Abstract
Background
Meningioma is the most frequent primary intracranial tumour. Surgical resection remains the main therapeutic option as pharmacological intervention is hampered by poor knowledge of their proteomic signature. There is an urgent need to identify new therapeutic targets and biomarkers of meningioma.
Methods
We performed proteomic profiling of grade I, II and III frozen meningioma specimens and three normal healthy human meninges using LC-MS/MS to analyse global proteins, enriched phosphoproteins and phosphopeptides. Differential expression and functional annotation of proteins was completed using Perseus, IPA® and DAVID. We validated differential expression of proteins and phosphoproteins by Western blot on a meningioma validation set and by immunohistochemistry.
Findings
We quantified 3888 proteins and 3074 phosphoproteins across all meningioma grades and normal meninges. Bioinformatics analysis revealed commonly upregulated proteins and phosphoproteins to be enriched in Gene Ontology terms associated with RNA metabolism. Validation studies confirmed significant overexpression of proteins such as EGFR and CKAP4 across all grades, as well as the aberrant activation of the downstream PI3K/AKT pathway, which seems differential between grades. Further, we validated upregulation of the total and activated phosphorylated form of the NIMA-related kinase, NEK9, involved in mitotic progression. Novel proteins identified and validated in meningioma included the nuclear proto-oncogene SET, the splicing factor SF2/ASF and the higher-grade specific protein, HK2, involved in cellular metabolism.
Interpretation
Overall, we generated a proteomic thesaurus of meningiomas for the identification of potential biomarkers and therapeutic targets.
Fund
This study was supported by Brain Tumour Research.
Keywords: Meningioma, Proteomics, Phosphoproteins, Differential expression, Grade-specific, RNA metabolism
Research in context.
Evidence before this study
There is currently no effective pharmacological intervention for meningioma, the most frequent primary intracranial tumour. There is now a wealth of genomic studies of meningioma that have been pivotal in elucidating the mutational profile of these tumours. However, the mechanistic involvement of these mutations in meningioma development or their potential as therapeutic targets has not yet been established. In the molecular landscape of meningioma, proteomics attempts to bridge the gap between the increasing expanse of genomic, epigenomic and transcriptomic knowledge of these tumours and the execution of these instructions at the proteome level. Previous proteomic studies have been limited by relatively small cohorts encompassing all WHO grades, have lacked an appropriate control or did not include tumour microenvironment.
Added value of this study
In this study, we performed extensive proteomic analyses of frozen tumour tissue covering all WHO grades of meningioma and healthy human meningeal tissue as a control, including to our knowledge, the largest sample size analysed to date of the rarest and most therapeutically challenging, grade III meningiomas. We did additional mutation screening of all samples and used a validation cohort. We describe the enrichment of numerous novel pathways including RNA processing and proteins in meningiomas, and oncoproteins commonly upregulated among all grades. Further, we also show upregulation of proteins including NEK9 and its activated phosphorylated form, previously undescribed in these tumours. We also demonstrate grade specific protein upregulation and activation of proteins.
Implications of all the available evidence
This study demonstrates that proteomic and phosphoproteomic analyses are instrumental in identifying new candidates for targeted therapies or biomarkers of this most common brain tumour. Indeed, our results confirm the significant overexpression of several new proteins and phosphoproteins in meningioma, including grade-specific candidates that with further investigation may have clinical importance when determining an effective treatment strategy.
Alt-text: Unlabelled Box
1. Introduction
Meningiomas are the most common primary intracranial tumour accounting for up to 36% of all primary central nervous system (CNS) tumours [1]. They are classified by the World Health Organization (WHO) as slow growing benign WHO grade I (~80%), atypical WHO grade II (18%) or malignant WHO grade III (1–3%) [1]. Many meningiomas can be treated effectively by surgical resection [2]. Excision is occasionally associated with morbidity and can be difficult to perform depending on tumour location, often preventing complete removal [2]. Radiotherapy is used as an adjunct, whilst current chemotherapies remain ineffective [2]. Prediction of tumour recurrence and prognosis is based primarily on histological grade and extent of surgical resection: 5 year recurrence rates following gross total resection, for grade I, II and III meningiomas are approximately 3%, 38% and 78%, respectively [2]. However, a recently described DNA methylation-based meningioma classification may improve future predictions of tumour recurrence and prognosis [3].
The genomic landscape of meningiomas is well characterised. Approximately 60% sporadic meningiomas harbour mutations in the Neurofibromatosis 2 (NF2, merlin) gene, while mutations in genes including TRAF7, KLF4, AKT1, SMO, PIK3CA, POLR2A, PRKAR1A, AKT3 and SUFU have been identified in the remaining 40% non-NF2 mutant meningiomas [[4], [5], [6]]. More recently, studies have defined distinct genetic profiles underlying primary and recurrent atypical meningioma as well as the presence of BAP1 mutations in a subset of WHO grade III rhabdoid meningiomas [7,8].
Potential molecular targets of meningiomas have previously been identified including the growth factor receptors EGFR, PDGFR and VEGFR, along with their signalling pathway activation, yet their pharmaceutical targeting has yielded mixed results in clinical trials [[9], [10], [11]]. Treatment of recurrent meningioma with the EGFR inhibitors erlotinib and gefitinib produced no significant response; whilst the VEGFR and PDGFR inhibitor sunitinib (Sutent®), demonstrated some efficacy in recurrent/progressive high-grade meningiomas but was associated with considerable toxicity [10,11]. Therefore, the identification of aberrantly activated pathways and the associated molecular targets/biomarkers remains crucial for developing novel therapeutic strategies for meningioma.
Proteomic approaches provide a powerful tool to detect differentially or uniquely expressed proteins and reveal alterations in signalling pathways. Previous studies have described differential proteomic profiles between meningioma grades. Okamoto et al. analysed pure populations of tumour cells, however not considering a control; whilst, Sharma et al. analysed tumour specimens but using glial in place of meningeal tissue as healthy control [12,13]. Most recently, Parada et al. investigated the phosphoproteome across WHO grades of meningiomas and identified reduced expression of AKAP12 as a potential prognostic marker of high-grade meningioma [14].
In this study, we present the comparative proteomic analyses of different WHO grades of meningiomas vs. healthy human meninges aiming to characterise the pathogenic signature of these tumours. We analysed the global proteome as well as enriched phosphoproteins and phosphopeptides of 22 meningiomas and three normal meninges and identified the differential expression of novel proteins like the oncoproteins SET and SF2/ASF among all meningioma grades. In addition, we revealed among others, the activation of phosphoproteins like phospho-AKT and phospho-NEK9. Grade-specific analysis allowed for the identification of upregulated proteins and phosphoproteins and their associated enriched biological processes in high-grade aggressive meningiomas. We validated differential expression using additional techniques and a validation cohort. Our results describe to our knowledge, the largest proteomic study of meningioma tissue and suggest several candidates with promise as therapeutic targets or biomarkers in meningioma.
2. Materials and methods
2.1. Clinical material
Anonymised meningioma samples under the ‘J’ series were provided by the BRain Archive and Information Network (BRAIN UK) under ethical approval by the South West research ethics committee (REC No: 14/SC/0098; IRAS project ID: 143874, BRAIN UK Ref: 15/011). Anonymised ‘MN’ meningioma samples were collected under ethical approval by the South West research ethics committee (REC No: 14/SW/0119; IRAS project ID: 153351) and local research and development approval (Plymouth Hospitals NHS Trust: R&D No: 14/P/056 and North Bristol NHS Trust: R&D No: 3458). Clinical and histological details about the specimens are presented in Supplementary Tables S1 and S2. Tumours were separated into a ‘discovery set’ for MS analysis consisting of 22 meningiomas (WHO grade I: n = 8, WHO grade II: n = 8, WHO grade III: n = 6) and a ‘validation set’ composed of 15 meningioma samples (WHO grade I: n = 5, WHO grade II: n = 5, WHO grade III: n = 5). Two frozen normal meninges were obtained from Analytical Biological Services Inc. and one human brain cerebral meninges was purchased from Novus Biologicals® (NB820-59183; lot B105014).
2.2. Phosphoprotein and phosphopeptide enrichment
For both phosphoprotein and phosphopeptide enrichment 2·5 mg of protein lysate was used as starting material. Phosphoproteins were enriched from frozen tissue using the commercially available Qiagen® PhosphoProtein Purification Kit (Qiagen) according to the manufacturer's instructions. Previous studies using this kit have reported an 88% elution recovery of phosphoproteins [15]. Protein concentration was determined as before [16].
For phosphopeptide enrichment, samples were homogenised from frozen in lysis buffer (8 M urea, 100 mM Tris-HCl, pH 8·0). Samples were frozen at −80 °C for 24 h, thawed on ice and centrifuged at 16,000 ×g for 15 min at 4 °C. Supernatant was collected in Eppendorf® Protein LoBind microcentrifuge tubes and protein concentration determined [16]. Prior to enrichment using titanium dioxide (TiO2) beads, 2·5 mg of protein lysate was subjected to in-solution digestion. Proteins were reduced and alkylated by incubation with 0·1 M dithiotheitol (DTT) for 30 min at room temperature (RT), followed by further incubation with 50 mM 2-iodoacetamide in the dark for 15 min at RT. Lys-C protease (Lysyl Endopeptidase®, Mass Spectrometry Grade, Wako) was then added at a protease: protein ratio of 1: 100 (w/w) in 50 mM ammonium bicarbonate (ABC) and incubated overnight (O/N) at 37 °C. Samples were diluted in 50 mM ABC to a final concentration of 2 M urea and incubated O/N at 37 °C with trypsin (Promega, Wisconsin, US), added at a protease: protein ratio of 1: 50 (w/w). Digested samples were acidified to a final concentration of 0·1% trifluoroacetic acid (TFA) and peptides desalted using HyperSep™ C18 Cartridges (Thermo Fisher Scientific, Massachusetts, US). Columns were washed with buffer A (1% TFA, 0·5% acetic acid), phosphopeptides eluted in buffer B (80% acetonitrile (ACN, LC-MS grade), 0·5% acetic acid, 1% TFA) and dried in a vacuum concentrator. Phosphopeptides were enriched by batch-wise incubation with Titansphere 10 μm TiO2 beads (GL Sciences) as described by Lasonder et al. [17] with the following modifications. Initially, TiO2 beads (1 mg beads per incubation) were incubated in wash buffer A (80% ACN, 5% TFA) followed by incubation in buffer B (60% ACN, 5% TFA, 5% glycerol) for 5 min at 1000 rpm, RT. TiO2 beads were sedimented by centrifugation at 2000 ×g for 1 min, buffer removed, sample peptide digests added in 1·5 ml Eppendorf® vials and incubated with the beads under continuous shaking for 1 h. Beads were washed three times with 100 μl buffer B, followed by three washes with 100 μl buffer A. Phosphopeptides were eluted following a two-step elution protocol by Fukuda et al. [18], acidified with TFA, purified by stop and go extraction (STAGE) tips [19] and stored at −20 °C prior to mass spectrometry (MS) analysis.
2.3. Protein fractionation and in-gel digestion
Proteins and phosphoproteins were separated using SDS-PAGE on 4–15% Mini-PROTEAN® TGX™ Precast Gels (Bio-Rad). For global proteome analysis 50 μg of protein lysate was separated. Gels were stained with Coomassie Blue R-350 (GE Healthcare Life Sciences) until lanes were visible. Destaining was performed using a destaining solution (50% LC/MS grade water, 40% MeOH and 10% acetic acid) O/N at RT. Sample lanes were excised from gels, sliced into 6 fractions that were cut into 1 × 1 mm pieces before in-gel digestion. In-gel digestion was performed following Shevchenko et al. [20]. Briefly, gel pieces were equilibrated with alternate incubation of 100% acetonitrile and 50 mM ABC. Proteins were reduced by incubation with 10 mM DTT in 50 mM ABC for 20 min at 56 °C in shaking at 700 rpm and then alkylated by incubation with 50 mM 2-iodoacetamide in 50 mM ABC for 20 min at RT in the dark. Proteins were digested in 12·5 ng/μl trypsin (Promega, Wisconsin, US) in 50 mM ABC O/N at 37 °C. Digested peptides were acidified with a final concentration of 2% TFA for 20 min at RT 1400 rpm. Peptides were then extracted by two 5 min incubations of gel pieces with buffer B (80% ACN, 0.5% acetic acid, 1% TFA) at 1400 rpm. Supernatant was pooled together for each sample and ACN was evaporated in a vacuum concentrator.
