Abstract
Background
Osteosarcoma (OS) is a malignant tumor mainly occurring in young people. Due to the limited effective therapeutic strategies, OS patients cannot achieve further survival improvement. G-protein-coupled receptors (GPCRs) constitute the largest family of cell membrane receptors and consequently hold the significant promise for tumor imaging and targeted therapy. We aimed to explore the biological functions of Sphingosine 1-phosphate receptor 3 (S1PR3), one of the members of GPCRs family, in OS and the possibility of S1PR3 as an effective target for the treatment of osteosarcoma.
Methods
The quantitative real time PCR (qRT-PCR) and western blotting were used to analyze the mRNA and protein expressions. Cell counting kit-8 (CCK8), colony formation assay and cell apoptosis assay were performed to test the cellular proliferation in vitro. Subcutaneous xenograft mouse model was generated to evaluate the functions of S1PR3 in vivo. RNA sequencing was used to compare gene expression patterns between S1PR3-knockdown and control MNNG-HOS cells. In addition, metabolic alternations in OS cells were monitored by XF96 metabolic flux analyzer. Co-immunoprecipitation (Co-IP) assay was used to explore the interaction between Yes-associated protein (YAP) and c-MYC. Chromatin immunoprecipitation was used to investigate the binding capability of PGAM1 and YAP or c-MYC. Moreover, the activities of promoter were determined by the luciferase reporter assay.
Findings
S1PR3 and its specific ligand Sphingosine 1-phosphate (S1P) were found elevated in OS, and the higher expression of S1PR3 was correlated with the poor survival rate. Moreover, our study has proved that the S1P/S1PR3 axis play roles in proliferation promotion, apoptosis inhibition, and aerobic glycolysis promotion of osteosarcoma cells. Mechanistically, the S1P/S1PR3 axis inhibited the phosphorylation of YAP and promoted the nuclear translocation of YAP, which contributed to the formation of the YAP–c-MYC complex and enhanced transcription of the important glycolysis enzyme PGAM1. Moreover, the S1PR3 antagonist TY52156 exhibited in vitro and in vivo synergistic inhibitory effects with methotrexate on OS cell growth.
Interpretation
Our study unveiled a role of S1P, a bioactive phospholipid, in glucose metabolism reprogram through interaction with its receptor S1PR3. Targeting S1P/S1PR3 axis might serve as a potential therapeutic target for patients with OS.
Fund
This research was supported by National Natural Science Foundation of China (81472445 and 81672587).
Keywords: Osteosarcoma, S1PR3, S1P, YAP, Warburg effect, Cell proliferation
Research in the context.
Evidence before this study
Growing evidence has suggested that G-protein-coupled receptors (GPCRs) hold the significant promise for tumor imaging and targeted therapy. The G-protein-coupled receptor sphingosine 1-phosphate receptor 3 (S1PR3) has been found to play critical roles in the development and progression of several tumors through interaction with its ligand Sphingosine 1-phosphate (S1P). However, the effects of S1P/S1PR3 axis on osteosarcoma (OS) still remain elusive.
Added value of this study
This study investigated the expression level, biological function, downstream regulation and clinical impact of S1PR3 on OS. We observed that increased S1PR3 expression predicted poor survival rate in OS patients and promoted cell proliferation by inducing the Warburg effect in OS. In addition, we also found that S1PR3 antagonist TY52156 exhibited the synergistic inhibitory effects with methotrexate on OS cell growth. Mechanistically, we uncovered that S1P/S1PR3 axis promoted the aerobic glycolysis in OS through inhibiting the phosphorylation of Yes-associated protein (YAP) and promoting the nuclear translocation of YAP, which contributed to the formation of the YAP–c-MYC complex and enhanced transcription of the important glycolysis enzyme PGAM1.
Implications of all the available evidence
Our findings delineated one of the instrumental molecular mechanisms underlying the glucose metabolism reprogram in the OS and indicated that S1P/S1PR3 axis was a potential therapeutic target for patients with OS.
Alt-text: Unlabelled Box
1. Introduction
Osteosarcoma (OS) is a malignant bone tumor and ranks the eighth top cancer among adolescents and young adults [1,2]. Due to its rapid growth and aggressiveness, OS typically extends beyond the bone into nearby musculoskeletal structures [3,4]. With the development and advancements in surgery, as well as various chemotherapy regimens, the 5-year overall survival rate has been significantly improved in OS. However, the survival rate has plateaued and has not significantly increased over the past three decades [5]. Thus, there is an urgent need for an increased understanding of the mechanisms underlying the progression of OS to develop more effective therapies for OS treatment.
G protein-coupled receptors (GPCRs) have been identified as one of the most popular drug target classes in pharmaceuticals, which was effective in treating various indications, such as inflammation, pain and metabolic dysfunctions [6]. Lots of previous studies have also demonstrated that GPCRs play a significant part in cancer growth and development [7,8]. Thus, we presume that there may be potential candidate targets for preventing OS progression in GPCRs.
Sphingosine 1-phosphate (S1P) is a bioactive phospholipid and is supposed to regulate many cellular physiology and pathology processes, such as proliferation, motility, survival, and cytoskeletal rearrangement, via binding to a family of five GPCRs, known as S1PR1–S1PR5 [[9], [10], [11], [12]]. Several lines of evidence have suggested that the S1P/S1PR3 axis was closely associated with proliferation, migration, and angiogenesis in various human cancer cells, such as breast cancer, nasopharyngeal carcinoma, ependymomas and ovarian cancer cells [[13], [14], [15], [16], [17]]. However, to our knowledge, the biological mechanism of this axis in OS still remains unclear.
In this study, we demonstrated that the S1P/S1PR3 axis enhanced the aerobic glycolysis and facilitated the OS growth. Further mechanistic studies showed that S1PR3 was a novel regulator of YAP and S1PR3-mediated YAP nuclear localization contributed to the aerobic glycolysis in OS growth. Moreover, S1PR3 antagonist TY52156 had shown a synergistic effect with methotrexate on tumor cell growth impairment in vitro and in vivo. Thus, targeting S1PR3 held promise as a novel anticancer strategy.
2. Materials and methods
2.1. Cell lines and reagents
The human osteosarcoma cell lines MG63, MNNG-HOS and Saos-2 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The human osteosarcoma cell line U-2OS was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). These cell lines were cultured following the ATCC protocols. Standardized culture conditions had been described previously [18].
The antibodies used were S1PR3 (ab126622; Abcam, Cambridge, UK), YAP (ab52771; Abcam, Cambridge, UK), p-YAP (ab76252; Abcam, Cambridge, UK), c-Myc (ab32072; Abcam), Ki67 (GB13030; Servicebio, Wuhan, China), β-actin (M1210-2; Hua'an Biology, Chuzhou, China), GAPDH (bsm-33033M; Bioss, Beijing, China), anti-rabbit IgG light chain (ab99697, Abcam), anti-rabbit IgG heavy chain (ab99702, Abcam), and anti-mouse IgG light chain (A25012, Abbkine, CA, USA).
2.2. Drugs and chemotherapeutic reagents
S1P (CAS26993-30-6; Cayman Chemical, Michigan, USA), Verteporfin (S1786; Selleckchem, Houston, Texas, USA), TY52156 (HY-19736; MedChem Express, Monmouth, NJ, USA) and Methotrexate (MTX) (S1210; Selleckchem, Houston, Texas, USA) were diluted and stored in accordance with manufacturer recommendations. As for in vitro experiments, drug stocks were diluted in the base media. While stocks were diluted in saline immediately prior to use in vivo experiments. 2 μM Verteporfin, 10 μM TY52156 and 1 μM MTX were used in in vitro experiments.
2.3. RNA sequencing
Total RNA from cell samples was isolated by Trizol reagent following the manufacturer's instructions. RNA quality was analyzed using an Agilent 2100 Bioanalyzer (Agilent). We purified the library fragments with the AMPure XP system (Beckman Coulter, Beverly, USA). The clustering of the index-coded samples was analyzed on a cBot Cluster Generation System by using the TruSeq PE Cluster Kit v3-cBot-HS (Illumia). After cluster generation, we sequence the library preparations on an Illumina Hiseq X Ten and generated 150 bp paired-end reads. Read numbers mapped to each gene were counted by using HTSeq v0.6.0. Then, the FPKM of each gene was calculated according to the length of the gene and reads count mapped to this gene.
2.4. Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) was performed using the GSEA software which was supported by the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). GSEA was analyzed for comparing the différential gene expression between sh-Control group and sh-S1PR3 group. In addition, the enrichment score was calculated.
2.5. Plasmid transfection
Plasmid transfection was done as previously described [19]. The short hairpin (sh)RNAs targeting S1PR3 sequences were as follows: sh-1, 5′-GCATCGCTTACAAGGTCAACA-3′, sh-2, 5′-GGAACTGCCTGCACAATCTCC-3′ and sh-CON, 5′-TTCTCCGAACGTGTCACGT-3′. Western blotting was used to verify the efficiency of the overexpression or knockdown.
2.6. RNA isolation and quantitative RT-PCR (qRT-PCR)
Total RNA extraction and RNA reverse transcription were done as described in our previously report [19]. Real-time PCR analyses were performed using a 7500 Real-time PCR system (Applied biosystems) as previously described [27]. β-actin was set as an internal control. All primers were listed in Supplementary Table 1.
2.7. Western blotting
Western blotting analysis was done as described in our previous report [20]. In brief, total cellular proteins were extracted from the target cells by RIPA lysis buffer (Beyotime, Shanghai, China) according to the manufacturer's instructions. Equal amounts of proteins were loaded onto 10% Tris-glycine sodium dodecyl sulfate-polyacrylimide gel electrophoresis gels (Bio-Rad Laboratories, CA, USA). Then the separated proteins were transferred onto nitrocellulose membranes (Millipore, MA, USA). After blocking with 5% non-fat milk, the membranes were incubated with a primary antibody at 4 °C overnight. The membranes were further incubated with secondary antibody and protein signals were detected under the ECL detection kit (Share-bio, Shanghai, China).
