Abstract
We seek to examine the use of an image processing pipeline on Magnetic Resonance Imaging (MRI) to identify features of Focal Cortical Dysplasia (FCD) in children who were suspected to have FCD on MRI (MRI-positive) and those with MRI-negative epilepsy. We aim to use a computer-aided diagnosis system to identify epileptogenic lesions with a combination of established morphometric features and textural analysis using Gray-Level Co-occurrence Matrices (GLCM) on MRI sequences. We implemented a modified version of the 2-Step Bayesian classifier method to a paediatric cohort with medically intractable epilepsy with MRI-positive and MRI-negative epilepsy, and evaluated the performance of the algorithm trained on textural features derived from T1-weighted (T1-w), T2-weighted (T2-w), and FLAIR (Fluid Attenuated Inversion Recovery) sequences. For MRI-positive cases, T1-w has the highest subjectwise sensitivity relative to T2-w and FLAIR (94% vs. 90% vs. 71% respectively), and also the highest lesional sensitivity (63% vs. 60% vs. 42% respectively), but the lowest lesional specificity (75% vs. 80% vs. 89% respectively). Combination of all three sequences improved the performance of the algorithm, with 97% subjectwise sensitivity. For MRI-negative cases, T1-w has the highest subjectwise sensitivity relative to T2-w and FLAIR (48% vs. 30% vs. 39% respectively), and also the highest lesional sensitivity (31% vs. 22% vs. 28% respectively). However, T2-w has the highest lesional specificity relative to T1-w and FLAIR (95% vs. 94% vs. 92% respectively) for MRI-negative cases. Combination of all three sequences improved the performance of the algorithm, with 70% subjectwise sensitivity. The 2-Step Naïve Bayes classifier correctly rejected 100% of the healthy subjects for all three sequences. Using combined morphometric and textural analysis in a 2-Step Bayesian classifier, applied to multiple MRI sequences, can assist with lesion detection in children with intractable epilepsy.
Keywords: Computer-aided diagnosis, Epilepsy, Focal cortical dysplasia, Morphometric and textural analysis
1. Introduction
In children with medically refractory focal epilepsy, a lesion can be identified on MRI in 30–85% of cases (Duncan, 1997; Colombo et al., 2003). Focal Cortical Dysplasia (FCD) is a brain malformation that is frequently responsible for epilepsy in children and accounts for approximately 26% of intractable epilepsy in children (Wyllie et al., 1998). Unlike tumors, FCD can be subtle and difficult to detect using standard MRI. The MRI features of FCD include abnormal cortical thickness, abnormal sulcal pattern, blurring of the gray-white matter junction, and abnormal signal in the white and/or gray matter.
FCD is one of the most challenging groups of lesions to detect on conventional MRI as the imaging features may be subtle and not infrequently missed. Additionally, the commonly recognized features of FCD may not all be present in a given case. Some cases of FCD may exhibit one or two of the identifiable features, while others may exhibit more (Antel et al., 2003). For these reasons, limiting assessment to one feature could inevitably lead to false negative. Those with FCD and normal MRI are known to have MRI-negative epilepsy, or non-lesional epilepsy. Cases with visible lesions on MRI have MRI-positive epilepsy (Antel et al., 2003). Up to 72% of cases of MRI negative epilepsy have FCD (Lee et al., 2005; Jeha et al., 2007; McGonigal et al., 2007; Krsek et al., 2007).
In studies conducted with adult cohorts, FCD detection was improved with computational techniques, but they have not been as effective when applied to developing brains (Adler et al., 2017). For an adult cohort, Antel et al. (Antel et al., 2003) implemented a 2-Step Bayesian classifier in which attribute data was split into morphometric and textural features, and the classifier is first trained on morphometry then using features derived from Gray-Level Co-occurrence Matrices (GLCMs). A two-classifier approach is justified because the two groups of attributes extract different types of information from the MRI volume. Their method produced high values of subjectwise sensitivity (colocalized lesion in 15/18 of the FCD subjects) and subjectwise specificity (rejected all Control subjects). Antel et al. (Antel et al., 2003) have used volumetric T1-weighted (T1-w) imaging to evaluate morphometric and textural features. However, different MRI acquisitions vary in terms of their ability to recognize certain features of FCD. For instance, studies have indicated T2-weighted (T2-w), and Fluid Attenuated Inversion Recovery (FLAIR) images are better at identifying abnormal signal in the white matter compared to sequences acquired from T1-w (Focke et al., 2008). On the other hand, volumetric T1-w images, due to their thinner slices, are superior for examining variations in cortical thickness (De Cocker et al., 2012). Hence, using different MRI acquisitions for textural analysis may contribute to improving lesion identification.
The paper by Antel et al. (Antel et al., 2003) served as a key motivation in our experimental setup. This method was effective in identifying FCD in adult cases and we aim to apply a similar approach to paediatric cases. We implemented a modified version of this method to perform classification using Naive Bayes to create a 2-Step classifier applied to paediatric subjects and evaluate the performance of this algorithm on developing brains. Naive Bayes classification is a machine learning technique based on Bayes' Theorem, and assumes that the value of a particular feature is independent of the value of any other features, given the class variable. The algorithm is called “naive” because it makes a naive assumption that each feature is independent of other features. Although it is a relatively simple idea, Naive Bayes can often outperform other more sophisticated algorithms and is extremely useful in common image classification applications. The algorithm allows us to predict a class, given a set of features using probabilities. By the class conditional independence assumption, all predictor variables contribute independently to class labels and any dependence between input variables is disregarded; this process has the advantage of being less computationally expensive. We applied the 2-Step Naive Bayes Classifier trained on morphometric and textural features, not only on volumetric T1-w, but also on T2-w and FLAIR sequences. In this study, we explored the development of a unified mathematical model that considers segmentation, morphometric analysis, and textural analysis in a joint formulation for the purpose of identifying FCD in children with focal intractable epilepsy.