Digested peptides and phosphopeptides were purified by STAGE tips [19]. Peptides were loaded onto C18-StageTips (Empore™ SPE Disks C18, 3 M) conditioned by 50 μl of methanol and equilibrated using 50 μl buffer B (80% ACN, 0·5% acetic acid, 1% TFA) followed by 50 μl buffer A (1% TFA, 0·5% acetic acid). Tips were washed with 50 μl buffer A and peptides eluted in 40 μl buffer B into LoBind microcentrifuge tubes. Eluted peptides were dried down completely in a vacuum concentrator and peptides resuspended in buffer A.
2.4. Liquid chromatography tandem mass spectrometry and protein identification
MS and protein identification was carried out as previously described [16], with the following modifications. MS data was analysed to identify proteins with the Andromeda peptide database search engine integrated into the computational proteomics platform MaxQuant version (1.5.0.30) [[21], [22], [23]]. Andromeda search parameters for protein identification specified a first search mass tolerance of 20 ppm and a main search tolerance of 4·5 ppm for the parental peptide. Minimal peptide length was set at six amino acids. Proteins were quantified with label free quantification (LFQ) values representing normalised summed peptide intensities correlating with protein abundances, where the ‘match between run’ option was permitted between runs with a 0·7 min elution time interval. Venn diagrams depicting the distribution of identified proteins and phosphoproteins were created with Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
2.5. Protein quantification analysis and functional annotation analysis
LFQ values were log2 transformed using the Perseus software suite 1.5.0.31 [22] to achieve normal data distribution, which was verified by visual inspection of histogram distribution plots of log2 transformed data generated in Perseus for each sample. Proteins identified in at least three runs were considered for LFQ and entries with an LFQ equal to zero were kept. Statistical significance of changes in abundance between sample groups was calculated by a two-tailed t-test, with p-values adjusted for multiple testing by a permutation-based FDR at 5%. Microsoft Excel was used to calculate ratios and fold changes (FC) followed by log2 transformation. A log2 FC ≥ 1·5 and log2 FC ≤ −1·5 with adjusted p-value < 0·05 were considered. Results are visualised by Volcano plots. Differentially expressed proteins (DEPs) for hierarchical clustering were obtained by submitting relative expression profiles identified in at least three runs to Perseus and performing a four-group one-way ANOVA (p-value < 0·05) on imputation supplemented data. Venn diagrams depicting upregulated proteins and phosphoproteins among all meningioma grades were created using euler APE (http://www.eulerdiagrams.org/eulerAPE/).
Pathway enrichment analysis was generated through the use of IPA® (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). Statistical significance of enriched pathways was assessed by right-tailed Fisher's exact test and considered significant for p (FET) < 0.05. GO enrichment analyses were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/) against a background of the H. sapiens proteome [24]. Enrichment of GO FAT terms was considered statistically significant when corrected for multiple testing by the Benjamini-Hochberg method with adjusted p-values < 0·05. Cytoscape plugin Enrichment Map (http://www.cytoscape.org/) was used to visualise enriched GO terms [25].
Molecular signatures for higher grade meningiomas were generated with the GeneSign module in BubbleGUM software (https://omictools.com/bubblegum-tool). Grade II and III samples were defined as subsets of interest (test classes), with NMT and grade I samples as references. The Mean (test)/ Mean method was applied for extracting molecular signatures.
Phosphorylation sites were classified as described by Villen et al. [26] with the following addition: pY at position 0 then classify as “Tyrosine”.
2.6. DNA purification and genotyping
DNA purification was performed with Qiagen® DNeasy® Blood & Tissue Kit (Qiagen) following manufacturer's instructions. DNA was screened for NF2 mutations by the Manchester Centre for Genomic Medicine using Next Generation sequencing (NGS) and Multiplex Ligation-dependent Probe Amplification (MLPA®) dosage test. Mutational screening of AKT1, KLF4, TRAF7, SMO, SMARCB1 and POLR2A was completed using KASP genotyping [27].
2.7. Cell culture and proliferation assay
The immortalised benign meningioma cell line, Ben-Men-1 (BM1) (DSMZ Cat# ACC-599, RRID:CVCL_1959) [28] and the malignant meningioma cell line KT21-MG1 (KT21) (RRID:CVCL_M429) [29] were cultured in meningioma medium (Dulbecco's Modified Eagle's Media (DMEM; Thermo Fisher Scientific), supplemented with 10% foetal bovine serum (FBS; Sigma-Aldrich®), 1% d-(+)-glucose (Sigma-Aldrich®), 100 U/ml penicllin/streptomycin (Thermo Fisher Scientific) and 2 mM l-glutamine (Thermo Fisher Scientific)) at 37 °C in humidified 5% CO2. Human meningeal cells (HMC) were obtained from ScienCell™ and routinely cultured in the manufacturer's recommended medium and growth supplements at 37 °C in humidified 5% CO2. BM1 and KT21 cell lines were plated in 96-well culture plates (Greiner Bio-One; #655088) at approximately 3000 cells per well. Cells were allowed to adhere and proliferate for 24 h. AZD5363 and Ku-0063794 (Selleckchem.com) were resuspended in dimethyl sulfoxide (DMSO; Sigma-Aldrich®) and serially diluted to the appropriate concentration in meningioma medium. Vehicle-treated control cells were grown with the addition of 0·1% DMSO. Cells were treated every 24 h by careful removal of media from wells, as not to detach cells, followed by addition of fresh drug during a 72 h incubation period at 37 °C in humidified 5% CO2. Cell viability was determined using the ‘CellTiter-Glo® Luminescent Cell Viability Assay’ (Promega; #G7570). Each drug concentration was tested in triplicate. Cell viability was calculated as a percentage of control cells, untreated cells as negative control and media alone as positive control. Graphs were generated using GraphPad Prism 5.
2.8. Cell lysate and tumour tissue preparation
Meningioma tissue samples of the validation set were manually homogenised from frozen in approximately 400 μl lysis buffer consisting of RIPA buffer (50 mM Tris-HCl pH 7.4, 0·1% SDS, 1% NP-40, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 0·5% sodium deoxycholate) protease inhibitor used at 1:20 (cOmplete™, EDTA-free Protease Inhibitor Cocktail, Sigma-Aldrich®; #11873580001) and phosphatase inhibitor cocktails used at 1:100 (Santa Cruz Biotechnology, Inc.; sc-45,045; sc-45,065). Cells were similarly lysed in a volume of lysis buffer appropriate for the size of cell culture dish. Lysates were placed at −80 °C for at least 24 h before being thawed on ice and centrifuged at 16,000 ×g for 15 min at 4 °C. Supernatant was transferred to fresh 1·5 ml microcentrifuge tubes and protein concentration determined as before using the Pierce™ BCA Protein Assay Kit.
2.9. Western blotting
Proteins were separated on 8% or 15% Laemmli SDS-PAGE, depending upon the molecular weight of the protein of interest and transferred to a polyvinylidene difluoride membrane (Immun-Blot® PVDF membrane, Bio-Rad). Membranes were blocked for 1 h at room temperature with 5% skimmed milk in phosphate buffered saline (PBS) with 0·05% Tween-20, before incubation with specific primary antibodies O/N at 4 °C. Primary antibody details are listed in Supplementary Table S3. Specific antigen-antibody interaction was detected with anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidise (Bio-Rad). Detection was achieved using the ECL or ECL Plus Western Blotting substrate (Pierce). Membranes were exposed to Amersham Hyperfilm ECL (GE Healthcare Life Sciences). Films were scanned at a resolution of 600 dpi using a HP Scanjet 2400.
2.10. Immunohistochemistry
Four-micrometer paraffin sections were de-waxed, rehydrated and incubated with primary antibodies at room temperature O/N using antigen retrieval methods as previously described [9]. Primary antibody details are listed in Supplementary Table S3. Proteins were visualised with the Vectastain Universal Elite ABC kit (Vector Laboratories Ltd). Slides were counterstained with haematoxylin (Sigma-Aldrich). As a control, sections were incubated with omission of the primary antibody. Immunohistochemical results were reviewed by neuropathologist, Hilton, DA. Semi-quantitative assessment of staining intensity was scored as follows: 0 if negative; 1 if low immunoreactivity; 2 if moderate and 3 if high.
2.11. Data availability
Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [30] with the following dataset identifiers and reviewer account details: global proteome PXD007073; phosphoproteome PXD007044; phosphopeptides PXD007125.
3. Results
3.1. Mutational screening of meningioma
To determine the presence of NF2 mutations in the discovery set we performed NGS and MLPA. In all but five tumours, we confirmed mutations in the NF2 gene; we further screened the remaining five for hot-spot mutations in AKT1, KLF4, TRAF7, SMO, SMARCB1 and POLR2A [4,5] using KASP, but we did not identify any mutation (Supplementary Table S4). We additionally used Western blotting to determine the expression of NF2 and pNF2-S518. In general, all 22 tumour lysates showed some expression of NF2 protein except for the grade III sample, J17 (Supplementary Fig. S1a). This finding may be seen to conflict with the NF2−/− genotype confirmed by NGS and MLPA of some tumours. However, it is likely NF2 expression originates from the tumour microenvironment. Indeed, meningiomas are described as heterogeneous tumours comprising mainly of tumour cells alongside a variable presence of infiltrating immune cells, most frequently macrophages [31]. Further, expression of pNF2-S518 could not be detected in any NF2−/− confirmed tumours and thus may be the better complementary technique in addition to mutational screening (Supplementary Fig. S1a). In contrast, four of the five NF2+/+ meningiomas could clearly be seen to show higher NF2 expression with concomitant pNF2-S518 expression (J8, J15, J1 and J2) compared to all other tumours and NMT in WB (Supplementary Fig. S1a).
3.2. Proteins identified in meningiomas
We performed in-depth global protein and phosphoprotein expression profiling of human meningiomas compared to normal human meninges. We analysed a total of 22 meningiomas; global proteome and phosphoprotein pull-down was performed on 14 meningiomas, whilst global proteome and phosphopeptide analysis was performed on the remaining 8 samples. Phosphoprotein and phosphopeptide analyses required large quantities of starting material and thus were mutually exclusive for tumour samples. In total, we sequenced and identified 3905 proteins and 3162 phosphoproteins with 1% FDR (Supplementary Table S5). We identified 3619 proteins within NMT, 3850 across grade I, 3852 across grade II and 3855 across grade III (Supplementary Fig. S2a). From the phosphoprotein dataset we identified 2793 phosphoproteins in NMT, 2965 across grade I, 2934 across grade II and 2868 across grade III (Supplementary Fig. S2b).
