Abstract
Background
Molecular subtyping of cancer aimed to predict patient overall survival (OS) and nominate drug targets for patient treatments is central to precision oncology. Owing to the rapid development of phosphoproteomics, we can now measure thousands of phosphoproteins in human cancer tissues. However, limited studies report how to analyse the complex phosphoproteomic data for cancer subtyping and to nominate druggable kinase candidates.
Findings
In this work, we reanalysed the phosphoproteomic data of high-grade serous ovarian cancer (HGSOC) from the Clinical Proteomic Tumour Analysis Consortium (CPTAC). Our analysis classified HGSOC into 5 major subtypes that were associated with different OS and appeared to be more accurate than that achieved with protein profiling. We provided a workflow to identify 29 kinases whose increased activities in tumours are associated with poor survival. The altered kinase signalling landscape of HGSOC included the PI3K/AKT/mTOR, cell cycle and MAP kinase signalling pathways. We also developed a “patient-specific” hierarchy of clinically actionable kinases and selected kinase inhibitors by considering kinase activation and kinase inhibitor selectivity.
Interpretation
Our study offered a global phosphoproteomics data analysis workflow to aid in cancer molecular subtyping, determining phosphorylation-based cancer hallmarks and facilitating nomination of kinase inhibition in cancer.
Keywords: Ovarian cancer, Phosphoproteomics, Druggable kinase
Research in context.
Evidence before this study
We searched the PubMed database according to the terms [(“phosphoproteomics” OR “phosphorylation”) AND (“cancer” OR “tumour”) among English-language articles before September 2, 2018. We found ten studies using quantitative phosphoproteomics to measure the human cancer clinical samples. These studies did not pay much attention to the potential clinical application of phosphoproteomics. Few studies report how to use these data to nominate kinase candidates for clinical intervention. Application of phosphoproteomics in translational research remains an urgent need.
Added value of this study
We took the high-grade serous ovarian cancer (HGSOC) datasets from the CPTAC as a case study to present a workflow to analyse large-scale phosphoproteomics data for cancer subtyping and to nominate druggable kinase candidates. A “patient-specific” hierarchy of clinically actionable kinases was developed by considering kinase activation and kinase inhibitor selectivity.
Implications of all the available evidence
We proposed a bioinformatics analysis workflow to distil information and knowledge from large-scale phosphoproteomics data. This work detailed the processes of how to subtype cancer with phosphorylation data to be associated with clinical outcome and nominate actionable kinase targets for clinical intervention. Our current study might provide a strategy and workflow to investigate kinase activation and then select the corresponding kinase targets for treatment for the particular patient.
Alt-text: Unlabelled Box
1. Introduction
Phosphoproteomics focuses on the identification and quantification of phosphorylated residues on proteins in biological specimens [1]. Owing to the recent technological advances, liquid chromatography-mass spectrometry (LC-MS)-based quantitative phosphoproteomics can measure thousands of phosphorylation events, from which kinase activities can be inferred and kinase-targeted therapies can be developed [[2], [3], [4]]. It has been successfully used to rationalize responses to kinase inhibitors, to identify drug targets, and for the development of new therapies against several diseases including cancer, diabetes, and neurodegenerative diseases [5]. In recent years, great efforts have been made towards the identification of biomarkers that can predict the clinical efficacy of kinase inhibitors and other drugs [4]. However, most were performed using cell lines or xenograft models that may not completely reflect the signalling events of human tissues.
Currently, several studies have used the LC-MS-based phosphoproteomics to focus on the measurement of signalling molecules and phosphoproteins in human tissues, including breast cancer [6], ovarian cancer [7], prostate cancer [8], acute myeloid leukaemia [9], hepatocellular carcinoma [10], lung cancer [11] and bladder cancer [12]. The main discoveries based on the phosphoproteomics are summarized in Supplementary Table 1. These studies mainly focus on the identification of alterations of signalling pathways and did not pay much attention to potential clinical application of phosphoproteomics. For example, the Clinical Proteomic Tumour Analysis Consortium (CPTAC) acquired extensive phosphoproteomics data and identified several altered signalling pathways including RhoA, PDGFRB, and the integrin-like pathways [7]. However, identifying and using phospho-proteins as cancer biomarkers is still in the early stage, and most studies did not associate phosphorylation with clinical outcomes such as overall survival (OS) and progression-free interval time (PFI). Few studies focused on how to use the phosphoproteomics to nominate kinase candidates for clinical intervention. Application of phosphoproteomics in translational research remains a challenge.
Ovarian cancer is among one of the most life-threatening malignancies for woman. High-grade serous ovarian cancer (HGSOC) accounts for 70–80% of ovarian cancer deaths, and overall survival has not changed significantly for several decades [13,14]. The standard clinical management for advanced-stage ovarian cancer includes surgery followed by adjuvant chemotherapy [13]. However, after initial response, tumour recurrence is encountered in approximately 70% of patients who will eventually die of a progressively chemo-resistant cancer [13]. Further studies for better understanding HGSOC-specific molecular carcinogenesis mechanisms and druggable targets are in urgent demand. The targeted therapy of ovarian cancer mainly focuses on anti-angiogenesis poly(ADP-ribose) polymerase inhibitors (PARPi), inhibition of DNA repair and cell cycle, inhibition of the PI3K/AKT/mTOR pathway and immunotherapeutic strategies [15,16]. Many of these innovative approaches already demonstrate promising activity in ovarian cancer. One of the most important issues is the selection of patients who could benefit from the therapies. The most successful example is that patients who carry germline mutations in either BRCA1 or BRCA2 are sensitive to PARPi [17]. However, except for TP53 (>90%) and BRCA1/BRCA2 (~15%), the mutation frequencies of other genes are quite rare (<5%) [18]. More drug targets and predictive markers are needed. In this work, we take the CPTAC HGSOC phosphoproteomics dataset as a case study to present a workflow to illustrate how phosphoproteomics profiling could be used to aid in the prediction of prognosis, determine phosphorylation-based cancer hallmarks and nominate kinase targets for clinical intervention.
2. Materials and methods
2.1. Dada acquisition and processing
The phosphoproteomic and proteomic data of HGSOC, which were carried out by the CPTAC program, were downloaded from the website [7]. The clinical information including the overall survival (OS) and progression-free interval time (PFI) were obtained from the TCGA Pan-Cancer Clinical Data Resource after mapping the barcodes [19] (Supplementary Table 2). Phosphorylation sites were retained when phosphopeptides showed an Ascore >19; otherwise, the precise modification site was deemed ambiguous [20]. We found that most phosphorylation sites whose maximum intensity in all experiments was less than the first quartile value were detected in fewer than 10 samples, as shown in Fig. S1a. We only used the phosphorylation sites with higher abundance, whose maximum intensity in all experiments ranked in the top 75% (Fig. S1b). The missing data were imputed with the minimum non-zero values of global phosphorylation sites. For the proteomics data, the same filter criteria were used. After these steps, the remaining phosphorylation sites and proteins were used for subsequent analysis.
2.2. Statistical analysis
Consensus clustering was performed using the R package ConsensusClusterPlus [21]. The phosphorylation sites were quantile normalized before clustering. The Kaplan-Meier method was used to perform the survival analysis, and the difference was tested using the log-rank test. Cox proportional hazards regression analysis which takes age, stage and grade into account were performed, and p-values were adjusted using the Benjamini and Hochberg method [22]. All statistical analyses were performed using R 3.31 and Python 3.6.2 (with Anaconda 5.1.0).
2.3. Construction of the kinase–substrate interaction dataset
The comprehensive kinase–substrate interaction dataset was constructed through integration of the data from the four public databases: PhosphoSitePlus (10,266 interactions) [23], Phospho.ELM (1479 interactions) [24], PhosphoNetworks (4402 interactions) [25] and UniprotKB (3092 interactions) [26]. To extract the sequences surrounding the phosphorylation sites in these interactions, we mapped the substrate sites to the human proteins in the Swiss-Prot database, and unmapped sites were removed. Finally, 15,384 non-redundant kinase substrate interactions remained. We then expanded the kinase-substrate relationships for other sites based on the position weight matrix (PWM) score and protein-protein interactions collected from databases.
PWMs were constructed for those kinases with >30 substrates and used to predict more kinase-substrate interactions. The PWM score of amino acid residue i at site j was defined as:
| (1) |
Here, pij represents the observed frequency of amino acid i at site j, and bi represented the background frequency of amino acid i in human phosphoproteomics. When counting the observed frequency, one pseudo count of every 20 amino acids was added, to make pi positive. An example of the calculation of the PWM score is displayed in the Fig. S2a.
Then if a phosphorylation site meets both of the following criteria, it will be predicted as the substrate site of a specific kinase: 1) similar sequence pattern with known substrate sites of the kinase; and 2) substrate protein interacted with the kinase. Here, the similar sequence pattern was evaluated based on the PWM scores. The PWM score of a phosphorylation site was defined as the sum of the PWM scores of residues from position −7 to +7, with the exception of the phosphosites. For each kinase, we took the 3rd quartile of PWM scores of its known substrates as the cut-off. Those sequences with PWM scores higher than the cut-off were defined as the potential substrates of the kinase. Those candidate substrates were further filtered by the known protein-protein interactions downloaded from eight public protein databases: BioGrid [27], PioPlex [28], CCSB [29], DIP [30], HPRD [31], IntAct [32], MINT [33], and PINA [34]. The types of interactions between proteins collected from databases included physical interaction, transcriptional regulation and sequential catalysis. Only those kinase-substrate relations with known protein-protein interaction support remained. In addition, the correlations of the PWM scores with different sequence lengths (from ±4 to ±6 positions) are shown in Fig. S2b. These results indicated that the PWM scores generated by different lengths correlated well and the kinase-substrate dataset generated by different sequence lengths should be similar.