2. Material and methods
2.1. Subjects
Fifty-four children with focal intractable epilepsy and have undergone epilepsy surgery were included in the study. Thirty-one children have subtle lesion on MRI that was suspected to be FCD (MRI-positive) and 23 children have MRI-negative focal epilepsy. The mean age of the MRI-positive patients was 11.63 years (standard deviation (SD) 3.38 years, age range of 6.45–17.46 years), with 18 males and 13 females. The mean age of the MRI-negative patients was 11.64 years (SD 3.84 years, age range of 5.17–17.16 years), with 13 males and 10 females. Healthy controls consisted of 13 children (mean age 12.97 years, SD 3.20 years, range 7.02–17.11 years), 8 males and 5 females, without neurological or psychiatric diagnosis. The patient demographics of MRI positive and MRI negative patients are presented in Table 1 and Table 2 respectively. There were no significant differences in the age of patients versus controls subjects (p > .05).
Table 1.
Patient demographics for MRI-positive patients.
| Age | Sex | Location of lesion | ILAE surgical outcome | Histology |
|---|---|---|---|---|
| 12.99 | f | L superior temporal & middle temporal & temporal pole | II | Non-specific gliosis |
| 11.80 | m | R inferior temporal & fusiform | I | FCD type IIA |
| 9.48 | m | L superior frontal | I | FCD type IIA |
| 15.76 | m | L inferior temporal & fusiform | I | FCD type IIB |
| 7.55 | f | R precentral | I | Non-specific gliosis |
| 7.67 | m | L insula | I | FCD type IIB |
| 16.39 | m | L parsopercularis | I | Non-specific gliosis |
| 14.45 | f | L temporal pole & superior temporal & middle temporal | III | Oligodendrocytosis |
| 7.21 | f | L superior frontal | II | Non-specific gliosis |
| 13.32 | m | L parahippocampal & fusiform | I | Oligodendrocytosis |
| 14.74 | f | R lateral orbital frontal & medial orbital frontal | I | Oligodendrocytosis |
| 13.65 | f | R inferior parietal & lateral occipital | I | FCD type IIB |
| 10.26 | m | L temporal pole & superior temporal & middle temporal | III | Non-specific gliosis |
| 13.05 | m | L temporal pole & superior temporal & middle temporal | I | Oligodendrocytosis |
| 10.30 | f | L superior frontal & rostral middle frontal & caudal middle frontal & parsopercularis & parsorbitalis & parstriangularis & lateral orbitofrontal | I | Oligodendrocytosis |
| 12.12 | m | L temporal pole & superior temporal & middle temporal | I | Non-specific gliosis |
| 6.45 | m | R precentral & rostral middle frontal & caudal middle frontal | I | FCD type IIB |
| 6.72 | m | L superior frontal | I | FCD type IIA |
| 8.73 | m | R superior frontal & rostral middle frontal & caudal middle frontal & parsopercularis & partriangularis & parstriangularis & lateral orbitofrontal | III | FCD type I |
| 7.95 | f | L postcentral | I | FCD type IIB |
| 13.84 | m | L paracentral & posterior cingulate | I | Non-specific gliosis |
| 9.18 | f | R superior frontal & rostral middle frontal & caudal middle frontal & parsopercularis & parstriangularis & lateral orbitofrontal | I | Oligodendrocytosis |
| 7.27 | f | R Transverse temporal & superior temporal & middle temporal & inferior temporal & inferior parietal & lateral occipital | III | FCD type I |
| 6.99 | m | L transverse temporal & superior temporal & middle temporal & inferior temporal & fusiform & parahippocampal gyrus & lingual & pericalcarine & cuneus & lateral occipital | I | FCD type IIA |
| 14.82 | m | L temporal pole & superior temporal & middle temporal | V | FCD type IB |
| 17.46 | f | L temporal pole & superior temporal & middle temporal | IV | Oligodendrocytosis |
| 16.21 | m | L superior frontal & rostral middle frontal & caudal middle frontal & parsopercularis & parstriangularis & lateral orbitofrontal | I | FCD type I |
| 16.21 | f | R precuneus | V | FCD type I |
| 12.34 | f | R superior frontal & paracentral | I | FCD type IIA |
| 12.30 | m | L temporal pole & superior temporal & middle temporal & inferior temporal & transverse temporal | I | FCD type IIA |
| 13.35 | m | L precentral & postcentral & superior parietal & supramarginal | I | FCD type IIB |
Table 2.
Patient demographics for MRI-negative patients.
