Abstract
Recent advancements in next generation sequencing (NGS) technology have led to the identification of the giant sarcomere gene, titin (TTN), as a major human disease gene. Truncating variants of TTN (TTNtv) especially in the A-band region account for 20% of dilated cardiomyopathy (DCM) cases. Much attention has been focused on assessment and interpretation of TTNtv in human disease; however, missense and non-frameshifting insertions/deletions (NFS-INDELs) are difficult to assess and interpret in clinical diagnostic workflow. Targeted sequencing covering all exons of TTN was performed on a cohort of 530 primary DCM patients from three cardiogenetic centres across Europe. Using stringent bioinformatic filtering, twenty-nine and two rare TTN missense and NFS-INDELs variants predicted deleterious were identified in 6.98% and 0.38% of DCM patients, respectively. However, when compared with those identified in the largest available reference population database, no significant enrichment of such variants was identified in DCM patients. Moreover, DCM patients and reference individuals had comparable frequencies of splice-region missense variants with predicted splicing alteration. DCM patients and reference populations had comparable frequencies of rare predicted deleterious TTN missense variants including splice-region missense variants suggesting that these variants are not independently causative for DCM. Hence, these variants should be classified as likely benign in the clinical diagnostic workflow, although a modifier effect cannot be excluded at this stage.
Subject terms: Genetic testing, Cardiovascular genetics
Introduction
With a prevalence of at least 1:2500, dilated cardiomyopathy (DCM) is a relatively common cause for heart failure and sudden cardiac death, and the most prevalent indication for heart transplantation1,2. DCM is characterized by progressive left ventricular dilatation and systolic dysfunction. Based on aetiology, the European Society of Cardiology (ESC) classified DCM into familial (genetic) and non-familial forms, with up to 30–50% of idiopathic DCM cases being familial3,4. Familial DCM exhibits genetic heterogeneity and follows generally an autosomal dominant inheritance. Most of the disease-causing DCM variants have been identified in genes encoding the components of the sarcomere, desmosome, cytoskeleton, nuclear lamina, mitochondria, and calcium-handling proteins5,6.
Variants leading to truncations of the giant sarcomeric protein titin (TTNtv) are the most frequent cause of DCM. Recently, we and others have identified clinically relevant TTNtv in about 20% of unselected and familial DCM cases7–10. Interestingly, DCM-causing TTNtv are clustered in the clinically relevant A-band region of the gene whereas TTNtv in reference and healthy individuals are clustered more in the I-band region where they may be spliced out, due to the usage of an alternative promoter, from mature long TTN isoforms resulting in minimal or no functional consequences11. Interpretation of TTNtv in clinical setting has been challenged by several factors including lack of knowledge of the true frequency of TTNtv in the general population, lack of understanding of different protein domains and alternatively spliced isoforms, and variable disease presentation hampering assessment of co-segregation. We and others have previously addressed these issues7,9,12–14. However, assessing the relevance of TTN missense variants in the pathogenesis of DCM has been even more difficult due to the high frequency of these variants in both reference individuals and DCM patients and the absence of large pedigrees where segregation can be studied. While few studies have reported modifier roles for TTN missense variants15,16, others have considered TTN missense variants as disease-causing variants in DCM, arrhythmogenic right ventricular cardiomyopathy (ARVC), and left ventricular non-compaction cardiomyopathy (LVNC) but these observations suggest recessive inheritance pattern and no convincing segregation has been demonstrated so far17–19. Furthermore, TTNtvs are also occasionally found in patients with SCD/aborted SCD who do not have diagnosis of DCM or DCM with LVNC20,21
In the current study, we focused on TTN missense and non-frameshifting insertion-deletion (NFS-INDELs) variants identified in DCM patients and reference population. By analyzing TTN missense and NFS-INDELs variants identified in 530 primary DCM patients, over 60,000 ExAC individuals, and for the first time, the largest available 123,136 gnomAD database reference individuals, we report a seemingly insignificant enrichment of TTN missense variants in DCM patients compared with reference individuals.
Results
Patient Cohort
DCM patients in this multi-centre study were recruited from three major cardiogenetic centres in Finland and the Netherlands. All patients fulfilled diagnostic criteria for DCM. Table 1 summarizes spectrum of pathogenic and likely pathogenic variants identified in the patients.
Table 1.
Distribution and spectrum of other pathogenic and likely pathogenic variants identified in the cohort.