3.3. Commonly upregulated proteins and phosphoproteins among all meningioma grades
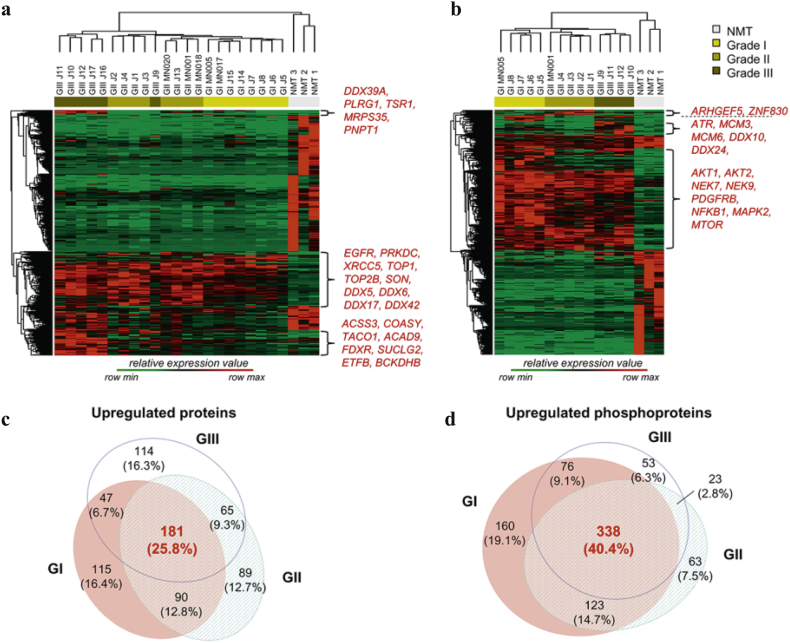
Proteins and phosphoproteins identified in at least 3 runs were quantified by LFQ and transformed into relative expression data across all grades resulting in expression profiles for 3888 proteins (Supplementary Table S6a) and 3074 phosphoproteins (Supplementary Table S6b). To get an overview we first grouped meningioma samples by hierarchical clustering based on differentially expressed proteins (DEPs) across all grades and healthy controls (Fig. 1a and b). Global proteome revealed a clear separation of tumour from control based on proteomic profiles, and meningiomas clustered into respective WHO grades, with the exception of the grade III sample J9, which was assigned to the grade II cluster (Fig. 1a). Three clusters of proteins displayed higher expression in at least one grade compared to NMT. The cluster having higher expression across all grades included EGFR, PRKDC, XRCC5, TOP1, TOP2B, SON and the DEAD box RNA helicases DDX5, DDX6, DDX17 and DDX42; whilst two smaller clusters with increased grade III-specific expression contained mainly mitochondrial related proteins (Fig. 1a). Similarly, the phosphoprotein dataset showed a marked separation of tumours vs. control, with meningiomas clustering into respective WHO grade (Fig. 1b). Again, we identified three main clusters of phosphoproteins displaying higher expression in at least one grade compared to NMT. Among phosphoproteins present in the largest cluster were AKT1 and 2, NEK7, NEK9, PDGFRB, NFKB1, MAPK2 and mTOR. The grade III-specific cluster contained the serine/threonine protein kinase ATR, the minichromosome maintenance proteins MCM3 and MCM6, previously identified in meningioma cell lines [32], and the DEAD box RNA helicases DDX10 and DDX24 (Fig. 1b).
Fig. 1.
Comparative analysis of the global proteome and phosphoproteins of different grades of meningiomas. (a) Unsupervised hierarchical clustering of 481 differentially expressed proteins (meningiomas n = 22; NMT n = 3). Tumours clustered separately from NMT and into respective WHO grades, with the exception of the grade III sample, J9. Three protein clusters are annotated demonstrating higher relative expression in at least one or more meningioma grade compared to NMT. (b) Unsupervised hierarchical clustering of 1296 differentially expressed phosphoproteins (meningiomas n = 14; NMT n = 3). Tumours clustered separately from NMT in their phosphoprotein profiles. Three phosphoprotein clusters are annotated showing higher relative expression in at least one or more meningioma grade compared to NMT. Clustering was generated using Perseus 1.5.0.31 software suite, based on four-group one-way ANOVA, p-value<.05, corrected for multiple testing. (c) Venn diagram depicting unique and common upregulated proteins among different meningioma grades vs. NMT. (d) Venn diagram depicting unique and common upregulated phosphoproteins among different meningioma grades vs. NMT. Upregulated: log2 FC ≥ 1.5; p-value < .05 (two-tailed t-test). Venn diagrams were created using Euler APE (http://www.eulerdiagrams.org/eulerAPE/). Proteomic data used to create this figure are provided in Supplementary Table S6a and b.
Comparative analysis of the global proteome and phosphoproteins of different grades of meningiomas. (a) Unsupervised hierarchical clustering of 481 differentially expressed proteins (meningiomas n = 22; NMT n = 3). Tumours clustered separately from NMT and into respective WHO grades, with the exception of the grade III sample, J9. Three protein clusters are annotated demonstrating higher relative expression in at least one or more meningioma grade compared to NMT. (b) Unsupervised hierarchical clustering of 1296 differentially expressed phosphoproteins (meningiomas n = 14; NMT n = 3). Tumours clustered separately from NMT in their phosphoprotein profiles. Three phosphoprotein clusters are annotated showing higher relative expression in at least one or more meningioma grade compared to NMT. Clustering was generated using Perseus 1.5.0.31 software suite, based on four-group one-way ANOVA, p-value<.05, corrected for multiple testing. (c) Venn diagram depicting unique and common upregulated proteins among different meningioma grades vs. NMT. (d) Venn diagram depicting unique and common upregulated phosphoproteins among different meningioma grades vs. NMT. Upregulated: log2 FC ≥ 1.5; p-value < .05 (two-tailed t-test). Venn diagrams were created using Euler APE (http://www.eulerdiagrams.org/eulerAPE/). Proteomic data used to create this figure are provided in Supplementary Table S6a and b.
Next, we focussed on proteins and phosphoproteins upregulated in meningiomas compared to control, enabling us to identify overexpressed proteins and differences in protein phosphorylation status. We determined significantly upregulated proteins and phosphoproteins common to all meningioma grades by overlapping upregulated DEPs from all grades, as depicted in the Venn diagrams (Fig. 1c and d; Supplementary Table S6). This enabled us to identify 181 proteins and 338 phosphoproteins to be commonly upregulated among all grades vs. NMT, some not yet described in meningiomas including SET, TRIP6 and the copper transport protein ATOX1. Partial lists of significantly upregulated proteins and phosphoproteins common to all grades are shown in Table 1, Table 2 respectively.
Table 1.
Partial list of proteins commonly and significantly upregulated among all grades of meningiomas compared to normal meningeal tissue (log2 fold-change ≥ 1.5; p-value < .05). Proteins specific to one of the two groups compared were assigned a fold change of infinity.
| Gene symbol | Protein name | UniProt Accession number | Log2 fold-change (GI vs. NMT) | Log2 fold-change (GII vs. NMT) | Log2 fold-change (GIII vs. NMT) |
|---|---|---|---|---|---|
| DNJC8 | DnaJ homolog subfamily C member 8 | O75937 | ∞ | ∞ | ∞ |
| SET | Protein SET | Q01105 | ∞ | ∞ | ∞ |
| STAT2 | Isoform 2 of Signal transducer and activator of transcription 2 | P52630-4 | ∞ | ∞ | ∞ |
| ATOX1 | Copper transport protein ATOX1 | O00244 | ∞ | ∞ | ∞ |
| SON | Isoform J of Protein SON | P18583-10 | ∞ | ∞ | ∞ |
| TRIP6 | Thyroid receptor-interacting protein 6 | Q15654 | ∞ | ∞ | ∞ |
| FOXP1 | Isoform 2 of Forkhead box protein P1 | Q9H334-2 | ∞ | ∞ | ∞ |
| EIF2A | Isoform 3 of Eukaryotic translation initiation factor 2A | Q9BY44-3 | ∞ | ∞ | ∞ |
| NAV1 | Isoform 5 of Neuron navigator 1 | Q8NEY1-5 | ∞ | ∞ | ∞ |
| NEK9 | Serine/threonine-protein kinase Nek9 | Q8TD19 | 4.83 | 4.33 | 2.89 |
| CENPV | Isoform 3 of Centromere protein V | Q7Z7K6-3 | 4.57 | 4.62 | 4.60 |
| S10AA | Protein S100-A10 | P60903 | 4.33 | 3.66 | 4.13 |
| SERPH | Serpin H1 | P50454 | 4.29 | 4.43 | 3.82 |
| ST1A1 | Sulfotransferase 1A1 | P50225 | 4.28 | 4.37 | 4.09 |
| H4 | Histone H4 | P62805 | 4.08 | 3.17 | 3.17 |
| SET | Isoform 2 of Protein SET | Q01105-2 | 3.74 | 3.99 | 4.18 |
| STAT1 | Signal transducer and activator of transcription 1-alpha/beta | P42224 | 3.67 | 3.67 | 4.46 |
| HDGF | Hepatoma-derived growth factor | P51858 | 3.53 | 3.54 | 3.53 |
| FLNB | Isoform 8 of Filamin-B | O75369-8 | 2.87 | 2.96 | 2.67 |
| NUCL | Nucleolin | P19338 | 2.85 | 2.97 | 3.20 |
| NAMPT | Nicotinamide phosphoribosyltransferase | P43490 | 2.78 | 3.67 | 3.56 |
| PAXI | Isoform Alpha of Paxillin | P49023-2 | 2.65 | 3.36 | 2.72 |
| CKAP4 | Cytoskeleton-associated protein 4 | Q07065 | 2.63 | 2.22 | 2.97 |
| EF1A1 | Elongation factor 1-alpha 1 | P68104 | 2.54 | 2.65 | 2.41 |
| SRSF1 | Serine/arginine-rich splicing factor 1 | Q07955 | 2.12 | 1.85 | 1 |
| PAK2 | Serine/threonine-protein kinase PAK 2 | Q13177 | 1.99 | 2.22 | 2.07 |
| SIAS | Sialic acid synthase | Q9NR45 | 1.95 | 2.32 | 2.28 |
| SPAG9 | Isoform 4 of C-Jun-amino-terminal kinase-interacting protein 4 | O60271-4 | 1.78 | 2.73 | 1.74 |
| CLIC1 | Chloride intracellular channel protein 1 | O00299 | 1.75 | 1.90 | 2.26 |
| EPS8L2 | Epidermal growth factor receptor kinase substrate 8-like protein 2 | Q9H6S3 | 1.66 | 3.26 | 3.04 |
Table 2.
Partial list of phosphoproteins commonly and significantly upregulated among all grades of meningiomas compared to normal meningeal tissue (log2 fold-change ≥ 1.5; p-value < .05). Phosphoproteins specific to one of the two groups compared were assigned a fold change of infinity.