2.8. Quantification of S1P
Enzyme-linked immunosorbent assay (ELISA) was performed to quantify the expression of S1P from cell culture supernatants using a Sphingosine 1-Phosphate ELISA Kit (K-1900; Echelon Biosciences, Salt Lake City, USA) following the manufacturer's instructions. In brief, approximately 1 × 106 cells were seeded in six-well plates and cultured with the medium and 10% FBS under standardized condition When cells were found adherent, serum-free medium was substituted for the culture medium. After 32 h seeding, cells and the culture medium were centrifuged at 3000g for 10 min, and then the supernatants were aliquoted for analysis.
2.9. Immunohistochemistry (IHC) staining
We examined the expression of S1PR3 with immunohistochemistry (IHC) using a tissue microarray containing 40 OS samples from Alena Biotechnology Co., Ltd. (Xi'an, China). The IHC assay protocol was described previously [19]. Anti- S1PR3 (1:100) antibodies were used. The intensity of the nuclei or cytoplasm staining of each tumor was assigned in line with the following criteria: 0–5%, 6–35%, 36–70% and >70% staining were scored of 0, 1, 2, 3 respectively. The final immunoreactive score was determined by multiplying the intensity scores, judged by two senior pathologists in a blinded manner. Tumors with scores < 2 were defined as negative, while scores ≥ 2 as negative.
2.10. Immunofluorescence (IF)
OS cells were planted in 12-well chambers (Ibidi, Germany) for IF. We fixed cells with 4% polyformaldehyde (30 min), permeabilized with 0.1% TritonX-100 (2 min) and blocked with 10% BSA (60 min) at room temperature. Following instructions include deparaffinization, rehydration, heat-mediated antigen retrieval in citric acid (pH 6.0) and blocking with 10% BSA. All cells and slides were incubated overnight with YAP antibodies at 4 °C and then labeled with Alexa Fluor-594-conjugated secondary antibody (1:200) at room temperature. DAPI was used to stain the nucleus for 2 min (Sigma, USA). Confocal microscopy (LSM 510, METALaser scanning microscope, Zeiss) was used to acquire the images.
2.11. Cell counting kit-8 (CCK-8) assay, Colony formation assay and cell apoptosis assay
CCK-8 assay, colony formation assay and cell apoptosis assay was performed as previously described [19].
2.12. Mouse xenograft model
6-weeks-old athymic male nu/nu mice were used in this study. Mice were manipulated and housed according to the criteria of Laboratory Animals. Animal studies were approved by the Research Ethics Committee of East China Normal University. 1.5 × 106 of the target cells in 100 μl PBS were injected subcutaneously into the mice to grow xenografts. Tumor diameters were monitored every 5 days. After 20 days, mice were scarificed, and the xenografts were stripped out and weighed for further analysis. For chronic administration, when animals bore visible tumors (50 mm3), they were randomly divided into four groups (Ctr group, MTX group, TY52156 group, and MTX plus TY52156 group). Each group was treated with different administration:100 μl of 0.9% NaCl, MTX (5 mg/kg), TY52156 (3 mg/kg) and MTX (5 mg/kg) plus TY52156 (3 mg/kg) every four days respectively. Tumor diameters were monitored every 4 days. Tumor volumes were calculated by volume = (Width2 × Length)/2.
2.13. Terminal deoxynucleotidyl transferase (TdT) dUTP nick-end labeling (TUNEL) assay
A TUNEL kit (Roche, Basel, Switzerland) was used to quantify the proportion of apoptotic cells in the xenograft tumors. We preformed this assay as previously described [19].
2.14. Oxidative phosphorylation and glycolysis analysis
The real-time glycolytic rate (ECAR) and oxygen consumption rate (OCR) of OS cells were analyzed by an XF96 metabolic flux analyzer (Seahorse Biosciences, Billerica, MA, USA) as previously described [19]. Briefly, 3 × 104 target cells were seeded into each well of a Seahorse XF 96 cell culture microplate. After the probes were calibrated, for OCR, 1 mM oligomycin, 1 mM p-trifluoromethoxy carbonyl cyanide phenylhydrazone (FCCP) and 2 mM antimycin A plus 2 mM rotenone (Rote/AA) were sequentially injected; and for ECAR, 10 mM glucose, 1 mM oligomycin, and 80 mM 2-deoxyglucose (2-DG) were sequentially injected. Data were assessed and analyzed by using Seahorse XF-96 Wave software.
2.15. Co-immunoprecipitation (Co-IP) assay
MNNG-HOS and U-2OS proteins were extracted through total protein extraction buffer (Beyotime, China). Protein A/G sepharose (Santa Cruz Biotechnology) was pre-incubated with anti-c-MYC or anti-YAP antibody for 30–60 min on a spinning wheel at 4 °C with two washes. The bead-antibody complexes were then suspended with protein lysate. All Co-IP was performed overnight on a spinning wheel at 4 °C. The beads were washed 3 times with extraction buffer, and were collected by centrifugation at 3000g. The immunoprecipitates were subjected to western blotting.
2.16. Chromatin immunoprecipitation (CHIP) assay
MNNG-HOS and U-2OS cells were fixed in 1% formaldehyde solution at 37 °C for 10 min. ChIP assay was performed using the Pierce™ agarose ChIP kit (Thermo Fisher Scientific). Premix Taq (Cell Signaling) was used to quantify the DNA-protein complexes formed by immunoprecipitating DNA with control IgG (Cell Signaling), anti-YAP and anti-c-MYC from the sonicated cell lysates. The specific primers used in the process of CHIP were listed in Supplementary Table 1.
2.17. Luciferase reporter assay
Luciferase reporter assay was performed as described in our previous report [19,21]. In brief, wild-type or YAP or c-MYC overexpressed MNNH-HOS and U-2OS cells were co-transfected with pGL4.10-Promotor vectors and pRL-TK Renilla plasmids. A Dual-Luciferase Reporter Assay System (Promega) was used to analyze the luciferase activity. The wild type and mutant PGAM1 promoters regions were list in Supplementary Table 1.
2.18. Database analysis
Survival rate analyzed by a Kaplan–Meier analysis of 88 OS patients were referenced from an online database (http://hgserver1.amc.nl). The data from GEO database (GSE 42352) were used to compare the gene expression levels of S1PR3 between the OS and mesenchymal stem cell (MSC).
2.19. Statistical analyses
All statistics were carried out using GraphPad Prism 7.0 and Excel for Windows (La Jolla, CA, USA). The chi-square test was used for proportion comparison. Comparisons between two groups were performed by two-tailed Student's t-test. All error bars in this study was represented the mean ± S.D. p values > .05 = ns, p values <.05 = *, p values < .01 = **, p values < .001 = ***.
2.20. Data availability
The data sets of MNNG-HOS cell line silenced of S1PR3, which support the findings of this study are available in the Sequence Read Archive (SRA) repository under accession NO.SRP165691. All other remaining data that support our findings are available from the authors upon reasonable request.
3. Results
3.1. S1P and S1PR3 were both upregulated in OS and higher expression of S1PR3 was closely related to the poor survival rate in OS
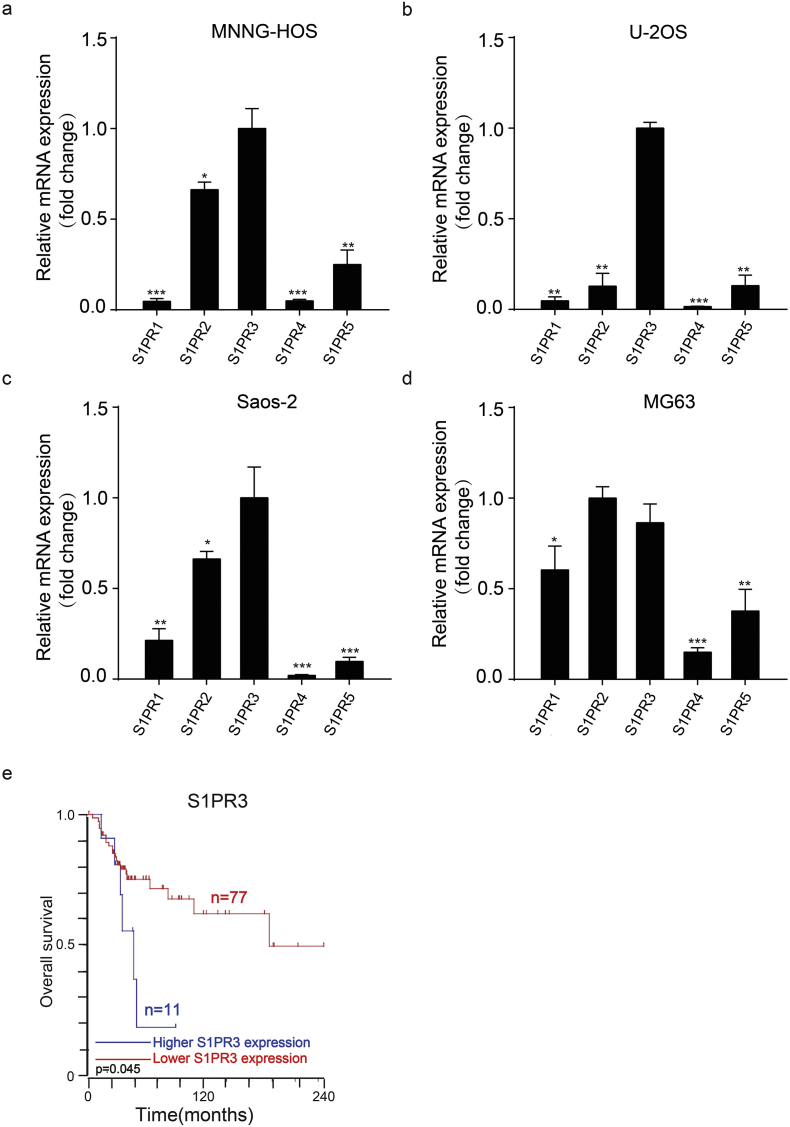
It has been reported that sphingosine kinase, resulting in the generation and secretion of S1P, was upregulated in OS cells and tissues [22]. Accordingly, we compared the S1P levels in the supernatants of hFOB1.19, a common osteoblast cell line, with that in human OS cell lines (MNNG-HOS, U-2OS, Saos-2 and MG63) through enzyme-linked immunosorbent assay (ELISA). As shown in Fig. 1a, OS cell lines secreted more S1P than osteoblast cell lines. We next compared the mRNA levels of all five S1PRs by quantitative RT-PCR in OS cell lines. Compared to the other four S1PRs, S1PR3 exhibited a higher expression in most OS cell lines (Supplementary Fig. 1a–d). Then we compared the expression level of S1PR3 between hFOB1.19 and OS cell lines via western blotting. OS cells displayed significantly higher S1PR3 levels compared to the osteoblast cell line, especially in MNNG-HOS and U-2OS cell lines (Fig. 1b). Furthermore, we detected the expression difference of S1PR3 between OS (n = 40) and osteochondroma (OC) (n = 20) tissue samples through immunohistochemistry (IHC) staining. S1PR3 exhibited higher expression in OS tissues than in OC tissues (Fig. 1c and d). We also found that the higher expression of S1PR3 was related to the advanced pathological staging in OS (Fig. 1e). Moreover, we downloaded the mRNA expression data from GEO database (GSE 42352) and analyzed the expression profiles of S1PR3 in 12 mesenchymal stem cell (MSC) and 84 OS tissues. The results also indicated that S1PR3 was upregulated in OS tissues (Supplementary Fig. 1e). We further analyzed the prognostic value of S1PR3 in OS patients according to an online database. As shown in Fig. 1f and Supplementary Fig. 1f, Kaplan–Meier analysis demonstrated that OS patients with a higher expression of S1PR3 had a lower overall survival rate (p = .045, Kaplan-meier analysis) and metastasis-free survival rate (p = .017, Kaplan-meier analysis). Collectively, we found that S1P and S1PR3 were both upregulated in OS. Simultaneously, the higher expression of S1PR3 was linked to a poor survival rate and might promote the progression of OS.
