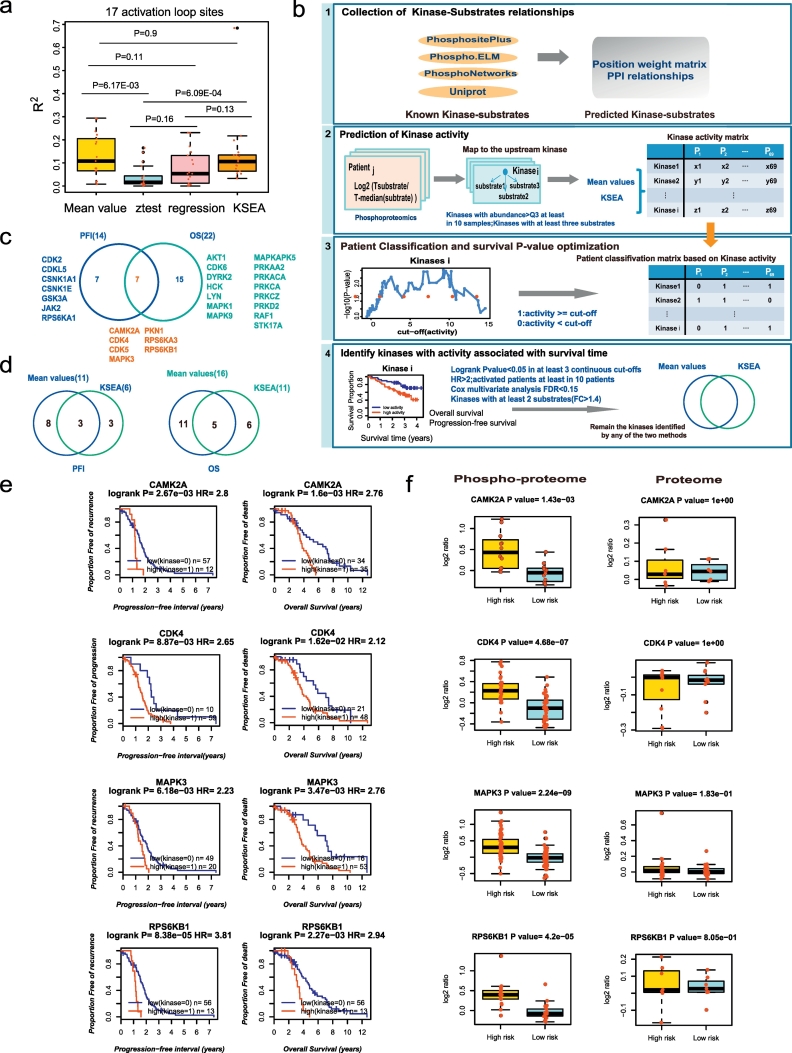
Fig. 3.
Nominating potential druggable kinases.
a. The correlations between the kinase activities predicted by the four methods with the intensities of the activation loop phosphorylation sites; b. The workflow of the identification of druggable kinases; c. Kinases whose increased activities in tumours are both associated with poor clinical outcome; d. Overlaps of the results for the two kinase activity prediction methods; e-f. Kinases whose increased activities in tumours are both associated with the progression-free interval and overall survival. e. Survival curves of the kinases and f. boxplot of the kinases substrates in the phosphoproteome and proteome data. Low(Kinase = 0)/high(Kinase = 1): patients with kinase activity lower than/above the cut-off. High risk/Low risk: patients with poor/better survival. HR: hazard ratio.