| Gene symbol | Protein name | UniProt Accession number | Log2 fold-change (GI vs. NMT) | Log2 fold-change (GII vs. NMT) | Log2 fold-change (GIII vs. NMT) |
|---|---|---|---|---|---|
| SSH3 | Protein phosphatase Slingshot homolog 3 | Q8TE77 | ∞ | ∞ | ∞ |
| ACSS2 | Acetyl-coenzyme A synthetase, cytoplasmic | Q9NR19 | ∞ | ∞ | ∞ |
| FAK2 | Isoform 2 of Protein-tyrosine kinase 2-beta | Q14289-2 | ∞ | ∞ | ∞ |
| MRCKB | Serine/threonine-protein kinase MRCK beta | Q9Y5S2 | ∞ | ∞ | ∞ |
| AKT2 | RAC-beta serine/threonine-protein kinase | P31751 | ∞ | ∞ | ∞ |
| RPS6KB1 | Ribosomal protein S6 kinase beta-1 | P23443-4 | ∞ | ∞ | ∞ |
| RTKN | Rhotekin | Q9BST9 | ∞ | ∞ | ∞ |
| NFKB1 | Nuclear factor NF-kappa-B p105 subunit | P19838 | ∞ | ∞ | ∞ |
| WASL | Neural Wiskott-Aldrich syndrome protein | O00401 | ∞ | ∞ | ∞ |
| IRF3 | Interferon regulatory factor 3 | Q14653 | ∞ | ∞ | ∞ |
| RB1 | Retinoblastoma-associated protein | P06400 | ∞ | ∞ | ∞ |
| AKT1 | RAC-alpha serine/threonine-protein kinase | P31749 | 7.00 | 5.74 | 4.97 |
| EPS8L2 | Epidermal growth factor receptor kinase substrate 8-like protein 2 | Q9H6S3 | 6.90 | 5.80 | 6.14 |
| CTNND1 | Isoform 1A of Catenin delta-1 | O60716-5 | 6.72 | 5.75 | 5.63 |
| ACACA | Acetyl-CoA carboxylase 1 | Q13085 | 5.67 | 5.84 | 5.54 |
| TUBB6 | Tubulin beta-6 chain | Q9BUF5 | 5.51 | 4.69 | 4.31 |
| PRKAA1 | Isoform 2 of 5-AMP-activated protein kinase catalytic subunit alpha-1 | Q13131-2 | 4.79 | 4.11 | 3.84 |
| S10AA | Protein S100-A10 | P60903 | 4.58 | 5.93 | 6.88 |
| MAPK2 | MAP kinase-activated protein kinase 2 | P49137 | 3.92 | 1.96 | 2.64 |
| TUBA1C | Tubulin alpha-1C chain | Q9BQE3 | 3.79 | 2.38 | 1.89 |
| SRC | Proto-oncogene tyrosine-protein kinase Src | P12931 | 3.78 | 3.63 | 3.73 |
| FLNB | Isoform 2 of Filamin-B | O75369-2 | 3.73 | 3.17 | 2.94 |
| PAXI | Isoform Alpha of Paxillin | P49023-2 | 3.70 | 4.26 | 8.09 |
| FAK1 | Isoform 7 of Focal adhesion kinase 1 | Q05397-7 | 3.64 | 2.00 | 2.67 |
| MP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 | P45985 | 2.66 | 1.88 | 1.72 |
| MP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | P36507 | 2.62 | 2.41 | 2.05 |
| SET | Protein SET | Q01105 | 2.40 | 3.29 | 3.18 |
| TUBB4B | Tubulin beta-4B chain | P68371 | 2.35 | 2.44 | 1.97 |
| PRKDC | DNA-dependent protein kinase catalytic subunit | P78527 | 2.14 | 2.30 | 2.97 |
| NUCL | Nucleolin | P19338 | 1.67 | 2.22 | 2.78 |
3.4. Functional annotation of commonly upregulated proteins and phosphoproteins in meningiomas
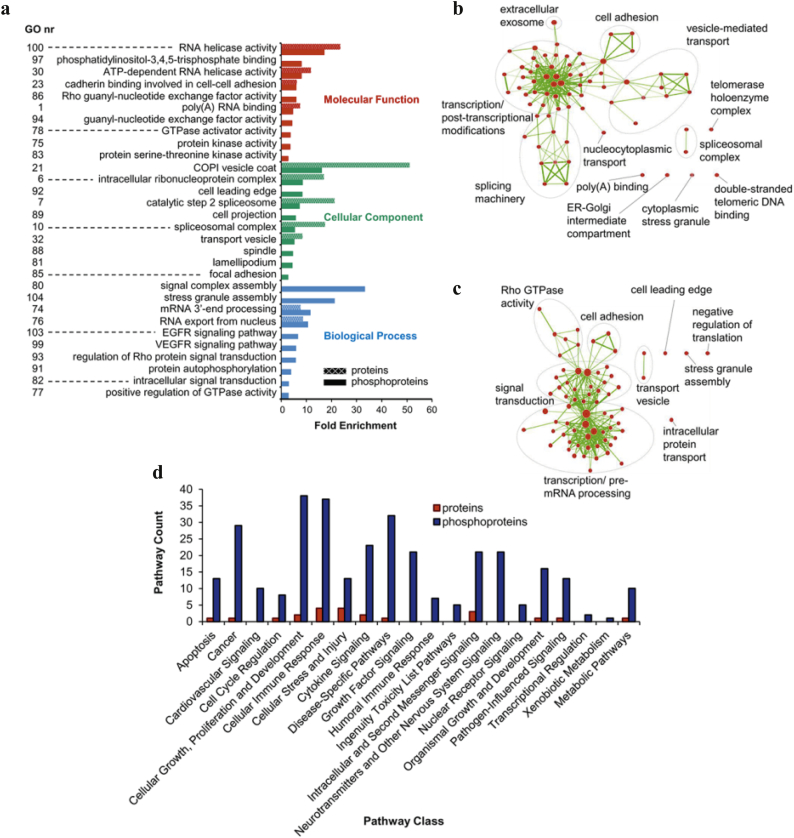
Functional annotation analyses were performed to establish signalling pathways, biological processes and molecular functions associated with DEPs. We annotated the 181 proteins and 338 phosphoproteins upregulated among all grades by GO enrichment analysis (Supplementary Table S7a and b), and compared significantly enriched terms of phosphoproteins with corresponding fold enrichments of proteins to identify GO terms predominantly targeted by phosphorylation. The most relevant enriched GO terms are presented in Fig. 2a; among the most upregulated proteins were RNA helicase activity (23-fold), COPI vesicle coat (51-fold) and the RNA export from nucleus (9-fold) in the ‘molecular function’, ‘cellular components’ and ‘biological process’ groups respectively. Several terms with the strongest enrichment for phosphoproteins were found associated with RNA helicase activity (17-fold) and COPI vesicle coat (16-fold), but we also identified specific phosphoprotein-enriched terms including the signal complex assembly (33-fold), EGFR (6-fold) and VEGFR (6-fold) signalling pathways, the last two already described in meningiomas [[9], [10], [11]] (Fig. 2a).
Fig. 2.
Functional annotation of proteins and phosphoproteins significantly upregulated and common to all meningioma grades. (a) Gene Ontology (GO) enrichment analysis by the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 and corresponding enrichment for proteins is shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. Supplementary Table S7a and b provide details of the GO enrichment analysis and the genes involved. (b) Enrichment map of GO terms enriched in upregulated proteins shown in an interaction network. (c) Enrichment map of GO terms enriched in upregulated phosphoproteins. Nodes represent enriched GO terms with colour intensities reflecting statistical significance and line thickness correlating to the degree of overlap. Numbers refer to descriptions provided in (a) and Supplementary Tables S7c and d. (d) Ingenuity Pathway Analysis of commonly upregulated proteins and phosphoproteins. Pathways significantly associated with two or more proteins or phosphoproteins (Fisher's exact test right-tailed, p-value < .05) were grouped into pathway classes according to Ingenuity Canonical Pathways. Supplementary Tables S7e and f provide details of IPA pathway analysis and pathway classification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Functional annotation of proteins and phosphoproteins significantly upregulated and common to all meningioma grades. (a) Gene Ontology (GO) enrichment analysis by the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 and corresponding enrichment for proteins is shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. Supplementary Table S7a and b provide details of the GO enrichment analysis and the genes involved. (b) Enrichment map of GO terms enriched in upregulated proteins shown in an interaction network. (c) Enrichment map of GO terms enriched in upregulated phosphoproteins. Nodes represent enriched GO terms with colour intensities reflecting statistical significance and line thickness correlating to the degree of overlap. Numbers refer to descriptions provided in (a) and Supplementary Tables S7c and d. (d) Ingenuity Pathway Analysis of commonly upregulated proteins and phosphoproteins. Pathways significantly associated with two or more proteins or phosphoproteins (Fisher's exact test right-tailed, p-value < .05) were grouped into pathway classes according to Ingenuity Canonical Pathways. Supplementary Tables S7e and f provide details of IPA pathway analysis and pathway classification. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To obtain a global view of the biological functions associated with upregulated proteins and phosphoproteins, enriched GO terms were visualised in an ‘enrichment map’ (Fig. 2b and c; Supplementary Table S7c and d). Six clusters of GO terms were identified in the protein enrichment map, the largest comprised of 36 nodes associated with transcription/post-transcriptional modifications, including nodes related to mRNA processing and the spliceosomal complex, which was also identified in two additional clusters (‘splicing machinery’ and ‘spliceosomal complex’) (Fig. 2b). The phosphoprotein enrichment map displayed five clusters of GO terms representing biological functions; the most represented were ‘transcription/pre mRNA processing’ and ‘signal transduction’, with 34 and 21 nodes respectively (Fig. 2c).
Finally, we used IPA to identify canonical pathways significantly associated with upregulated proteins and phosphoproteins. IPA revealed 22 and 198 pathways associated with proteins and phosphoproteins, respectively (Supplementary Table S7e and f). Pathways containing ≥2 proteins or phosphoproteins were grouped under broader functional pathway classes according to Ingenuity Canonical Pathways (Fig. 2d). Classes associated with upregulated proteins included ‘cellular immune response’ and ‘intracellular and second messenger signaling’, with no more than four pathways in a class at any time. Upregulated phosphoproteins were most frequently associated with pathways related to ‘cellular growth, proliferation and development’, ‘cellular immune response’, ‘disease-specific pathways’, ‘cancer’ as well as ‘transcriptional regulation’; within these groups we identified the integrin, EIF2, FAK, ERK/MAPK, paxillin, Rho Family GTPases and G-protein coupled receptor signalling pathways (Supplementary Table S7f).
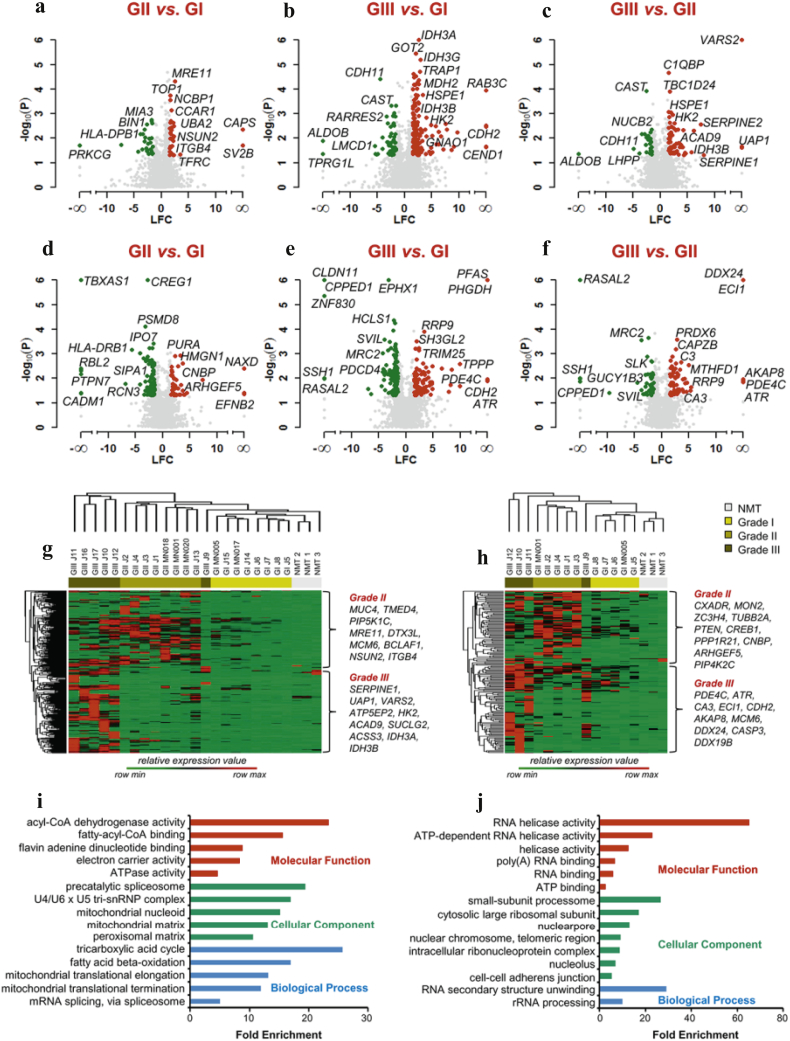
3.5. Differential protein and phosphoprotein expression between meningioma grades
Next, we performed comparative analysis to identify DEPs between grades. Significant differences were assessed by Student's t-tests and DEPs visualised in Volcano plots (Fig. 3; Supplementary Table S8). We identified 260 proteins to be significantly differentially expressed between grade II vs. grade I (Fig. 3a), 424 proteins between grade III vs. grade I (Fig. 3b) and 243 proteins between grade III vs. grade II (Fig. 3c). Proteins significantly upregulated in grade II meningiomas vs. grade I included the DNA damage response protein MRE11, TOP1 involved in DNA replication and NCBP1, a component of the cap-binding complex that binds the 5′-cap of pre-mRNAs and plays a role in RNA biogenesis (Fig. 3a). Within higher-grade specific proteins, we identified the glycolytic enzyme HK2 (hexokinase-2), known for its key role in aerobic glycolysis [33], and the mitochondrial related proteins HSPE1 and IDH3B (Fig. 3b and c).
