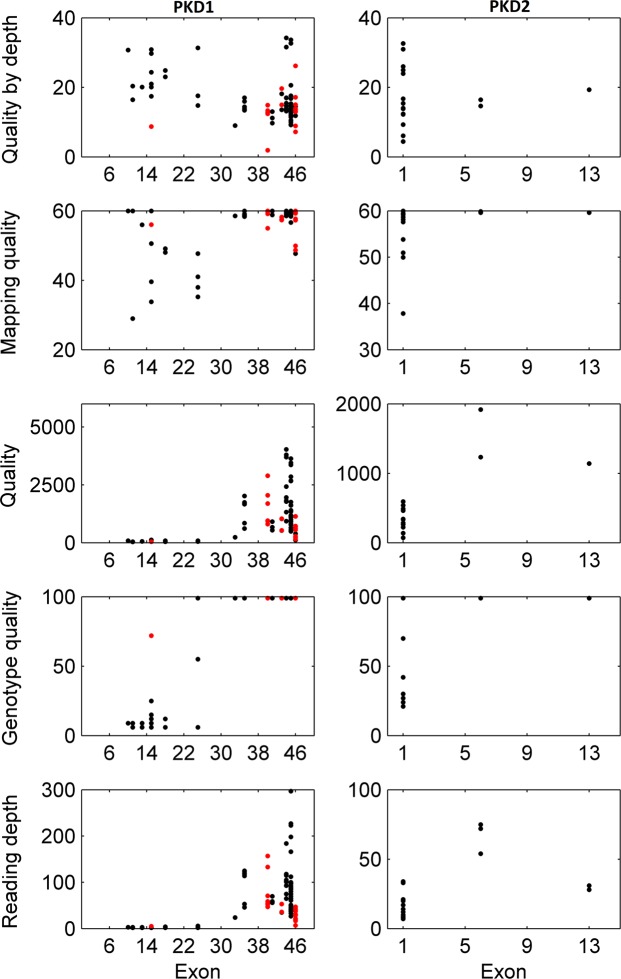
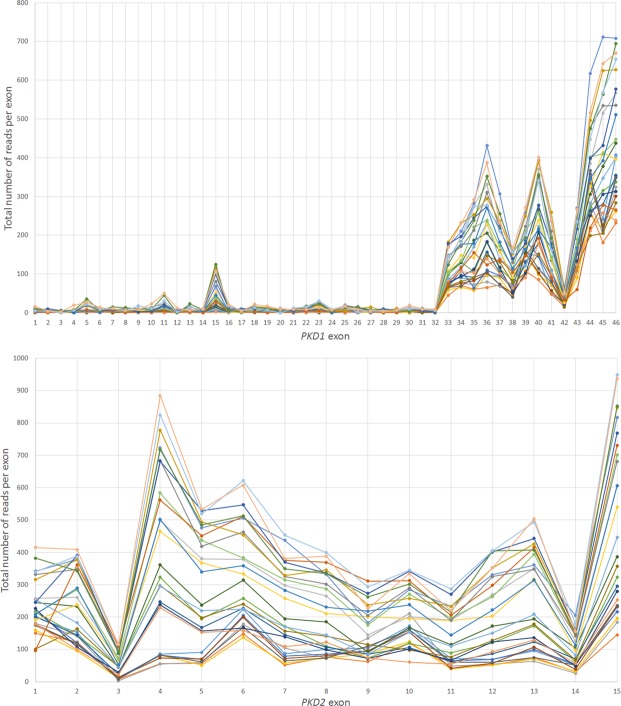
All variants detected in exons 1–32 of PKD1 showed low mean reading depth (RD) ranging from 2 to 5 (Figs 2 and 3). Despite a low RD of 2, the mutation found in family 1 (16:2158441-SNV) was called and detected, but only in one of the 6 patients from this family (Tables 2 and 4). We also calculated the total number of reads per PKD1 exon in each sample, and all samples showed a similar trend (Fig. 4). Exons 1–32 had a noticeably lower total number of reads in comparison to exons 33–46. Mean Genotype Quality (GQ) and mean quality were also low for variants called in exons 1–32 in comparison to variants called in exons 33–46. However, the mean Mapping Quality (MQ) was roughly the same for all variants called across the whole PKD1 gene. When the quality was normalised by depth, called variants in exons 1–32 showed a relatively similar range of scores to those variants called in exons 33–46 (Fig. 2).
Figure 2.
Quality assessment of called variants in PKD1 and PKD2 using WES. Exons 1–32 of PKD1 had low reading depth, genotype quality and quality. ADPKD mutations shown in red.
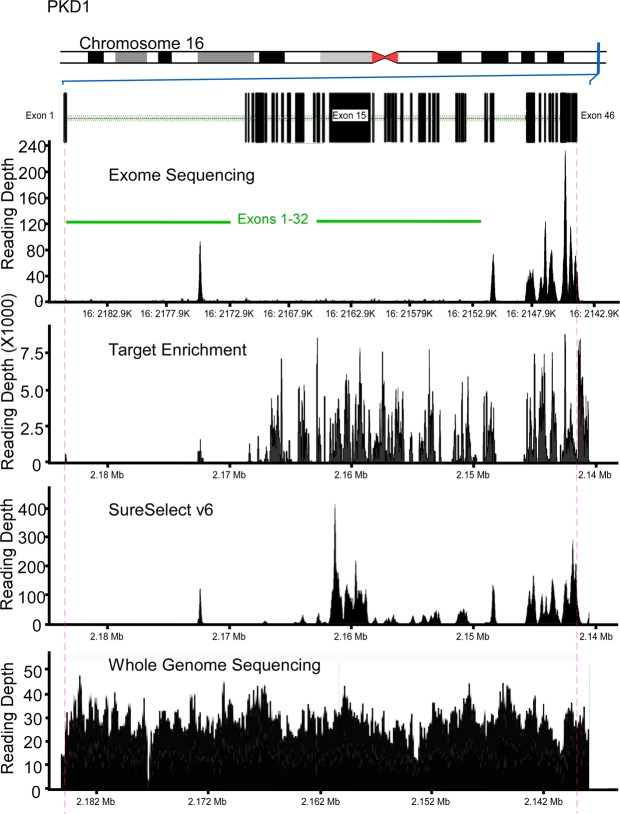
Figure 3.