| Age | Sex | Location of surgical resection | ILAE Surgical outcome | Histology |
|---|---|---|---|---|
| 5.17 | m | R precentral, post central, R parsopercularis & parstriangularis, R middle & inferior temporal, | I | Non-specific gliosis |
| 15.03 | f | L postcentral | I | Non-specific gliosis |
| 9.39 | f | R superior frontal | I | No abnormality |
| 13.58 | m | L middle temporal, inferior temporal, L postcentral, superior parietal | I | FDC type I |
| 11.15 | f | R inferior temporal, R fusiform | I | Oligodendrocytosis |
| 13.89 | m | L superior frontal | I | Non-specific gliosis |
| 16.86 | f | L temporal pole, L superior temporal | IV | Heterotopic neurons |
| 15.60 | m | R temporal pole, R superior temporal, R middle temporal | I | Non-specific gliosis |
| 8.6 | m | R lateral occipital, R lingual | IV | FCD type I |
| 15.68 | m | L precentral | I | FCD type I |
| 13.06 | m | L superior frontal | I | FCD type IIA |
| 12.18 | f | L superior frontal and L paracentral | I | FCD type IIA |
| 9.51 | m | R inferior temporal | I | FCD type IIA |
| 10.14 | f | R precentral, postcentral, R caudal and rostral middle frontal, R parsopercularis, R superior temporal | I | FCD type I |
| 6.95 | m | L precentral | I | FCD type IIA |
| 7.26 | f | L caudal middle frontal gyrus, L precentral | I | Oligodendrocytosis |
| 5.25 | m | L superior frontal, L rostral middle frontal | I | Melanocytic cells in leptomeninges |
| 17.16 | f | R superior frontal, R caudal middle frontal, R rostral middle frontal | IV | FCD type I |
| 14.47 | f | R precentral, R postcentral, R superior parietal | V | Non-specific gliosis |
| 9.99 | m | L middle temporal | V | Non-specific gliosis |
| 16.89 | f | L precentral, L superior frontal, L paracentral | II | Non-specific gliosis |
| 6.46 | m | L superior frontal, L caudal middle frontal, L rostral middle frontal, L medial orbitofrontal, L lateral orbitofrontal, L pars orbitalis, L pars triangularis, L pars opercularis | III | Increased telangiectatic vessels in white matter |
| 13.46 | m | L temporal pole, L middle temporal gyrus | I | Oligodendrocytosis |
2.2. MRI
Patients and controls underwent MRI on Philips 3 T scanner (Philips Medical System, Best, Netherlands) using 8-channel head coil with the same imaging parameters. Patients underwent high-resolution epilepsy protocol, which included volumetric T1-w (TR/TE = 4.9/2.3 msec, ST = 0.9 mm, FOV = 22 cm, matrix = 220 × 220), axial and coronal FLAIR (TR/TE = 10,000/140, ST = 3 mm, FOV = 22 cm, matrix = 316 × 290) and axial and coronal proton density (PD)/T2-w (TR/TE = 4200/40/80, ST = 3 mm, FOV = 22 cm, matrix = 400 × 272). All patients underwent epilepsy surgery resection and had post-operative Computed tomography (CT) or volumetric T1-w MRI on the same scanner. All controls underwent volumetric T1-w, axial T2-w, and axial FLAIR imaging, using the same parameters as for the patients.
The MR images were reviewed by a neuroradiologist with expertise in epilepsy imaging, who identified the FCD. The location of the FCD was determined using anatomical labels from FreeSurfer. All the MRI identified FCD were concordant with the surgical resection site. In patients with MRI-negative epilepsy, the surgical resection location, as assessed on post-operative CT or MRI, was used as the reference standard.
2.3. Software
Research was conducted on an Ubuntu operating system using R (v3.4.1), MATLAB (R2017a) in combination with SPM12 (Statistical Parametric Mapping), and FreeSurfer software packages. FreeSurfer v5.3.0 (Dale et al., 1999; Dale and Sereno, 1993; Fischl and Dale, 2000; Fischl et al., 1999; Fischl et al., 2004) was employed for preprocessing T1-w images for all cases and controls in order to perform cortical reconstruction using Surface Based Morphometry (SBM), produce anatomical labelling and structural details. FreeSurfer can directly process T1-w acquisitions, as opposed to T2-w and FLAIR. In order to incorporate T2-w and FLAIR sequences into the analysis, the SPM package was used with MATLAB and FreeSurfer output for T2-w and FLAIR acquisitions to prepare the images for analysis.
2.4. Methods of measuring performance
To measure performance of the algorithm we will use the measures subjectwise sensitivity, subjectwise specificity, lesional sensitivity and lesional specificity. Subjectwise sensitivity is the sum of all epilepsy subjects in which a lesion is correctly identified (true positive) divided by the total number of epilepsy subjects. Subjectwise specificity is the sum of all control subjects in which no lesion is identified (true negative) divided by the total number of control subjects. A structure is considered lesional if it was specified in the surgical resection site on the post-operative MRI or CT for the subject. Lesional sensitivity is measured as the sum of all FreeSurfer defined cortical segments labeled lesional by the classifier for all subjects divided by the total segments labeled lesional for all subjects in pre-surgical reports in MRI-positive cases or surgical resection site in MRI-negative cases. Lesional specificity is the sum of all segments labeled non-lesional by the classifier for FCD subjects divided by the total segments labeled non-lesional for FCD subjects in pre-surgical reports in MRI-positive cases or surgical resection site in MRI-negative cases. We will consider lesional sensitivity and lesional specificity as our key performance measures, and use the subjectwise sensitivity and subjectwise specificity as supplementary information for analysis. Mathematically defining these measures, for NF number of FCD patients and NC number of healthy control subjects,
In these equations (TP)i is the true positive relating to the i-th subject. The other quantities are similarly defined. We are predominantly interested in subjectwise performance, that is to say, a technique's ability to not select any lesional structures in control cases and partially/wholly colocalize lesions in FCD patients. However the extent of the lesion can differ vastly from case to case. Fig. 1 depicts a case where the lesion was contained within a single Freesurfer defined cortical segment, however in most other cases it was contained with multiple segments. For this reason, we incorporate lesional measures as additional measures to overcome the subjectwise measures limitations in localization.