Fig. 3.
Differential protein and phosphoprotein expression between meningioma grades. Volcano plots of protein and phosphoprotein abundance between different grades of meningiomas. Differential expression of proteins is shown: (a) grade II vs. grade I; (b) grade III vs. grade I and (c) grade III vs. grade II. Differential expression of phosphoproteins is shown: (d) grade II vs. grade I; (e) grade III vs. grade I and (f) grade III vs. grade II. Depicted in the plots are the comparisons of log2 fold changes (LFC) versus p-values (Student's t-test between replicate measurements). Red dots: Upregulated proteins and phosphoproteins (log2 fold change ≥ 1.5; p-value < .05). Green dots: Downregulated proteins and phosphoproteins (log2 fold change ≤ 1.5; p-value < .05). Grey dots: Proteins and phosphoproteins that did not meet these criteria. Proteins and phosphoproteins specific to one of the two groups compared were assigned a fold change of infinity. See also Supplementary Table S8a and b. (g) Hierarchical clustering of the protein molecular signature of grade II and III meningiomas. (h) Hierarchical clustering of the phosphoprotein molecular signature of grade II and III meningiomas. Molecular signatures were generated by the computational tool BubbleGUM (https://omictools.com/bubblegum-tool). Hierarchical clustering created using Perseus 1.5.0.31 software suite. Supplementary Table S9a–d provide details of all molecular signatures. (i) GO enrichment analysis of proteins derived from the molecular signature of GIII meningiomas. (j) GO enrichment analysis of phosphoproteins derived from the molecular signature of GIII meningiomas. GO enrichment analysis was performed using the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 and corresponding enrichment for proteins is shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. Proteins and phosphoproteins associated with these terms are provided in Supplementary Table S9e and f. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Differential protein and phosphoprotein expression between meningioma grades. Volcano plots of protein and phosphoprotein abundance between different grades of meningiomas. Differential expression of proteins is shown: (a) grade II vs. grade I; (b) grade III vs. grade I and (c) grade III vs. grade II. Differential expression of phosphoproteins is shown: (d) grade II vs. grade I; (e) grade III vs. grade I and (f) grade III vs. grade II. Depicted in the plots are the comparisons of log2 fold changes (LFC) versus p-values (Student's t-test between replicate measurements). Red dots: Upregulated proteins and phosphoproteins (log2 fold change ≥ 1.5; p-value < .05). Green dots: Downregulated proteins and phosphoproteins (log2 fold change ≤ 1.5; p-value < .05). Grey dots: Proteins and phosphoproteins that did not meet these criteria. Proteins and phosphoproteins specific to one of the two groups compared were assigned a fold change of infinity. See also Supplementary Table S8a and b. (g) Hierarchical clustering of the protein molecular signature of grade II and III meningiomas. (h) Hierarchical clustering of the phosphoprotein molecular signature of grade II and III meningiomas. Molecular signatures were generated by the computational tool BubbleGUM (https://omictools.com/bubblegum-tool). Hierarchical clustering created using Perseus 1.5.0.31 software suite. Supplementary Table S9a–d provide details of all molecular signatures. (i) GO enrichment analysis of proteins derived from the molecular signature of GIII meningiomas. (j) GO enrichment analysis of phosphoproteins derived from the molecular signature of GIII meningiomas. GO enrichment analysis was performed using the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 and corresponding enrichment for proteins is shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. Proteins and phosphoproteins associated with these terms are provided in Supplementary Table S9e and f. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Among the phosphoproteins we found 404 DEPs between grade II vs. grade I (Fig. 3d), 423 DEPs between grade III vs. grade I (Fig. 3e) and 346 DEPs between grade III vs. grade II (Fig. 3f). Significantly upregulated phosphoproteins in grade II meningiomas included EFNB2, a ligand for the EPH family of RTKs, recently identified to be activated in NF2-deficient meningioma (Fig. 3d) [34]. Among the downregulated phosphoproteins was the MAP kinase-specific phosphatase PTPN7, a negative regulator of ERK2 in K562 leukaemia cells [35] (Fig. 3d). Grade III-specific upregulated phosphoproteins included the cell cycle checkpoint protein ATR, often critical for tumour cell survival during increased levels of DNA replication stress and described as a therapeutic target for advanced solid tumours (Fig. 3e and f) [36].
To better characterise the proteomic landscape of aggressive meningiomas by considering also NMT as control, we extracted individual molecular signatures for grade II and grade III using the computational tool BubbleGUM. A grade II signature was generated by comparison with NMT and grade I, and a grade III signature by comparison with NMT, grade I and grade II, keeping the FC > 2. Hierarchical clustering of relative expression of proteins and phosphoproteins revealed clustering of tumour samples into their respective grades, again with the exception of J9 (Fig. 3g and h). We identified grade II and III signatures of 191 and 192 proteins respectively (Fig. 3g), and grade II and III signatures of 88 and 84 phosphoproteins respectively (Fig. 3h). Among these, were upregulated proteins already visualised in volcano plots (Fig. 3a–f); yet, additional grade-specific proteins were established like the minichromosome maintenance protein MCM6 in grade II meningioma and the grade III-specific mitochondrial proteins ATP5EP2 and SUCLG2 (Fig. 3g). Grade II-specific phosphoproteins included the tumour suppressor PTEN, suggested to contribute to oncogenesis when inactivated by phosphorylation [37] (Fig. 3h). In grade III meningiomas we identified increased expression of the RNA helicase phosphoprotein DDX19B that upon phosphorylation by CHEK1 functions in the DNA damage response in an ATR/CHEK1-dependent manner [38] (Fig. 3h); indeed, ATR was identified as a grade III-specific phosphoprotein in our previous analyses (Fig. 1b; 3e and f). Complete lists of proteins and phosphoproteins are shown in Supplementary Tables S9a–d.
Lastly, GO enrichment analysis was performed to functionally annotate the grade III-specific signature of proteins and phosphoproteins (Fig. 3i and j; Supplementary Table S9e and f). The most enriched protein terms were acyl-CoA dehydrogenase activity (23-fold), precatalytic spliceosome (19-fold) and tricarboxylic acid cycle (26-fold) in the three groups respectively; overall, the majority of enriched terms were associated with mitochondrial function (Fig. 3i). Among grade III-specific phosphoproteins, terms with the strongest enrichments included RNA helicase activity (65-fold), small-subunit processome (26-fold) and RNA secondary structure unwinding (29-fold) in the three groups respectively, with terms mainly linked to RNA activity and ribosome biogenesis (Fig. 3j).
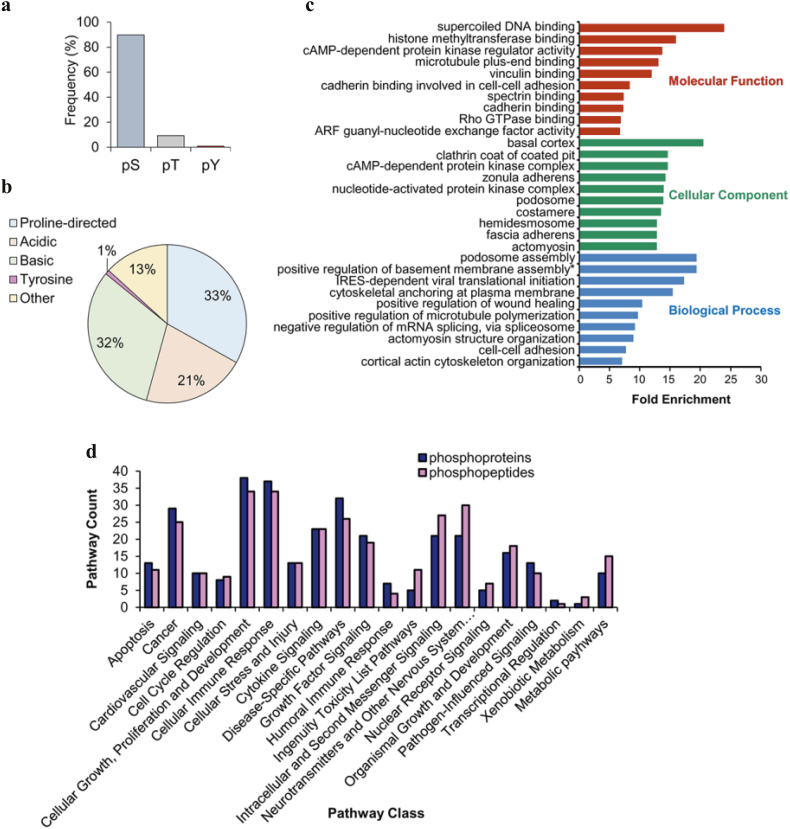
3.6. Compendium of protein phosphorylation sites in meningiomas
In order to identify specific phosphorylation sites, we enriched and analysed phosphorylated peptides by MS. Overall, we sequenced and identified 3622 phosphopeptides, of which 2729 (75%) were identified with a phosphosite post-translational modification (PTM) localization probability score > 0.75 (Supplementary Table S10a). Our distribution of serine (~90%), threonine (~9%) and tyrosine (~1%) phosphorylation sites (Fig. 4a) were similar to previous studies [39]. We further classified phosphopeptides based on their chemical properties using a binary decision tree [26]. The majority of sites were classified as either proline-directed (33%) or basic (32%), followed by acidic (21%), other (13%) and tyrosine (1%) (Fig. 4b). The 3622 identified phosphopeptides covered 1320 proteins, of which 56% overlapped with the phosphoprotein dataset. GO analysis of the overlap showed a strong enrichment in terms related to the cellular membrane and its adhesion properties (e.g. basal cortex, zonula adherens, podosome assembly), microtubule regulation, cytoskeletal organization and DNA binding (Fig. 4c; Supplementary Table S10b). Additionally, we analysed the 1320 proteins associated with the enriched phosphopeptides using IPA and identified 211 significantly associated canonical pathways (Fig. 4d; Supplementary Table S10c). Pathways were grouped under broader functional classes as before. Proteins identified by phosphopeptides showed a similar trend of pathway classification when compared to the identified upregulated phosphoproteins (Fig. 4d); the majority of pathways were associated with ‘cellular growth, proliferation and development’ (34 pathways) and ‘cellular immune response’ (34 pathways).
Fig. 4.