| Patients | All (N = 530) | Helsinki cohort (N = 145) | Groningen cohort (N = 169) | Amsterdam cohort (N = 216) |
|---|---|---|---|---|
| Gender distribution, n (%) | ||||
| Males | 311 (58.7) | 107 (73.8) | 95 (56.2) | 109 (50.5) |
| Females | 219 (41.3) | 38 (26.2) | 74 (43.8) | 107 (49.5) |
| Diagnostic yield, n (%) | ||||
| Mutation positive | 157 (29.6) | 51 (35.2) | 37 (21.9) | 69 (31.9) |
| Mutation negative | 373 (70.4) | 94 (64.8) | 137 (81.1) | 147 (68.1) |
| Causative gene, n (%) | ||||
| Sarcomeric | 108 (20.4) | 28 (19.3) | 22 (13.0) | 58 (26.9) |
| TPM1 | 1 (0.2) | — | — | 1 (0.5) |
| TCAP | 1 (0.2) | 1 (0.7) | — | — |
| TNNT2 | 2 (0.4) | 1 (0.7) | — | 1 (0.5) |
| TNNI3 | 2 (0.4) | — | — | 2 (0.9) |
| MYH7 | 5 (0.9) | 1 (0.7) | 2 (1.2) | 2 (0.9) |
| TTN | 97 (18.3) | 25 (17.2) | 20 (11.8) | 52 (24.1) |
| Non-sarcomeric | 49 (9.2) | 23 (15.7) | 15 (8.9) | 11 (5.1) |
| DMD | 1 (0.2) | 1 (0.7) | — | — |
| RBM20 | 10 (1.9) | 2 (1.4) | 3 (1.8) | 5 (2.3) |
| PLN | 11 (2.1) | — | 7 (4.1) | 4 (1.9) |
| DSP | 12 (2.3) | 8 (5.5) | 2 (1.2) | 2 (0.9) |
| LMNA | 15 (2.8) | 12 (8.3) | 3 (1.8) | — |
Deriving Allele Frequency for Potentially Deleterious DCM Variant
To set a threshold for filtering TTN missense variants identified in DCM cohorts, we obtained the gnomAD allele frequency of the most common pathogenic variant associated with DCM with a wide consensus of the pathogenicity in the field. Covering 100% of the total gnomAD alleles, the PLN c.40_42delAGA, p.Arg14del (NM_002667.4) variant is the most common “certainly pathogenic” DCM variant22–26. This variant is absent in the ExAC individuals (allele count [AC] = 0), while it is present in two individuals (AC = 2) in the gnomAD database. Based on these observations, the minor allele frequency (MAF) cut-offs were set to 0.0016% (AC = 2) and 0.0016% (AC = 4) for ExAC and gnomAD datasets respectively, and variants with higher frequency in the reference populations would not be expected to be pathogenic.
TTN NFS-INDELs and Missense Variants in DCM Cohort
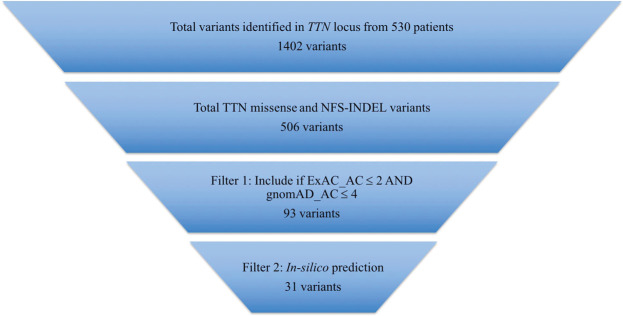
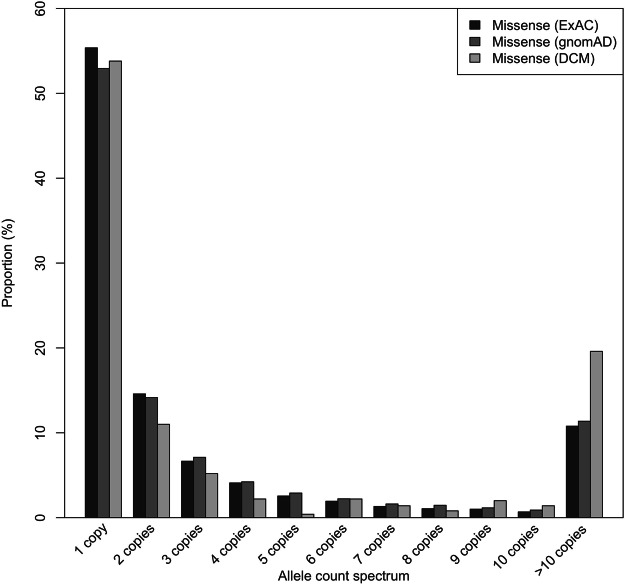
Initial variant calling with GATK yielded a total of 1402 unique TTN variants identified in 530 primary DCM patients, of which 506 were missense and NFS-INDELs (Fig. 1), 39 frameshift variants, 27 nonsense variants, 14 splice-site variants, 205 silent variants, 4 untranslated region (UTR) variants, and the remaining 607 variants being intronic. Of the total variants, 52.7% were private (Fig. 2). Applying the ExAC and gnomAD MAF cut-offs yielded a total of 87 missense and 5 NFS-INDEL variants. Taken together, 18.9% and 1.9% of DCM patients carried rare heterozygous missense and NFS-INDELs, respectively. Given that many rare and even private variants are believed to be innocent bystanders, variant pathogenicity potential was further assessed by using three in-silico prediction tools. Implementing this, 33.3% (29/87) of the rare missense variants were predicted deleterious. Interestingly, 69.0% (20/29) of the predicted deleterious missense variants were in the A-band region of TTN. Of note, 55.2% (16/29) of the rare missense variants were absent in both ExAC and gnomAD reference individuals.
Figure 1.
Bioinformatic filtering strategy to identify “deleterious” variants. Total TTN variants were filtered using allele frequency (0.0016%) threshold derived from the frequency of known pathogenic DCM variants identified in gnomAD reference individuals. Variants more frequent than derived thresholds in respective sources were not expected to be deleterious.
Figure 2.
Allele frequency spectrum of TTN missense variants in DCM and reference individuals. More than 50% of TTN missense variants in DCM patients as well as in reference individuals were private. Potentially pathogenic private and/or rare TTN missense variants were not enriched in DCM patients compared with ExAC and gnomAD reference individuals.