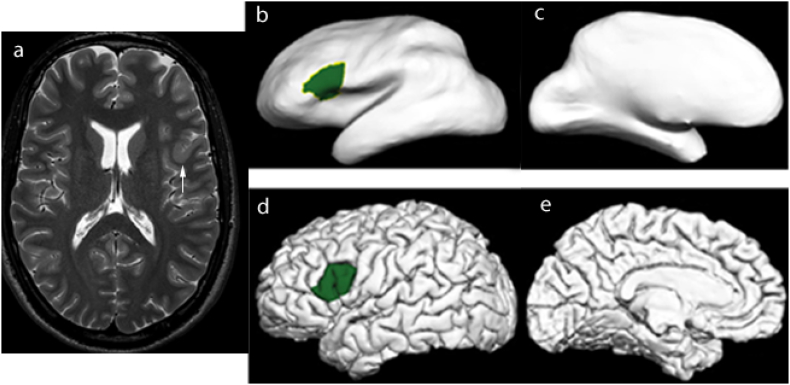
Fig. 1.
(a) There is thickening of the cortex in the left inferior frontal gyrus (arrow), in keeping with focal cortical dysplasia. Lesion labelling on (b and c) inflated and (d and e) pial surfaces.
2.5. Common features of T1-weighted MRI - cortical thickness and blurring of gray-white matter interface
Abnormal cortical thickness is one of the features of FCD (Lüsebrink et al., 2013). Cortical thickness measurements were acquired using the FreeSurfer software. The FreeSurfer constructs models to represent the pial surface and white matter surface with the surfaces generated in the form of triangular meshes. First, the shortest distance from the white matter surface to the pial surface was measured. Next, the distance from the pial surface to the white matter surface was computed. The average of the two values was taken to be the cortical thickness. Cortical thickness measures were saved as a vector of values, and the thickness vectors were sampled into a volume. To improve detection, a 3D-Gaussian smoothing filter (sigma = 1.9, filter size = 17) was applied to the cortical volume followed by a gamma correction.
Blurring at the gray-white matter interface is another MRI feature of FCD. To identify the location of the interface the FreeSurfer produced ribbon.mgz volumes was used. The ribbon.mgz volume is a binary mask of the cortical ribbon that maps the left/right hemisphere gray matter and white matter. To detect the interface, at each voxel the neighboring 8 voxels were examined. If at least 30% were contained in the gray matter and at least 30% were contained in the white matter, the voxel was considered to fall on the interface (Antel et al., 2003). A binary mask volume of the interface was composed for each case. To model the blur we employed the gradient magnitude, a measure of intensity change at each pixel of an image. Following this, gradient magnitude can be computed by . The gradient magnitude was computed for each case using the original volume following intensity normalization from FreeSurfer. The interface binary mask was used to segment the interface regions of the gradient magnitude. Blurring is indicated by regions with lower values of gradient magnitude.
2.6. Textural analysis
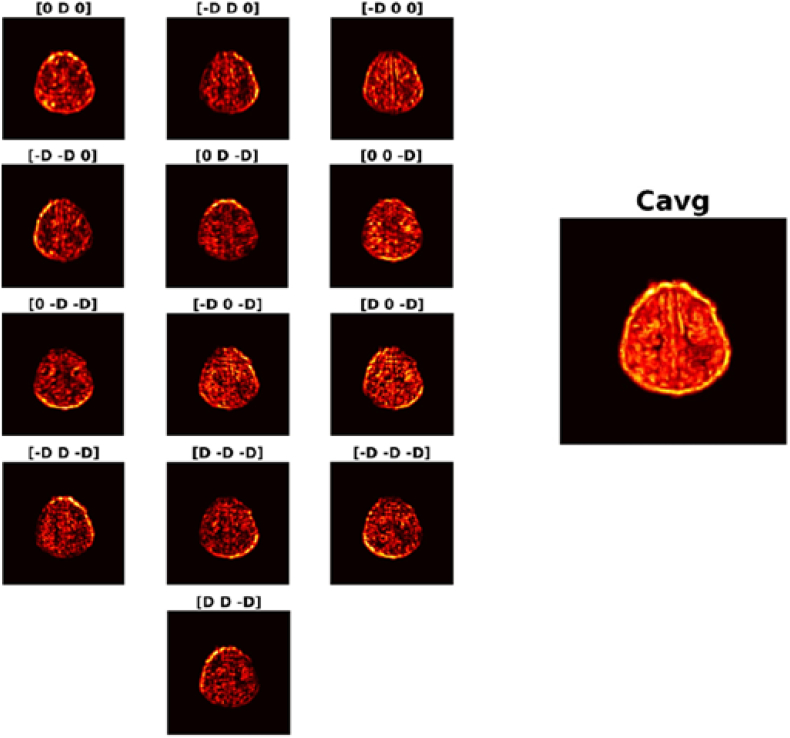
Quantitatively representing texture was performed using Gray-Level Co-occurrence Matrices (GLCM). Symmetric GLCMs were computed 3-dimensionally on T1-w acquisitions with the following parameters: a distance of 3 voxels, and an intensity range of 32 gray levels such that 13 co-occurrence matrices were produced per voxel. From each generated matrix 12 textural descriptor features were computed: Angular Second Momentum, Contrast, Entropy, Correlation, Homogeneity, Variance, Sum Mean, Inertia, Cluster Shade, Cluster Tendency, Max Probability, and Inverse Variance. For each feature, the average of the 13 was taken to be the value mapped back to the initial position. Fig. 2 depicts a sample axial slice using the 13 directions to produce the average contrast volume.
Fig. 2.
Contrast volumes using D = 3 for 13 directions and resulting Cavg. To generate symmetric 3-D GLCMs, 13 directions are considered. For each direction a matrix is generated and the contrast is computed. This value gets mapped back to the original voxel around which the GLCMs were generated. The average of the 13 values produced considering each direction gets mapped to the same location in Cavg. Doing so for all points produces the average Contrast volume. A sample axial slice of each direction and the generated average for Contrast is displayed.