WES and Target enrichment coverage map of PKD1. WES of exons 1 to 32 of PKD1 showed low coverage while PKD2 coverage showed proper depth of all exons. Target Enrichment showed proper coverage for coding regions of PKD1 and PKD2. SureSelect v6 improved the coverage of PKD1 in comparison to WES TruSeq v3 but exons 1 to 14 remained poorly covered. WGS showed proper covering of the entire PKD1.
Table 4.
Quality assessment of variants detected by WES.
| WES Variant | Exon | Mean Reading Depth ( ± SD) | Mean Genotype Quality (GQ) ( ± SD) | Mean Mapping Quality (MQ) ( ± SD) | Mean Quality by Depth (QD) ( ± SD) | Mean Quality ( ± SD) | |
|---|---|---|---|---|---|---|---|
| PKD1 (NM_001009944.2) | |||||||
| 16:2140009-Ins | 46 | 14.0 (±7) | 99 (±0) | 48.2 (±0.73) | 13.19 (±1.86) | 255.23 (±193.04) | |
| 16:2140010-MIX | 46 | 29.33 (±11.68) | 99 (±0) | 56.08 (±4.71) | 14.12 (±5.09) | 524.63 (±312.54) | |
| 16:2140294-SNV | 45 | 114.71 (±46.76) | 99 (±0) | 59.01 (±1.57) | 11.34 (±2.09) | 1924.45 (±1142.22) | |
| 16:2140321-SNV | 45 | 216.40 (±44.17) | 99 (±0) | 59.05 (±0.69) | 12.74 (±1.86) | 2824.37 (±730.75) | |
| 16:2140454-SNV | 45 | 53.60 (±19.97) | 99 (±0) | 59.41 (±0.49) | 19.09 (±7.81) | 1155.87 (±346.94) | |
| 16:2140554-SNV | 45 | 55.80 (±11.33) | 99 (±0) | 59.37 (±0.61) | 12.63 (±1.82) | 708.17 (±135.65) | |
| 16:2140680-SNV | 44 | 105 (±29.11) | 99 (±0) | 59.45 (±0.49) | 18.99 (±8.04) | 2420.89 (±1146.87) | |
| 16:2141025-Ins | 43 | 44.0 (±9) | 99 (±0) | 57.88 (±0.51) | 15.86 (±3.22) | 781.73 (±356.38) | |
| 16:2141028-SNV | 43 | 44.5 (±8.5) | 99 (±0) | 57.81 (±0.61) | 17.32 (±0.3.32) | 790.77 (±353.31) | |
| 16:2141795-SNV | 41 | 61.67 (±6.01) | 99 (±0) | 59.42 (±0.82) | 11.32 (±1.65) | 709.77 (±186.27) | |
| 16:2142112-Del | 40 | 169 (NA) | 99 (NA) | 60 (NA) | 17.12 (NA) | 2894.73 (NA) | |
| 16:2142113-SNV | 40 | 67.14 (±27.77 | 99 (±0) | 58.47 (±2.33) | 13.17 (±1.02) | 1290.31 (±550.12) | |
| 16:2144176-SNV | 35 | 108 (±27.61) | 99 (±0) | 59.03 (±0.71) | 14.99 (±1.44) | 1612.17 (±446.16) | |
| 16:2144182-SNV | 35 | 80 (±34) | 99 (±0) | 58.66 (±0.25) | 13.88 (±0.67) | 1141.27 (±741.75) | |
| 16:2147399-SNV | 33 | 24 (NA) | 99 (NA) | 58.55 (NA) | 9.78 (NA) | 234.77 (NA) | |
| 16:2152387-SNV | 25 | 3.25 (±1.64) | 41.5 (±44.75) | 40.45 (±6.45) | 21.25 (±8.87) | 68.09 (±18.58) | |
| 16:2152388-SNV | 25 | 3.25 (±1.64) | 41.5 (±44.75) | 40.45 (±6.45) | 21.25 (±8,87) | 68.09 (±18.58) | |
| 16:2156447-SNV | 18 | 3.0 (±1) | 9 (±4.3) | 48.14 (±0.79) | 23.94 (±1.31) | 70.88 (±29.90) | |
| 16:2158441-SNV† | 15 | 2.0 (NA) | 6 (NA) | 60 (NA) | 30.87 (NA) | 61.74 (NA) | |
| 16:2158570-SNV | 15 | 5.0 (NA) | 72 (NA) | 56.04 (NA) | 8.75 (NA) | 43.77 (NA) | |
| 16:2159405-SNV | 15 | 3.0 (NA) | 9 (NA) | 60 (NA) | 17.43 (NA) | 52.28 (NA) | |
| 16:2159996-SNV | 15 | 4.67 (±0.47) | 17.3 (±6.8) | 41.33 (±8.53) | 25.03 (±4.42) | 115.23 (±9.05) | |
| 16:2160503-SNV | 15 | 3.0 (NA) | 9 (NA) | 60 (NA) | 20.09 (NA) | 60.28 (NA) | |
| 16:2162887-SNV | 13 | 3.0 (NA) | 9 (NA) | 56 (NA) | 20.09 (NA) | 60.28 (NA) | |
| 16:2164294-SNV | 11 | 3.0 (NA) | 9 (NA) | 60 (NA) | 16.43 (NA) | 49.28 (NA) | |
| 16:2164808-SNV | 11 | 2.0 (NA) | 6 (NA) | 29 (NA) | 20.35 (NA) | 40.74 (NA) | |