2.4. An unbiased bioinformatics approach to nominate kinases as potential therapeutic targets
2.4.1. Predicting kinase activities for each patient
Based on the assumption that the activation of a kinase is reflected by the phosphorylation state of its substrates, the mean values method was used to estimate the kinase activity after normalization of phosphopeptide abundance by protein abundance [35]. The Mean value method calculated the average fold change difference for all detected substrate sites from the same kinase as a measurement of the kinase activation/inhibition. Three additional methods, including the multiple linear regression (MLR) model [36], KSEA algorithm and Z-test [37], were also applied to estimate the kinase activity.
The MLR model was constructed as follows:
| (2) |
where is the coefficient of regression; xij=1 if phosphosite j is a substrate of kinase i, otherwise 0. i ranges from 1 to n (n = 192 for all 192 kinases), yj is the relative value of phosphosite j, and is computed as follows:
| (3) |
Tj and Mj are quantitative values of phosphosite j in the tumour and median of all the tumours; TPj and MPj are the quantitative values of the protein corresponding to phosphosite j in the tumour and the median of all the tumours. Tj + 1 instead of Tj was used to calculate yj. Adding 1 was used to avoid obtaining fold changes that are too large considering some phosphorylation sites/proteins with low expression levels. Ridge regression, defined as below, was used to solve the regression problem.
| (4) |
Here, y is the vector of yj and X is the matrix of xij; β is the vector of coefficients βi, taken as the estimation of kinase activity; λ‖β‖2 is ridge penalty, and was applied to reduce the over-fitting of the MLR model and multi-collinearity of the interactome. For each patient, λ was set to 0.0001. Scikit-learn package (http://scikit-learn.org/stable/) was used to compute the ridge regression.
The KSEA package from https://github.com/evocellnet/ksea uses a modified weight Kolmogorov-Smirnov test to search for kinases with significant enrichment of upregulated or downloaded substrates. The relative values of phosphosites and related kinases-substrate interactions were taken as the input. The significance was estimated with a null distribution of 1000 permutations. The resulting P-values were log10- transformed and signed based on the average sign of all substrates, which was taken as the final kinase activity [37].
Z-test compares the mean fold change of substrate sites of a kinase to that of all substrate sites:
| (5) |
where S is the mean fold change of substrate sites of a kinase in one patient, Phos is the mean fold change of all substrate sites in the patient, m is the number of substrate sites of the kinase, and σ is the standard deviation of fold changes of substrate sites.
2.4.2. Classification of patients based on kinase-patient matrix
We wanted to find out kinases whose high activities in tumours were associated with overall poor survival compared with low activity. Thus, we need to define the cut-off value that separate the high and low activity group for each kinase. Firstly, patients were grouped into high and low activity groups by an arbitrary cut-off value, then significance of the clinical outcome difference between the two groups was tested by the log-rank test and then a log-rank P value were calculated. Secondly, we iterated the arbitrary cutoff value until we obtained the lowest log-rank P value to find the cut-off value that would be used to define the high and low activity group.
2.5. Nomination of kinase inhibitors for each patient
The preferences of each kinase inhibitor in one patient were measured by the following preference score:
| (6) |
where PScoreij is the preference score of drug i in patient j, βjk is the activity of kinase k in patient j. Here the kinase activity was measured by the Mean value method. Cik was the concentration and target dependent selectivity (CATDS) value of inhibitor i to kinase k. Citotal was the sum of CATDS of inhibitor i to its all targeted kinases. CATDS could measure the engagement of a specific protein target at a particular drug concentration relative to all target protein engagements of that drug at the same concentration. More details of the CATDS have been reported in a previous study [38].
3. Results
3.1. Kinase activity pattern in HGSOC
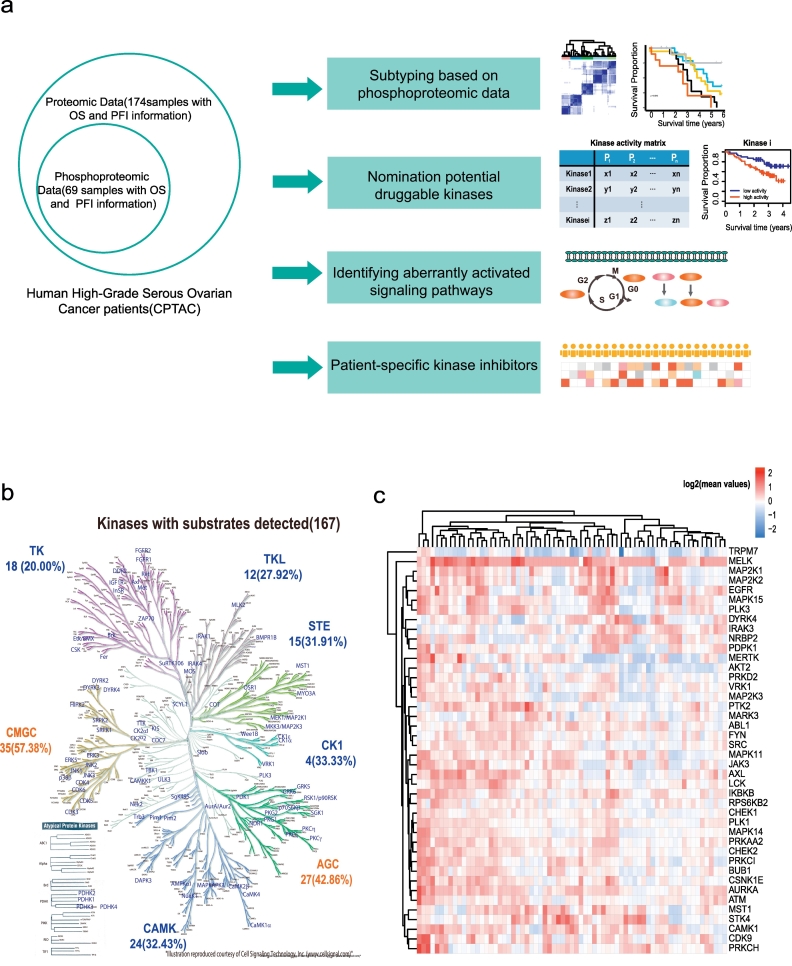
The CPTAC HGSOC datasets contained both phosphoproteomic and proteomic data for 69 patients with clinical outcomes [7] (Fig. 1a). After data pre-processing (see Methods), 10,171 phosphorylation sites and 6878 proteins were remained for subsequent analysis (Fig. S3a-b), which included 8481 (83.38%) phosphoserine sites, 1444 (14.2%) phosphothreonine sites and 246 (2.42%) phosphotyrosine sites (Fig. S3a).
Fig. 1.
The main workflow in this study and kinase activity patterns in HGSOC.
a. Application of phosphoproteomic data in ovarian cancer; b. Kinome tree annotated using Kinome Render from Cell Signalling Technology, Inc. (www.cellsignal.com); Each kinase had at least two substrates. CMGC contains cyclin-dependent kinase, MAPK, glycogen synthase kinase 3 and CDC2-like (cell division control 2, A-type cyclin-dependent kinase); STE consists of the MAPK cascade families; TKL (tyrosine kinase-like) consists of the MLK (mixed-lineage kinase), LISK (LIMK/TESK), and IRAK; AGC contains protein kinases A, G and C; CAMK (calcium/calmodulin- dependent protein kinase); CK1 (casein kinase 1) contains the CK1, TTBK (tau tubulin kinase), and VRK (vaccinia-related kinase) families; TK (tyrosine kinase) c. Kinase activity pattern in HGSOC.
As the activation status of a kinase can be inferred by its substrates [9,39], we built 17,833 kinase-substrate relationships from public databases (see Methods, Supplementary Table 3). In the ovarian cancer dataset, 1913 substrates and 170 kinases with at least two substrates were mapped, of which 167 (98.23%) were annotated in the kinome tree [40] (Fig. 1b). The main kinase groups were CMGC, which contains CDK kinases and MAPK families, and AGC, which consists of protein kinases A, G and C. For each kinase, the average number of substrates was 11 and the median number of substrates was 4 (Fig. S4a, Supplementary Table 4). The enhanced phosphorylation of a substrate site can be interpreted by the enhancement of both kinase activity and substrate protein level. In this study, we are interested in those phosphorylation changes caused by the kinase activity. We then calculated the normalized value of p-site-intensityphosphorylation/protein-intensityprofiling to correct for altered protein expression [35] and then computed a fold change difference of the phosphorylation site in the tumour for each patient divided by the median value of the 69 tumours. We used the average fold difference for all detected substrate sites for the same kinase as a measurement of the kinase activation/inhibition (Supplementary Table 5). Forty-two kinases with the top 1/4 ranked values of average fold difference across the patients were shown in Fig. 1c. The kinase activity pattern for each patient was diverse. The highly activated kinases included MELK, MAP2K1/2, PTK2, STK4, and CDK9.