2.7. Experimental setup
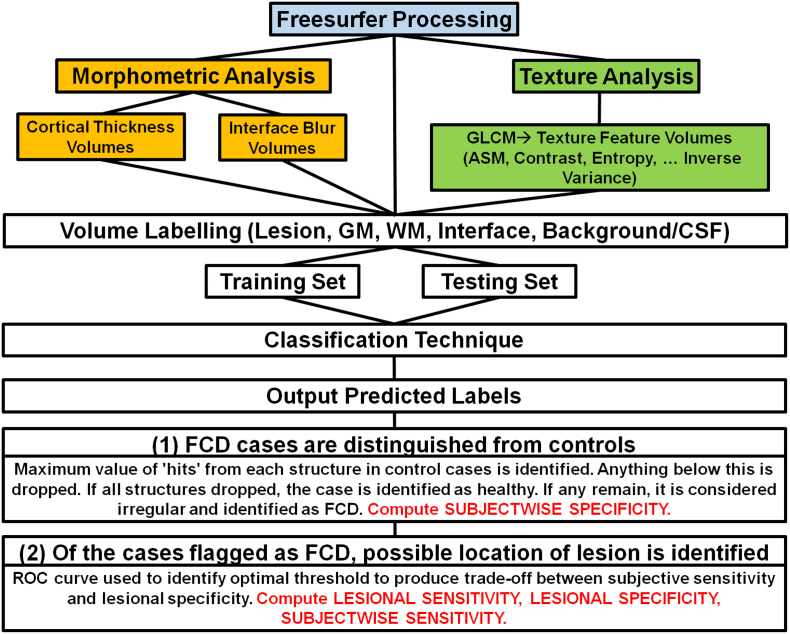
The complete pipeline is presented in Fig. 3. Z-score volumes were computed for each textural descriptor attribute average volume. Once all the volumes for all cases were produced and within-subject z-scores computed, they were normalized by computing between-subject z-scores using the overall mean and standard deviation relative to each feature. We employed the computational models for two of the common FCD traits cortical thickness, and blurring of the gray-white matter interface, as described above. These volumes too were standardized by computing z-scores. For classification we considered the z-score volumes of the established morphometric MRI features and the textural features. A voxel in a volume could be labeled by five possible tags: lesion, gray matter, white matter, gray-white matter interface and background or cerebrospinal fluid (CSF). Two datasets were constructed for classification, a training and testing set, with sampling rates differing between the two. We seek to train the classifier on a training set balanced with respect to label classes, and test performance on a testing set unbalanced in this respect. Sampling frequencies of the training set data are selected to achieve roughly equal numbers of voxels in each label. For the testing set, voxels are uniformly selected from all label classes.
Fig. 3.
Experimental setup.
2.8. 2-Step Naïve Bayes classifier
Classification was performed using the 2-Step method following the approach described in Antel et al. (Antel et al., 2003). In the 2-step method, data is first trained solely on morphometric features. Data that is classified as lesional is then reclassified using textural features in the second step. A 2-classifier approach is required because the two groups of attributes extract different types of information from the MRI volume. The computational models of MRI morphometric characteristics of FCD (i.e. cortical thickness and blurring of the gray-white matter interface) measure first-order statistical properties of individual voxels. The texture features measure second-order properties of the volume by assessing spatial relationships amongst voxels of varying intensities using the associated GLCMs. The 2-Step classifiers allows training in distinct regions within feature space.
We can generalize the approach as follows: given the training and testing data set, we classify the test data as lesional or normal using a leave-one-out (N − 1) approach for each subject N ϵ 1, 2, 3, … , 44 (31 MRI-Positive cases, 13 healthy controls). Results of experiments for each classification technique are written to .csv files containing voxel information and predicted class labels to be read into MATLAB. These results are used to implement a 2-Step pipeline in which cases are first identified as either control or FCD, and then the structural location of the lesion is identified. To do so, control cases are examined for the maximum cluster size within each of the structures to introduce structural thresholds. Lesional clusters smaller than these thresholds within each structure are deemed a result of random noise and omitted from the lesion map produced by the classifier. Thus, for each case, the thresholds are applied to drop structures. If all structures remain unflagged, the case is identified as a control. If any region is flagged, the case is deemed irregular and identified as an FCD case. For all cases identified as FCD, the next step is to find the possible location of the lesion. To do this we select an appropriate threshold using criterion selection via ROC curves. The Youden index (J) can be used as a means of selecting the optimal parameter of a ROC curve (Ekelund, 2012). Due to the nature of our pipeline for classification, subjectwise sensitivity and subjectwise specificity are computed in isolated steps. Thus we employ a modified Youden's J index that aims to produce a balance between subjectwise sensitivity and lesional specificity. To select the optimal threshold for each algorithm implementation we utilized this modified J index. We can express this index relative to subjectwise sensitivity and subjectwise specificity in terms of the (TP)i, (TN)i, (FP)i, and (FN)i relating to the i-th subject as
The 2-Step Naive Bayes classifier written in R code is available on GitHub at: https://github.com/ImagingLab/2StepBayesianClassifier. Future upgrades to the code will be publicly available on this repository.