| 16:2165470-SNV | 10 | 3.0 (NA) | 9 (NA) | 60 (NA) | 30.09 (NA) | 92.28 (NA) | |
| PKD2 (NM_000297.3) | |||||||
| 4:88928968-SNV | 1 | 12.6 (±5.35) | 67.9 (±35.45) | 53.31 (±7.45) | 19.86 (±8.51) | 322.69 (±125.66) | |
| 4:88929305-SNV | 1 | 27 (±11.26) | 99 (NA) | 59.32 (±0.16) | 13.91 (±2.14) | 567.04 (±38.47) | |
| 4:88929453-SNV | 1 | 11 (±1.41) | 64.5 (±48.79) | 59.64 (±0.51) | 12.16 (±12.01) | 179.42 (±150.83) | |
| 4:88967919-SNV | 6 | 67 (±11.35) | 99 (NA) | 59.755 (±0.17) | 15.54 (±1.27) | 1576.08 (±483.92) | |
| 4:88989089-SNV | 13 | 31 (NA) | 99 (NA) | 59.63 (NA) | 36.77 (NA) | 1140.41 (NA) | |
Standard deviation (SD) is noted as non-applicable (NA) when there is only one value indicating that the variant was detected in one sample.
Figure 4.
Total number of reads per exon for PKD1 and PKD2 using WES. Upper image shows that PKD1 exons 1 to 32 have relatively lower number of total reads in comparison with PKD1 exons 33–46 while PKD2 exons “lower image” shows better distribution of readings per exon in comparison with PKD1.
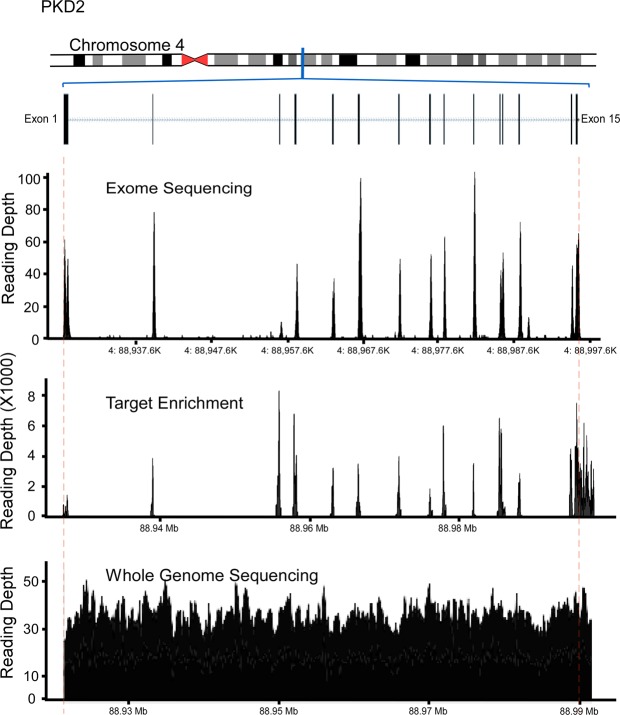
For PKD2, all exons were properly covered, and all called variants showed a mean RD > 10, mean GQ was >60, mean MQ was close to 60 and mean quality ranged between 179.42 and 1576.08. Exon 3 showed the lowest total number of reads in all samples (Figs 2, 4 and 5).
Figure 5.
WES, Target enrichment coverage map and WGS of PKD2. PKD2 was covered properly using all methods.
WGS detected PKD1 mutations located in the duplicated region
DNA samples of 4 ADPKD patients were selected for WGS analysis: 2 patients with PKD1 mutations in the duplicated region of PKD1 and 2 patients with mutations outside the duplicated region of PKD1 (mutations were previously identified using LR-PCR). The coverage analysis of WGS for PKD1 and PKD2 showed good and uniform coverage, including exons 1–32 of PKD1 (Figs 3 and 4). Three out of 4 mutations were successfully detected using WGS, including the 2 mutations located in the PKD1 duplicated region (exon 15), c.5014_5015delAG and c.6727 C > T, whereas c.12627_12655dup29 (located in exon 46) was missed (Table 5).
Table 5.
PKD1 mutations detected by WGS*.
| cDNA Change | Protein | Mean Depth | Quality |
|---|---|---|---|
| c.11863dupC | p.Q3955fs | 25 | 40.3 |
| c.5014_5015delAG | p.Arg1672Glyfs*98 | 34 | 34.9 |
| c.6727 C > T | p.Q2243X | 37 | 40.1 |
*The genomes of 4 ADPKD with known mutations where analyzed using WGS. 3 out 4 mutations were detected successfully while the c.12627_12655dup29 was missed.
Targeted enrichment system detected mutations in PKD1 duplicated regions
Samples with PKD1 mutations from families 5 (c.5014_5015delAG) and 6 (c.6727 C > T) both located in exon 15, which were found using LR-PCR and Sanger sequencing but could not be detected with WES, were reanalysed using targeted enrichment arrays. These specific mutations were identified in all the samples analysed using targeted enrichment of PKD1. Coverage maps of targeted enrichment of PKD1 and PKD2 showed good coverage and depth in the coding regions of both genes (Figs 3 and 5). The newer version of SureSelect capture arrays shows significant improvement in the representation of exons 15–32 of the PKD1 gene (Fig. 3). However, capture of exons 1–14 remains poor.