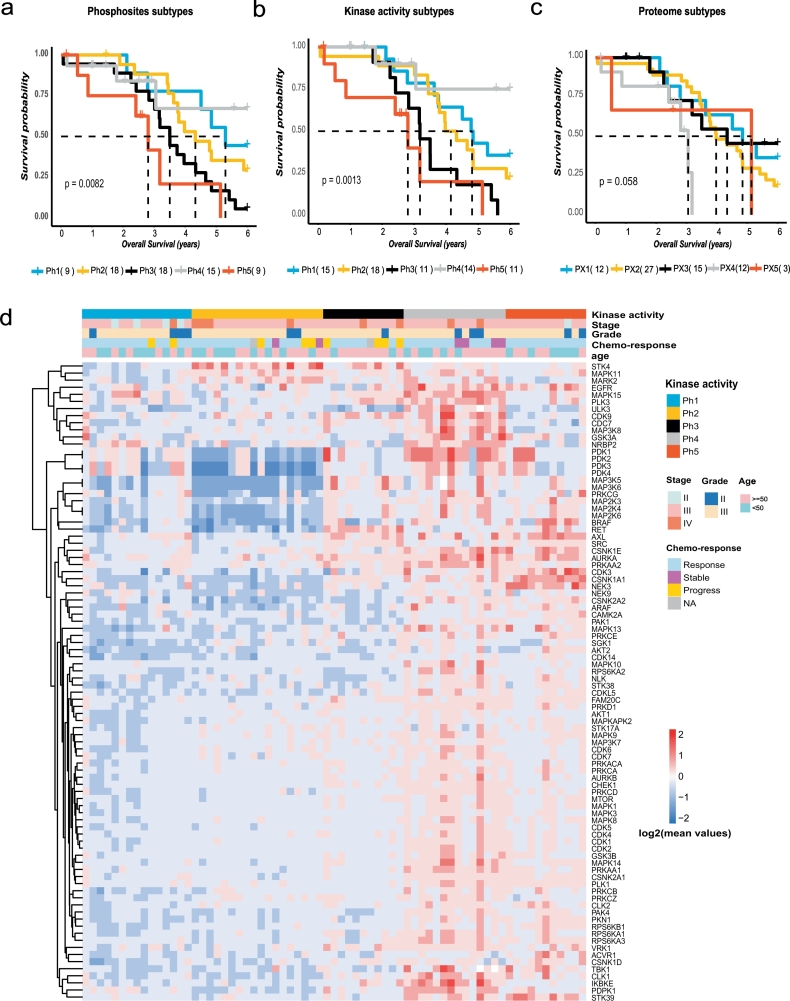
3.2. Phosphoproteomic subtyping of HGSOC and association with OS
We then explored whether the HGSOC patients could be further classified into subtypes based on the phosphorylation sites or kinase activities. Using the values of 10,172 phosphorylation sites in the tumours, we employed consensus clustering [21] to identify HGSOC subtypes. Five clusters (Ph1–5) were apparent (Fig. S5a) and associated with different OS (Log-rank test, P = .0082, Fig. 2a). When using the 48 kinase activities as input for consensus clustering, we also obtained five subtypes (Fig. S5b) with similar OS associations (Log-rank test, P = .0013, Fig. 2b). Interestingly, proteome subtyping also generated 5 subtypes, but the p-value for association of subtypes with OS was larger than those obtained from phosphoproteomics data (Log-rank test, P = .058, Fig. 2c, Fig. S5c). The kinase activity-based subtyping appeared to be better than the other two methods to extract patients with the best OS. These results suggest that phosphoproteome subtyping is better correlated with OS than proteome subtyping. None of the subtypes correlated with the progression-free interval time. Other clinical characteristics, such as age, stage, grade, and chemo-response were also annotated in Fig. 2d, and the frequencies of these characteristics within the five subtypes are shown in Supplementary Table 6. Compared with Ph1-Ph4, Ph5 responded to chemotherapy, and Ph2-Ph3 appeared to display disease progress after chemotherapy (Fisher-test, P < .05).
Fig. 2.
Phosphoproteome subtyping of HGSOC with different overall survival.
The association of molecular subtypes based on a. phosphosites, b. kinases activity, and c. proteins with overall survival of patients. d. Kinase activity and other clinical parameters across 69 patients in Ph1–5. Kaplan-Meier analysis, P value from the log-rank test;
Ninety kinases were identified as exhibiting differential activities among the subtypes (ANOVA, FDR < 0.01). As shown in Fig. 2d, Ph1-Ph3 were characterized by few activated kinases. The most activated kinases in Ph1 included PDK3/4, which contributed to the regulation of glucose metabolism. Ph2-Ph3 had STK4 activated, which promoted apoptosis. Ph4 and Ph5 carried the most activated kinases. PLK3, ULK3, CDK9, CDC7, MAP3K8, IKBKE, and PDPK1 were activated most frequently in Ph4. Ph5 had BRAF, AXL, CDK3, CSNK1A1, and NEK3 activated, which might account for the worse survival of Ph5. Among these kinases, there were 8 kinases whose activities significantly correlated with clinical outcome (P value <.05, Cox regression taking age, stage, grade and chemo-response into account, Supplementary Table 6). These kinases were mainly the features of Ph4 and Ph5 subtypes such as CDK9, CDC7, BRAF and AXL. Ph4 had the best survival and Ph5 had the worst survival, which might have more apparent characteristics than other subtypes.
3.3. Nomination of kinases as potential therapeutic targets
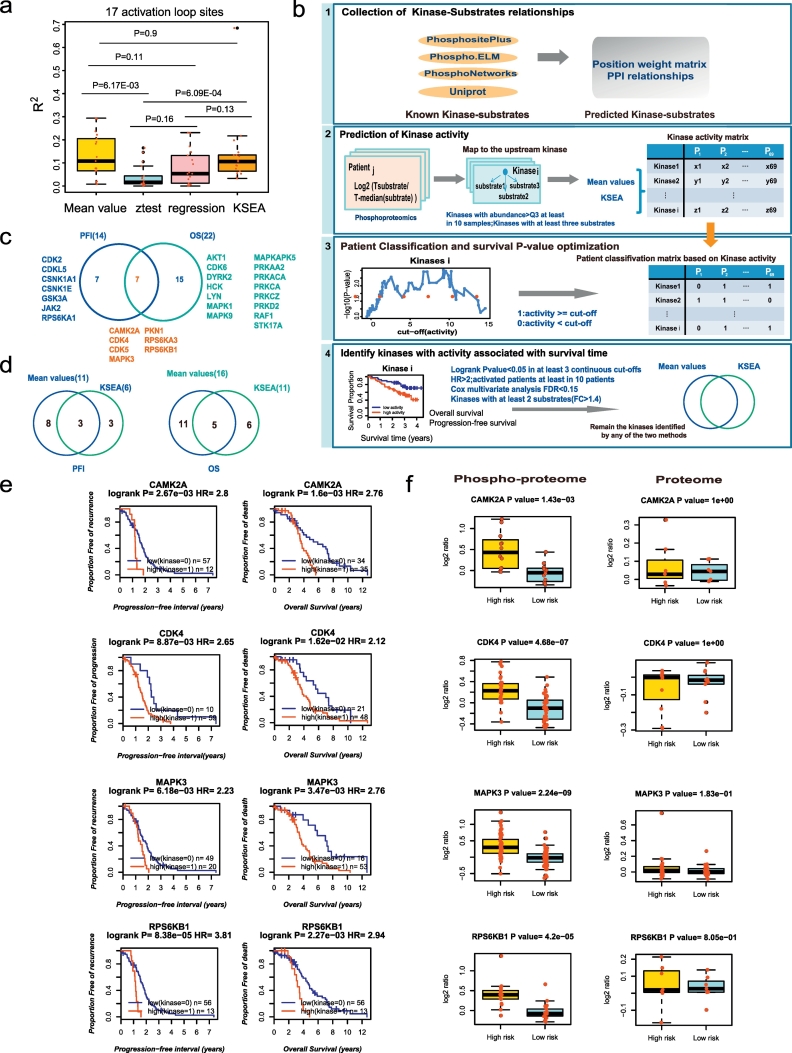
We then postulated that activated kinases might be therapeutic targets if their activities are negatively correlated with poor survival. In addition to Mean value method, we also adapted the Z-test, the KSEA [37] and the multiple linear regression (MLR) model to measure the kinase activity based on the kinase-substrate relationships. With each of these four methods, we could compute the “kinase activity” for every patient and generate a kinase-patient matrix. Then four kinase activity matrices were obtained; each row represents the activity of a specific kinase across different patients, and each column represents the activities of different kinases for a patient. It is known that one of the most common methods for regulating kinase activity is the phosphorylation of the activation loop. The activation loop is a site of protein-protein interactions that can be critical in controlling the localization and regulation of a kinase and its binding partners [41]. To evaluate the accuracy of the four kinase activity prediction methods, we collected 618 activation loop phosphorylation sites from the PhosphoSitePlus database, and 17 sites were detected in the dataset (Supplementary Table 7). The correlations of the predicted kinases activities with the intensity of the activation loop phosphorylation sites were performed by the linear regression. We found that the kinase activities predicted by the four methods were positively correlated with the intensity of the activation loop phosphorylation sites (Fig. 3a, Fig. S6, Supplementary Table 7). The Mean value and KSEA methods performed significantly better than the other two methods (Wilcox test P < .05, Fig. 3a) and were retained for subsequent analysis.