Twenty-nine rare heterozygous TTN missense variants predicted deleterious were identified in 6.8% (36/530) of DCM patients, with only one patient harbouring two rare heterozygous TTN missense variants. Heterozygous NFS-INDELS affecting at least five transcripts of the TTN (Transcript Count Index, TCI ≥ 5) were identified in 0.37% (2/530) of DCM patients. Of note, all the NFS-INDELS were in the I-band region of TTN (Tables 2 and 3). When considering mutation status, gene elusive DCM patients and mutation positive DCM patients had comparable frequencies of rare TTN missense variants further suggesting that these variants may not be monogenic cause of DCM (Supplemantary Table S1).
Table 2.
TTN missense and non-frameshifting INDELs variants in DCM vs. general population.
| Total TTN NFS-INDEL allele | Total TTN missense allele | |||||||
|---|---|---|---|---|---|---|---|---|
| DCM (n = 530) | gnomAD (n = 123,136) | OR | p-value | DCM (n = 530) | gnomAD (n = 123,136) | OR | p-value | |
| Raw variant (before assessment) | ||||||||
| Unique variant count | 9 | 289 | 497 | 17726 | ||||
| After assessment using derived thresholds | ||||||||
| Unique variant count | 5 | 226 | 87 | 13808 | ||||
| Number of individuals (n) | 10 | 337 | 100 | 21102 | ||||
| Prevalence (%) | 1.887 | 0.273 | 7.0 | 3.2E-06 | 18.867 | 17.137 | 1.1 | 0.3187 |
| Distribution by sarcomere domain, n (%) | ||||||||
| Z-disc | 0 (0.00) | 32 (0.026) | NA | 2 (0.377) | 1074 (0.872) | 0.4 | 0.3400 | |
| I-band | 10 (1.887) | 104 (0.084) | 22.7 | 8.7E-11 | 46 (8.679) | 6720 (5.457) | 1.6 | 0.0015 |
| A-band | 0 (0.00) | 137 (0.111) | NA | 58 (10.943) | 10725 (8.709) | 1.3 | 0.0815 | |
| M-band | 0 (0.00) | 53 (0.043) | NA | 2 (0.377) | 1488 (1.208) | 0.3 | 0.1000 | |
| After assessment using derived thresholds and pathogenicity prediction | ||||||||
| Unique variant count | 2 | 172 | 29 | 4657 | ||||
| Number of individuals (n) | 2 | 255 | 37 | 7071 | ||||
| Prevalence (%) | 0.377 | 0.207 | 1.8 | 0.3000 | 6.981 | 5.74 | 1.2 | 0.2200 |
| Distribution by sarcomere domain, n (%) | ||||||||
| Z-disc | 0 (0.00) | 31 (0.025) | NA | 1 (0.188) | 243 (0.197) | 0.95 | 1.0000 | |
| I-band | 2 (0.377) | 30 (0.024) | 15.5 | 0.0083 | 10 (1.887) | 1486 (1.206) | 1.6 | 0.1581 |
| A-band | 0 (0.00) | 137 (0.111) | NA | 25 (4.717) | 4760 (3.865) | 1.2 | 0.3100 | |
| M-band | 0 (0.00) | 53 (0.043) | NA | 0 | 540 (0.438) | NA | NA | |
Table 3.
Rare TTN missense variant classified as deleterious by in-silico prediction.
| SYMBOL | Position | Nucleotide Change | AA Change | EXON | DCM (n) | ExAC (n) | gnomAD (n) |
|---|---|---|---|---|---|---|---|
| Z-disk | |||||||
| TTN | 179664242 | c.886 G > A | Val296Met | 6 | 1 | 0 | 0 |
| I-band | |||||||
| TTN | 179640476 | c.6115 C > T | Leu2039Phe | 28 | 1 | 0 | 0 |
| TTN | 179634837 | c.8591 A > G | Tyr2864Cys | 36 | 1 | 0 | 0 |
| TTN | 179634555 | c.8753 G > C | Gly2918Ala | 37 | 1 | 1 | 2 |
| TTN | 179632835 | c.9211 G > C | Glu3071Gln | 39 | 1 | 2 | 4 |
| TTN | 179588312 | c.21515 G > T | Arg7172Ile | 74 | 3 | 0 | 0 |
| TTN | 179576946 | c.27611 C > T | Pro9204Leu | 96 | 1 | 1 | 2 |
| TTN | 179574299 | c.28747 A > T | Ile9583Phe | 99 | 1 | 0 | 0 |
| TTN | 179494979 | c.44270 T > A | Leu14757His | 239 | 2 | 0 | 0 |
| A-band | |||||||
| TTN | 179477634 | c.49814 T > G | Val16605Gly | 265 | 6 | 0 | 0 |
| TTN | 179476201 | c.50755 G > T | Val16919Phe | 269 | 1 | 0 | 0 |
| TTN | 179472191 | c.53224 T > A | Tyr17742Asn | 277 | 1 | 0 | 0 |
| TTN | 179452778 | c.63356 G > A | Cys21119Tyr | 305 | 1 | 1 | 1 |
| TTN | 179441303 | c.69668 G > C | Gly23223Ala | 325 | 1 | 0 | 0 |
| TTN | 179425479 | c.85380 A > G | Ile28460Met | 326 | 1 | 1 | 3 |
| TTN | 179429090 | c.81769 C > A | Pro27257Thr | 326 | 0 | 0 | 0 |
| TTN | 179432751 | c.78108 G > C | Leu26036Phe | 326 | 1 | 0 | 1 |
| TTN | 179435376 | c.75483 G > C | Lys25161Asn | 326 | 1 | 0 | 1 |
| TTN | 179436260 | c.74599 A > G | Thr24867Ala | 326 | 1 | 0 | 0 |