Fig. 1.
S1P and S1PR3 expression level are both increased in osteosarcoma (OS). a. Quantitative analysis of the secretion of Sphingosine 1-phosphate (S1P) in normal osteoblast cell line (hFOB1.19) and osteosarcoma (OS) cell lines (MNNG-HOS, U-2OS, Saos-2 and MG63) by enzyme-linked immunosorbent assay (ELISA). Values are means ± SD, *p < .05, **p < .01 (Student's t-test). b. The expression patterns of S1P receptor 3 (S1PR3) in hFOB1.19, a normal osteoblast cell line, and OS cell lines (MNNG-HOS, U-2OS, Saos-2 and MG63) by western blotting. c. The protein expression of S1PR3 in osteohondroma (OC), a commom type of benign bone tumor, and OS tissues by immunohistochemical (IHC) staining. Scale bars = 100 μm. d. Statistical analysis of IHC staining results based on the expression level of S1PR3 in OS (n = 40) and OC (n = 20) tissues. **p < .01 (chi-square test). e. Statistical analysis of IHC staining results based on the expression level of S1PR3 in T1 stage (n = 9) and T2 stage (n = 20) OS tissues. *p < .05 (chi-square test). f. Kaplan-Meier analysis of the correlations between S1PR3 expression and metastasisfree survival rate of 88 OS patients based on an online database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
Supplementary Fig. 1.
a–d. The expression of S1PRs (S1PR1, S1PR2, S1PR3, S1PR4 and S1PR5) were detected via quantitative real time PCR (qRT-PCR) in OS cell lines (MNNG-HOS, U-2OS, Saos-2 and MG63). Values are means ± SD, *p < .05, **p < .01, ***p < .001 (Student's t-test). e. Relative expression levels of S1PR3 in 12 mesenchymal stem cell (MSC) and 84 OS tissues in GEO database (GSE 42352), The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. ***p < .001 (Student's t-test). f. Kaplan-Meier analysis of the correlations between S1PR3 expression and overall survival rate of 88 OS patients based on an online database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi).
3.2. S1P/S1PR3 axis promoted the proliferation and inhibited the apoptosis of OS cells
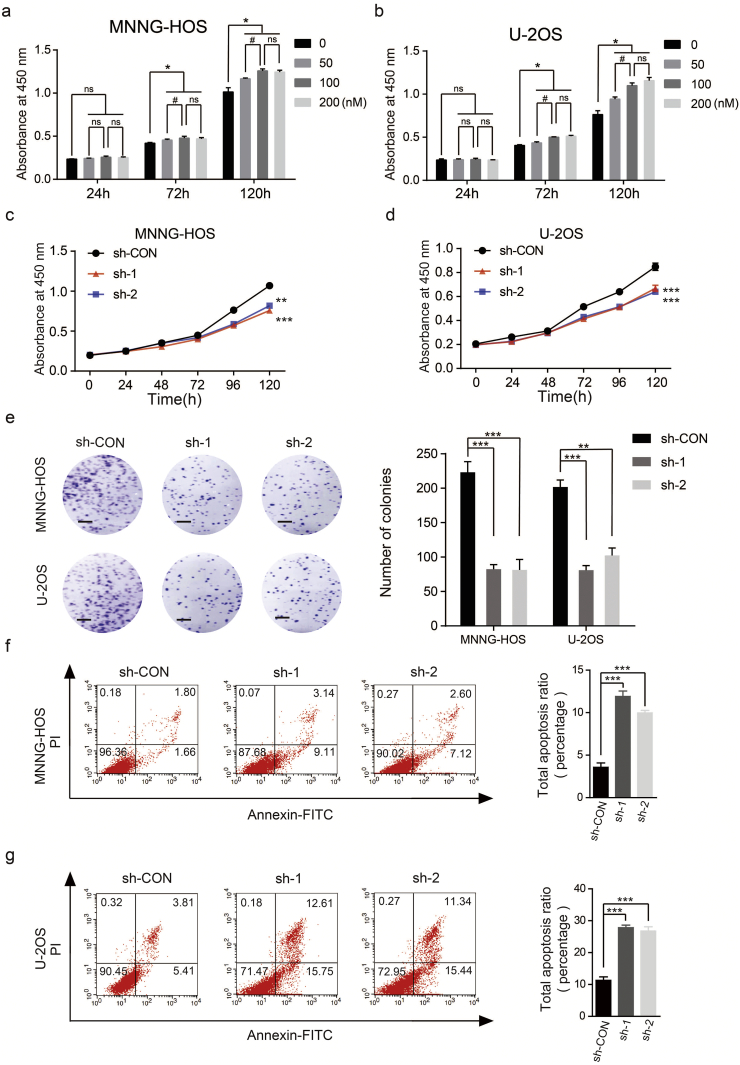
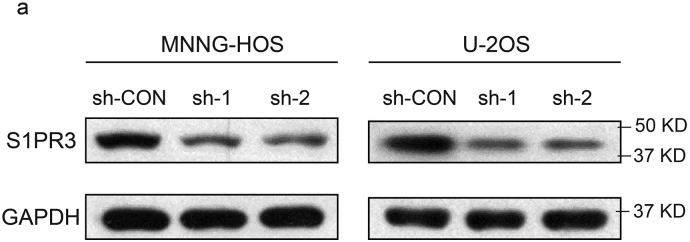
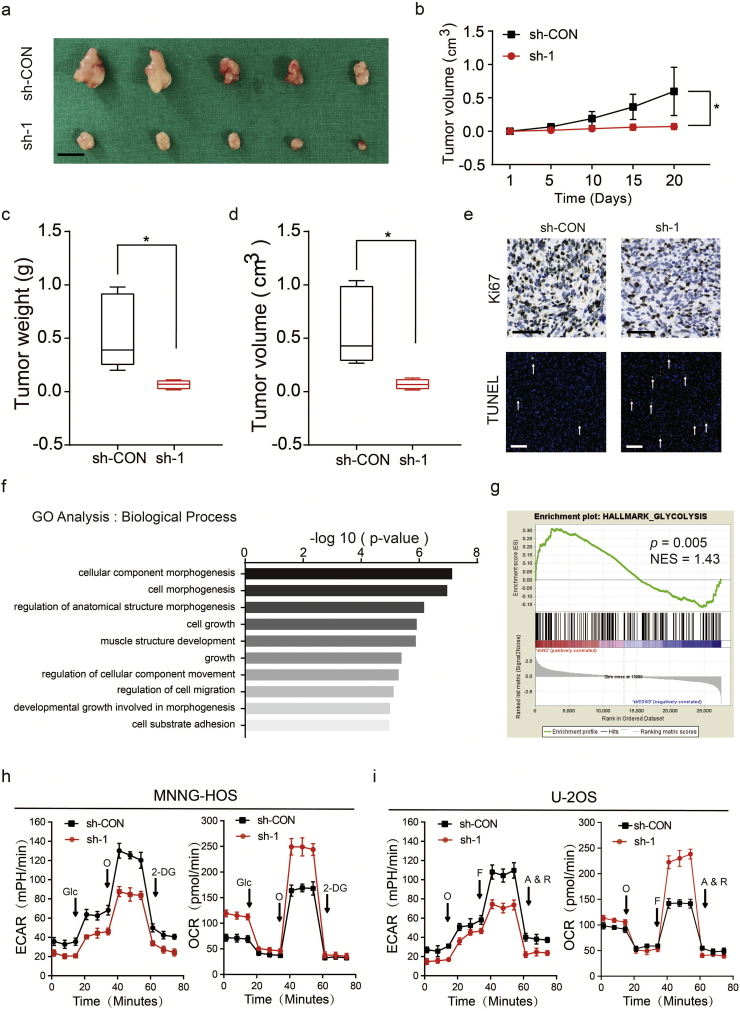
In order to study the biological functions of S1P/S1PR3 axis in OS, we first stimulated OS cells by adding S1P to the medium exogenously. Stimulation with S1P promoted the proliferation of OS cells, with a maximal response observed at 100 nM (Fig. 2a and b). Next, we blocked the S1P/S1PR3 axis by shRNAs against S1PR3. While a non-targeting shRNA was taken as a control, and western blotting were performed to test the efficiency of knockdown (Supplementary Fig. 2a). We first studied the influence of S1PR3 knockdown in OS growth in vitro. The silencing of S1PR3 partly suppressed the proliferation of OS cells through using CCK-8 assay (Fig. 2c and d) and colony formation assay (Fig. 2e). We then explored the effects of S1PR3 knockdown on cell apoptosis in OS. As shown in Fig. 2f and g, knockdown of S1PR3 could significantly promote the apoptosis of OS cells. Moreover, we analyzed whether S1PR3 could enhance proliferation and inhibit apoptosis in vivo. The stable S1PR3 knockdown and control MNNG-HOS cells were injected into the nude mice subcutaneously. An obvious delay in the growth speed of tumors was seen in the stable S1PR3-knockdown group, as well as a reduced tumor weight and volume (Fig. 3a–d). The results of IHC staining and TUNEL assay revealed declining expression of Ki67 and rising rate of apoptosis in the xenografted tumors of S1PR3-knockdown group (Fig. 3e). Furthermore, we performed RNA sequencing to compare gene expression patterns between S1PR3-knockdown and control MNNG-HOS cells. In order to ensure the accurateness of data analysis, differentially expressed genes were defined as an average fold-change > 2.0 and p-value < .05. As displayed in Fig. 3f, the results of GO analysis (biological process) were in consistent with our previous study that S1PR3 promoted OS cell growth. These data pointed out that the S1P/S1PR3 axis was supposed to promote the proliferation and inhibit the apoptosis of OS cells.