Fig. 3.
Nominating potential druggable kinases.
a. The correlations between the kinase activities predicted by the four methods with the intensities of the activation loop phosphorylation sites; b. The workflow of the identification of druggable kinases; c. Kinases whose increased activities in tumours are both associated with poor clinical outcome; d. Overlaps of the results for the two kinase activity prediction methods; e-f. Kinases whose increased activities in tumours are both associated with the progression-free interval and overall survival. e. Survival curves of the kinases and f. boxplot of the kinases substrates in the phosphoproteome and proteome data. Low(Kinase = 0)/high(Kinase = 1): patients with kinase activity lower than/above the cut-off. High risk/Low risk: patients with poor/better survival. HR: hazard ratio.
The above two kinase activity matrices allowed us to stratify patients into two groups according to high and low kinase activity. The cut-off value was individually determined to find the lowest P value of survival time according to a log-rank test (Fig. 3b, panel 3). To reduce the false positive rate, we only retained the kinases with a log-rank P value <.05 in at least 3 continuous cut-off values. In addition, based on the kinase activities predicted by the Mean value method, we performed Cox regression and multiple testing p-value correction, taking age, stage, grade and chemo-response into account (Fig. 3b, panel 4). Fourteen and twenty-two kinases were identified, whose activities were significantly associated with poor PFI or OS, respectively (Fig. 3c, Fig. S7, Fig. S8, Supplementary Table S8, Supplementary Table S9). The two methods (Mean value and KSEA) had 3 kinases that overlapped for PFI association and 5 kinases that overlapped for OS, both of which account for around 50% of KSEA results. (Fig. 3d). Finally, to expand the list of kinase targets, we nominated the union of the two methods, including 29 non-overlap kinases that were associated with PFI or OS as potential therapeutic targets (Table 1, Fig. S3b).
Table 1.
Kinases associated with survival of HGSOC patients.
| Kinases | Method | Survival time | HR | Logrank P value | Summary |
|---|---|---|---|---|---|
| PI3K/AkT/mTOR pathway | |||||
| AKT1 | KSEA | OS | 4.23 | 9.70E-03 | Cell grow and proliferation |
| PRKAA2 | Mean values; KSEA | OS | 3.19;2.65 | 6.20E-03; 0.010 | Cellular energy metabolism |
| PRKACA | KSEA | OS | 4.68 | 5.30E-03 | Differentiation, proliferation, and apoptosis. |
| PRKCA | KSEA | OS | 2.06 | 4.80E-02 | Cell adhesion, cell transformation, cell cycle checkpoint |
| PRKCZ | Mean values | OS | 3.12 | 2.60E-03 | Tight junction |
| RPS6KA1 | KSEA | PFI | 5.20 | 3.00E-03 | Cell growth and differentiation |
| RPS6KA3 | KSEA | PFI/OS | 2.07;2.90 | 0.030; 9.26E-04 | Cell grow and proliferation |
| RPS6KB1 | Mean values | PFI/OS | 3.81;2.94 | 8.38E-05; 2.27E-03 | Cell grow and proliferation |
| PKN1 | Mean values; KSEA | PFI;PFI/OS | 3.50;2.35; 2.19 | 4.80E-03; 1.11E-03; 0.025 | Cell migration, tumour cell invasion |
| Cell cycle | |||||
| CDK2 | KSEA | PFI | 4.38 | 1.05E-05 | G1 to S phase transition |
| CDK4 | Mean values; KSEA | OS;PFI/OS | 2.11;2.65; 2.06 | 1.60E-02; 8.87E-03; 9.40E-03 | G(1)/S transition |
| CDK5 | Mean values | PFI/OS | 2.19;3.40 | 4.0E-03; 6.3E-03 | Cell cycle, cell proliferation |
| DNA damage | |||||
| CDK6 | Mean values | OS | 2.02 | 1.20E-02 | G1 phase progression and G1/S transition |
| CDKL5 | Mean values | PFI | 2.79 | 4.00E-04 | Cell proliferation |
| DYRK2 | Mean values | OS | 2.62 | 1.30E-03 | Cellular growth and/or development |
| MAPK pathway | |||||
| MAPK1 | Mean values; KSEA | OS | 3.20;2.76 | 0.020;0.012 | Cell growth, adhesion, survival and differentiation |
| MAPK3 | Mean values | PFI/OS | 2.23;2.76 | 6.2E-03; 3.4E-03 | Cell growth, adhesion, survival and differentiation |
| MAPK9 | Mean values | OS | 2.65 | 1.10E-02 | |
| MAPKAPK5 | Mean values | OS | 2.40 | 1.40E-03 | Stress and inflammatory responses, nuclear export, and cell proliferation |
| RAF1 | Mean values | OS | 2.80 | 2.10E-03 | Activate the dual specificity protein kinases MEK1 and MEK2 |
| Wnt signalling pathway | |||||
| CSNK1A1 | Mean values | PFI | 2.67 | 1.90E-03 | DNA repair, cell division, nuclear localization and membrane transport |
| CSNK1E | Mean values | PFI | 2.02 | 7.60E-03 | DNA replication and repair |
| GSK3A | Mean values; KSEA | PFI | 3.03;2.40 | 9.54E-05; 9.70E-04 | Cell cycle progression, differentiation, and apoptosis |
| CAMK2A | Mean values; KSEA | PFI/OS;OS | 2.80;2.76; 3.48 | 2.70E-03; 1.60E-03; 3.00E-04 | Ca(2+)/calmodulin-dependent protein kinases |
| Immune associated targets | |||||
| JAK2 | Mean values | PFI | 2.03 | 5.50E-03 | Cytokine receptor signalling pathway; responses to gamma interferons |
| HCK | Mean values; KSEA | OS | 4.20;3.73 | 2.13E-05; 2.41E-05 | A member of the Src family of tyrosine kinases |
| LYN | KSEA | OS | 3.10 | 1.50E-03 | A member of the Src family of tyrosine kinases |
| Others | |||||
| STK17A | Mean values | OS | 2.70 | 4.20E-04 | Apoptosis |
| PRKD2 | Mean values | OS | 2.31 | 5.40E-03 | Cell migration and differentiation |
Seven kinases (CAMK2A, CDK4, CDK5, MAPK3, PKN1, RPS6KA3 and RPS6KB1) were found to be associated with both PFI and OS (Fig. 3c). As shown in Fig. 3e, higher kinase activities of CAMK2A, CDK4, MAPK3 and RPS6KB1 were significantly correlated with OS and PFI (Log-rank test, P < .05). In contrast, the abundance of substrates measured from protein profiling showed a smaller difference between the high- -risk (shorter OS) and the low-risk group (longer OS) (Fig. 3f). Other kinases showed similar results (Fig. S9, Fig. S10, Fig. S11, Fig. S12, Fig. S13, Fig. S14, Fig. S15). Interestingly, we only identified 6 kinases (TNIK, PRKCE, PRKD3, STK36, STK4 and PIK3CB) whose increase in protein abundance correlated with poor PFI or OS (Supplemental Table 8, Fig. S7b). There was no overlap between the kinase list based on activity and abundance. Therefore, it is necessary to take the activity of kinases into consideration when identifying kinase targets.