| TTN | 179439256 | c.71603 G > A | Arg23868Gln | 326 | 1 | 1 | 2 |
| TTN | 179439440 | c.71419 T > C | Tyr23807His | 326 | 1 | 0 | 0 |
| TTN | 179419452 | c.88622 T > C | Ile29541Thr | 332 | 1 | 1 | 4 |
| TTN | 179416971 | c.90656 T > A | Val30219Asp | 335 | 1 | 0 | 0 |
| TTN | 179417593 | c.90034 G > A | Glu30012Lys | 335 | 1 | 0 | 0 |
| TTN | 179418010 | c.89617 T > C | Trp29873Arg | 335 | 1 | 0 | 0 |
| TTN | 179414844 | c.91721 A > T | Glu30574Val | 337 | 2 | 0 | 1 |
| TTN | 179414390 | c.92059 G > A | Gly30687Ser | 338 | 1 | 2 | 4 |
| TTN | 179411891 | c.94361 T > C | Leu31454Pro | 340 | 1 | 0 | 0 |
| TTN | 179403949 | c.98713 G > A | Glu32905Lys | 353 | 1 | 1 | 3 |
Variant positions were reported for the RefSeq transcript NM_001267550.
TTN Missense and NFS-INDELs Variants in Reference Population
Applying the derived allele frequency thresholds to TTN variants reported in ExAC, we identified 8265 and 82 rare missense and NFS-INDELs variants respectively in 16.5% (10035/60707) and 0.15% (93/60707) of the ExAC individuals. Of the ExAC rare missense variants, 24% (1990/8265) were predicted deleterious by the three prediction tools used in this study. In total, 3.98% of ExAC individuals carried a rare TTN missense variant predicted deleterious. In addition, 0.11% (65/60706) of ExAC cohort harboured rare heterozygous NFS-INDELs affecting at least five transcripts of TTN.
Given that gnomAD cohort is almost twice the size of ExAC cohort and ExAC individuals are in gnomAD cohort, we set the threshold to four and considered variants more frequent than this in gnomAD as likely benign. With this threshold, we identified 13808 rare missense variants present in 17.1% (21102/123136) of gnomAD cohort. However, rare heterozygous TTN missense variants predicted deleterious were observed in 5.7% (7071/123136) of the gnomAD (Table 2). Notably, 54.6% and 52.2% of TTN missense and NFS-INDELS identified in ExAC and gnomAD cohorts respectively were private (Fig. 2).
Multiple Heterozygous TTN Variants
To estimate the frequency of reference individuals with multiple heterozygous TTN variants, the phase 3 call set of the 1000 Genomes project was utilized since it provides sample-level genotype data. Only TTN missense variants with allele frequency less than or equal the derived thresholds were used for this analysis. We identified 8.2% (205/2504) of the 1000 Genomes Project phase 3 cohorts with a rare heterozygous TTN missense variant. Of these, 5.4% (11/205) harboured two heterozygous rare TTN missense variants.
When considering eleven individuals in 1000 Genomes Project phase 3 cohorts with TTN truncations (TTNtv) as published previously12, 18.2% (2/11) of these individuals, both from South Asian population also harboured a rare heterozygous TTN missense variant, thus, being heterozygous for both rare TTNtv and TTN missense variant. However, it is unclear whether they are in cis (at the same allele) or in trans (at different allele)
Interestingly, only one DCM patient in this study harboured two rare heterozygous TTN missense variants, one in the A-band and the other at the distal I-band TTN region. Five out of thirty-six (13.9%) DCM patients with rare TTN missense variants predicted deleterious harbored a TTNtv (Table 4).
Table 4.
DCM patients with both predicted deleterious TTN missense variant and TTN truncation (TTNtv).
| Gene | Deleterious TTN missense variants | Other known P/LP TTNtv | ||
|---|---|---|---|---|
| Nucleotide Change | AA Change | Nucleotide Change | AA Change | |
| TTN | c.44270 T > A | p.Leu14757His | c.45322 C > T | p.Arg15108* |
| TTN | c.91721 A > T | p.Glu30574Val | c.76373delC | p.Pro25458Glnfs*9 |
| TTN | c.98713 G > A | p.Glu32905Lys | c.8426dup | p.Asn2809Lysfs*7 |
| TTN | c.69668 G > C | p.Gly23223Ala | c.69671delC | p.Pro23224HisfsTer10 |
| TTN | c.49814 T > G | p.Val16605Gly | c.75391delG | p.V25131Lfs*16 |
Variant positions were reported for the RefSeq transcript NM_001267550.