Discussion
Mutations in either PKD1 or PKD2 may cause ADPKD. While genetic analysis of PKD2 is relatively easy, genetic analysis of PKD1 is more complex because of its highly polymorphic nature, large size and pseudogene regions. LR-PCR and Sanger sequencing are the current gold standard methods for genetic analysis of PKD1. However, these methods are labour intensive and require a substantial amount of time to analyse the large number of amplicons. In the current study, we evaluated the efficiency of WES, WGS and target enrichment as genetic diagnostic tools for ADPKD and as potential replacement methods for the LR-PCR and Sanger sequencing techniques.
ADPKD is diagnosed using ultrasonographic age-related cyst number criteria29. It has also been shown that computed tomography and magnetic resonance imaging may be effectively utilised for the same purpose30. These image-based diagnostic approaches are highly reliable in older patients (aged >30 years) but not in younger adult patients, which limits their utility in kidney donations to exclude disease status in young kidney donors11. Genetic analysis of ADPKD genes provides an alternative reliable diagnosis tool for younger patients. In the current study, these methods demonstrated high sensitivity and specificity, as they detected all the pathogenic mutations that segregated with the disease in all patients.
Recent advances in genetic sequencing, including NGS platforms such as WES, are considered revolutionising tools for genetic diagnostic research. The ability of WES to generate accurate, efficient, fast and cost-effective genetic data is important when considering implementation in medical practice to improve diagnosis and disease treatments in general. For ADPKD, WES is considered an effective tool to explore and identify potential disease modifier genes, particularly where our current knowledge of these modifiers is poor31–33. In this study, we assessed the effectiveness of utilising WES as a diagnostic tool for ADPKD and whether it can replace the current gold standard methods for ADPKD genetic diagnosis: LR-PCR and Sanger sequencing. To achieve this objective, we avoided filtering the called variants and analysed the quality scores for all variants called in PKD1 and PKD2. WES showed major limitations when used to detect mutations in PKD1 exons 1–32, as test sensitivity for this particular region was only 7.14%, whereas it demonstrated high sensitivity for the remaining coding regions of PKD1. This was also reflected by the low RDs observed, particularly over PKD1 exons 1–32 (Figs 2 and 3). The low RD concurred with the significantly lower number of total reads per exon in this region compared to the PKD1 exons 33–46 and PKD2 (Fig. 5). Moreover, the mean GQ for the called variants in PKD1 exons 1–32 was correlated with the read depth pattern, as they scored low in comparison with the variants called elsewhere in PKD1 and PKD2. GQ indicates the likelihood of the called genotype being correct; the higher the value, the more accurate the genotype calls. Normally, a GQ > 20 is considered acceptable33; however, in our WES results, the mean GQ for called variants in PKD1 exons 1–32 ranged from 6 to 72 (Table 4). Despite the low GQ score of 6 and a RD of only 2 for the called mutation found in family 1 (16:2158441-SNV, c.6727 C > T), the WES results were confirmed by LR-PCR and Sanger sequencing, indicating its validity. However, the mean MQ scores across PKD1 and PKD2 were high in general, indicating that all the reads were informative and correctly mapped. Although there is no clear cut off MQ value34, a score > 20 is usually acceptable according to the GATK tool35 (Table 4). These poor results may be attributed to the PKD1 pseudogenes, as designing capture oligonucleotide probes (65 nt) that specifically target PKD1 genuine regions and avoid its pseudo-regions poses a considerable challenge. As a result, pseudogenes are captured in parallel with the genuine PKD1 regions, complicating data analysis, compromising result reliability and often yielding false-positive results, as mutations in pseudogenes are detected, resulting in reduced test specificity21,27,36. However, it is possible that readings mapped to multiple genome sites tend to be avoided, as demonstrated by our results. In general, the low coverage and RD of the PKD1 exons 1–32 could represent a major diagnostic limitation, as some mutations may be missed and consequently compromise the sensitivity and efficacy of the test. In both WES and WGS, mutation detection sensitivity can be improved by increasing the depth of sequencing and by the utilization of newer capture arrays. The newer version of SureSelect V6 capture arrays shows significant improvement in the representation of the exons 15–32 of the PKD1 gene (Fig. 3). For example, SureSelect V6 arrays covers all exons with a target size of 60 MB and 758,086 probes, while V4 of SureSelect capture arrays have a target size of 51 MB and covers 334,378 exons.
WGS showed a more uniform representation of entire PKD1 and PKD2, including exons 1–32 of PKD1. It has been suggested that longer read lengths and avoidance of capture bias enhanced the ability of WGS to detect pathological ADPKD mutations, including those in the duplicated region of PKD137,38. However, detecting small to medium size indels (10–1000 bases) remains a challenge (Table 5). We are currently utilizing three algorithms in parallel; Pindel, UnifiedGenotyper and HaplotypeCaller to call indels39.