Fig. 2.
S1P/S1PR3 signaling facilitates proliferation and suppresses apoptosis of OS cells in vitro. a and b. Dose-dependent effects of S1P in the proliferation capability of MNNG-HOS and U-2OS cells using the cell counting kit (CCK)-8 assay. Values are means ± SD, *p < .05, (50 nM, 100 nM and 200 nM vs 0 nM); #p < .05 (100 nM vs 200 nM); non sensual (ns), p > .05 (Student's t-test). c and d. Knockdown of S1PR3 suppressed proliferation capability of MNNG-HOS and U-2OS cells using the CCK-8 assay. Values are means ± SD, **p < .01, ***p < .001 (Student's t-test). e. Knockdown of S1PR3 suppressed proliferation of OS cells (MNNG-HOS and U-2OS) using colony formation assay. Representative photographs of the colony formation assay were shown in the left panel. Scale bars = 5 mm. Values are means ± SD, **p < .01, ***p < .001 (Student's t-test). f and g. Knockdown of S1PR3 significantly induces apoptosis of MNNG-HOS and U-2OS cells. Values are means ± SD, ***p < .001 (Student's t-test).
Supplementary Fig. 2.
a. Interference efficacy of sh-RNA targeting of S1PR3 in MNNG-HOS and U-2OS cells was determined by western blotting.
Fig. 3.
S1PR3 facilitates OS proliferation in vivo and promotes the aerobic glycolysis in OS cells. a. Morphologic characteristics of xenograft tumors from MNNG-HOS/sh-Control group and MNNG-HOS/sh-S1PR3 group (n = 5). Scale bars = 1 cm. b. The tumor volumes were measured with calipers every 5 days. Values are means ± SD, *p < .05 (Student's t-test). c. Tumor weights in 20 day were measured in each group. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. n = 5, **p < .01 (Student's t-test). d. Tumor volumes in 20 day were measured in each group. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. n = 5, *p < .05 (Student's t-test). e. Representative images of Ki67 and TUNEL staining in xenograft tumors from sh-S1PR3 and sh-Control mice. A TUNEL positive cell is indicated (arrow). f. Representative GO (GO: biological process) categories affected by S1PR3 knock-down in MNNG-HOS cells. g. Gene set enrichment analysis (GSEA) using hallmark gene sets was performed to compare the MNNG-HOS sh-NC group and sh-S1PR3 group. NES, normalized enrichment score. h and i. Extracellular acidification rate (ECAR) and O2 consumption rate (OCR) of MNNG-HOS and U-2OS cells in sh-Control and sh-S1PR3 group was detected. O: Oligomycin, F: FCCP, A&R: antimycin A/rotenone, Glc: glucose, Oligo: oligomycin, 2-DG: 2-deoxy-d-glucose. Values are means ± SD.
3.3. S1P/S1PR3 axis promotes the aerobic glycolysis in OS cells
To further explore the role of S1PR3 in OS, we set enrichment analysis (GSEA) based on the results of RNA sequencing for gene. Our data implied that genes involved in the glycolysis pathway were particularly enriched in the control MNNG-HOS group, suggesting the potential regulatory roles of S1PR3 in glucose metabolism (Fig. 3g). The Warburg effect was the phenomenon in which cancer cells predominantly generate energy through aerobic glycolysis rather than by oxidative phosphorylation (OXPHOS) [23]. It had been widely approbated that the Warburg effect leads to the promotion of survival and apoptosis inhibition in tumors [24]. We then explored if S1P/S1PR3 axis regulated the aerobic glycolysis in OS. As shown in Fig. 3h and i, we observed a reduction in the glycolytic rate (ECAR) following the S1PR3-shRNA treatment of MNNG-HOS and U-2OS cells. In contrast, the mitochondrial respiration (OCR) increased when S1PR3 was depleted in OS cells (Fig. 3h and i). Taken together, we suggested that the S1P/S1PR3 axis devoted to the activation of the Warburg effect in OS cells.
3.4. S1P/S1PR3 inhibits the Hippo pathway by promoting nuclear translocation of YAP
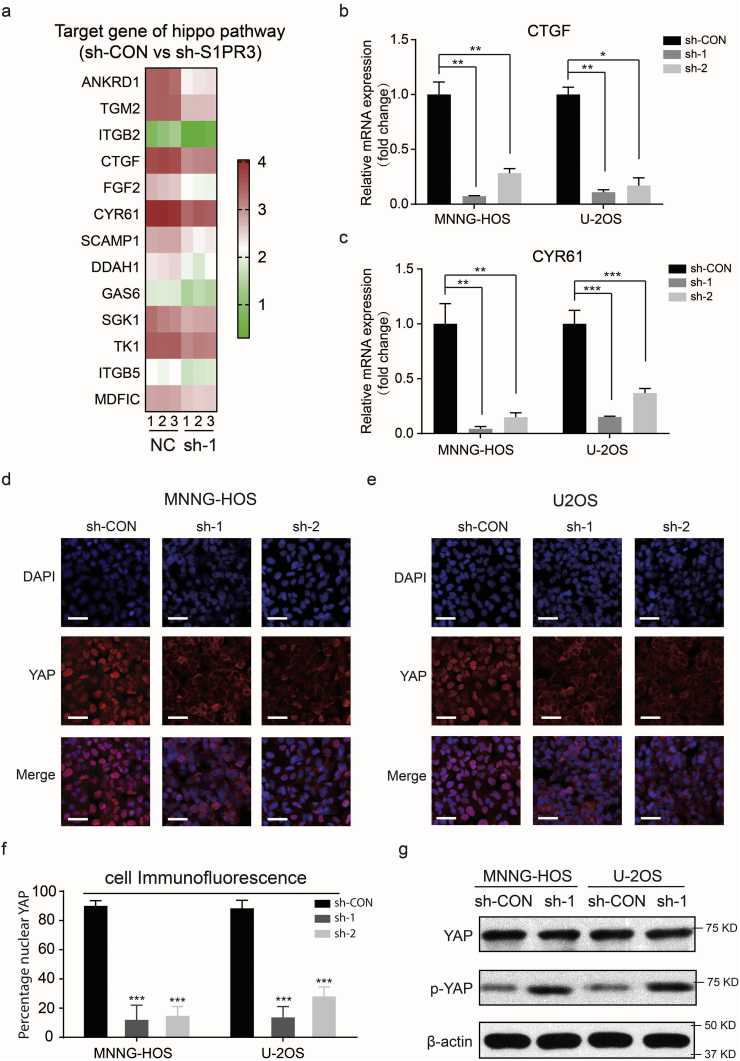
In mammalian Hippo pathway, YAP is inactivated by Large Tumor Suppressor (LATS) through retaining their cytoplasmic localization and protein degradation. Otherwise, when the Hippo pathway is inactive, the dephosphorylated YAP will enter the nucleus and interact with some key transcription factors to promote the transcription of some genes that involved in cell proliferation, apoptosis and reprogramming of glucose metabolism [[25], [26], [27], [28]]. We next asked whether S1PR3 could devote to the aerobic glycolysis in OS through the regulation of YAP. Comparison analysis of results of RNA sequencing exhibited that a large number of the YAP target genes, which were evolutionarily conserved and had been summarized by Cordenonsi M et al. [29], were downregulated in S1PR3-knockdown cells (Fig. 4a). The mRNA expression level of YAP typical target genes, CTGF (Connective Tissue Growth Factor) and CYR61 (Cysteine Rich Angiogenic Inducer 61), were significantly decreased in S1PR3-knockdown cells, which was in line with the RNA sequencing results (Fig. 4b and c). Furthermore, the results of immunofluorescence (IF) staining revealed that YAP localized mainly in the nucleus of sh-Control OS cells but in cytoplasm of sh-S1PR3 OS cells at middle cell density (Figs. 4d–f). Moreover, in both MNNG-HOS and U-2OS cells, S1PR3 suppressed the Hippo pathway activation based on reduced p-YAP levels (Fig. 4g). Above results indicated that the S1P/S1PR3 axis inhibits the Hippo pathway by suppressing the phosphorylation of YAP and promoting nuclear translocation of YAP.
Fig. 4.