3. Results
The results of the 2-Step Bayesian classifier on MRI-positive cases are presented in Table 3. Considering solely the performance on MRI-positive cases using T1-w data, the resulting subjectwise specificity, subjectwise sensitivity, lesional specificity and lesional sensitivity respectively, with surgical resection site as the reference standard for the lesion, were 100%, 94%, 75%, and 63% (Table 3). Thus the method correctly rejected 13/13 of healthy subjects. It also colocalized lesions in 29/31 of the FCD cases with the location of the lesions as visualized by a neuroradiologist, with 63% coverage of the complete extent of the lesion (see Fig. 4 for example of MRI-positive cases). The 2-Step Naive Bayes classifier method was also implemented on volumetric T2-w and FLAIR modalities. Using T2-w images, the subjectwise specificity, subjectwise sensitivity, lesional specificity and lesional sensitivity were 100%, 90%, 80%, and 60%. With FLAIR images, the subjectwise specificity, subjectwise sensitivity, lesional specificity and lesional sensitivity were 100%, 71%, 89%, and 42%. In one subject, T1-w and T2-w imaging did not identify a lesion but FLAIR detected the lesion. In both cases where T1-w imaging failed, T2 also failed. However, there were 8 subjects where FLAIR were normal and T2 identified a lesion. Combining the T1-w, T2-w, and FLAIR improved the subjectwise sensitivity to 97% while the subjectwise specificity remained at 100%. The number of false positive cases, that is, the abnormality identified by the classifier that did not correspond to the FCD location, on T1-w, FLAIR and T2-w were 1 case, 6 cases, and 1 case respectively.
Table 3.
Evaluating 2-Step Naive Bayes performances using T1, T2, FLAIR on MRI-positive cases.
| T1 | T2 | FLAIR | |
|---|---|---|---|
| Subjectwise specificity (%) | 100 | 100 | 100 |
| Subjectwise sensitivity (%) | 94 | 90 | 71 |
| Lesional specificity (%) | 75 | 80 | 89 |
| Lesional sensitivity (%) | 63 | 60 | 42 |
Fig. 4.
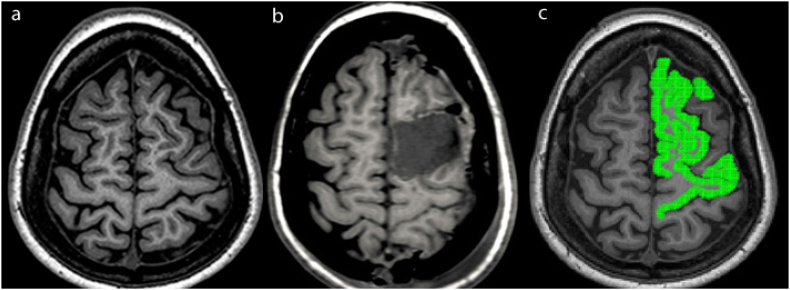
MRI-positive case. (a) Axial T1 image shows the lesion in the right parietal lobe, with increased T1 signal and blurring of the gray-white matter junction. (b) Axial T1 slice with FreeSurfer regions using 2-Step Naive Bayes Classification method with all selected structures in green.
We seek to extend the methods used on MRI-positive cases to MRI-negative cases, where the FCD features were likely more subtle and difficult to detect. 23 MRI negative cases were classified using the 2-Step Naive Bayes classifier. Considering the 23 MRI-negative cases, the threshold was selected using ROC curves in order to perform with the same precision as the MRI-positive cases. The resulting subjectwise specificity, subjectwise sensitivity, lesional specificity, and lesional sensitivity (with surgical resection site as the reference standard for the lesion) were 100%, 48%, 94% and 31% respectively (Table 4). Thus, using classification techniques, lesions were selected in 11 of the 23 subjects that were not identified using traditional visual assessment of MRI by neuroradiologist (see Fig. 5 for an example of MRI-negative case). Using T2-w images, the subjectwise specificity, subjectwise sensitivity, lesional specificity and lesional sensitivity were 100%, 30%, 95% and 22% respectively. With FLAIR images, the subjectwise specificity, subjectwise sensitivity, lesional specificity and lesional sensitivity were 100%, 39%, 92% and 28%. There were 3 subjects where T2-w imaging identified an abnormality but T1-w and FLAIR did not identify a lesion, and 1 subject where FLAIR identified an abnormality but T1-w and T2-w did not identify a lesion. Combining the T1-w, T2-w, and FLAIR improved the subjectwise sensitivity to 70% while the subjectwise specificity remained at 100%. The number of false positive cases, that is, the abnormality identified by the classifier that did not correspond to the surgical location, on T1-w, FLAIR and T2-w were 10 cases, 14 cases and 15 cases respectively.
Table 4.
Evaluating 2-Step Naive Bayes performances using T1, T2, FLAIR on MRI-negative cases.
| T1 | T2 | FLAIR | |
|---|---|---|---|
| Subjectwise specificity (%) | 100 | 100 | 100 |
| Subjectwise sensitivity (%) | 48 | 30 | 39 |
| Lesional specificity (%) | 94 | 95 | 92 |
| Lesional sensitivity (%) | 31 | 22 | 28 |
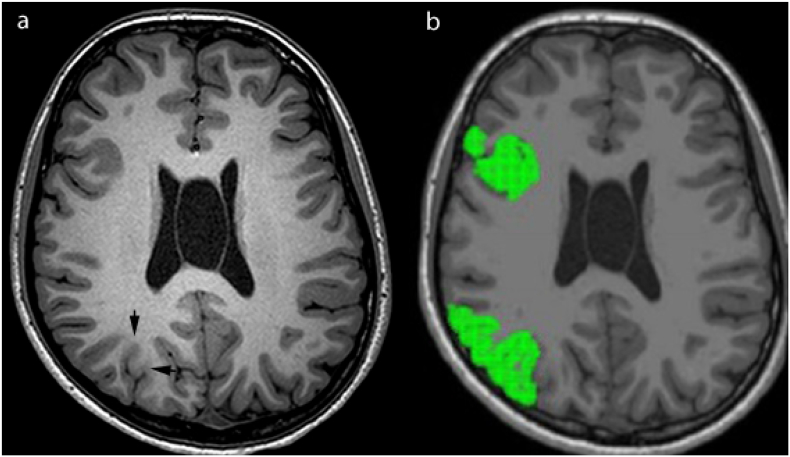
Fig. 5.