Distribution of TTN NFS-INDELs and Missense Variants in DCM and Reference Cohorts
The proportion of DCM patients harbouring rare TTN missense and NFS-INDEL variants before and after variant assessment was compared with that identified in the reference population. 18.9% (100/530) of DCM patients harboured rare TTN missense variants as against 17.3% in gnomAD reference individuals (OR = 1.1; 95% CI: 0.90–1.35; P = 0.32). Moreover, DCM patients had a slightly higher frequency of TTN missense in the TTN I-band region (8.7% vs. 5.5%; OR = 1.1; 95% CI: 1.2–2.1; P = 0.0015) and A-band region (10.9% vs. 8.7%; OR = 1.3; 95% CI: 1.0–1.6; P = 0.08). Rare NFS-INDELs was detected in 1.9% of DCM patients as against 0.3% in gnomAD reference individuals (OR = 7.0; P = 3.2 × 10−6). Of note, all NFS-INDELs identified in DCM patients clustered in the TTN I-band region. When considering rare TTN missense and NFS-INDELs together, there was no statistically significant enrichment of these variants in DCM patients and within TTN domains compared to the observed frequency and distribution in gnomAD database reference individuals (Table 2 and Fig. 3). Moreover, distribution of TTN missense variants was similar in patients with a known pathogenic or likely pathogenic DCM mutation (Mutation positive) compared to those without a mutation (Mutation negative) when the variant classification follows ACMG criteria (Supplementary Table S1).
Figure 3.
Spatial distribution of rare TTN missense variants in DCM cohort and reference population. Titin is linearly depicted with its 152 Ig-like domains in purple and 132 fibronectin type III domains in green. TTN missense variants identified in patients as well as ExAC and gnomAD reference individuals are shown as coloured bars. Bisque bar represents the number of transcripts (TCI). Variants are shown relative to the Meta transcript position. The dark grey bars indicate exons unique to the Meta transcript. The dashed lines below the number of transcripts schematic indicate the location of variants within the sarcomere.
Splice-Region Missense Variants with Predicted Splicing Effect are not enriched in DCM
Given that variants that affect splicing can directly cause disease or contribute to disease severity, we analysed missense variants located near exon-intron boundaries (splice-regions). A total of five rare splice-region missense variants affecting 3.6% (19/530) of DCM patients was identified. Of note, only two out of five were predicted to potentially alter natural splice site or activate exonic cryptic splice-sites. When compared with ExAC reference individuals (DCM: 3.6% vs. ExAC: 2.9%; P = 0.4624) and gnomAD reference individuals (DCM: 3.6% vs. gnomAD: 3.0%; P = 0.4808), there was no significant enrichment of variants with high potential for defective splicing in DCM patients.
Discussion
Recent advancement in high-throughput sequencing technology has unravelled TTN as a major human disease gene, mutated in both human skeletal and cardiac muscles disease. Inclusion of the TTN gene in genetic screening for DCM has led to an increase in diagnostic rate for adult DCM by at least 18% and 25% respectively in sporadic and familial DCM cases8–10,27,28. Much attention has been focused on the assessment and clinical interpretation of frameshift, nonsense and splice-site variants, leading to a truncation of TTN, leaving missense and NFS-INDELs variants in this major human disease gene unassessed and unexplored. Consequently, these variants are difficult to assess and interpret in the clinical diagnostic workflow.
DCM is a genetically heterogeneous disorder and multiple genes have been implicated in the pathogenesis of DCM. Truncations of the giant sarcomeric protein, TTN, are to date, the most common genetic cause of DCM, accounting for about 20% of cases, and are overrepresented in the A-band region of the protein. Such A-band and/or distal I-band TTNtv have been estimated to have nearly 98% chance of being disease causing when identified in unselected DCM patients7. Interestingly, a recent study showed a significant association of TTNtv in constitutive exons throughout TTN protein with DCM29. Furthermore, it has been shown that TTNtv rarely cause pediatric DCM but the gene typically expresses the disease at middle-age30. In addition, we and others have addressed several issues and challenges associated with the clinical assessment and interpretation of TTNtv in both research and clinical diagnostic setting7,9,12–14. In contrast, the role and contribution of missense and NFS-INDELs variants in this gene has been poorly addressed so far.
In this European multi-centre DCM study, we assessed the possible role of TTN missense and NFS-INDELs variants in DCM. By analyzing TTN variants in: 1) a large primary DCM cohort (n = 530), 2) the widely used ExAC reference individuals, and 3) the largest available gnomAD database reference individuals, we showed that there is a statistically insignificant enrichment of rare TTN missense and NFS-INDEL variants in DCM patients compared with reference individuals. This observation holds even when considering different TTN regions, suggesting that TTN missense and NFS-INDEL variants do not cause DCM and should therefore be classified as “likely benign” in clinical diagnostic settings. Using stringent bioinformatic filtering criteria, we identified rare TTN missense variants predicted deleterious in 7.0% of primary DCM cases as against 5.7% in gnomAD reference individuals (OR = 1.2; P = 0.22). When considering different TTN domains (Z-disk, I-band, A-band and M-band), we identified no enrichment of these variants in DCM patients compared to reference individuals.