Compendium of protein phosphorylation sites in meningiomas. (a) Distribution of amino acid phosphorylation for serine, threonine and tyrosine across meningiomas. See Supplementary Table S10a for all phosphorylation sites. (b) Distribution of phosphorylation site classes in meningiomas. The phosphorylated residue is located at the central position within a sequence window of 13 amino acids. Classes were defined by the chemical properties of the sequence window peptide as acidic, basic, proline-directed, tyrosine or other by a binary decision tree method [26]. (c) GO enrichment analysis of 738 phosphoproteins common to the phosphoprotein and phosphopeptide datasets performed by the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 are shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. *positive regulation of basement membrane assembly = positive regulation of basement membrane assembly involved in embryonic body morphogenesis. Phosphoproteins associated with these terms are provided in Supplementary Table S10b. (d) Ingenuity Pathway Analysis of commonly upregulated phosphoproteins and proteins associated with identified phosphopeptides. Pathways significantly associated with two or more phosphoproteins (Fisher's exact test right-tailed, p-value < .05) were grouped into pathway classes according to Ingenuity Canonical Pathways. Supplementary Tables S7f and S10c provide details of IPA pathway analysis and pathway classification. Grade I n = 3; grade II n = 3 and grade III n = 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Compendium of protein phosphorylation sites in meningiomas. (a) Distribution of amino acid phosphorylation for serine, threonine and tyrosine across meningiomas. See Supplementary Table S10a for all phosphorylation sites. (b) Distribution of phosphorylation site classes in meningiomas. The phosphorylated residue is located at the central position within a sequence window of 13 amino acids. Classes were defined by the chemical properties of the sequence window peptide as acidic, basic, proline-directed, tyrosine or other by a binary decision tree method [26]. (c) GO enrichment analysis of 738 phosphoproteins common to the phosphoprotein and phosphopeptide datasets performed by the web tool DAVID v6.8. Terms containing at least three phosphoproteins with Benjamini-Hochberg adjusted p < .05 are shown. Gene Ontologies representing Molecular Function are presented in red, Cellular Component in green and Biological Processes in blue. Fold enrichment relative to the H. sapiens proteome is displayed at the x-axis. *positive regulation of basement membrane assembly = positive regulation of basement membrane assembly involved in embryonic body morphogenesis. Phosphoproteins associated with these terms are provided in Supplementary Table S10b. (d) Ingenuity Pathway Analysis of commonly upregulated phosphoproteins and proteins associated with identified phosphopeptides. Pathways significantly associated with two or more phosphoproteins (Fisher's exact test right-tailed, p-value < .05) were grouped into pathway classes according to Ingenuity Canonical Pathways. Supplementary Tables S7f and S10c provide details of IPA pathway analysis and pathway classification. Grade I n = 3; grade II n = 3 and grade III n = 2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. Validation of identified proteins and phosphoproteins
We validated our proteomic results with a subset of proteins and phosphoproteins identified as upregulated in meningiomas compared to NMT. We validated by WB on a selection of the discovery set samples as well as on a validation set of 15 meningiomas (Fig. 5a; Supplementary Fig. S3). NF2 expression of the validation set is shown in Supplementary Fig. S1b.
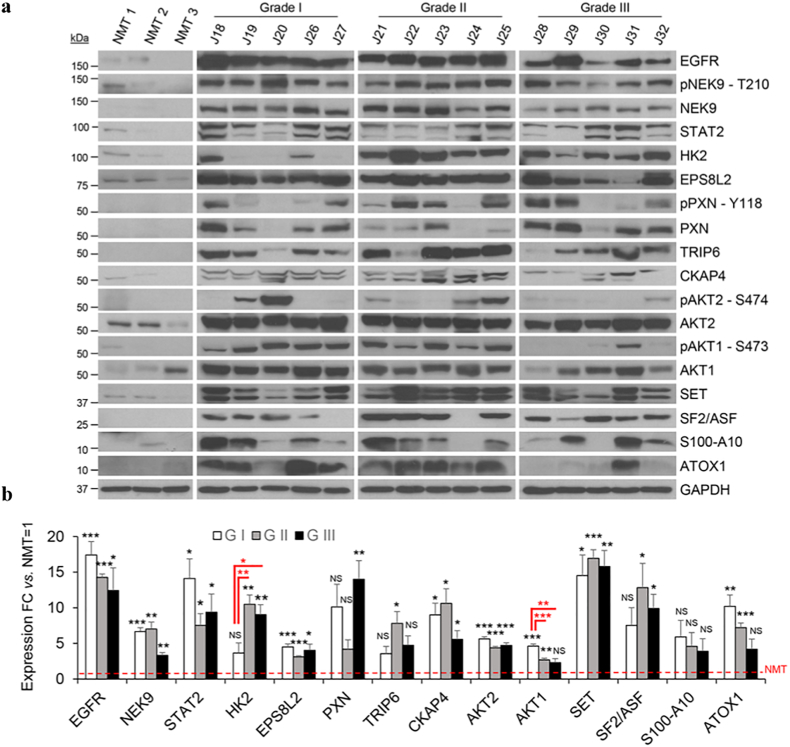
Fig. 5.
Western blot validation of proteins and phosphoproteins identified as significantly upregulated in meningiomas. (a) Western blot analysis of EGFR, pNEK9-T210, NEK9, STAT2, HK2, EPS8L2, pPXN-Y118, PXN, TRIP6, CKAP4, pAKT2-S474, pAKT-S473, SET, SF2/ASF, S100-A10, ATOX1 across different grades of meningioma and NMT. GAPDH was used as loading control. Clinical details of the validation set used in this figure are presented in Supplementary Table S2. (b) Histogram representing Western blot quantification of EGFR, NEK9, STAT2, HK2, EPS8L2, PXN, TRIP6, CKAP4, AKT2, AKT1, SET, SF2/ASF, S100-A10 and ATOX1 shown in (a) compared to levels in NMT. Statistical significance is shown by: *p ≤ .05; **p ≤ .01; ***p ≤ .001; NS: not significant. NMT n = 3; grade I n = 5, grade II n = 5 and grade III n = 5.
We confirmed the significant overexpression of EGFR, NEK9, STAT2, EPS8L2, CKAP4 and SET in all grades compared to NMT by WB with an average of ~15 folds for EGFR, ~6 folds for NEK9, ~10 folds for STAT2, ~4 folds for EPS8L2 ~9 folds for CKAP4 and ~16 folds for SET (Fig. 5a and b). Further, WB quantification established HK2 as statistically overexpressed in grade II and III vs. grade I meningiomas; indeed, HK2 was identified as a higher-grade specific protein in our earlier analyses (Fig. 5b; 3b and c). By WB, we additionally confirmed the overexpression of PXN, TRIP6, S100-A10, SF2/ASF and ATOX1; all the proteins listed above showed immunoreactivity in the majority of meningiomas with weak or absent expression in NMT (Fig. 5a and b).
Among the phosphoproteins, we confirmed the overexpression of pPXN-Y118, although variable across meningioma samples and grades, and pNEK9-T210, which was phosphorylated across all samples and grades with weak expression in NMT (Fig. 5a). Lastly, pAKT2-S474 demonstrated variable expression in grade I and II meningiomas and was not detected in grade III, whilst the phosphorylation of AKT1 on S473 was detected predominantly in grade I and II meningiomas (Fig. 5a). Total AKT1 and AKT2 were identified in all grades (Fig. 5a).
As a proof of principle we further investigated the potential of pAKT1-S473 as a therapeutic target in low-grade meningioma in vitro using the benign meningioma cell line, BM1, and the malignant meningioma cell line, KT21 [28,29]. First, we established activation of the PI3K/AKT/mTOR pathway in the meningioma validation set by the phosphorylation of two downstream targets, GSK3β and PRAS40 [40,41]. Expression of pGSK3β-S9 and pPRAS40-T246 followed a trend similar to pAKT1-S473, displaying lower expression in grade III compared to grade I and II meningiomas (Supplementary Fig. S4a and b). Next, we confirmed that whilst both BM1 and KT21 showed increased pAKT1-S473 levels compared to normal human meningeal cells (HMC), KT21 demonstrated a significant reduction in pAKT1-S473 compared to BM1 by WB (Supplementary Fig. S4c and d). Cells were then treated with the pan-AKT kinase inhibitor, AZD5363, previously shown to be effective in the treatment of metastatic grade I meningothelial meningioma [42]. Treatment of BM1 and KT21 cells with AZD5363 reduced phosphorylation of GSK3β and PRAS40 30 min post-treatment, yet following treatment with increasing concentrations of AZD5363 the IC50 of BM1 and KT21 cells were high at 33·1 μM and 67·8 μM, respectively (Supplementary Fig. S4e and f). Next, we sought to determine if inhibition of the mTOR complex, previously described to be activated in meningioma, may be a more promising target [43]. We selected the dual mTOR inhibitor Ku-0063794 that has demonstrated preclinical efficacy in reducing the viability of renal cell carcinoma cell lines [44,45]. Following treatment of BM1 and KT21 cells with Ku-0063794, pGSK3β-S9 levels in KT21 cells decreased to less than half that of vehicle-treated cells but not in BM1, whilst pPRAS40-T246 displayed similar reduction in BM1 and KT21 cells (Supplementary Fig. S4g). Following treatment of BM1 and KT21 cells with Ku-0063794, BM1 cells showed a greater reduction in cell viability with an IC50 of 1·1 μM, whereas KT21 cells reached IC50 at 3·6 μM, over double that of BM1 (Supplementary Fig. S4h).
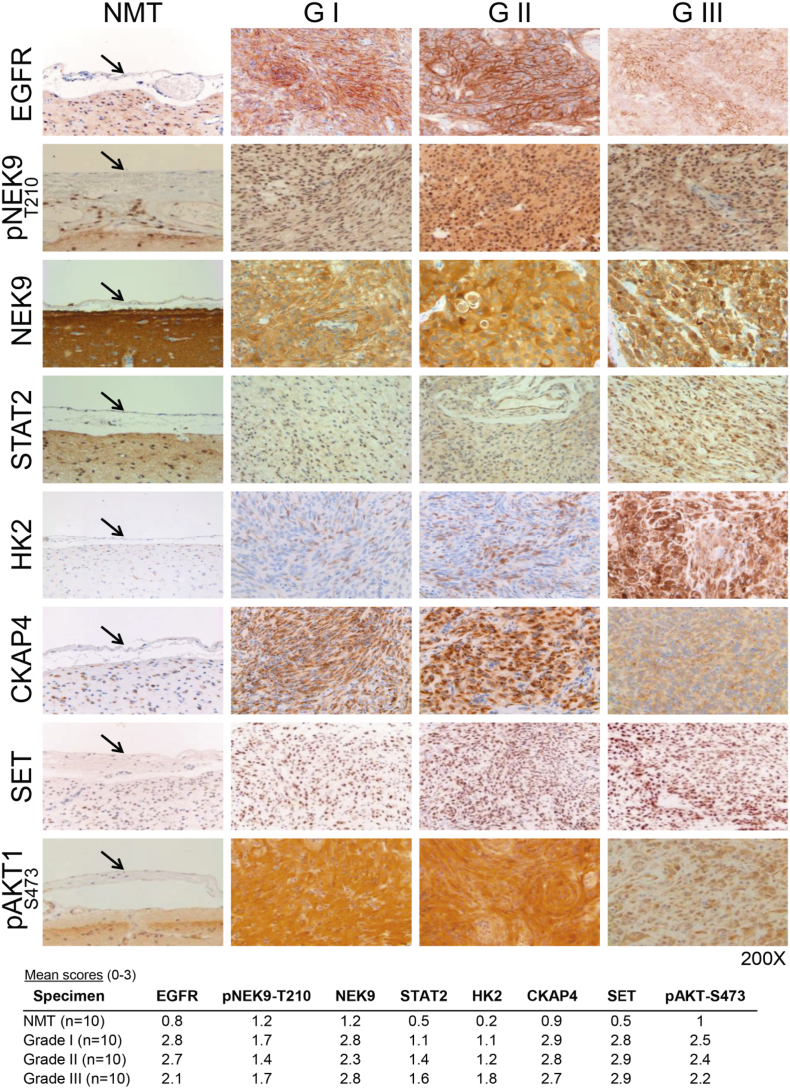
In addition to validation by WB, we performed IHC studies comprising 10 sections per sample group (NMT, grade I, II and III) to examine the expression levels of selected proteins and phosphoproteins. Importantly, this was not just another validation set but allowed expansion of the control cohort. Subsequent to immunostaining, we found EGFR, pNEK9-T210, NEK9, STAT2, HK2, CKAP4, SET and pAKT1-S473 exhibited similar patterns of expression across meningioma grades as those seen by WB (Fig. 6). In the majority of cases, immunostaining was stronger in meningiomas than in control meninges with relatively little difference between grades (Fig. 6; Supplementary Table S11). Localisation of NEK9, STAT2, HK2 and CKAP4 appeared cytoplasmic; SET was found to be predominantly nuclear, while EGFR was localised to the cytoplasm and plasma membrane (Fig. 6). In contrast to the cytoplasmic immunostaining of NEK9, we observed a strong nuclear presence of pNEK9-T210 (Fig. 6).