S1PR3 suppresses the Hippo pathway in OS cells. a. A heat map showing the expression of the YAP target genes across MNNG-HOS NC samples and S1PR3 knockdown with shRNA samples. b and c. Relative mRNA levels of YAP target genes (CTGF and CYR61) in S1PR3-knockdown or sh-Control MNNG-HOS and U-2OS cells. Values are means ± SD, *p < .05, **p < .01, ***p < .001 (Student's t-test). d and e. Immunofluorescence of YAP in sh-Control and sh-S1PR3 MNNG-HOS and U-2OS cells. YAP is shown by red fluorescence, and the cell nuclei were stained with DAPI (blue fluorescence), Scale bars = 100 μm. f. Analysis of the percentage of cells with predominantly nuclear YAP. n = 5 randomly chosen fields. Values are means ± SD, ***p < .001 (Student's t-test). g. The level of phospho-YAP and total-YAP in sh-Control versus sh-S1PR3 MNNG-HOS and U-2OS cells. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.5. YAP is responsible for S1PR3-mediated cell growth and the Warburg effect
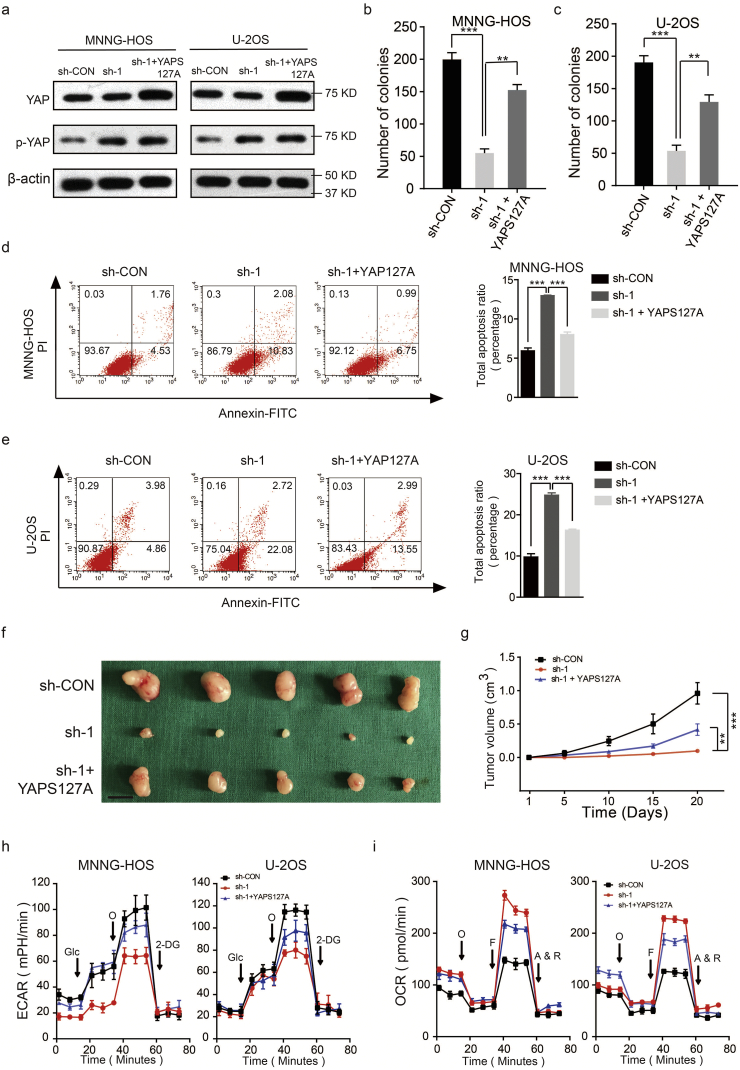
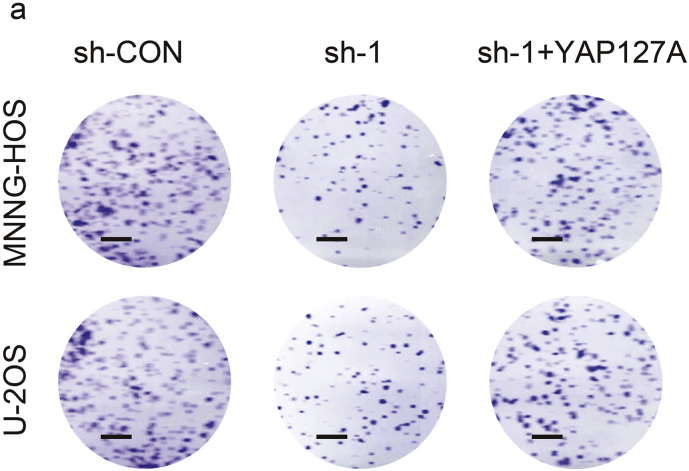
We further assessed whether the overexpression of YAP could reversed the inhibition of S1PR3 knockdown on the pro-growth and the Warburg effect of OS cells. We first overexpressed YAPS127A (constitutively active YAP that cannot be phosphorylated by LATS kinases) in wild-type and S1PR3 knockdown OS cell lines (MNNG-HOS and U-2OS). The western blot was used to confirm the efficiency of overexpression (Fig. 5a). As shown in Fig. 5b–e and Supplementary Fig. 3a, overexpression of YAPS127A partly reversed the suppressed effects of S1PR3 knockdown on the pro-growth properties of OS cells. In the in vivo experiment, the anti-tumorigenic effect on S1PR3-knockdown group was partly reversed by YAPS127A overexpression (Fig. 5f and g). Consistently, YAPS127A overexpression also partly restored the Warburg effect in MNNG-HOS and U-2OS (Fig. 5h and i). These results validated that S1PR3 promotes OS growth and the Warburg effect by inhibiting the Hippo/YAP pathway.
Fig. 5.
Overexpression of YAP partly reverses the effect of S1PR3 knockdown in OS. a. Overexpression efficacy of YAPS127A (constitutively active YAP that cannot be phosphorylated by LATS kinases) in sh-S1PR3 OS cells (MNNG-HOS and U-2OS) was detected by western blotting. b and c. Overexpression of YAP partly reversed the suppressed effects of S1PR3-knockdown on the colony formation capability of MNNG-HOS and U-2OS cells, values are means ± SD, **p < .01, ***p < .001 (Student's t-test). d and e. Overexpression of YAP partly reversed the induce effect of S1PR3-knockdown on the apoptosis of OS cells (MNNG-HOS and U-2OS). Values are means ± SD, ***p < .001 (Student's t-test). f. Morphologic characteristics of xenograft tumors from MNNG-HOS/sh-Control group, MNNG-HOS/sh-S1PR3 group and MNNG-HOS/sh-S1PR3 + YAPS127A group (n = 5). Scale bars = 1 cm. g. Overexpression of YAP partly rescued the inhibitory effects of S1PR3-knockdown on the growth rate of MNNG-HOS cells in vivo. The volumes of tumors were measured every 5 days, values are means ± SD, **p < .01, ***p < .001 (Student's t-test). h. The ECAR in OS cells (MNNG-HOS and U-2OS) in different groups (sh-Control, sh-S1PR3 and sh-S1PR3 + ov-YAP) were determined. Values are means ± SD. i. The OCR in OS cells (MNNG-HOS and U-2OS) in different groups (sh-Control, sh-S1PR3 and sh-S1PR3 + ov-YAP) were determined. Values are means ± SD.
Supplementary Fig. 3.
a. Representative images of OS colony formation assay in sh-Control, sh-S1PR3 and sh-S1PR3 + ov-YAP groups are shown. Scale bars = 5 mm.
3.6. S1PR3 regulates the expression of PGAM1 via the YAP/c-YMC complex in OS cells
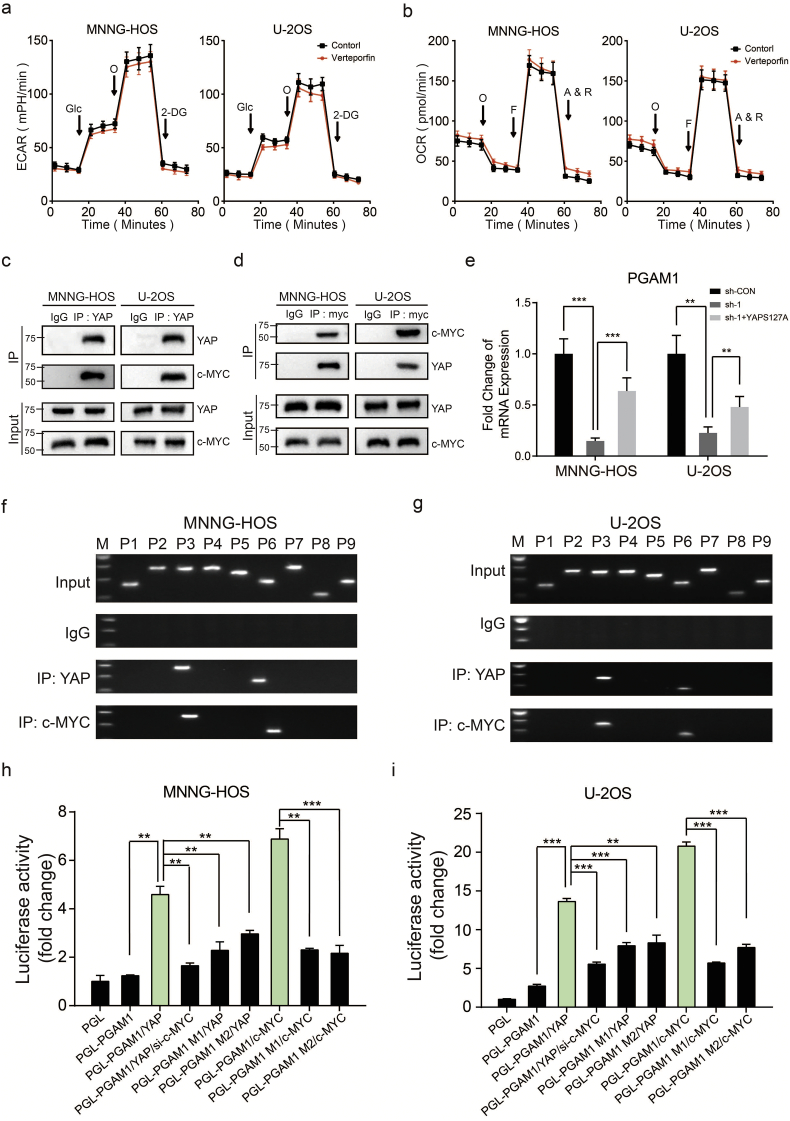
Due to the lack of a DNA-binding domain in YAP, it requires for the interactions with other transcriptional factors for recruitment to chromatin, especially the TEAD family members [30]. We first tested whether the combination of YAP and TEAD had an influence on the aerobic glycolysis in OS cells. However, administration of Verteporfin (2 μM), a specific inhibitor targeting YAP-TEAD association, exerted little inhibitory impact on aerobic glycolysis of OS cells (Fig. 6a and b). Many studies have illustrated that c-MYC was concerned with modulating key enzymes involved in aerobic glycolysis as well as important processes demanding for the Warburg effect [31]. We then explored if S1PR3/YAP axis regulated Warburg effect in OS through regulating the expression or function of c-MYC. As exhibited in Supplementary Fig. 4a, compared with the control group, the expression of c-MYC was not significantly altered by the depletion of S1PR3 or overexpression of YAP in OS cells. Notably, as shown in Fig. 6c and d, the interaction between YAP and c-MYC were uncovered in both MNNG-HOS and U-2OS cells. According to the above results, we speculated that S1PR3-mediated Warburg effect in OS cells might depend on YAP mediated promotion of transcriptional activity on c-MYC.