A recent study assessing the role of TTN missense variants identified “severe” missense variants in 25.2% (37/147) of DCM probands15. We randomly selected five “severe” missense variants as published by the authors and applied our bioinformatic filtering strategy. Of note, 40% (2/5) of these “severe” variants were found in at least seven gnomAD database reference individuals, and as such would not be considered “deleterious” using our approach. Interestingly, five probands in the study were double heterozygous for a “severe” TTN missense variant and a pathogenic (P)/likely pathogenic (LP) variant in LMNA, MYH7 and SCN5A. Three (3/5) of these patients harboured a P/LP LMNA variant in addition to “severe” TTN variant. Two of the “severe” TTN missense variants were later reclassified by the authors as “unlikely” disease associated, possibly due to lack of segregation, and third patient harbouring a P/LP LMNA variant co-occurring with a “severe” TTN missense variant had undergone heart transplantation15. In another study of an extended DCM family with 14 affected subjects, four had severe DCM requiring heart transplantation in early adulthood31. In all affected patients, the P/LP LMNA p.(Lys219Thr) variant was identified. In addition to this variant, a TTN missense variant, p.(Leu4855Phe), was identified in all four patients with severe phenotypes, all of whom had transplantation at a younger age compared to those with only LMNA p.(Lys219Thr) variant. Although this may suggest that TTN missense variants can modify disease severity, the TTN p.(Leu4855Phe) is found heterozygous in 15 individuals in gnomAD reference population and the position is multi-allelic, thus stronger evidence is needed before scientific interpretation of potential modifier role can be inferred.
Disruption of natural splice sites by point mutations can result in exon skipping, activation of cryptic splice-site, and rarely intron retention32, with activation of cryptic splice-site being the second most frequent consequence of such mutations. Such point mutations, put together, account for 10–15% of all mutations causing human inherited disease33,34. By analyzing missense variants located near exon-intron boundaries (splice-region) of TTN, we identified five rare splice-region missense variants in DCM patients. Two (40%) of these five splice-region missense variants were predicted to disrupt natural splice-site. To test for possible enrichment of this event in DCM patients, we randomly and iteratively selected five rare splice-regions variants from gnomAD and tested their potential to alter splicing. Surprisingly, two (40%) of the five randomly chosen splice-region missense variants were predicted to alter splicing, suggesting that splice-region missense variants identified in our DCM cohort do not carry higher risk of splicing defect than any other splice-region missense variants in the population.
DCM cohorts in this study were sequenced using high coverage sequencing assays yielding more uniform coverage even across difficult-to-sequence regions that may alone increase likelihood of finding slightly higher variant counts from DCM cohorts compared to the population databases. If we overestimate that DCM has a prevalence of 1 per 500 and further speculate that TTN missense variants would account 10% of all DCM cases leading to a frequency of 1 per 5000 (0.02%) in TTN missense positive DCM. However, 5.7% of gnomAD reference population have rare missense variant predicted deleterious by in-silico tools, thus 99.7% of these missense variants are not expected to cause DCM in monogenic manner. Thus, these variants should be classified as likely benign in clinical and diagnostic workflow as far as more understanding have been gathered of them individually and as a group.
Rare NFS-INDELs affecting at least five transcripts of TTN (TCI ≥ 5) were identified in 0.37% (2/530) of DCM patients as against 0.21% (255/123136) in gnomAD reference individuals (OR = 1.8; P = 0.3). Failure to show significant enrichment of NFS-INDELs in our DCM cohort suggests that the vast majority of these variants are non-contributory to the monogenic etiology of DCM.
Limitations
We acknowledge the following limitations: The health status of reference individuals used is not known even though curators of reference population databases have made every effort to exclude individuals with severe pediatric diseases from these cohorts. Moreover, we had limited evidence of the effect of predicted deleterious TTN missense variants in patients with other P/LP variants including TTNtv. Functional analysis of predicted deleterious TTN missense and NFS-INDELs was beyond the scope of this study; and we recommend that such should be combined with segregation information and de novo status to reclassify the variants.
Conclusion
The current data revealed that predicted deleterious TTN missense and NFS-INDEL variants are not significantly enriched in a large group of DCM patients compared to control populations, and should therefore be classified as likely benign in a clinical diagnostic setting. Nevertheless, it cannot be excluded that some rare TTN missense variants might have modifier effect for the phenotype.
Methods
Patient Cohorts and Clinical Evaluation
This European multi-centre study was approved by institutional ethical committees of the University of Helsinki, Academic Medical Centre (Amsterdam, the Netherlands), and University Medical Centre (Groningen, the Netherlands) and informed consent was obtained from all subjects. The DCM cohort in this study comprised of 530 unrelated primary DCM patients of European origin. The patients were recruited at three major cardiogenetic centres in Finland (Helsinki; n = 145) and the Netherlands (Amsterdam; n = 216, and Groningen; n = 169). All patients fulfilled the diagnostic criteria for DCM (LV size/volume >117% or >2 SD of the reference value and LV-EF <45%)35.
Genetic Studies of DCM
Genomic DNA was extracted from patients’ blood samples by standard procedures and guidelines. TTN was sequenced in a total of 530 primary DCM patients using targeted panels of increasing sizes as previously described8,27. Irrespective of the sequencing panel used, sequence data from each patient was processed uniformly following GATK best practices recommendation to identify genetic variants36. Identified variants were annotated using Ensembl’s Variant Effect Predictor (VEP v87) tool37, and SIFT38, PolyPhen39, and MutationTaster40 pathogenicity scores and predictions were obtained for each missense variant via the database of nonsynonymous SNP functional prediction (dbNSFPv2.9.2)41. Rare TTN missense variants predicted deleterious by the three prediction tools were considered candidate risk variants. NFS-INDELs were assessed using the Transcript Count Index (TCI) as previously published7.