Target enrichment of PKD1 resulted in successful detection of mutations located in the duplicated regions (c.6727 C > T and c.5014_5015delAG, both located in exon 15), which could not be detected efficiently using WES. This coincides with the good coverage and RD produced using target enrichment over all coding regions of PKD1, including the problematic exons 1–32 and PKD2. One of the key differences between WES and target enrichment is gDNA library preparation and enrichment. For WES, sequence enrichment is achieved through an oligonucleotide probe-based capture strategy, whereas a PCR-based method is used for target enrichment40–43. While the oligonucleotide probe-based capture strategy for larger DNA regions is preferred over PCR-based methods for time and cost reasons, its capture efficiency is between 70–80%, as exons with high GC or AT content have reduced hybridisation and amplification efficiency19,44. In addition, a common limitation for capture-based enrichment methods is the presence of pseudogenes, which results in complications in variant calling and data interpretation of the targeted regions19. However, PCR enrichment methods are more efficient and reliable when analysing genes with pseudo-regions such as PKD1, as they have shown high specificity, sensitivity and reproducibility45. In addition, the ability to modify the design of probes in the PCR method provides another advantage, as this allows specific targeting of the functional gene rather than the pseudogenes, avoiding diagnostic limitations, including false negative results caused by low coverage of PKD1 duplicated regions (as seen in WES) and false-positive results caused by the inability to avoid pseudogenes and mutations in these regions. Target enrichment of PKD1 appears to overcome the limitations of WES and perhaps is more suitable, now, for ADPKD diagnostic applications.
In conclusion, the ability to effectively implement WES in current medical practices will improve care of patients by enhancing disease diagnosis and treatment planning. WES provides a rapid diagnostic tool for many genetic diseases and disorders, as it allows identification of common and novel genetic variants that may then be evaluated for their pathological impact. As for ADPKD, although the WES platform successfully identified novel PKD1 mutations, it showed low sensitivity and RD of the duplicated regions, which represent a challenge for effective and reliable genetic diagnosis of ADPKD. WES enrichment strategies must be improved to solve the low sensitivity problem in the PKD1 duplicated regions. Such enhancements would allow more rapid and accurate genetic analysis of ADPKD patients, which in turn would contribute to better disease management and improve our understanding of the molecular pathology underlying the disease.
Methods
Patient inclusion criteria
Six families with a history of ADPKD were selected from the Nephrology unit database at Mubarak Al-Kabeer Hospital, Kuwait for inclusion in the current study. ADPKD patients showed typical clinical presentations of ADPKD, including multiple renal cysts and impaired kidney function. Total 51 individuals from 6 families with typical ADPKD, including 26 ADPKD patients and 25 at-risk individuals were enrolled in the current study, which was reviewed and approved by the Joint Committee for The Protection of Human Subjects in Research of the Health Sciences Center (HSC) at Kuwait University and the Kuwait Institute for Medical Specialization (KIMS) (Reference: VDR/JC/690). Written informed consent was obtained from all patients before their enrollment in the study. All methods were performed in accordance with the guidelines of the joint HSC and KIMS ethical committee.
Clinical evaluation
Clinical evaluation of all 51 subjects from the 6 families was performed. Individuals were evaluated using ultrasonographic analysis and renal function tests to confirm their disease status, including those showing negative results in mutation screening. Healthy individuals were later used as negative controls for comparative analysis with ADPKD patients.
Abdominal ultrasound
All subjects enrolled in the current study except those who had undergone kidney transplants were instructed to fast for 6 h prior to abdominal ultrasound examinations, which were performed using a Logic 7 GE ultrasound with a curvilinear 3.5 MHZ probe. Focused ultrasound was performed to assess the kidneys, liver, pancreas and ovaries in female subjects. Each kidney was assessed in multiple views. Renal cysts in each kidney were examined and counted for diagnostic purposes according to the unified criteria for ultrasonographic diagnosis of ADPKD22. Each total kidney volume (TKV) was calculated using the ellipsoid formula: volume = length × lateral diameter × anterior-posterior diameter × π/6. TKV was calculated automatically by the machine in cubic centimetres (cc) and adjusted for height (htTKV expressed as cc/m)46. Each patient’s liver and pancreas were also screened for the presence or absence of cysts.
Renal function test
From each subject, 5-mL blood samples were taken and used to conduct renal function test (RFT), which were performed at the Mubarak Al-Kabeer Hospital, Jabriya, Kuwait. Serum creatinine levels were determined for each patient and expressed as µ mol/L. Estimated glomerular filtration rates (eGFR) were calculated using the CKD-EPI creatinine equation (2009) developed by Levey et al.47. Calculations were performed using the GFR calculator provided on The National Kidney Foundation website www.kidney.org.
The RFT was not performed for ADPKD patients who reached ESRD, were on dialysis or had undergone a kidney transplant. For these patients, only abdominal ultrasound and genetic testing were performed to confirm ADPKD diagnosis, and their eGFR and htTKV were not included in the analysis.
DNA isolation
From each subject, 10-mL blood samples were collected at the Nephrology Department of the Mubarak Al-Kabeer Hospital, Jabriya, Kuwait and genomic DNA was isolated from blood samples using a Gentra Puregene Blood Kit (Qiagen, 158467) following the manufacturer’s protocol.
Mutation screening and classification of variants
Long-Range PCR
Mutations were screened in the genomic DNA of all subjects using locus-specific amplification and direct sequencing of exonic and flanking intronic regions of PKD1 and PKD211. Segregation was tested in family members using sequence analysis of the relevant genomic fragments. The significance of missense variants was assessed using the ADPKD Mutation Database (http://pkdb.mayo.edu), multi-sequence alignments and substitution assessment tools (SIFT, PolyPhen2 and Align GVGD), as previously described11,20 Novel frame-shifting insertions and deletions were defined as pathogenic.