Fig. 6.
Immunohistochemistry validation of proteins and phosphoproteins identified as significantly upregulated in meningiomas. Representative images showing immunohistochemical staining of EGFR, pNEK9-T210, NEK9, STAT2, HK2, CKAP4, SET and pAKT-S473 in all meningioma grades compared to NMT (see black arrow for normal meninges). Mean intensity scores are presented below for meningioma specimens and normal meninges examined. Supplementary Table S11 details the full list of specimens and corresponding semi-quantitative assessment. NMT n = 10; grade I n = 10, grade II n = 10 and grade III n = 10.
Immunohistochemistry validation of proteins and phosphoproteins identified as significantly upregulated in meningiomas. Representative images showing immunohistochemical staining of EGFR, pNEK9-T210, NEK9, STAT2, HK2, CKAP4, SET and pAKT-S473 in all meningioma grades compared to NMT (see black arrow for normal meninges). Mean intensity scores are presented below for meningioma specimens and normal meninges examined. Supplementary Table S11 details the full list of specimens and corresponding semi-quantitative assessment. NMT n = 10; grade I n = 10, grade II n = 10 and grade III n = 10.
4. Discussion
Meningiomas are the most frequent primary CNS tumours and currently there is no effective pharmacological therapy. We performed proteomic analyses of frozen meningiomas of all grades compared to healthy meninges to decipher the molecular signature of these tumours. In addition to global proteome profiling, we used a dual enrichment approach; employing affinity chromatography to enrich phosphoproteins and TiO2 beads to enrich phosphopeptides for a comprehensive analysis of the meningioma phosphoproteome. The two datasets shared 56% of the identified phosphoproteins and several canonical pathways. The number of phosphoproteins identified by phosphoprotein enrichment was more than double that identified by phosphopeptide enrichment, thus we performed in-depth bioinformatics on the phosphoprotein dataset first to elucidate dysregulated protein activity and biological functions that underlie meningioma pathogenesis.
Initially, we analysed data by unsupervised hierarchical clustering based on DEPs across all grades meningioma and NMT as control. For both global proteome and enriched phosphoproteins, we had a clear separation of tumours vs. controls, as well as a separation between tumour grades. The only exception was the grade III sample J9, which was grouped together with grade II tumours; however, since meningiomas can progress from a grade II to a grade III, we cannot exclude that this tumour still retains some grade II features.
We identified 181 proteins and 338 phosphoproteins commonly and significantly upregulated among all meningioma grades compared to NMT. Among the commonly upregulated proteins that were further validated, we identified both isoforms of the nuclear proto-oncogene SET (I2PP2A), which participates in many cellular processes including cell-cycle regulation, gene transcription, epigenetic regulation and cell migration [46]. SET is overexpressed in other cancers, contributing to tumourigenesis in part through its inhibition of the tumour suppressor phosphatase PP2A, which acts as a negative regulator of cellular growth and survival pathways [46]. Both SET isoforms are components of the inhibitor of acetyltransferases (INHAT) complex shown to bind histones, predominantly histone H4, and inhibit histone acetyltransferase activity thus regulating chromatin modification and transcriptional activity [47]. Interestingly, histone H4 was another commonly upregulated protein identified by our global proteome analysis. SET is also part of a complex that localises to the endoplasmic reticulum but was found in the nucleus inhibiting apoptosis after cytotoxic T lymphocyte attack [48]. This protein has never been studied in meningioma, but deciphering the causes of its overexpression and identifying the binding partners could clarify which role it plays in the pathogenesis and whether this could lead to a therapeutic intervention.
Another commonly and significantly upregulated protein among all grades was STAT2, a STAT-family member involved in immune response through activation of the JAK/STAT pathway after stimulation with type I IFNs (IFN-alpha and IFN-beta) [49]. Activated STAT2 forms heterodimers with STAT1, also found to be commonly upregulated (Table 1) and previously identified by our group as a common target in NF2-negative schwannomas and meningioma BM1 cells, and further validated as overexpressed in meningioma [16] (Ferluga et al. unpublished). These results, together with the identification of ‘cellular immune response’ and ‘cytokine signalling’ as two of the most upregulated pathway classes identified by both phosphoprotein and phosphopeptide enrichment, suggest that the tumour microenviroment and immunomodulatory molecules could have a pivotal role in meningioma development and progression, and present the STAT protein family as plausible targets.
In our previous study we also identified EPS8L2 [16], which here was again found commonly overexpressed in all meningioma grades as both total and phosphorylated protein. EPS8L2 is a member of the EPS8 family, comprising also EPS8L1 and EPS8L3; the role of the –like forms is still unclear, although it was proposed that they can substitute EPS8 function [50]. High levels of EPS8 were able to induce nuclear translocation and activation of YAP, a member of the Hippo pathway that is usually active in NF2-negative meningiomas [51]. EPS8 is also a substrate for EGFR and it participates in both EGFR signalling through Rac, and vesicular trafficking through Rab5 [52], which is required for the fusion of plasma membranes and early endosomes [53]. ‘Cortical actin cytoskeleton organization’ and ‘Cytoskeletal anchoring at plasma membrane’ are two GO terms identified in the phosphopeptide dataset, and ‘Transport vesicle’ is a common GO term for both total and phosphoproteins, so the signalling downstream EGFR and the role of EPS8L2 in meningioma cells certainly warrant further investigation.
One additional validated protein that associates with EGFR and we find commonly upregulated was CKAP4. This protein is a high-affinity epithelial cell surface receptor for the antiproliferative factor secreted by bladder epithelial cells [54] and plays an important role in mediating the anchoring of the ER to microtubules, but its biological function in meningioma is unknown. Again, this protein connects the ‘EGFR signalling’ and ‘positive regulation of microtubule polymerization’ GO terms and its role should be further deciphered.
Comparing our global proteome dataset with the previously generated proteomes by Sharma et al. [13], who similarly analysed all meningioma grades, we identified an overlap of 1428 proteins. Among our 181 upregulated DEPs common to all grades, 25 were also found by Sharma et al., including validated proteins like CKAP4 and S100-A10. Further, we identified MX1, ANP32E, SUPT16H, NEK9 and DDX42 as commonly upregulated among all grades, which were previously shown by Saydam et al. to be expressed in a human benign meningioma cell line vs. human primary arachnoidal cells [32].
Among the phosphoproteins that were commonly and significantly upregulated in all grades compared to NMT we identified phosphoproteins that are well characterised in cancer like phopsho-RB1, phospho-SRC or phospho-FAK, and others that are novel. Validation studies on phosphoproteins are more challenging as they rely on availability of phospho-specific antibodies. Here, we validated the overexpression and activation of PXN across all grades that works as a scaffold at the focal adhesions sites interacting with structural and signalling proteins like FAK and SRC. Following phosphorylation in response to growth factors and integrin-mediated cell adhesion, phospho-PXN regulates cell proliferation, adhesion and motility [55]. Further to identification of the enriched GO term ‘focal adhesion’, we observed phospho-PXN was expressed in two-thirds of the meningiomas analysed but not in NMT, so the therapeutic potential of targeting this signalling should be further investigated.
One additional validated protein, which was significantly overexpressed and phosphorylated across all grades but not in NMT was NEK9, a serine/threonine-protein kinase of the NIMA family protein kinases that together with other members of the family functions in the regulation of mitosis [56]. In particular, NEK9 controls G1/S transition and S phase progression when phosphorylated on T210 and associated with the heterodimeric complex FACT, formed by the proteins SPT16 and SSRP1 [57]; indeed, both proteins were also identified in our proteome dataset as upregulated in all grades compared to NMT, like the validated phospho-NEK9. Although NEK9 has been previously identified to be exclusively expressed in a benign meningioma cell line compared to primary arachnoidal cells, its phosphorylated active form has not been shown in meningioma until now [32]. Furthermore, NEK9 activates the family members NEK6 and NEK7, and together the three proteins function in mitotic progression with a role in mitotic spindle formation, a GO term established as enriched among phosphoproteins identified as commonly upregulated among all grades in our dataset [58]. Phosphorylated NEK7 was also identified as commonly overexpressed in all grades compared to NMT and detected in our phosphopeptide analysis to be phosphorylated on S195, the phosphorylation of which is catalysed by NEK9 [58]. Recent studies have identified NEK9 as a potent target of the FDA-approved kinase inhibitor, dabrafenib, inhibiting growth of NRAS- and KRAS-mutant cancer cells [59]. Our findings thus present NEK9, as well as NEK7 and the FACT complex as potential novel drug targets in meningioma, proposing their further validation.
In addition to identifying common molecular signatures between grades, we wanted to characterise signatures specific to grade II and III as these tumours are more therapeutically challenging. Among the 191 proteins defining a grade II signature we found mucin-4 (MUC4), its overexpression enhanced proliferation and invasive potential of GBM cells by upregulating EGFR expression [60]; indeed, silencing EGFR in pancreatic cancer cells decreased MUC4 expression by reducing the phosphorylation of STAT1 [61]. Within the grade III-specific proteins we identified the glycolytic enzyme HK2, known for its key role in aerobic glycolysis and thus, the “Warburg effect” in rapidly growing cancer cells [33]. HK2 has previously been associated with the promotion of tumour growth, migration and metastasis in GBM and pancreatic cancer [62,63]. HK2 was further validated, demonstrating significantly higher expression in grade II and III meningiomas compared to NMT and grade I as well as clearly displaying a much stronger immunoreactivity in IHC in grade III meningiomas. Indeed, the inhibition of HK2 in cancer by administration of small-molecule inhibitors such as 3-bromopyruvate and the development of selective HK2 inhibitors indicate the potential of targeting this enzyme in high-grade meningioma [64,65].
We also validated expression and activation of AKT1 and AKT2 as possible therapeutic targets. Clark et al. described a constitutively active mutation of AKT1 (AKT1E17K) in NF2-positive meningiomas [5], and other studies have shown that inhibition of phospho-AKT1 by the Integrin-Linked Kinase inhibitor OSU-T315 in BM1 cells was able to arrest cell cycle and induce cell death [66]. Further, expression of AKT2 in meningioma was previously described as a potential prognostic marker [67]. Based on our validation, AKT1 and AKT2 were both overexpressed in all grades, and pAKT1-S473 was persistently identified in grade I and II. We conducted preliminary functional validation experiments as proof of principle to determine if the inhibition of pAKT1-S473 was a more effective therapeutic target in low rather than high-grade meningioma. However, we observed AKT inhibition did not produce a strong reduction in cell viability independent of grade. Instead, we demonstrated inhibition with a dual mTOR inhibitor may prove to have increased efficacy in lower, compared to higher-grade meningioma. An additional possibility for aberrant AKT1 activation in meningioma could be CKAP4, a protein also identified as consistently overexpressed in grade I and II meningiomas in our data, where pAKT1 was mainly detected. Recent studies of pancreatic and lung cancers have related the activation of AKT1 to CKAP4 and its binding to the secretory protein Dickkopf1, leading to the downstream activation of PI3K and enhanced cellular proliferation [68].