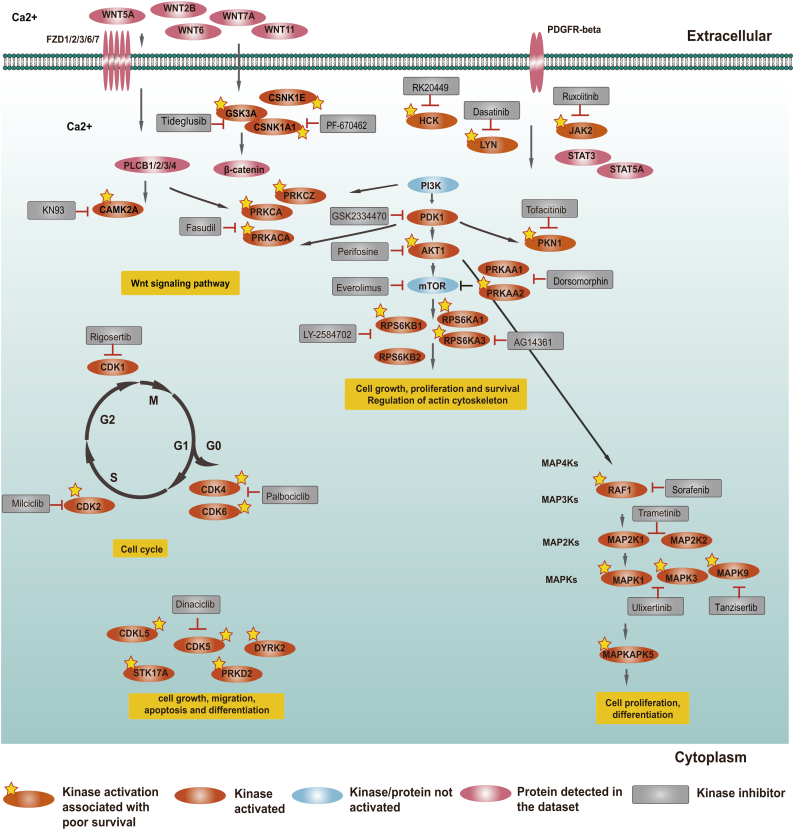
3.4. Major dysregulated kinase pathways in HGSOC
The 29 potential therapeutic targets could be classified into 5 pathways within the conceptual framework of hallmark cancer signalling: (1) the PI3K/AKT/mTOR network, (2) Wnt signalling, (3) MAPK network, (4) cell cycle and (5) other pathways, such as cell migration and PDGFRB pathways (Fig. 4, Table 1). In addition, in order to make the pathways more intact, 6 kinases (PDK1, CDK1, PRKAA1, RPS6KB2, MAP2K1 and MAP2K2), whose maximum activity across the patients ranked in the top 1/4, were also mapped in Fig. 4. The PI3K/AKT/mTOR signalling pathway contained the most potential therapeutic targets such as AKT1, PRKAA2, PRKACA, PRKCA, PRKCZ, RPS6KA1, RPS6KA3, RPS6KB1 and PKN1 (Fig. 4). Higher AKT1 expression has been reported to correlate with poor prognosis in ovarian cancer and could be a biomarker of treatment response in ovarian cancer [[42], [43], [44]]. Clinical trials of several AKT inhibitors in ovarian cancer have been conducted [45]. Elevated of RPS6KA1 abundance could promote the metastasis of ovarian cancer cells [46]. The cell cycle had 6 potential therapeutic targets (Fig. 4). In the cell cycle pathways, all of the cyclin-dependent kinases identified as potential therapeutic targets were validated in cell models of ovarian cancer. Targeting CDK2 was a novel therapeutic strategy in CCNE1-amplified ovarian cancer [47]. CDK4/6 inhibitors have shown significant activity against several solid tumours such as breast cancer, non-small cell lung cancer and melanoma [48]. These inhibitors also have shown promising preclinical activity in ovarian cancer [49]. CDK5 is important for neuronal development [50]; increased expression of CDK5 in human ovarian cancers correlates with poor overall survival, and inhibition of CDK5 could increase paclitaxel sensitivity [51]. In the MAPK signalling pathway, MAPK1 was associated with frequent somatic copy number amplification in HGSOC [18]. Overexpression of MAP2K1 correlated to progression free survival of ovarian cancer [52]. MAPK9 represents an attractive targets in cancer [53]. In addition, three kinases (JAK2, HCK and LYN) have been reported as therapeutic targets in immune and cancer cells [[54], [55], [56]]. Summaries of other kinases are provided in Table 1 and Supplemental Table 10. In the CPTAC paper, proteins and phosphoproteins abundance of the RhoA-regulatory, PDGFRB, and integrinlike kinase pathways were found to be associated with survival. Three kinases (JAK2, RAF1 and PRKCA) in these pathways were also found in our study. In addition, four kinases (RAF1, MAPK1, MAPK3 and AKT1) showed statistically significantly increased phosphorylation by a reverse-phase protein array (RPPA) in short-surviving patients compared with long-surviving patients [7]. These four kinases have also been identified in our study. The comparisons between the CPTAC study and our work are shown in Table 2.
Fig. 4.
Major pathways mapped by the kinases activated in HGSOC.
Here, if a kinase meets one of the following criteria, it would be defined as activated: (1) whose activity was associated with poor overall survival; and (2) maximum activity across the patients ranked in the top 1/4. If a kinase mapped in Fig.4 did not meet one of the above criteria, it was denoted as “not activated”. If a protein was not detected in the dataset, we also denoted it as “not activated”.
Table 2.
The comparisons between CPTAC study and our work.
| CPTAC study | Our study | |
|---|---|---|
| Subtyping with phosphoproteome data | No | Yes, the subtypes were significantly associated with different overall survival based on kinase activity(P = .0013). |
| Subtyping with proteome data | Yes, but the subtypes were not associated with clinical outcome(P > .5). | Yes, the subtypes were tend to be associated with different overall survival(P = .058). |
| Method for determing kinase activity | They did not predict kinase activity. | Two methods: Mean values and Kinase substrate enrichment analysis (KSEA). |
| Nomination potential druggable kinases | No | Yes, we identified 35 potential druggable kinases. |
| Identifying aberrantly activated signalling pathways |
Yes, they used proteins and phosphoproteins whose abundance were associated with survival (a two-sided t-test) to identify pathways: The RhoA-regulatory, PDGFRB, and integrinlike kinase pathways. | Yes, we used kinases whose increased activities in tumours are associated with poor survival (log-rank test) to paint the altered signalling, which were centered on the PI3K/AkT/mTOR pathway, cell cycle and MAP kinase signalling pathways. |
| Development of patient-specific kinase inhibitors | No | Yes, we developed a patient-specific hierarchy of clinically actionable kinases and selected kinase inhibitors by considering kinase activation and kinase inhibitor selectivity. |
| Integrating proteomic data with the genomic data |
Yes | No |
| The main significance of the study | Layering proteomic and genomic data from ovarian tumours provides insights into how signalling pathways correspond to specific genome rearrangements. | This work detailed the processes of how to subtype cancer with phosphorylation data to be associated with clinical outcome, and nominate actionable kinase targets for clinical intervention. |
3.5. A “patient-specific” hierarchy of clinically actionable kinases and inhibitors
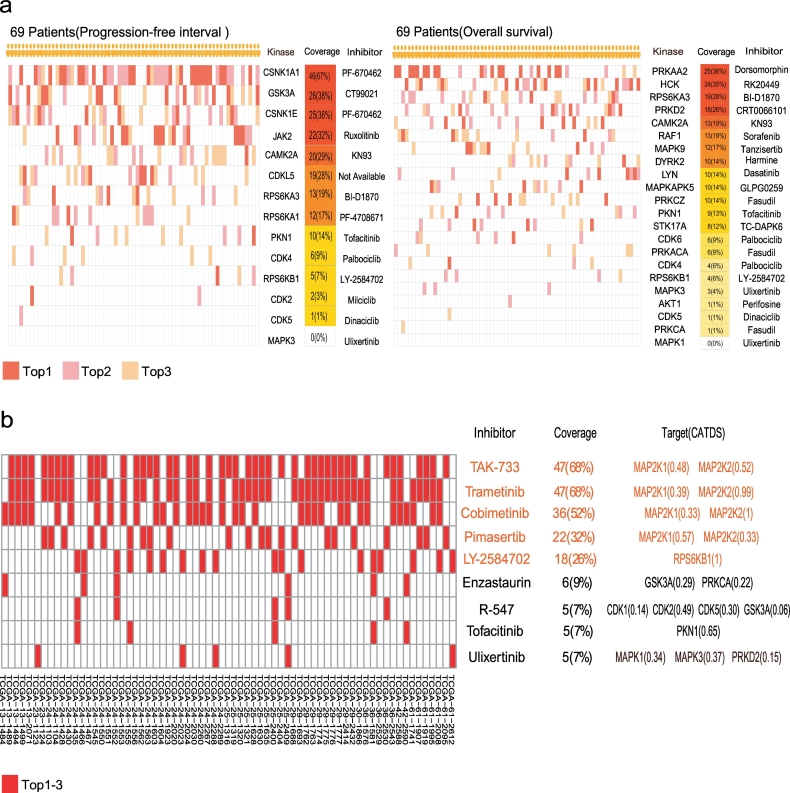
We sought to nominate kinase targets for each individual patient and drew the top three potential therapeutic target activity patterns for every patient based on kinase activity derived from the mean values of all measured corresponding substrates (Fig. 5a). It was evident that each patient had a unique top three potential therapeutic target activation pattern. For the PFI-associated kinases, the top five frequently activated potential therapeutic targets included CSNK1A1, GSK3A, CSNK1E, JAK2 and CAMK2A (Fig. 5a-left panel). The most frequently activated CSNK1A1 was identified in 46/69 (67%) of the patients. For OS-associated kinases, the top five frequently activated potential therapeutic targets included PRKAA2, HCK, RPS6KA3, PRKD2 and CAMK2A (Fig. 5a-right panel). The most frequently activated PRKAA2 was identified in 25/69 (36%) of the patients, while the frequency for CDK activation was low. A “patient-specific” hierarchy of the top three most activated kinases and the corresponding inhibitors is displayed in Fig. 5a, which provides an entry point for selecting truly individualized kinase inhibition therapy.
Fig. 5.
“Patient-specific” kinase inhibitors.
a. “Patient-specific” kinase inhibitors. The kinases were ranked based on the mean values of the corresponding substrates; b. Selectivity of inhibitors targeting at candidate kinases shown in Fig. 4. The inhibitors were ranked based on the preference score. Preference score: the sum of inhibition of its target kinase activities weighted by CATDS. CATDS: the concentration and target dependent selectivity.