Fig. 6.
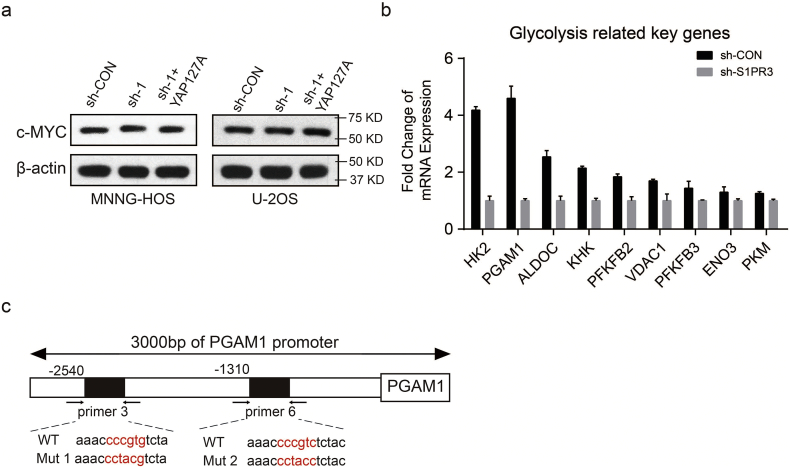
Interplay between S1PR3, YAP, c-MYC and PGAM1 contributes to Warburg effect in OS. a and b. Altered level of ECAR and OCR in OS cells (MNNG-HOS and U-2OS) with treatment of Verteporfin (2 μM) or DMSO. Values are means ± SD. c and d. Interactions between c-MYC and YAP demonstrated by CO-IP assay. The input was performed as control. e. Relative mRNA levels of PGAM1 in OS cells (MNNG-HOS and U-2OS) in different groups (sh-Control, sh-S1PR3 and sh-S1PR3 + ov-YAP). Values are means ± SD, **p < .01, ***p < .001 (Student's t-test). f and g. A ChIP assay was used to verify the potential c-MYC and YAP co-binding site in the PGAM1 promoter region in MNNG-HOS and U-2OS cell lines. Input fractions and IgG were used as controls. h and i. Luciferase activities of YAP-overexpressing, c-MYC-overexpressing, YAP-overexpressing and c-MYC-knockdown or control OS cells (MNNG-HOS and U-2OS) in luciferase reporter plasmid containing wild-type and mutant 1(M1) or mutant 2 (M2) PGAM1 promoter. The data shown are mean ± SD, **p < .01, ***p < .001 (Student's t-test).
Supplementary Fig. 4.
a. The level of c-MYC in sh-Control, sh-S1PR3 and sh-S1PR3 + ov-YAP groups are shown. b. mRNA expression of the key glycolytic genes were determined by qRT–PCR in S1PR3-knockdown or sh-Control MNNG-HOS. c. The mutated sequences of the PGAM1 promotor used for luciferase activities of OS cells (mutation site: red).
To further explore the target genes that co-regulated by YAP and c-MYC, we first compared the mRNA level of glycolytic genes between control group and S1PR3-knockdown group in MNNG-HOS cell through RNA sequencing. As revealed in Supplementary Fig. 4b, knockdown of S1PR3 significantly suppressed mRNA expression of several key glycolytic genes in MNNG-HOS cells, especially Phosphoglycerate mutase-1 (PGAM1) (Supplementary Fig. 4b). PGAM1, a glycolytic gene, catalyzing the conversion of 3-phosphoglycerate to 2-phosphoglycerate, which promotes a metabolic switch toward glycolysis to support cell proliferation and survival [32]. Accordingly, targeting PGAM1 appears to predict a promising strategy for some tumors [[33], [34], [35]]. Furthermore, downregulation of PGAM1 in S1PR3 knockdown MNNG-HOS cells could be restored by overexpression of YAPS127A, suggesting that PGAM1 might be a potential target gene of YAP-c-MYC (Fig. 6e). Hence, a ChIP-PCR assay was performed to verify our hypothesis. As shown in Fig 4f and g, we displayed that the PGAM1 promoter was co-occupied by YAP and c-MYC in several regions flanked by primers 3 and 6 in MNNG-HOS and U-2OS cells (Fig. 6f and g). Furthermore, the wild-type PGAM1 promoter but not mutant constructs could be activated by YAP or c-MYC overexpression. (Fig. 6h, i and Supplementary Fig. 4c). In addition, YAP-activated PGAM1 transcription was obviously suppressed by c-MYC knockdown (Fig. 6h and i). These results indicated that S1PR3 regulated the expression of PGAM1 via the YAP/c-YMC complex in OS cells.
3.7. Specific antagonist of S1PR3 combined with methotrexate suppresses the growth of OS cells in vitro and in vivo
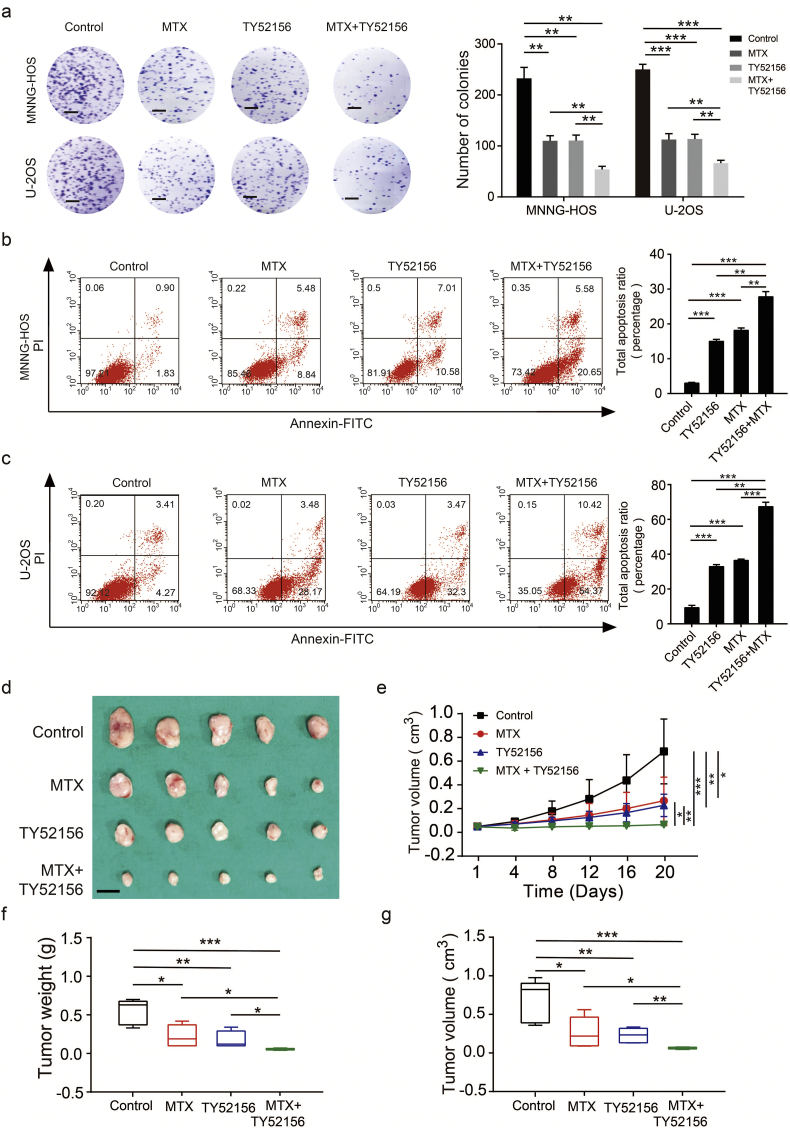
Methotrexate (MTX), a deoxycytidine analog that inhibits DNA replication and thus prevents tumor growth, is a widely used chemotherapy for OS. TY52156 is an antagonist of S1PR3 and has a high degree of selectivity for the S1PR3 receptor [36,37]. Next, we evaluated whether the function of MTX on the anti-tumorigenic ability of OS could be enhanced by TY52156. Treatment of MTX or TY52156 inhibited the proliferation of OS cells and facilitated their apoptosis in vitro. Furthermore, the combination of MTX and TY52156 could enhance these functions (Fig. 7a–c). In an in vivo assay, both MTX (50 mg/kg) and TY52156 (10 mg/kg) were treated every 4 days to implanted mice, which resulted in smaller tumors compared to the MTX or TY52156 only treatment groups (Fig. 7d–g). Collectively, these data indicate that the combination of MTX and TY52156 may provide an additional treatment benefit.