Whole exome sequencing
DNA samples from 26 ADPKD patients from the 6 families were prepared and enriched using TruSeq v3, SureSlect v4 or v6 following the manufacturer’s protocol. Agilent’s QPCR NGS Library Quantification Kit (G4880A) was used to determine the DNA concentration of each library prepared. Enriched samples were pooled at a final concentration of 10 nM. Exome sequencing was performed using the Illumina HiSeq2000 platform. For mapping and alignment, read files (Fastq) were generated and obtained from the HiSeq2000 platform using the manufacturer’s proprietary software. Burrows-Wheeler Aligner package version 0.6.148 was used to locate reads in the most recent map of the human genome (hg38/GRCh38). To ensure a minimum number of mismatching bases across the reads obtained, which in turn reduces false-positive SNP calls (indels), we utilised Genome Analysis Tool Kit (GATK) version 1.635, which locally realigned mapped reads around potential insertion/deletion (indel) sites. Picard version 1.62 was used to label duplicate reads so as to remove those likely resulting from PCR bias. Generated BAM files were further manipulated using Samtools version 0.1.1849. To improve the quality of variants calls, GATK’s covariance recalibration was used to recalibrate base quality (Phred scale) scores. GATK Unified Genotyper was used to call SNP and indel variants in each sample50. Variant novelty was determined using dbSNP.
For variant calling and analysis, we used Golden Helix SNP & Variation Suite version 8.3.4 for Win64. We performed multiple sample variant calling to reduce calling sequencing errors and to enhance the accuracy and sensitivity of calling51–53. Filtered and unfiltered VCF files of all samples were uploaded and Homo sapiens (Human), GRCh37 g 1k (Feb2009) was used as the default genome assembly. PKD1 (NM_001009944.2) and PKD2 (NM_000297.3) variant annotations and analyses were performed using the dbNSFP Functional Predictions and Scores 2.9, GHI database54–56, which provided pathological prediction scores from prediction algorithms, including SIFT, Polyphen2, LRT, FATHMM and MetaLR; and conservation scores from phyloP100way_vertebrate, phastCons100way vertebrate, GERP++ and SiPhy and other function annotations, which were used to assess pathological impact. All non-synonymous and insertion/deletion variants were tested and validated in healthy and affected members of the ADPKD families using Sanger sequencing.
No filters were applied when calling variants to assess the quality of called variants in PKD1 and PKD2. The quality was assessed by analysing the reading depth (RD), mapping quality (MQ), genotype quality (GQ), phred scale quality and quality by depth for each called variant. When the same variant was called in more than one sample, the mean of the quality measure was obtained and the standard deviation (STD) was calculated.
The PKD1 and PKD2 regions captured using WES were obtained by plotting BAM files against RefSeq genes 105v2, NCBI to highlight the gene regions. In addition, the total reads per exon in both genes were calculated for each sample and presented graphically.
Whole genome sequencing and analysis
DNA was extracted from blood samples using Qiagen Kits according to the manufacturer’s protocol. DNA quality and concentration were determined, and then 1 ug of the DNA was used for WGS using the Illumina TruSeq DNA sample preparation guide to obtain a final library of 300–400 bp average insert size. Covaris systems, which produces dsDNA fragments with 3′ and 5′ overhangs, was used to fragment genomic DNA that was then converted to have blunt ends using an end repair mix; the 3′ overhangs were removed using 3′ to 5′ exonuclease, and the 5′ overhangs were filled by the polymerase. The library size was selected using different ratios of the sample purification beads. Ligation was prevented by adding a single adenine nucleotide to the 3′ end of the blunted fragments, and the corresponding thymine nucleotide provided a complementary overhang on the 3′ end of the adapter. Multiple indexing adapters were ligated to both ends of the fragments in preparation for hybridisation in the Illumina flow cell (Illumina Hiseq2500). Isaac aligner software was used to align NGS sequencing data57. Resulting VCF files were analysed using golden helix software for the WES data.
Targeted enrichment arrays
Four samples with mutations in exons 1–32 of PKD1 that were detected by LR-PCR but not WES were selected for this analysis. HaloPlex Custom kits (Agilent Genomics) were used to design panels covering PKD1 and PKD2 coding exons, such as 5′ UTR and 3′ UTR. The design was made for the Illumina platform using RefSeq, Ensembl, CCDS, Gencode, VEGA and SNP databases, and the designed panels were used according to the manufacturer’s protocol (G9900–90020). Generated BAM and VCF files were analysed using Golden Helix SNP & Variation Suite version 8.3.4 for Win64. Homo sapiens (Human), GRCh37 g 1k (Feb2009) was used as the default genome assembly. We eliminated all filters for the analysis to evaluate the quality of all called variants, which was assessed by analysing the RD, MQ, GQ, phred scale quality and quality by depth for each called variant.
Statistical analysis
To test the hypothesis that the slope for predicting htTKV levels from subject age is greater for ADPKD patients than for healthy subjects, linear regression analyses were performed on htTKV-age data from ADPKD patients and healthy subjects. This was followed by a comparison of the resulting regression slopes using Student’s t-test to evaluate the difference between the slopes. Similar analyses were also performed on the eGFR-age data.
Kaplan–Meier renal survival analysis
Survival times were calculated as the time of onset of ESRD, and a Kaplan–Meier product-limit survival curve was constructed using MATLAB and Statistics Toolbox, Release 2012b (The MathWorks, Inc., Natick, MA, USA). Twenty-three patients had already reached ESRD by the time of the analysis (some of the ESRD patients were part of the 6 families enrolled in the current study, but were excluded from the genetic testing as they died prior to screening). The median survival time was calculated as the smallest survival time for which the estimated probability of renal survival was ≤0.5. The mean survival time was calculated as the area under the Kaplan–Meier survival curve58.
Supplementary information
Acknowledgements
We would like to thank the staff of the radio-diagnosis department in Mubarak Al-Kabeer Hospital for their help with the clinical evaluation of the patients. This study was funded by Kuwait University Research Grant SRUL02/13, Kuwait Foundation for the Advancement of Science grant PR17-13MM-07 and Mayo PKD center DK090728.