MRI-negative case. (a) Axial T1 image shows no abnormality seen by visual assessment, and (b) the surgical resection site. (c) Axial T1 slice with selected FreeSurfer regions using 2-Step Naive Bayes classification method with all selected structures in green, which colocalize to the surgical resection site.
4. Discussion
The method described by Antel et al. (Antel et al., 2003) assessed 18 MRI-visible FCD and 14 control cases from an adult cohort. The true location of lesions was manually segmented by an expert to be used in the verification of performance. An automated classifier was developed using a 2-step Bayesian classifier trained on recognized MRI FCD features and textural features derived from GLCMs to classify voxels as lesional or non-lesional. In adult cohorts the resulting subjectwise specificity = 100% and subjectwise sensitivity = 83% using a 2-step Naive Bayes classifier (Antel et al., 2003). The technique explored in this study differs from the one developed by Antel et al. (Antel et al., 2003) in terms of the textural features used, as well as incorporating the impact of using T2-w and FLAIR sequences. In this study, we evaluated children with medically intractable epilepsy, including those with MRI-positive and MRI-negative epilepsy. We showed that in children with MRI-positive epilepsy, volumetric T1-w had the highest subjectwise sensitivity (94%) and lesional sensitivity (63%), and lowest lesional specificity (75%) relative to T2-w and FLAIR using the 2-step Bayesian classifier. In a study conducted by Adler et al. using a neural network classifier and surface features trained on a paediatric dataset, the method detected MRI-visible FCDs in 16 out of 22 subjects with a sensitivity of 73% (Adler et al., 2017).
Of the MR sequences assessed, FLAIR failed to identify the largest number of lesions (9 cases) in patients with MRI-positive epilepsy, and T2-w failed to identify the largest number of lesions (16 cases) in patients with MRI-negative epilepsy. There was one case that neither T1-w nor T2-w sequences detected a lesion, but FLAIR identified the lesion in MRI-positive epilepsy. There were 3 cases where T2-w identified an abnormality, but T1-w and FLAIR did not identify a lesion in MRI-negative epilepsy. While the implementation on volumetric T1-w images was found to offer the best results for both MRI-positive and MRI-negative epilepsy, the argument can be made for combining information from multiple modalities into a single method in future work. In the study by Adler et al. (Adler et al., 2017) T1-w and FLAIR scans were incorporated into analysis. To examine brightening at the gray-white matter boundary, FLAIR intensity was sampled at this region and the surrounding area. This differs from the approach in this study which samples from the complete brain.
Patients with MRI-negative cases have lower subjectwise sensitivity and lesional sensitivity (48% and 31% respectively) compared to MRI-positive cases (subjectwise sensitivity and lesional sensitivity of 94% and 63% respectively). Patients with MRI-negative epilepsy likely have subtle changes on MRI that were not readily identified by visual assessment. The features could also be more difficult to detect using morphometric and textural analyses. Jin et al. (Jin et al., 2018) have also found that the sensitivity of MRI-negative cases was significantly lower than MRI-positive cases (52.9% vs. 81.8% respectively). For morphometric analysis, we have assessed cortical thickness and blurring of the gray-white matter interface, as these are two relatively common features of FCD (Alshafai et al., 2014). Additional measures such as curvature, sulcal depth and signal intensity could be extracted with FreeSurfer. It is possible that by extracting these additional features, we may improve the sensitivity of identifying an abnormality in MRI-negative cases. The false positive cases were higher in patients with MRI-negative than MRI-positive epilepsy. We have developed the algorithm on MRI-positive epilepsy cases and then applied the algorithm to MRI-negative cases, which may have resulted in less optimal threshold selection for MRI-negative cases, resulting in higher number of false positives in MRI-negative epilepsy. Future research focusing on training the classifier on combined MRI-positive and MRI-negative cases simultaneously, rather than training the algorithm on MRI-positive cases and then testing the algorithm on MRI-negative cases may be more optimal. In practice, any abnormality identified by computer aided tool including classifier should be correlated with the epileptogenic zone as assessed using video electroencephalography (EEG) and other functional imaging such as fluoro-deoxyglucose (FDG)-PET.
There are several limitations of this study. Fist, using the classification method, the lesions have coverage but were not localized. Thus, the technique performed well in terms of selecting the correct structure but the issue was in over-selection. In cases where the lesion is not restricted within a single FreeSurfer cortical segment, the method failed to identify the lesion in some cortical segments. This also affects subjects with small lesions as it will overestimate their size due to the defined structures of FreeSurfer labels. Manual lesion selection could improve results, particularly in regards to lesional sensitivity. However, manual lesion selection is a time consuming task, as an expert would need to go through the images slice by slice to select the lesion. Vertex wise classification serves as the natural extension of our future work as a means of improving lesional sensitivity. An alternate approach to this issue would be to incorporate some measure of spatial spread of selected lesional voxels. Second, to identify gray-white matter interface, at each voxel, we assessed the 8 neighboring voxels to determine if they are in the gray or white matter. It is possible that this approach may under- or over-estimate gray-white junction blurring relative to an approach that evaluates the intensity contrast in a perpendicular direction to the cortical interface. Another limitation is the relatively small number of healthy controls, which did not permit the data to be split into unique training and testing datasets, but instead utilize a leave one out method of training. This likely contributes to the high subjectwise specificity seen across techniques. Future work can explore the inclusion of additional control cases.
In summary, computational techniques using a 2-Step Bayesian classifier trained on morphometric and textural analysis, applied to multiple MRI sequences, can assist with detecting a lesion on MRI in children with focal intractable epilepsy. By demonstrating the potential location of the abnormality, the neuroradiologists can perform a second review of these areas on MRI and correlate the changes with the epileptogenic zone as identified on video EEG and other functional imaging.