It is now commonly accepted now that most kinase inhibitors target the intended kinase as well as other kinases as off-target effects [38]. Recently, Bernhard Kuster et al. measured the on-target and “off-target” spectra of 243 clinically evaluated kinase inhibitors [38], in which quantitative inhibition values for on-target and off-target kinases were measured and presented as concentration and target dependent selectivity (CATDS) scores. Taking advantage of this resource, we tried to select the most appropriated kinase inhibitors for each patient by summarizing CATDS weighted inhibition effects of each inhibitor to the 35 kinases in hallmark cancer signalling pathways in Fig. 4 (see Methods). Surprisingly, we found that the top three inhibitors, TAK-733, trametinib and cobimetinib, all targeting MAP2K1/2, may be applied to 52%–68% of the patients (Fig. 5b). TAK-733 is a potent and selective MEK allosteric site inhibitor for MEK. It has demonstrated suppression of tumour growth in a wide range of tumour types, including melanoma and colorectal, lung, pancreatic and breast cancer [57,58]. Trametinib has been approved for the unresectable or metastatic melanoma with BRAF V600E or V600K mutation [59]. While the frequency of BRAF mutation is rare in ovarian cancer[18], its downstream effector MAP2K1/2 kinase activity is elevated in the CPTAC cohort, rendering them amenable to trametinib treatment. In fact, it has been reported that the high MAPK activity, as measured by phospho-MAPK intensity, could independently predict poor survival for HGSOC [60]. A “patient-specific” hierarchy of the top kinase inhibitors is displayed in Fig. 5b. It seemed that Uprosertib, R-547 and ruboxistaurin were the top three inhibitors that may be applied to the entire HGSOC cohort.
4. Discussion
Phosphoproteomics is playing a significant role in aiding our understanding of the molecular mechanisms governing human cancers [2,61]. Most of the studies applied this technique to generate data covering thousands of phosphorylation sites, yet use selected data most of the time to reveal activation of oncogenic pathways. Our case study of reanalysing the CPTAC HGSOC phosphoproteomics dataset demonstrated that phosphoproteomics data could be used to subtype HGSOC into 5 major classes (Ph1–5) associated with clinical outcomes. These subtypes were associated with patient OS and chemo-sensitivity, and they may be used after validation in clinics to assist clinicians in determining treatment plans. It is interesting to note that owing to its ability to regulate biological activities, protein phosphorylation-based molecular subtyping appears to be able to achieve more accurate result than subtyping with protein abundance. We compared the classifications by us and by the CPTAC group as well as the transcriptome-based classification and the subtyping comparisons were added to Fig. S16a. The subtype that overlapped best among the three kinds of classifications was the Ph4/Immunoreactive subtype, which had the best survival. The stromal subtype in the CPTAC proteome-based classification tended to be enriched in Ph5, which had the worst survival. The other three subtypes (Ph1-Ph3) had little overlaps. One major reason might be that the small sample size and multiple subtypes might limit the consistent comparison. A larger cohort is needed to further compare the multi-omics subtypes.
In this study, we also provided a workflow to nominate suitable kinases for further clinical evaluation. We found 29 kinases, functioning in the PI3K/AKT/ mTOR pathway, cell cycle and MAP kinase signalling pathways, whose elevated kinase activities were significantly associated with poor survival of the HGSOC patients. In our study, we applied the Mean value method, MLR, KSEA and Z-test, to infer kinase activities [36]. We found that the kinase activities predicted by the four methods were positively correlated with the phosphorylation intensity of the activation loop sites. Especially, the Mean value and KSEA methods performed significantly better than the other two methods. The KSEA uses a rank-based permutation and measures the activity of one kinase by calculating its significance relative to other kinases in the same sample. The Mean value method calculates the kinase activities by directly sum of downstream phosphosites. As displayed in Table 1, there were 15 kinases that only the Mean value method could identify compared with KSEA, fourteen of these kinases have been validated as potential targets in cancer(Supplementary Table 10). This suggests that the Mean value method could identify additional reasonable kinases. KSEA is complementary to the Mean value method, and was able to identify seven kinases that the Mean value method failed to capture (Table 1), all of these kinases have been validated as potential targets in cancer (Supplementary Table 10). Therefore, we suggest the use of the union of the two methods.
It is noteworthy that the HGSOC patients exhibited quite diverse and individualized kinase activation spectra. We thus developed a patient-specific hierarchy of clinically actionable kinases and inhibitors for precision oncology in kinase inhibition of HGSOC. While individualized precision oncology is elegant and conceptually appealing, it may be cost-prohibitive in a real world situation. To circumvent this problem, we propose to take advantage of the fact that most kinase inhibitors are not truly specific and have on-target kinase and the off-target kinase effects. Thus, the inhibitors have inherent drug selectivities and can thus be used as “pan-inhibitors” for certain combinations of activated kinases. Using this concept, we found that trametinib was designed as the MAP2K1/2 inhibitor and may be applied in half of the patients, making this inhibitor a likely actionable kinase inhibitor in fighting HGSOC. Logically, trametinib was approved for treating melanoma patients with BRAF mutations that lead to hyper-activation of MAP2K1/2. HGSOC patients without BRAF activation, but whose MAP2K1/2 was found to be activated in phosphoproteomics analysis, they might benefit from trametinib treatment, which warrants further investigation. It will be interesting to carry out a clinical trial for HGSOC to test the idea generated from this data reanalysis.
Some limitations of the study should be noted. Firstly, only 69 patients had both the phosphoproteomic and proteomic data. Considering the heterogenous nature of HGSOC, a larger cohort should be utilized to validate the subtypes and the corresponding specific features. Second, the kinase activity predicted in this study is based on the current databases of kinase substrate relationships, which could limit the analytical depth. We repeated the Mean value method with well-annotated kinase-substrate relations and found that the predicted kinase-substrate relationships could help identify additional reasonable kinases (Fig. S16b). The rapid accumulation of MS-based experiments could be used in the future to extend the databases of kinase-substrate relationships. Finally, the nomination of kinases for drug intervention is group based and the “patient-specific” hierarchy of clinically actionable kinases and inhibitors reported here was a proof of concept and needs further investigations such as performing a phosphoproteomic analysis of an independent panel of patient-derived tumour xenograft (PDX) models to validate the therapeutic effects. Specific phosphorylation sites should be selected to measure which patients have the higher kinase activity. The adjacent tissue from the patient could be used as the reference to predict kinase activity for precision therapy.
In summary, our results indicate an association between the molecular subtypes based on phosphoproteomics and clinical outcomes, suggesting that molecular characteristics of HGSOC reflected in phosphoproteomic patterns might dictate clinical outcomes. Currently, it is still difficult to select patients who are the most likely to respond to a given kinase inhibition treatment. Our current study might provide a strategy and workflow to investigate kinase activation and then select the corresponding kinase targets for treatment of a the particular patient. Future work is needed to translate our current finding and the use of phosphoproteomics as a vital technique to assist in the prediction of prognosis and the treatment of cancer for kinase inhibition therapy.
The following are the supplementary data related to this article.
Sample frequency of the phosphosites ranked at the bottom 1/4(a) and ranks of the phosphorylation sites based on the intensity(b).
An example for the calculation of the position weight matrix(a) and the correlations of the ±7 positions-based PWM scores with different sequence lengths (from±4 to ±6 positions) (b).
Overview of the phosphoproteome and proteome data. a. The number of phosphorylation sites and phosphorylation proteins detected in 69 patients; b. Numbers of proteins detected in 69 patients.
Numbers of substrates detected for each kinase.
Consensus clustering based on the phosphoproteome data. Consensus matrices and the cumulative distribution function (CDF) plots for the a. phosphorylation sites, b. kinases activity, and c. protein abundance.
Linear regression results of the activation loop phosphorylation sites. a. Comparisons of the correlations between the kinases activity predicted by the Mean value (left) and KSEA(right) methods with the intensity of the activated/not activated loop phosphorylation sites; Examples for the correlations between the kinases activities predicted by the b. Mean values/ c. KSEA methods with the intensities of the activation loop phosphorylation sites. R2: r square in the linear regression analysis.
Survival curves of the kinases based on activities and abundance. a. Survival curves of the kinases whose increased activities in tumours are both associated with progression-free interval and overall survival; b. Survival curves of six kinases whose overexpressed protein abundance as measured in the profiling experiments associated with clinical outcome. Low(Kinase = 0)/high(Kinase = 1): patients with kinase activity/abundance lower/above than cut-off.
Survival curves of the kinases whose increased activities in tumours are associated with progression-free interval or overall survival. Low (Kinase = 0)/high(Kinase = 1): patients with kinase activity lower/above than cut-off.
Boxplots of kinase substrates phosphorylation difference in two prognostic group (PI3K/AkT/mTOR pathway).
Boxplots of kinase substrates phosphorylation difference in two prognostic group (Cell cycle and WNT pathways).
Boxplots of kinase substrates phosphorylation difference in two prognostic group (MAPK and other pathways).
Heatmaps of substrates phosphorylation difference in two prognostic group (PI3K/AkT/mTOR pathway).
Heatmaps of substrates phosphorylation difference in two prognostic group (Cell cycle pathway).
Heatmaps of substrates phosphorylation difference in two prognostic group (WNT pathway and other pathways).
Heatmaps of substrates phosphorylation difference in two prognostic group (MAPK pathway).
Subtyping comparisons between this study and the CPTAC group (a); Overlaps of the nominated kinase targets based on known and expanded kinase-substrate relationship sets (b). PFI: progression-free interval time; OS: overall survival; Known: 15,384 well- annotated kinase-substrate relations; Expanded: 17,833 kinase-substrate relations (15,384 well-annotated kinase-substrate relations combined with 2449 expanded relations).
Applications of LC/MS phosphoproteomic analysis of clinical samples.
Clinical characteristics of 69 patients.
Kinase-substrate relationships from public databases.
Numbers of substrates for each kinase.
Kinases with average log2ratios of sustrates from 69 HGSOC patients.