Analysis of TTN Missense and NFS-INDELs Variants in the Population
Exonic boundaries of TTN in build 37 of the human genome (hg19) were downloaded from the Ensembl Genome Browser (www.ensembl.org), and were used to query ExAC and gnomAD databases (accessed 23 July 2017) for TTN exonic variants. Extracted variants were filtered for quality (Phred Quality Score 29) and coverage (15.0X), and calls that passed all quality filters were used for downstream analysis. The two datasets were re-annotated using VEP v87, processed uniformly to identify spectrum and distribution of missense and NFS-INDELS mutations in the reference population.
Variant Mapping to Uniprot Protein Domain
To allow inclusion of variants located in exons not present in the principal cardiac long isoform (N2BA), otherwise known as the canonical transcript with the Uniprot ID Q8WZ42-1, the location of variants identified in DCM patients as well as in the reference population was reported with respect to the Meta transcript annotation - (ENST00000589042/NM_001267550). Protein domain annotation for the Uniprot consensus sequence Q8WZ42-1 was transferred to the inferred Meta transcript as described earlier7,12.
In-silico Splicing Defect Prediction
The effect of splice-region missense variants on splicing efficiency were predicted with five different tools – Human Splicing Finder42,43, Splice Site Prediction by Neural Network (NNSPLICE)44, MaxEntScan45, GeneSplicer46, and Splice Site Finder, via Alamut splicing software v2.0 (Interactive Biosoftware, France) using default settings in all predictions. Controls for each rare splice-region missense variant were selected from gnomAD from near-by genomic location and with similar allele frequency.
Statistical Analysis
Between groups comparisons for categorical variables were performed using χ2 test if appropriate, otherwise, Fisher’s exact test. For non-parametric and continuous variables, between groups comparisons were done using Mann–Whitney U tests and independent samples t-tests combined with Levene’s tests respectively. Bonferroni correction was performed on all analyses to adjust for multiple testing. All statistical analyses were done using R statistical software (version 3.3.3).
Supplementary information
Acknowledgements
We would like to thank the Exome Aggregation Consortium and Genome Aggregation Database; and the groups that provided exome and genome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about and http://gnomad.broadinstitute.org/about. This work was supported by grants from the Sigrid Jusélius Foundation (T-PA, JK), Finnish Foundation for Cardiovascular Research (T-PA, JK, TH), Aarne Koskelo Foundation (TH), Special Governmental Subsidy (EVO) grants (TH), Ida Montin Foundation (OA), Orion Research Foundation (OA), Oskar Öflund foundation (OA), and the Netherlands Cardiovascular Research Initiative, an initiative with support of the Dutch Heart Foundation: CVON2012-10 PREDICT and CVON2014-40 DOSIS.
Author Contributions
Conception and study design: O.A., T.H., M.P.B., Y.M.P., T.P.A., S.M., K.S.Z., J.P.T., P.A.Z., J.K. Acquisition of data: O.A., T.H., R.H.L.D., J.D.H.J., L.G.B., M.P.B., Y.M.P., T.P.A., S.M., K.S.Z., J.P.T., P.A.Z., J.K. Analysis and interpretation of data: O.A., T.H., R.H.L.D., J.D.H.J., L.G.B., M.P.B., Y.M.P., T.P.A., K.S.Z., J.P.T., P.A.Z., J.K. Drafting the manuscript: O.A., T.H., K.S.Z., J.P.T., P.A.Z., J.K. Critical revision of the manuscript: All authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Karin van Spaendonck-Zwarts, J. Peter van Tintelen, Paul A. van der Zwaag and Juha Koskenvuo jointly supervised this work.
Change history
10/9/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
Oyediran Akinrinade, Email: oyediran.akinrinade@yahoo.com.
Juha Koskenvuo, Email: juha.koskenvuo@blueprintgenetics.com.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-39911-x.
References
- 1.Codd MB, Sugrue DD, Gersh BJ, Melton LJ., 3rd Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy. A population-based study in Olmsted County, Minnesota, 1975–1984. Circulation. 1989;80:564–572. doi: 10.1161/01.CIR.80.3.564. [DOI] [PubMed] [Google Scholar]
- 2.John R, et al. Long-term outcomes after cardiac transplantation: an experience based on different eras of immunosuppressive therapy. The Annals of thoracic surgery. 2001;72:440–449. doi: 10.1016/S0003-4975(01)02784-9. [DOI] [PubMed] [Google Scholar]
- 3.Elliott P, et al. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. European heart journal. 2008;29:270–276. doi: 10.1093/eurheartj/ehm342. [DOI] [PubMed] [Google Scholar]
- 4.Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annual review of genomics and human genetics. 2005;6:185–216. doi: 10.1146/annurev.genom.6.080604.162132. [DOI] [PubMed] [Google Scholar]
- 6.Dellefave L, McNally EM. The genetics of dilated cardiomyopathy. Current opinion in cardiology. 2010;25:198–204. doi: 10.1097/HCO.0b013e328337ba52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akinrinade O, Alastalo TP, Koskenvuo JW. Relevance of truncating titin mutations in dilated cardiomyopathy. Clin Genet. 2016;90:49–54. doi: 10.1111/cge.12741. [DOI] [PubMed] [Google Scholar]
- 8.Akinrinade O, et al. Genetics and genotype-phenotype correlations in Finnish patients with dilated cardiomyopathy. European heart journal. 2015;36:2327–2337. doi: 10.1093/eurheartj/ehv253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herman DS, et al. Truncations of titin causing dilated cardiomyopathy. The New England journal of medicine. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugh TJ, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genetics in medicine: official journal of the American College of Medical Genetics. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 11.Deo RC. Alternative Splicing, Internal Promoter, Nonsense-Mediated Decay, or All Three: Explaining the Distribution of Truncation Variants in Titin. Circulation. Cardiovascular genetics. 2016;9:419–425. doi: 10.1161/CIRCGENETICS.116.001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinrinade O, Koskenvuo JW, Alastalo TP. Prevalence of Titin Truncating Variants in General Population. PloS one. 2015;10:e0145284. doi: 10.1371/journal.pone.0145284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jansweijer JA, et al. Truncating titin mutations are associated with a mild and treatable form of dilated cardiomyopathy. European journal of heart failure. 2017;19:512–521. doi: 10.1002/ejhf.673. [DOI] [PubMed] [Google Scholar]