Acknowledgements
This research was conducted with the support of EpLink – The Epilepsy Research Program of the Ontario Brain Institute (OBI). The OBI is an independent non-profit corporation, funded partially by the Ontario government. The opinions, results and conclusions are those of the authors and no endorsement by the Ontario Brain Institute is intended or should be inferred. This research was also supported in part by an NSERC Discovery Grant for M.E.
References
- Adler S., Wagstyl K., Gunny R., Ronan L., Carmichael D., Cross J.H.…Baldeweg T. Novel surface features for automated detection of focal cortical dysplasias in paediatric epilepsy. NeuroImage Clin. 2017;14:18–27. doi: 10.1016/j.nicl.2016.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alshafai L., Ochi A., Go C., McCoy B., Hawkins C., Otsubo H.…Widjaja E. Clinical, EEG, MRI, MEG, and surgical outcomes of pediatric epilepsy with astrocytic inclusions versus focal cortical dysplasia. Epilepsia. 2014;55(10):1568–1575. doi: 10.1111/epi.12756. [DOI] [PubMed] [Google Scholar]
- Antel S.B., Collins D.L., Bernasconi N., Andermann F., Shinghal R., Kearney R.E.…Bernasconi A. Automated detection of focal cortical dysplasia lesions using computational models of their MRI characteristics and texture analysis. NeuroImage. 2003;19(4):1748–1759. doi: 10.1016/s1053-8119(03)00226-x. [DOI] [PubMed] [Google Scholar]
- Colombo N., Tassi L., Galli C., Citterio A., Russo G.L., Scialfa G., Spreafico R. Focal cortical dysplasias: MR imaging, histopathologic, and clinical correlations in surgically treated patients with epilepsy. Am. J. Neuroradiol. 2003;24(4):724–733. [PMC free article] [PubMed] [Google Scholar]
- Dale A.M., Sereno M.I. Improved localizadon of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. J. Cogn. Neurosci. 1993;5(2):162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dale A.M., Fischl B., Sereno M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. NeuroImage. 1999;9(2):179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- De Cocker L., D'Arco F., Demaerel P., Smithuis R. Role of MRI in epilepsy. Radiol. Assist. 2012 www.radiologyassista.nt.nl/en (Accessed, 1) [Google Scholar]
- Duncan J.S. Imaging and epilepsy. Brain J. Neurol. 1997;120(2):339–377. doi: 10.1093/brain/120.2.339. [DOI] [PubMed] [Google Scholar]
- Ekelund S. ROC Curves—what are they and how are they used? Point Care. 2012;11(1):16–21. [Google Scholar]
- Fischl B., Dale A.M. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc. Natl. Acad. Sci. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B., Sereno M.I., Dale A.M. Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. NeuroImage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B., Van Der Kouwe A., Destrieux C., Halgren E., Ségonne F., Salat D.H.…Kennedy D. Automatically parcellating the human cerebral cortex. Cereb. Cortex. 2004;14(1):11–22. doi: 10.1093/cercor/bhg087. [DOI] [PubMed] [Google Scholar]
- Focke N.K., Symms M.R., Burdett J.L., Duncan J.S. Voxel-based analysis of whole brain FLAIR at 3T detects focal cortical dysplasia. Epilepsia. 2008;49(5):786–793. doi: 10.1111/j.1528-1167.2007.01474.x. [DOI] [PubMed] [Google Scholar]
- Jeha L.E., Najm I., Bingaman W., Dinner D., Widdess-Walsh P., Lüders H. Surgical outcome and prognostic factors of frontal lobe epilepsy surgery. Brain. 2007;130(2):574–584. doi: 10.1093/brain/awl364. [DOI] [PubMed] [Google Scholar]
- Jin B., Krishnan B., Adler S., Wagstyl K., Hu W., Jones S., Ding M. Automated detection of focal cortical dysplasia type II with surface-based magnetic resonance imaging postprocessing and machine learning. Epilepsia. 2018;59(5):982–992. doi: 10.1111/epi.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsek P., Hajek M., Dezortova M., Jiru F., Skoch A., Marusic P.…Komarek V. 1H MR spectroscopic imaging in patients with MRI-negative extratemporal epilepsy: correlation with ictal onset zone and histopathology. Eur. Radiol. 2007;17(8):2126–2135. doi: 10.1007/s00330-007-0594-1. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Lee S.Y., Kim K.K., Hong K.S., Lee D.S., Chung C.K. Surgical outcome and prognostic factors of cryptogenic neocortical epilepsy. Ann. Neurol. 2005;58(4):525–532. doi: 10.1002/ana.20569. [DOI] [PubMed] [Google Scholar]
- Lüsebrink F., Wollrab A., Speck O. Cortical thickness determination of the human brain using high resolution 3 T and 7 T MRI data. NeuroImage. 2013;70:122–131. doi: 10.1016/j.neuroimage.2012.12.016. [DOI] [PubMed] [Google Scholar]
- McGonigal A., Bartolomei F., Régis J., Guye M., Gavaret M., Fonseca A.T.-D.…Péragut J.-C. Stereoelectroencephalography in presurgical assessment of MRI-negative epilepsy. Brain. 2007;130(12):3169–3183. doi: 10.1093/brain/awm218. [DOI] [PubMed] [Google Scholar]
- Wyllie E., Comair Y.G., Kotagal P., Bulacio J., Bingaman W., Ruggieri P. Seizure outcome after epilepsy surgery in children and adolescents. Ann. Neurol. 1998;44(5):740–748. doi: 10.1002/ana.410440507. [DOI] [PubMed] [Google Scholar]