Author Contributions
H.A. was involved in the conception and design of the study, acquisition of genetic data, analysis and interpretation of data, writing the manuscript and final approval of the version to be published. F.M. was involved in the conception and design of the study, acquisition and analysis of data and writing some sections of the manuscript. N.H., M.N. and A.A1. were involved in patient recruitment and clinical diagnosis. A.M.A. performed the radiological assessment and diagnosis. A.A2. generated the figures. M.A. performed clinical data analysis and associated statistical analysis and organised the database. J.A. was involved in manuscript critical revision. S.A. was involved in manuscript critical revision and overall conception and design of the study. P.H. was involved in the acquisition and analysis of data and critical manuscript revision. All authors contributed to the manuscript revision, read and approved the submitted version. A.A1.: Ali AlSahow A.A2.: Ashraf Al Madhoun.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hamad Ali, Email: hamad.ali@hsc.edu.kw.
Fahd Al-Mulla, Email: fahd@al-mulla.org.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40761-w.
References
- 1.Ong AC, Harris PC. Molecular pathogenesis of ADPKD: the polycystin complex gets complex. Kidney international. 2005;67:1234–1247. doi: 10.1111/j.1523-1755.2005.00201.x. [DOI] [PubMed] [Google Scholar]
- 2.Gabow PA. Autosomal dominant polycystic kidney disease. The New England journal of medicine. 1993;329:332–342. doi: 10.1056/NEJM199307293290508. [DOI] [PubMed] [Google Scholar]
- 3.Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nature reviews. Nephrology. 2010;6:197–206. doi: 10.1038/nrneph.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris PC, Torres VE. Polycystic kidney disease. Annu Rev Med. 2009;60:321–337. doi: 10.1146/annurev.med.60.101707.125712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris PC, Rossetti S. Determinants of renal disease variability in ADPKD. Adv Chronic Kidney Dis. 2010;17:131–139. doi: 10.1053/j.ackd.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossetti S, Harris PC. Genotype-phenotype correlations in autosomal dominant and autosomal recessive polycystic kidney disease. Journal of the American Society of Nephrology: JASN. 2007;18:1374–1380. doi: 10.1681/ASN.2007010125. [DOI] [PubMed] [Google Scholar]
- 7.Hateboer N, et al. Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 Study Group. Lancet. 1999;353:103–107. doi: 10.1016/S0140-6736(98)03495-3. [DOI] [PubMed] [Google Scholar]
- 8.Rossetti S, et al. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney international. 2009;75:848–855. doi: 10.1038/ki.2008.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris PC, Hopp K. The mutation, a key determinant of phenotype in ADPKD. Journal of the American Society of Nephrology: JASN. 2013;24:868–870. doi: 10.1681/ASN.2013040417. [DOI] [PubMed] [Google Scholar]
- 10.Audrezet MP, et al. Autosomal dominant polycystic kidney disease: comprehensive mutation analysis of PKD1 and PKD2 in 700 unrelated patients. Human mutation. 2012;33:1239–1250. doi: 10.1002/humu.22103. [DOI] [PubMed] [Google Scholar]
- 11.Rossetti S, et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. Journal of the American Society of Nephrology: JASN. 2007;18:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 12.Vouk K, et al. PKD1 and PKD2 mutations in Slovenian families with autosomal dominant polycystic kidney disease. BMC Med Genet. 2006;7:6. doi: 10.1186/1471-2350-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Obeidova L, et al. Novel mutations of PKD genes in the Czech population with autosomal dominant polycystic kidney disease. BMC Med Genet. 2014;15:41. doi: 10.1186/1471-2350-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang T, et al. Identification of novel mutations of PKD1 gene in Chinese patients with autosomal dominant polycystic kidney disease by targeted next-generation sequencing. Clinica chimica acta; international journal of clinical chemistry. 2014;433:12–19. doi: 10.1016/j.cca.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Yu C, et al. Identification of novel mutations in Chinese Hans with autosomal dominant polycystic kidney disease. BMC medical genetics. 2011;12:164. doi: 10.1186/1471-2350-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. The European Polycystic Kidney Disease Consortium. Cell78, 725 (1994). [PubMed]
- 17.Hughes J, et al. The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet. 1995;10:151–160. doi: 10.1038/ng0695-151. [DOI] [PubMed] [Google Scholar]
- 18.Bogdanova N, et al. Homologues to the first gene for autosomal dominant polycystic kidney disease are pseudogenes. Genomics. 2001;74:333–341. doi: 10.1006/geno.2001.6568. [DOI] [PubMed] [Google Scholar]
- 19.Claes KB, De Leeneer K. Dealing with pseudogenes in molecular diagnostics in the next-generation sequencing era. Methods in molecular biology. 2014;1167:303–315. doi: 10.1007/978-1-4939-0835-6_21. [DOI] [PubMed] [Google Scholar]
- 20.Watnick TJ, et al. An unusual pattern of mutation in the duplicated portion of PKD1 is revealed by use of a novel strategy for mutation detection. Hum Mol Genet. 1997;6:1473–1481. doi: 10.1093/hmg/6.9.1473. [DOI] [PubMed] [Google Scholar]
- 21.Rossetti S, et al. Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. Journal of the American Society of Nephrology: JASN. 2012;23:915–933. doi: 10.1681/ASN.2011101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y, et al. Unified criteria for ultrasonographic diagnosis of ADPKD. Journal of the American Society of Nephrology: JASN. 2009;20:205–212. doi: 10.1681/ASN.2008050507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shendure J, Ji H. Next-generation DNA sequencing. Nature biotechnology. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 24.Renkema KY, Stokman MF, Giles RH, Knoers NV. Next-generation sequencing for research and diagnostics in kidney disease. Nature reviews. Nephrology. 2014;10:433–444. doi: 10.1038/nrneph.2014.95. [DOI] [PubMed] [Google Scholar]