Baseline Characteristics and kinase signatures of the Ph1,Ph2,Ph3,Ph4,and Ph5 subtypes(n=69).
Activation loop sites and the linear regression results between kinase activities and the corrsponding intensity of loop sites.
Nomination of kinases as potential therapeutic targets.
Cox regression and multiple testing p-value correction with taking age, stage and grade into account for kinases associated with clinical outcome.
The validation results of candidate kinase targets from published paper.
Authors' contributions
T.L., J.Q. and F.H. directed and designed research; M.T., C·Y., D.Z. and M.Z. performed analyses of mass spectrometry data and adapted algorithms for data analysis; T.L, J.Q., Y.W., W.Z., C.W., M.T. and C.Y wrote the manuscript.
Competing interests
The authors have declared no competing interests.
Funding sources
This work was supported by the National Program on Key Basic Research Project (973 Program, 2014CBA02000), National International Cooperation Grant (2014D FB30010 and 2014DFA33160), Beijing Municipal Science and Technology “Frontier Project” (Z131100005213003), National Key Research and Development Program of China (2017YFC0908404, 2017YFC0908400 and 2018YFA0507504), National Natural Science Foundation of China (61773025), The interdisciplinary medicine Seed Fund of Peking University (BMU2017MB001), National Institute of Health (Illuminating Druggable Genome, U01MH105026), Shanghai Municipal Science and Technology Major Project (2017SHZDZX01), and a grant from the State Key Laboratory of Proteomics (SKLP-YA201401). The funders had no roles in study design, data collection, data analysis and interpretation, or writing the manuscript.
Contributor Information
Fuchu He, Email: hefc@nic.bmi.ac.cn.
Jun Qin, Email: jqin1965@126.com.
Tingting Li, Email: litt@hsc.pku.edu.cn.
References
- 1.Nilsson C.L. Advances in quantitative phosphoproteomics. Anal Chem. 2012;84:735–746. doi: 10.1021/ac202877y. [DOI] [PubMed] [Google Scholar]
- 2.Casado P., Hijazi M., Britton D., Cutillas P.R. Impact of phosphoproteomics in the translation of kinase-targeted therapies. Proteomics. 2017:17. doi: 10.1002/pmic.201600235. [DOI] [PubMed] [Google Scholar]
- 3.Wu X., Xing X., Dowlut D., Zeng Y., Liu J., Liu X. Integrating phosphoproteomics into kinase-targeted cancer therapies in precision medicine. J Proteomics. 2019 Jan 16;191:68–79. doi: 10.1016/j.jprot.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 4.Yu L.R., Issaq H.J., Veenstra T.D. Phosphoproteomics for the discovery of kinases as cancer biomarkers and drug targets. Proteomics Clin Appl. 2007;1:1042–1057. doi: 10.1002/prca.200700102. [DOI] [PubMed] [Google Scholar]
- 5.Thygesen C., Boll I., Finsen B., Modzel M., Larsen M.R. Characterizing disease-associated changes in post-translational modifications by mass spectrometry. Expert Rev Proteomics. 2018;15:245–258. doi: 10.1080/14789450.2018.1433036. [DOI] [PubMed] [Google Scholar]
- 6.Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H., Liu T., Zhang Z., Payne S.H., Zhang B., McDermott J.E. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drake J.M., Paull E.O., Graham N.A., Lee J.K., Smith B.A., Titz B. Phosphoproteome integration reveals patient-specific networks in prostate cancer. Cell. 2016;166:1041–1054. doi: 10.1016/j.cell.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casado P., Rodriguez-Prados J.C., Cosulich S.C., Guichard S., Vanhaesebroeck B., Joel S. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 10.Dazert E., Colombi M., Boldanova T., Moes S., Adametz D., Quagliata L. Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc Natl Acad Sci U S A. 2016;113:1381–1386. doi: 10.1073/pnas.1523434113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 12.Khadjavi A., Mannu F., Destefanis P., Sacerdote C., Battaglia A., Allasia M. Early diagnosis of bladder cancer through the detection of urinary tyrosine-phosphorylated proteins. Br J Cancer. 2015;113:469–475. doi: 10.1038/bjc.2015.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borley J., Wilhelm-Benartzi C., Brown R., Ghaem-Maghami S. Does tumour biology determine surgical success in the treatment of epithelial ovarian cancer? A systematic literature review. Br J Cancer. 2012;107:1069–1074. doi: 10.1038/bjc.2012.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowtell D.D., Bohm S., Ahmed A.A., Aspuria P.J., Bast R.C., Jr., Beral V. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–679. doi: 10.1038/nrc4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunewald T., Ledermann J.A. Targeted therapies for ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:139–152. doi: 10.1016/j.bpobgyn.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Vetter M.H., Hays J.L. Use of targeted therapeutics in epithelial ovarian cancer: a review of current literature and future directions. Clin Ther. 2018;40:361–371. doi: 10.1016/j.clinthera.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Lord C.J., Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., Lichtenberg, T., Hoadley, K. A., Poisson, L. M., Lazar, A. J., Cherniack, A. D., Kovatich, A. J., Benz, C. C., Levine, D. A., Lee, A. V., Omberg, L., Wolf, D. M., Shriver, C. D., Thorsson, V., Cancer Genome Atlas Research. 2018. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics Cell 173, 400-416 e11. [DOI] [PMC free article] [PubMed]
- 20.Beausoleil S.A., Villen J., Gerber S.A., Rush J., Gygi S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 21.Wilkerson M.D., Hayes D.N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 23.Hornbeck P.V., Zhang B., Murray B., Kornhauser J.M., Latham V., Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43:D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinkel H., Chica C., Via A., Gould C.M., Jensen L.J., Gibson T.J. Phospho.ELM: a database of phosphorylation sites--update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J., Rho H.S., Newman R.H., Zhang J., Zhu H., Qian J. PhosphoNetworks: a database for human phosphorylation networks. Bioinformatics. 2014;30:141–142. doi: 10.1093/bioinformatics/btt627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., Bridge A.J. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol Biol. 2016;1374:23–54. doi: 10.1007/978-1-4939-3167-5_2. [DOI] [PubMed] [Google Scholar]
- 27.Stark C., Breitkreutz B.J., Chatr-Aryamontri A., Boucher L., Oughtred R., Livstone M.S. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huttlin E.L., Ting L., Bruckner R.J., Gebreab F., Gygi M.P., Szpyt J. The BioPlex network: a systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rolland, T., Tasan, M., Charloteaux, B., Pevzner, S. J., Zhong, Q., Sahni, N., Yi, S., Lemmens, I., Fontanillo, C., Mosca, R., Kamburov, A., Ghiassian, S. D., Yang, X., Ghamsari, L., Balcha, D., Begg, B. E., Braun, P., Brehme, M., Broly, M. P., Carvunis, A. R., Convery-Zupan, D., Corominas, R., Coulombe-Huntington, J., Dann, E., Dreze, M., Dricot, A., Fan, C., Franzosa, E., Gebreab, F., Gutierrez, B. J., Hardy, M. F., Jin, M., Kang, S., Kiros, R., Lin, G. N., Luck, K., Macwilliams, A., Menche, J., Murray, R. R., Palagi, A., Poulin, M. M., Rambout, X., Rasla, J., Reichert, P., Romero, V., Ruyssinck, E., Sahalie, J. M., Scholz, A., Shah, A. A., Sharma, A., Shen, Y., Spirohn, K., Tam, S., Tejeda, A. O., Trigg, S. A., Twizere, J. C., Vega, K., Walsh, J., Cusick, M. E., Xia, Y., Barabasi, A. L., Iakoucheva, L. M., Aloy, P., De Las Rivas, J., Tavernier, J., Calderwood, M. A., Hill, D. E., Hao, T., Roth, F. P. & Vidal, M. 2014. A proteome-scale map of the human interactome network. Cell, 159, 1212–1226. [DOI] [PMC free article] [PubMed]
- 30.Xenarios I., Salwinski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S. Human protein reference database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermjakob H., Montecchi-Palazzi L., Lewington C., Mudali S., Kerrien S., Orchard S. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licata L., Briganti L., Peluso D., Perfetto L., Iannuccelli M., Galeota E. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–D861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowley M.J., Pinese M., Kassahn K.S., Waddell N., Pearson J.V., Grimmond S.M. PINA v2.0: mining interactome modules. Nucleic Acids Res. 2012;40:D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R., Dephoure N., Haas W., Huttlin E.L., Zhai B., Sowa M.E. Correct interpretation of comprehensive phosphorylation dynamics requires normalization by protein expression changes. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.009654. (M111009654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hernandez-Armenta C., Ochoa D., Goncalves E., Saez-Rodriguez J., Beltrao P. Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics. 2017;33:1845–1851. doi: 10.1093/bioinformatics/btx082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ochoa D., Jonikas M., Lawrence R.T., El Debs B., Selkrig J., Typas A. An atlas of human kinase regulation. Mol Syst Biol. 2016;12:888. doi: 10.15252/msb.20167295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klaeger S., Heinzlmeir S., Wilhelm M., Polzer H., Vick B., Koenig P.A. The target landscape of clinical kinase drugs. Science. 2017;358 doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drake J.M., Graham N.A., Stoyanova T., Sedghi A., Goldstein A.S., Cai H. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci U S A. 2012;109:1643–1648. doi: 10.1073/pnas.1120985109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 41.Nolen B., Taylor S., Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 42.Altomare D.A., Wang H.Q., Skele K.L., De Rienzo A., Klein-Szanto A.J., Godwin A.K. AKT and mTOR phosphorylation is frequently detected in ovarian cancer and can be targeted to disrupt ovarian tumor cell growth. Oncogene. 2004;23:5853–5857. doi: 10.1038/sj.onc.1207721. [DOI] [PubMed] [Google Scholar]