- 14.van Spaendonck-Zwarts KY, et al. Titin gene mutations are common in families with both peripartum cardiomyopathy and dilated cardiomyopathy. European heart journal. 2014;35:2165–2173. doi: 10.1093/eurheartj/ehu050. [DOI] [PubMed] [Google Scholar]
- 15.Begay, R. L. et al. Role of Titin Missense Variants in Dilated Cardiomyopathy. Journal of the American Heart Association4, 10.1161/JAHA.115.002645 (2015). [DOI] [PMC free article] [PubMed]
- 16.Golbus JR, et al. Population-based variation in cardiomyopathy genes. Circulation. Cardiovascular genetics. 2012;5:391–399. doi: 10.1161/CIRCGENETICS.112.962928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinson JT, et al. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–986. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings R, et al. Combination of Whole Genome Sequencing, Linkage, and Functional Studies Implicates a Missense Mutation in Titin as a Cause of Autosomal Dominant Cardiomyopathy With Features of Left Ventricular Noncompaction. Circulation. Cardiovascular genetics. 2016;9:426–435. doi: 10.1161/CIRCGENETICS.116.001431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor M, et al. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation. 2011;124:876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tayal U, et al. Truncating Variants in Titin Independently Predict Early Arrhythmias in Patients With Dilated Cardiomyopathy. Journal of the American College of Cardiology. 2017;69:2466–2468. doi: 10.1016/j.jacc.2017.03.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapoor A, et al. Rare coding TTN variants are associated with electrocardiographic QT interval in the general population. Scientific reports. 2016;6:28356. doi: 10.1038/srep28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh R, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in medicine: official journal of the American College of Medical Genetics. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DeWitt MM, MacLeod HM, Soliven B, McNally EM. Phospholamban R14 deletion results in late-onset, mild, hereditary dilated cardiomyopathy. Journal of the American College of Cardiology. 2006;48:1396–1398. doi: 10.1016/j.jacc.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Haghighi K, et al. A mutation in the human phospholamban gene, deleting arginine 14, results in lethal, hereditary cardiomyopathy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1388–1393. doi: 10.1073/pnas.0510519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van der Zwaag PA, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. European journal of heart failure. 2012;14:1199–1207. doi: 10.1093/eurjhf/hfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Zwaag PA, et al. Recurrent and founder mutations in the Netherlands-Phospholamban p.Arg14del mutation causes arrhythmogenic cardiomyopathy. Netherlands heart journal: monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation. 2013;21:286–293. doi: 10.1007/s12471-013-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haas, J. et al. Atlas of the clinical genetics of human dilated cardiomyopathy. European heart journal, 10.1093/eurheartj/ehu301 (2014). [DOI] [PubMed]
- 28.Roberts AM, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Science translational medicine. 2015;7:270ra276. doi: 10.1126/scitranslmed.3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schafer S, et al. Titin-truncating variants affect heart function in disease cohorts and the general population. Nature genetics. 2017;49:46–53. doi: 10.1038/ng.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatkin D, et al. Titin truncating mutations: A rare cause of dilated cardiomyopathy in the young. Progress in pediatric cardiology. 2016;40:41–45. doi: 10.1016/j.ppedcard.2016.01.003. [DOI] [Google Scholar]
- 31.Roncarati R, et al. Doubly heterozygous LMNA and TTN mutations revealed by exome sequencing in a severe form of dilated cardiomyopathy. European journal of human genetics: EJHG. 2013;21:1105–1111. doi: 10.1038/ejhg.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 33.Krawczak M, et al. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 34.Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Human genetics. 1992;90:41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- 35.Mestroni L, et al. Guidelines for the study of familial dilated cardiomyopathies. Collaborative Research Group of the European Human and Capital Mobility Project on Familial Dilated Cardiomyopathy. European heart journal. 1999;20:93–102. doi: 10.1053/euhj.1998.1145. [DOI] [PubMed] [Google Scholar]
- 36.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic acids research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Current protocols in human genetics/editorial board, Jonathan L. Haines… [et al.] Chapter 7, Unit720, 10.1002/0471142905.hg0720s76 (2013). [DOI] [PMC free article] [PubMed]
- 40.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nature methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Jian X, Boerwinkle E. dbNSFPv2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desmet FO, et al. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubbard TJ, et al. Ensembl 2007. Nucleic acids research. 2007;35:D610–617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 45.Eng L, et al. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 46.Pertea M, Lin X, Salzberg SL. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.