- 25.Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. Journal of human genetics. 2014;59:5–15. doi: 10.1038/jhg.2013.114. [DOI] [PubMed] [Google Scholar]
- 26.Majewski J, Schwartzentruber J, Lalonde E, Montpetit A, Jabado N. What can exome sequencing do for you? Journal of medical genetics. 2011;48:580–589. doi: 10.1136/jmedgenet-2011-100223. [DOI] [PubMed] [Google Scholar]
- 27.Eisenberger T, et al. An efficient and comprehensive strategy for genetic diagnostics of polycystic kidney disease. PloS one. 2015;10:e0116680. doi: 10.1371/journal.pone.0116680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali H, et al. A novel PKD1 variant demonstrates a disease-modifying role in trans with a truncating PKD1 mutation in patients with Autosomal Dominant Polycystic Kidney Disease. BMC nephrology. 2015;16:26. doi: 10.1186/s12882-015-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravine D, et al. Evaluation of ultrasonographic diagnostic criteria for autosomal dominant polycystic kidney disease 1. Lancet. 1994;343:824–827. doi: 10.1016/S0140-6736(94)92026-5. [DOI] [PubMed] [Google Scholar]
- 30.Grantham JJ, et al. Volume progression in polycystic kidney disease. The New England journal of medicine. 2006;354:2122–2130. doi: 10.1056/NEJMoa054341. [DOI] [PubMed] [Google Scholar]
- 31.Fain PR, et al. Modifier genes play a significant role in the phenotypic expression of PKD1. Kidney international. 2005;67:1256–1267. doi: 10.1111/j.1523-1755.2005.00203.x. [DOI] [PubMed] [Google Scholar]
- 32.Emond MJ, et al. Exome sequencing of extreme phenotypes identifies DCTN4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nature genetics. 2012;44:886–889. doi: 10.1038/ng.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lettre, G. The search for genetic modifiers of disease severity in the beta-hemoglobinopathies. Cold Spring Harbor perspectives in medicine2, 10.1101/cshperspect.a015032 (2012). [DOI] [PMC free article] [PubMed]
- 34.Park MH, et al. Comprehensive analysis to improve the validation rate for single nucleotide variants detected by next-generation sequencing. PloS one. 2014;9:e86664. doi: 10.1371/journal.pone.0086664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trujillano D, et al. Diagnosis of autosomal dominant polycystic kidney disease using efficient PKD1 and PKD2 targeted next-generation sequencing. Molecular genetics & genomic medicine. 2014;2:412–421. doi: 10.1002/mgg3.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mallawaarachchi AC, et al. Whole-genome sequencing overcomes pseudogene homology to diagnose autosomal dominant polycystic kidney disease. European journal of human genetics: EJHG. 2016;24:1584–1590. doi: 10.1038/ejhg.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan P, Sung WK. Structural variation detection using next-generation sequencing data: A comparative technical review. Methods. 2016;102:36–49. doi: 10.1016/j.ymeth.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 39.Ghoneim DH, Myers JR, Tuttle E, Paciorkowski AR. Comparison of insertion/deletion calling algorithms on human next-generation sequencing data. BMC research notes. 2014;7:864. doi: 10.1186/1756-0500-7-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asan, et al. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome biology. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bodi K, et al. Comparison of commercially available target enrichment methods for next-generation sequencing. Journal of biomolecular techniques: JBT. 2013;24:73–86. doi: 10.7171/jbt.13-2402-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark MJ, et al. Performance comparison of exome DNA sequencing technologies. Nature biotechnology. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mertes F, et al. Targeted enrichment of genomic DNA regions for next-generation sequencing. Briefings in functional genomics. 2011;10:374–386. doi: 10.1093/bfgp/elr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Rawe J, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome medicine. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valencia CA, et al. Assessment of target enrichment platforms using massively parallel sequencing for the mutation detection for congenital muscular dystrophy. The Journal of molecular diagnostics: JMD. 2012;14:233–246. doi: 10.1016/j.jmoldx.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Breau RH, et al. A simple method to estimate renal volume from computed tomography. Canadian Urological Association journal = Journal de l’Association des urologues du Canada. 2013;7:189–192. doi: 10.5489/cuaj.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbard T, et al. The Ensembl genome database project. Nucleic acids research. 2002;30:38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng AY, Teo YY, Ong RT. Assessing single nucleotide variant detection and genotype calling on whole-genome sequenced individuals. Bioinformatics. 2014;30:1707–1713. doi: 10.1093/bioinformatics/btu067. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, Han S, Wang Z, Gelernter J, Yang BZ. Variant callers for next-generation sequencing data: a comparison study. PloS one. 2013;8:e75619. doi: 10.1371/journal.pone.0075619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Q, et al. Steps to ensure accuracy in genotype and SNP calling from Illumina sequencing data. BMC genomics. 2012;13(Suppl 8):S8. doi: 10.1186/1471-2164-13-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Human mutation. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Human mutation. 2013;34:E2393–2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Human molecular genetics. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raczy C, et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041–2043. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 58.Jager KJ, van Dijk PC, Zoccali C, Dekker FW. The analysis of survival data: the Kaplan-Meier method. Kidney international. 2008;74:560–565. doi: 10.1038/ki.2008.217. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.