Fig. 7.
Targeting S1PR3 combined with methotrexate suppresses the growth of OS cells in vitro and in vivo. a. Colony formation assays using MNNG-HOS and U-2OS cells treat with DMSO, TY52156 (10 μM), methotrexate (MTX) (1 μM) or combination of TY52156 and MTX. Representative photographs of the colony formation assay were shown in the left panel. Scale bars = 5 mm. Values are means ± SD, **p < .01, ***p < .001 (Student's t-test). b and c. Cell apoptosis assay using MNNG-HOS and U-2OS cells treat with DMSO, TY52156 (10 μM), methotrexate (MTX) (1 μM) or combination of TY52156 and MTX. Representative images of apoptosis assay were shown in the left panel. Values are means ± SD, **p < .01, ***p < .001 (Student's t-test). d. Subcutaneous xenografts transplanted with MNNG-HOS cells treat with 0.9% NaCl, TY52156 (3 mg/kg), methotrexate (MTX) (5 mg/kg) or combination of TY52156 and MTX every 4 days. e. Tumor volumes were measured with calipers every 4 days. n = 5, *p < .05, **p < .01, ***p < .001 (Student's t-test). f. Tumor weights in 20 day were measured in each group. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. n = 5, *p < .05, **p < .01 (Student's t-test). g. Tumor volumes in 20 day were measured in each group. The median, upper and lower quartiles were plotted, and the whiskers that extend from each box indicate the range values that were outside of the intra-quartile range. n = 5, *p < .05, **p < .01, ***p < .001 (Student's t-test).
4. Discussion
Recent reports have discovered the important roles of S1PRs family in the modulation of tumor growth, tumorigenesis and metastasis. The elevation of S1PR1 in T lymphoblastic lymphoma cells increased the expression of intercellular adhesion molecule 1 (ICAM1) and augmented cell-cell adhesion, which was concerned with the blockade of intravasation and hematological dissemination [38]. In cancer cells, S1PR2 induced acute myelogenous leukemia growth and was demonstrated to activate ezrin–radixin–moesin (ERM) proteins to promote movement and invasion of HeLa cells in culture [[39], [40], [41]]. A clinical study has reported that increased S1PR4 expression was linked with lower disease-free survival rate in a sample of 140 patients with estrogen receptor (ER)-negative breast cancer [42]. A recent study revealed that S1P/S1PR5 axis promoted mitotic progression in HeLa cells, generating chromosome segregation defects via S1PR5-dependent activation of the PI3K-AKT pathway [43]. S1PR3 also has been reported to have cancer-promoting properties and correlated with poor prognosis in intestinal tumors [17,37,[44], [45], [46]]. The levels of S1PR3 were upregulated in human lung adenocarcinomas and promotes the progression of human lung adenocarcinomas [46].The S1P/S1PR3 axis regulates cancer stemness by activating Notch signaling [37]. However, the expression and function of the S1P/S1PR3 axis in OS has not been studied. Interestingly, S1PR3 was necessary to promote bone formation in response to S1P, indicating that dysregulation of S1P or S1PR3 might bring about development of OS [47].We demonstrate that S1P and S1PR3 were both upregulated in OS; inhibiting S1P/S1PR3 axis by silencing of S1PR3, which produced an anti-tumor effect. In addition, a Kaplan–Meier analysis showed that poor overall survival and metastasis-free survival rates were associated with higher expression of S1PR3 in OS patients.
Increased aerobic glycolysis (the Warburg effect) emerges as an enlightening hallmark for cancers. This metabolic shift promotes cancer cell proliferation and survival, particularly by supplying more intermediates for some biosynthetic pathways and an adaptation to hypoxic conditions [[48], [49], [50]]. Recently, our team also reported that the Warburg effect existed in OS and promoted its proliferation [19]. However, the implication of the Warburg effect in the progression of OS remains to be investigated. In this study, we identified an S1PR3-dependent regulatory network involving YAP, c-MYC, and modulation the Warburg effect during the growth of OS.
The Hippo pathway participates in controlling organ size, and its dysregulation contributes to the tumorigenesis. Previous studies indicated that the Hippo-YAP pathway was a critical signaling branch downstream of GPCR. Yu et al. showed that S1P acted through G12/13-coupled receptors to inhibit the Hippo pathway by activating YAP and TAZ transcription coactivators [51]. The Hippo pathway also played an important role in regulating the energy metabolism of cancer cells [[52], [53], [54]]. However, the signaling connection between them is still largely unknown, especially in OS. In this study, we demonstrated that S1P/S1PR3-mediated YAP nuclear translocation promoted the aerobic glycolysis in OS cells. Knockdown of the S1PR3 gene in OS cells recapitulates growth inhibition, resulting in deficits in aerobic glycolysis and YAP cytoplasm translocation. Functionally, our study indicated that YAP could combine with c-MYC in the OS cells. Furthermore, we identified PGAM1, a key regulator in glycolysis, as a direct target gene of S1P/S1PR3 signaling.
In this study, we established a subcutaneous xenograft mouse model to evaluate the functions of S1PR3 in vivo. Patient-derived tumor xenograft (PDX) model and orthotopic xenograft model would be generated in future studies to further explore the possibility of S1PR3 as an effective target for the treatment of osteosarcoma. Furthermore, considering the low incidence of OS, it was difficult for us to make a more thorough evaluation of clinical significance of the S1PR3 in a large cohort.
In summary, our studies gave a new enlightenment into the molecular mechanisms for the glucose metabolism reprogram in the development of OS. Our results highlighted a previously unknown role for S1P/S1PR3 axis, which shed light on how S1P/S1PR3 axis initiated the Warburg effect by activating the YAP/c-MYC/PGAM1 pathway. Furthermore, our data demonstrated that S1PR3 specific antagonist TY52156 had synergistic inhibitory effects with methotrexate on OS cell growth, which also provided new ideas for clinical treatment of osteosarcoma. Thus, targeting S1P/S1PR3 axis might constitute a new approach for OS treatment.
The following are the supplementary data related to this article.
Primer sequences.
Funding
This work was supported by National Natural Science Foundation of China (Nos. 81472445 and 81672587).
Author contributions
D.Z., J.L., R.Z. and Y.F.S. were responsible for the concept and experimental design. Y.F.S., S.J.Z., S.Y.W., X.H.P., Q.L. and L.P.H. performed the experiments, data analysis and statistical analysis. Y.Q.J., Q.Z., Y.K.Z., Y.Z., J.W.X, J.B.G., H.B.L., S.H.J., H.L.Y. and R.Z. provided technical and material support. Y.F.S., S.J.Z., S.Y.W. and X.H.P. were involved in drafting and revision of the manuscript. R.Z., J.L. and D.Z. supervised this study. All authors discussed the results and commented on the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Rong Zhang, Email: Rongzhang@163.com.
Jun Li, Email: Junli@shsci.org.
Dong Zhou, Email: zhoudong1012@163.com.