- 43.Etemadmoghadam D., Bowtell D. AKT1 gene amplification as a biomarker of treatment response in ovarian cancer: mounting evidence of a therapeutic target. Gynecol Oncol. 2014;135:409–410. doi: 10.1016/j.ygyno.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Huang J., Zhang L., Greshock J., Colligon T.A., Wang Y., Ward R. Frequent genetic abnormalities of the PI3K/AKT pathway in primary ovarian cancer predict patient outcome. Genes Chromosomes Cancer. 2011;50:606–618. doi: 10.1002/gcc.20883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheaib B., Auguste A., Leary A. The PI3K/Akt/mTOR pathway in ovarian cancer: therapeutic opportunities and challenges. Chin J Cancer. 2015;34:4–16. doi: 10.5732/cjc.014.10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torchiaro E., Lorenzato A., Olivero M., Valdembri D., Gagliardi P.A., Gai M. Peritoneal and hematogenous metastases of ovarian cancer cells are both controlled by the p90RSK through a self-reinforcing cell autonomous mechanism. Oncotarget. 2016;7:712–728. doi: 10.18632/oncotarget.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etemadmoghadam D., Au-Yeung G., Wall M., Mitchell C., Kansara M., Loehrer E. Resistance to CDK2 inhibitors is associated with selection of polyploid cells in CCNE1-amplified ovarian cancer. Clin Cancer Res. 2013;19:5960–5971. doi: 10.1158/1078-0432.CCR-13-1337. [DOI] [PubMed] [Google Scholar]
- 48.Goel S., Decristo M.J., Watt A.C., Brinjones H., Sceneay J., Li B.B. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548:471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Konecny G.E. Cyclin-dependent kinase pathways as targets for women's cancer treatment. Curr Opin Obstet Gynecol. 2016;28:42–48. doi: 10.1097/GCO.0000000000000243. [DOI] [PubMed] [Google Scholar]
- 50.Wei F.Y., Tomizawa K. Cyclin-dependent kinase 5 (Cdk5): a potential therapeutic target for the treatment of neurodegenerative diseases and diabetes mellitus. Mini Rev Med Chem. 2007;7:1070–1074. doi: 10.2174/138955707782110114. [DOI] [PubMed] [Google Scholar]
- 51.Zhang S., Lu Z., Mao W., Ahmed A.A., Yang H., Zhou J. CDK5 regulates paclitaxel sensitivity in ovarian cancer cells by modulating AKT activation, p21Cip1- and p27Kip1-mediated G1 cell cycle arrest and apoptosis. PLoS One. 2015;e0131833:10. doi: 10.1371/journal.pone.0131833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Penzvalto Z., Lanczky A., Lenart J., Meggyeshazi N., Krenacs T., Szoboszlai N. MEK1 is associated with carboplatin resistance and is a prognostic biomarker in epithelial ovarian cancer. BMC Cancer. 2014;14:837. doi: 10.1186/1471-2407-14-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bubici C., Papa S. JNK signalling in cancer: in need of new, smarter therapeutic targets. Br J Pharmacol. 2014;171:24–37. doi: 10.1111/bph.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ajayi S., Becker H., Reinhardt H., Engelhardt M., Zeiser R., Von Bubnoff N. Ruxolitinib. Recent Results Cancer Res. 2018;212:119–132. doi: 10.1007/978-3-319-91439-8_6. [DOI] [PubMed] [Google Scholar]
- 55.Nguyen P.H., Fedorchenko O., Rosen N., Koch M., Barthel R., Winarski T. LYN kinase in the tumor microenvironment is essential for the progression of chronic lymphocytic leukemia. Cancer Cell. 2016;30:610–622. doi: 10.1016/j.ccell.2016.09.007. [DOI] [PubMed] [Google Scholar]
- 56.Poh A.R., Love C.G., Masson F., Preaudet A., Tsui C., Whitehead L. Inhibition of hematopoietic cell kinase activity suppresses myeloid cell-mediated colon cancer progression. Cancer Cell. 2017;31(563–575) doi: 10.1016/j.ccell.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong Q., Dougan D.R., Gong X., Halkowycz P., Jin B., Kanouni T. Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer. Bioorg Med Chem Lett. 2011;21:1315–1319. doi: 10.1016/j.bmcl.2011.01.071. [DOI] [PubMed] [Google Scholar]
- 58.Micel L.N., Tentler J.J., Tan A.C., Selby H.M., Brunkow K.L., Robertson K.M. Antitumor activity of the MEK inhibitor TAK-733 against melanoma cell lines and patient-derived tumor explants. Mol Cancer Ther. 2015;14:317–325. doi: 10.1158/1535-7163.MCT-13-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Long G.V., Stroyakovskiy D., Gogas H., Levchenko E., De Braud F., Larkin J. Dabrafenib and trametinib versus dabrafenib and placebo for Val600 BRAF-mutant melanoma: a multicentre, double-blind, phase 3 randomised controlled trial. Lancet. 2015;386:444–451. doi: 10.1016/S0140-6736(15)60898-4. [DOI] [PubMed] [Google Scholar]
- 60.Hew K.E., Miller P.C., El-Ashry D., Sun J., Besser A.H., Ince T.A. MAPK activation predicts poor outcome and the MEK inhibitor, Selumetinib, reverses antiestrogen resistance in ER-positive high-grade serous ovarian cancer. Clin Cancer Res. 2016;22:935–947. doi: 10.1158/1078-0432.CCR-15-0534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harsha H.C., Pandey A. Phosphoproteomics in cancer. Mol Oncol. 2010;4:482–495. doi: 10.1016/j.molonc.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample frequency of the phosphosites ranked at the bottom 1/4(a) and ranks of the phosphorylation sites based on the intensity(b).
An example for the calculation of the position weight matrix(a) and the correlations of the ±7 positions-based PWM scores with different sequence lengths (from±4 to ±6 positions) (b).
Overview of the phosphoproteome and proteome data. a. The number of phosphorylation sites and phosphorylation proteins detected in 69 patients; b. Numbers of proteins detected in 69 patients.
Numbers of substrates detected for each kinase.
Consensus clustering based on the phosphoproteome data. Consensus matrices and the cumulative distribution function (CDF) plots for the a. phosphorylation sites, b. kinases activity, and c. protein abundance.
Linear regression results of the activation loop phosphorylation sites. a. Comparisons of the correlations between the kinases activity predicted by the Mean value (left) and KSEA(right) methods with the intensity of the activated/not activated loop phosphorylation sites; Examples for the correlations between the kinases activities predicted by the b. Mean values/ c. KSEA methods with the intensities of the activation loop phosphorylation sites. R2: r square in the linear regression analysis.
Survival curves of the kinases based on activities and abundance. a. Survival curves of the kinases whose increased activities in tumours are both associated with progression-free interval and overall survival; b. Survival curves of six kinases whose overexpressed protein abundance as measured in the profiling experiments associated with clinical outcome. Low(Kinase = 0)/high(Kinase = 1): patients with kinase activity/abundance lower/above than cut-off.
Survival curves of the kinases whose increased activities in tumours are associated with progression-free interval or overall survival. Low (Kinase = 0)/high(Kinase = 1): patients with kinase activity lower/above than cut-off.
Boxplots of kinase substrates phosphorylation difference in two prognostic group (PI3K/AkT/mTOR pathway).
Boxplots of kinase substrates phosphorylation difference in two prognostic group (Cell cycle and WNT pathways).
Boxplots of kinase substrates phosphorylation difference in two prognostic group (MAPK and other pathways).
Heatmaps of substrates phosphorylation difference in two prognostic group (PI3K/AkT/mTOR pathway).
Heatmaps of substrates phosphorylation difference in two prognostic group (Cell cycle pathway).
Heatmaps of substrates phosphorylation difference in two prognostic group (WNT pathway and other pathways).
Heatmaps of substrates phosphorylation difference in two prognostic group (MAPK pathway).
Subtyping comparisons between this study and the CPTAC group (a); Overlaps of the nominated kinase targets based on known and expanded kinase-substrate relationship sets (b). PFI: progression-free interval time; OS: overall survival; Known: 15,384 well- annotated kinase-substrate relations; Expanded: 17,833 kinase-substrate relations (15,384 well-annotated kinase-substrate relations combined with 2449 expanded relations).
Applications of LC/MS phosphoproteomic analysis of clinical samples.
Clinical characteristics of 69 patients.
Kinase-substrate relationships from public databases.
Numbers of substrates for each kinase.
Kinases with average log2ratios of sustrates from 69 HGSOC patients.
Baseline Characteristics and kinase signatures of the Ph1,Ph2,Ph3,Ph4,and Ph5 subtypes(n=69).
Activation loop sites and the linear regression results between kinase activities and the corrsponding intensity of loop sites.
Nomination of kinases as potential therapeutic targets.
Cox regression and multiple testing p-value correction with taking age, stage and grade into account for kinases associated with clinical outcome.
The validation results of candidate kinase targets from published paper.