References
- 1.Bonuccelli G., Avnet S., Grisendi G., Salerno M., Granchi D., Dominici M. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget. 2014;5(17):7575–7588. doi: 10.18632/oncotarget.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moriarity B.S., Otto G.M., Rahrmann E.P., Rathe S.K., Wolf N.K., Weg M.T. A Sleeping Beauty forward genetic screen identifies new genes and pathways driving osteosarcoma development and metastasis. Nat Genet. 2015;47(6):615–624. doi: 10.1038/ng.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Group ESESNW Bone sarcomas: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl. 3):iii113–23. doi: 10.1093/annonc/mdu256. [DOI] [PubMed] [Google Scholar]
- 4.Savage S.A., Mirabello L. Using epidemiology and genomics to understand osteosarcoma etiology. Sarcoma. 2011;2011:548151. doi: 10.1155/2011/548151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 6.Nieto Gutierrez A., McDonald P.H. GPCRs: emerging anti-cancer drug targets. Cell Signal. 2018;41:65–74. doi: 10.1016/j.cellsig.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Lynch J.R., Wang J.Y. G protein-coupled receptor signaling in stem cells and cancer. Int J Mol Sci. 2016;17(5) doi: 10.3390/ijms17050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sever R., Brugge J.S. Signal transduction in cancer. Cold Spring Harb Perspect Med. 2015;5(4) doi: 10.1101/cshperspect.a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takabe K., Paugh S.W., Milstien S., Spiegel S. “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev. 2008;60(2):181–195. doi: 10.1124/pr.107.07113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yester J.W., Tizazu E., Harikumar K.B., Kordula T. Extracellular and intracellular sphingosine-1-phosphate in cancer. Cancer Metastasis Rev. 2011;30(3–4):577–597. doi: 10.1007/s10555-011-9305-0. [DOI] [PubMed] [Google Scholar]
- 11.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18(1):33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendoza A., Fang V., Chen C., Serasinghe M., Verma A., Muller J. Lymphatic endothelial S1P promotes mitochondrial function and survival in naive T cells. Nature. 2017;546(7656):158–161. doi: 10.1038/nature22352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai L., Liu Y., Xie L., Wu X., Qiu L., Di W. Sphingosine kinase 1/sphingosine-1-phosphate (S1P)/S1P receptor axis is involved in ovarian cancer angiogenesis. Oncotarget. 2017;8(43):74947–74961. doi: 10.18632/oncotarget.20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson C., Long J.S., Orange C., Tannahill C.L., Mallon E., McGlynn L.M. High expression of sphingosine 1-phosphate receptors, S1P1 and S1P3, sphingosine kinase 1, and extracellular signal-regulated kinase-1/2 is associated with development of tamoxifen resistance in estrogen receptor-positive breast cancer patients. Am J Pathol. 2010;177(5):2205–2215. doi: 10.2353/ajpath.2010.100220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E.S., Kim J.S., Kim S.G., Hwang S., Lee C.H., Moon A. Sphingosine 1-phosphate regulates matrix metalloproteinase-9 expression and breast cell invasion through S1P3-Galphaq coupling. J Cell Sci. 2011;124(Pt 13):2220–2230. doi: 10.1242/jcs.076794. [DOI] [PubMed] [Google Scholar]
- 16.Magrassi L., Marziliano N., Inzani F., Cassini P., Chiaranda I., Skrap M. EDG3 and SHC3 on chromosome 9q22 are co-amplified in human ependymomas. Cancer Lett. 2010;290(1):36–42. doi: 10.1016/j.canlet.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 17.Lee H.M., Lo K.W., Wei W., Tsao S.W., Chung G.T.Y., Ibrahim M.H. Oncogenic S1P signalling in EBV-associated nasopharyngeal carcinoma activates AKT and promotes cell migration through S1P receptor 3. J Pathol. 2017;242(1):62–72. doi: 10.1002/path.4879. [DOI] [PubMed] [Google Scholar]
- 18.Weng Y., Shen Y., He Y., Pan X., Xu J., Jiang Y. The miR-15b-5p/PDK4 axis regulates osteosarcoma proliferation through modulation of the Warburg effect. Biochem Biophys Res Commun. 2018;503(4):2749–2757. doi: 10.1016/j.bbrc.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 19.Zhao S.J., Shen Y.F., Li Q., He Y.J., Zhang Y.K., Hu L.P. SLIT2/ROBO1 axis contributes to the Warburg effect in osteosarcoma through activation of SRC/ERK/c-MYC/PFKFB2 pathway. Cell Death Dis. 2018;9(3):390. doi: 10.1038/s41419-018-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S.J., Jiang Y.Q., Xu N.W., Li Q., Zhang Q., Wang S.Y. SPARCL1 suppresses osteosarcoma metastasis and recruits macrophages by activation of canonical WNT/beta-catenin signaling through stabilization of the WNT-receptor complex. Oncogene. 2018;37(8):1049–1061. doi: 10.1038/onc.2017.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X.M., Cao X.Y., He P., Li J., Feng M.X., Zhang Y.L. Overexpression of Rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology. 2018;155(4):1233–1249. doi: 10.1053/j.gastro.2018.07.010. [e1222] [DOI] [PubMed] [Google Scholar]
- 22.Xu D., Zhu H., Wang C., Zhao W., Liu G., Bao G. SphK2 over-expression promotes osteosarcoma cell growth. Oncotarget. 2017;8(62):105525–105535. doi: 10.18632/oncotarget.22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koppenol W.H., Bounds P.L., Dang C.V. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 24.Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5 doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Z., He F., Chen M., Hua L., Wang W., Jiao S. DNA-binding mechanism of the Hippo pathway transcription factor TEAD4. Oncogene. 2017;36(30):4362–4369. doi: 10.1038/onc.2017.24. [DOI] [PubMed] [Google Scholar]
- 26.Yu F.X., Zhao B., Guan K.L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163(4):811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enzo E., Santinon G., Pocaterra A., Aragona M., Bresolin S., Forcato M. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. EMBO J. 2015;34(10):1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deran M., Yang J., Shen C.H., Peters E.C., Fitamant J., Chan P. Energy stress regulates hippo-YAP signaling involving AMPK-mediated regulation of angiomotin-like 1 protein. Cell Rep. 2014;9(2):495–503. doi: 10.1016/j.celrep.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordenonsi M., Zanconato F., Azzolin L., Forcato M., Rosato A., Frasson C. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147(4):759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 30.Pobbati A.V., Hong W. Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther. 2013;14(5):390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang S.H., Li J., Dong F.Y., Yang J.Y., Liu D.J., Yang X.M. Increased serotonin signaling contributes to the warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology. 2017;153(1):277–291. doi: 10.1053/j.gastro.2017.03.008. [e219] [DOI] [PubMed] [Google Scholar]
- 32.Jiang X., Sun Q., Li H., Li K., Ren X. The role of phosphoglycerate mutase 1 in tumor aerobic glycolysis and its potential therapeutic implications. Int J Cancer. 2014;135(9):1991–1996. doi: 10.1002/ijc.28637. [DOI] [PubMed] [Google Scholar]
- 33.Sun Q., Li S., Wang Y., Peng H., Zhang X., Zheng Y. Phosphoglyceric acid mutase-1 contributes to oncogenic mTOR-mediated tumor growth and confers non-small cell lung cancer patients with poor prognosis. Cell Death Differ. 2018;25(6):1160–1173. doi: 10.1038/s41418-017-0034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang D., Jin N., Sun W., Li X., Liu B., Xie Z. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene. 2017;36(20):2900–2909. doi: 10.1038/onc.2016.446. [DOI] [PubMed] [Google Scholar]
- 35.Hitosugi T., Zhou L., Fan J., Elf S., Zhang L., Xie J. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murakami A., Takasugi H., Ohnuma S., Koide Y., Sakurai A., Takeda S. Sphingosine 1-phosphate (S1P) regulates vascular contraction via S1P3 receptor: investigation based on a new S1P3 receptor antagonist. Mol Pharmacol. 2010;77(4):704–713. doi: 10.1124/mol.109.061481. [DOI] [PubMed] [Google Scholar]
- 37.Hirata N., Yamada S., Shoda T., Kurihara M., Sekino Y., Kanda Y. Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nat Commun. 2014;5:4806. doi: 10.1038/ncomms5806. [DOI] [PubMed] [Google Scholar]
- 38.Feng H., Stachura D.L., White R.M., Gutierrez A., Zhang L., Sanda T. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell. 2010;18(4):353–366. doi: 10.1016/j.ccr.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell J.A., Lewis A.C., Zhu W., Toubia J., Pitman M.R., Wallington-Beddoe C.T. Targeting sphingosine kinase 1 induces MCL1-dependent cell death in acute myeloid leukemia. Blood. 2017;129(6):771–782. doi: 10.1182/blood-2016-06-720433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adada M.M., Canals D., Jeong N., Kelkar A.D., Hernandez-Corbacho M., Pulkoski-Gross M.J. Intracellular sphingosine kinase 2-derived sphingosine-1-phosphate mediates epidermal growth factor-induced ezrin-radixin-moesin phosphorylation and cancer cell invasion. FASEB J. 2015;29(11):4654–4669. doi: 10.1096/fj.15-274340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ponnusamy S., Selvam S.P., Mehrotra S., Kawamori T., Snider A.J., Obeid L.M. Communication between host organism and cancer cells is transduced by systemic sphingosine kinase 1/sphingosine 1-phosphate signalling to regulate tumour metastasis. EMBO Mol Med. 2012;4(8):761–775. doi: 10.1002/emmm.201200244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohotski J., Long J.S., Orange C., Elsberger B., Mallon E., Doughty J. Expression of sphingosine 1-phosphate receptor 4 and sphingosine kinase 1 is associated with outcome in oestrogen receptor negative breast cancer (vol 106, pg 1453, 2012) Br J Cancer. 2012;107(4):756. doi: 10.1038/bjc.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrieu G., Ledoux A., Branka S., Bocquet M., Gilhodes J., Walzer T. Sphingosine 1-phosphate signaling through its receptor S1P5 promotes chromosome segregation and mitotic progression. Sci Signal. 2017:10(472). doi: 10.1126/scisignal.aah4007. [DOI] [PubMed] [Google Scholar]
- 44.Wang H., Huang H., Ding S.F. Sphingosine-1-phosphate promotes the proliferation and attenuates apoptosis of Endothelial progenitor cells via S1PR1/S1PR3/PI3K/Akt pathway. Cell Biol Int. 2018;42(11):1492–1502. doi: 10.1002/cbin.10991. [DOI] [PubMed] [Google Scholar]
- 45.Wang S., Liang Y., Chang W., Hu B., Zhang Y. Triple negative breast cancer depends on sphingosine kinase 1 (SphK1)/sphingosine-1-phosphate (S1P)/sphingosine 1-phosphate receptor 3 (S1PR3)/notch signaling for metastasis. Med Sci Monit. 2018;24:1912–1923. doi: 10.12659/MSM.905833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J., Liu J., Lee J.F., Zhang W., Kandouz M., Vanhecke G.C. TGF-beta/SMAD3 pathway stimulates sphingosine-1 phosphate receptor 3 expression: implication of sphingosine-1 phosphate receptor 3 in lung adenocarcinoma progression. J Biol Chem. 2016;291(53):27343–27353. doi: 10.1074/jbc.M116.740084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meshcheryakova A., Mechtcheriakova D., Pietschmann P. Sphingosine 1-phosphate signaling in bone remodeling: multifaceted roles and therapeutic potential. Expert Opin Ther Targets. 2017;21(7):725–737. doi: 10.1080/14728222.2017.1332180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 49.Lu J., Tan M., Cai Q. The Warburg effect in tumor progression: mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015;356(2 Pt A):156–164. doi: 10.1016/j.canlet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altman B.J., Stine Z.E., Dang C.V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16(11):749. doi: 10.1038/nrc.2016.114. [DOI] [PubMed] [Google Scholar]
- 51.Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng X., Han H., Liu G.P., Ma Y.X., Pan R.L., Sang L.J. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017;36(22):3325–3335. doi: 10.15252/embj.201797609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen R., Zhu S., Fan X.G., Wang H., Lotze M.T., Zeh H.J., III High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology. 2018;67(5):1823–1841. doi: 10.1002/hep.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin C., Xu X. YAP1-TEAD1-Glut1 axis dictates the oncogenic phenotypes of breast cancer cells by modulating glycolysis. Biomed Pharmacother. 2017;95:789–794. doi: 10.1016/j.biopha.2017.08.091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer sequences.
Data Availability Statement
The data sets of MNNG-HOS cell line silenced of S1PR3, which support the findings of this study are available in the Sequence Read Archive (SRA) repository under accession NO.SRP165691. All other remaining data that support our findings are available from the authors upon reasonable request.