Interestingly, our data highlighted an enrichment of GO terms related to RNA metabolism such as RNA helicase activity and splicing, not yet described in meningioma. We repeatedly identified many members of the DEAD-box family of RNA helicases by both global proteome and phosphoprotein analysis including DDX5, DDX6, DDX10, DDX17, DDX19B DDX24, DDX39A and DDX42 to be significantly upregulated in one or more grades of meningioma compared to NMT. Furthermore, we identified several components of the spliceosome as upregulated in meningioma including SNRPD3, SNRPF, SNRPE, SF3B2, SF3B5 as well as the splicing factors SRSF3, SRSF7 and SF2/ASF, the latter of which we validated as overexpressed in meningioma compared to NMT. Upregulation of SF2/ASF has been shown in many different human cancers and its overexpression causes oncogenic transformation of immortal rodent fibroblasts that can form sarcomas in nude mice [69]. SF2/ASF may contribute to tumourigenesis through aberrant splicing of its target genes including the tumour suppressor BIN1, identified as downregulated in our analysis of grade II vs. grade I meningioma. Aberrant splicing by SF2/ASF promotes an alternative BIN1 isoform lacking its tumour suppressive pro-apoptotic function [69]. Taken together, these results propose further investigation into RNA metabolism in meningioma may reveal targetable pathways and protein families, in addition to increasing our knowledge of the underlying molecular pathogenesis of these tumours.
In summary, we performed a comprehensive analysis of proteins and phosphoproteins in meningioma tissue of all grades compared to healthy meninges. We identified many relevant differentially expressed and aberrantly phosphorylated proteins common to all or grade-enriched. We validated a panel of candidates that following further studies incorporating different tumours may be potential biomarkers for all grades, like SET, STAT2, NEK9 and pNEK9, AKT1 and AKT2; and proteins/phosphoproteins enriched in grade I-II, like ATOX1, CKAP4 and pAKT1 or in grade III like HK2. Finally, we discussed pathways that appear to have a pivotal role in meningioma pathogenesis, like RNA metabolism, EGFR signalling, integrin-mediated cellular adhesion and cellular immune response.
The following are the supplementary data related to this article.
Supplementary material
Proteins and phosphoproteins identified by LC-MS/MS in meningiomas and meninges.
Differentially expressed proteins and phosphoproteins of each meningioma grade vs. normal meningeal tissue.
Functional annotation enrichment analysis results from DAVID and IPA.
Statistical analysis of quantified proteins and phosphoproteins visualised in Volcano plots.
Molecular signature and GO enrichment analysis of grade II and III meningiomas.
Protein phosphorylation sites in meningiomas and functional annotation of phosphopeptides by DAVID and IPA.
Semi-quantitative scoring of immunohistochemical staining for validation of proteins and phosphoproteins.
Acknowledgments
Acknowledgements
COH was supported by the charity Brain Tumour Research, SF by Eric Reid Fund. Tissue samples were obtained from NHS archives as part of the UK Brain Archive Information Network (BRAIN UK) which is funded by the Medical Research Council and Brain Tumour Research.
Funding sources
This study was supported by a grant from Brain Tumour Research. The funders had no role in study design, data collection, data analysis and interpretation, writing of the manuscript or decision to publish.
Declaration of interests
No conflicts of interest.
Author contributions
Conception and design: CO Hanemann.
Development of methodology: S Ferluga, J Dunn.
Acquisition of data: J Dunn, S Ferluga, V Sharma, D Hilton, CL Adams.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J Dunn, S Ferluga, M Futschik, CO Hanemann, E Lasonder.
Writing, review, and/or revision of the manuscript: J Dunn, S Ferluga, CO Hanemann, E Lasonder.
Study supervision: CO Hanemann.
References
- 1.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Marosi C., Hassler M., Roessler K., Reni M., Sant M., Mazza E. Meningioma. Crit Rev Oncol Hematol. 2008;67(2):153–171. doi: 10.1016/j.critrevonc.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 3.Sahm F., Schrimpf D., Stichel D., Jones D.T.W., Hielscher T., Schefzyk S. DNA methylation-based classification and grading system for meningioma: a multicentre, retrospective analysis. Lancet Oncol. 2017;18(5):682–694. doi: 10.1016/S1470-2045(17)30155-9. [DOI] [PubMed] [Google Scholar]
- 4.Clark V.E., Harmanci A.S., Bai H., Youngblood M.W., Lee T.I., Baranoski J.F. Recurrent somatic mutations in POLR2A define a distinct subset of meningiomas. Nat Genet. 2016;48(10):1253–1259. doi: 10.1038/ng.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark V.E., Erson-Omay E.Z., Serin A., Yin J., Cotney J., Ozduman K. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–1080. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuzawa S., Nishihara H., Tanaka S. Genetic landscape of meningioma. Brain Tumor Pathol. 2016;33(4):237–247. doi: 10.1007/s10014-016-0271-7. [DOI] [PubMed] [Google Scholar]
- 7.Harmanci A.S., Youngblood M.W., Clark V.E., Coskun S., Henegariu O., Duran D. Integrated genomic analyses of de novo pathways underlying atypical meningiomas. Nat Commun. 2017;8:14433. doi: 10.1038/ncomms14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar G.M., Abedalthagafi M., Vaubel R.A., Merrill P.H., Nayyar N., Gill C.M. Germline and somatic BAP1 mutations in high-grade rhabdoid meningiomas. Neuro Oncol. 2017;19(4):535–545. doi: 10.1093/neuonc/now235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton D.A., Shivane A., Kirk L., Bassiri K., Enki D.G., Hanemann C.O. Activation of multiple growth factor signalling pathways is frequent in meningiomas. Neuropathology. 2016;36(3):250–261. doi: 10.1111/neup.12266. [DOI] [PubMed] [Google Scholar]
- 10.Norden A.D., Raizer J.J., Abrey L.E., Lamborn K.R., Lassman A.B., Chang S.M. Phase II trials of erlotinib or gefitinib in patients with recurrent meningioma. J Neurooncol. 2010;96(2):211–217. doi: 10.1007/s11060-009-9948-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaley T.J., Wen P., Schiff D., Ligon K., Haidar S., Karimi S. Phase II trial of sunitinib for recurrent and progressive atypical and anaplastic meningioma. Neuro Oncol. 2015;17(1):116–121. doi: 10.1093/neuonc/nou148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto H., Li J., Vortmeyer A.O., Jaffe H., Lee Y.S., Glasker S. Comparative proteomic profiles of meningioma subtypes. Cancer Res. 2006;66(20):10199–10204. doi: 10.1158/0008-5472.CAN-06-0955. [DOI] [PubMed] [Google Scholar]
- 13.Sharma S., Ray S., Mukherjee S., Moiyadi A., Sridhar E., Srivastava S. Multipronged quantitative proteomic analyses indicate modulation of various signal transduction pathways in human meningiomas. Proteomics. 2015;15(2–3):394–407. doi: 10.1002/pmic.201400328. [DOI] [PubMed] [Google Scholar]
- 14.Parada C.A., Osbun J., Kaur S., Yakkioui Y., Shi M., Pan C. Kinome and phosphoproteome of high-grade meningiomas reveal AKAP12 as a central regulator of aggressiveness and its possible role in progression. Sci Rep. 2018;8(1):2098. doi: 10.1038/s41598-018-19308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meimoun P., Ambard-Bretteville F., Colas-des Francs-Small C., Valot B., Vidal J. Analysis of plant phosphoproteins. Anal Biochem. 2007;371(2):238–246. doi: 10.1016/j.ab.2007.08.022. [DOI] [PubMed] [Google Scholar]
- 16.Bassiri K., Ferluga S., Sharma V., Syed N., Adams C.L., Lasonder E. Global proteome and phospho-proteome analysis of merlin-deficient meningioma and schwannoma identifies PDLIM2 as a novel therapeutic target. EBioMedicine. 2017;16:76–86. doi: 10.1016/j.ebiom.2017.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lasonder E., Green J.L., Camarda G., Talabani H., Holder A.A., Langsley G. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase a signaling. J Proteome Res. 2012;11(11):5323–5337. doi: 10.1021/pr300557m. [DOI] [PubMed] [Google Scholar]
- 18.Fukuda I., Hirabayashi-Ishioka Y., Sakikawa I., Ota T., Yokoyama M., Uchiumi T. Optimization of enrichment conditions on TiO2 chromatography using glycerol as an additive reagent for effective phosphoproteomic analysis. J Proteome Res. 2013;12(12):5587–5597. doi: 10.1021/pr400546u. [DOI] [PubMed] [Google Scholar]
- 19.Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 20.Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 21.Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 23.Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 24.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Merico D., Isserlin R., Bader G.D. Visualizing gene-set enrichment results using the Cytoscape plug-in enrichment map. Methods Mol Biol. 2011;781:257–277. doi: 10.1007/978-1-61779-276-2_12. [DOI] [PubMed] [Google Scholar]
- 26.Villen J., Beausoleil S.A., Gerber S.A., Gygi S.P. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104(5):1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timpson N.J., Walter K., Min J.L., Tachmazidou I., Malerba G., Shin S.Y. A rare variant in APOC3 is associated with plasma triglyceride and VLDL levels in Europeans. Nat Commun. 2014;5:4871. doi: 10.1038/ncomms5871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puttmann S., Senner V., Braune S., Hillmann B., Exeler R., Rickert C.H. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab Invest. 2005;85(9):1163–1171. doi: 10.1038/labinvest.3700307. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka K., Sato C., Maeda Y., Koike M., Matsutani M., Yamada K. Establishment of a human malignant meningioma cell line with amplified c-myc oncogene. Cancer. 1989;64(11):2243–2249. doi: 10.1002/1097-0142(19891201)64:11<2243::aid-cncr2820641110>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 30.Vizcaino J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingues P.H., Teodosio C., Ortiz J., Sousa P., Otero A., Maillo A. Immunophenotypic identification and characterization of tumor cells and infiltrating cell populations in meningiomas. Am J Pathol. 2012;181(5):1749–1761. doi: 10.1016/j.ajpath.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 32.Saydam O., Senol O., Schaaij-Visser T.B., Pham T.V., Piersma S.R., Stemmer-Rachamimov A.O. Comparative protein profiling reveals minichromosome maintenance (MCM) proteins as novel potential tumor markers for meningiomas. J Proteome Res. 2010;9(1):485–494. doi: 10.1021/pr900834h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lis P., Dylag M., Niedzwiecka K., Ko Y.H., Pedersen P.L., Goffeau A. The HK2 dependent “Warburg effect” and mitochondrial oxidative phosphorylation in cancer: targets for effective therapy with 3-bromopyruvate. Molecules. 2016;21(12) doi: 10.3390/molecules21121730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angus S.P., Oblinger J.L., Stuhlmiller T.J., DeSouza P.A., Beauchamp R.L., Witt L. EPH receptor signaling as a novel therapeutic target in NF2-deficient meningioma. Neuro Oncol. 2018;20(9):1185–1196. doi: 10.1093/neuonc/noy046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettiford S.M., Herbst R. The MAP-kinase ERK2 is a specific substrate of the protein tyrosine phosphatase HePTP. Oncogene. 2000;19(7):858–869. doi: 10.1038/sj.onc.1203408. [DOI] [PubMed] [Google Scholar]
- 36.Karnitz L.M., Zou L. Molecular pathways: targeting atr in cancer therapy. Clin Cancer Res. 2015;21(21):4780–4785. doi: 10.1158/1078-0432.CCR-15-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z., Yuan X.G., Chen J., Luo S.W., Luo Z.J., Lu N.H. Reduced expression of PTEN and increased PTEN phosphorylation at residue Ser380 in gastric cancer tissues: a novel mechanism of PTEN inactivation. Clin Res Hepatol Gastroenterol. 2013;37(1):72–79. doi: 10.1016/j.clinre.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Hodroj D., Recolin B., Serhal K., Martinez S., Tsanov N., Abou Merhi R. An ATR-dependent function for the Ddx19 RNA helicase in nuclear R-loop metabolism. EMBO J. 2017;36(9):1182–1198. doi: 10.15252/embj.201695131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou H., Di Palma S., Preisinger C., Peng M., Polat A.N., Heck A.J. Toward a comprehensive characterization of a human cancer cell phosphoproteome. J Proteome Res. 2013;12(1):260–271. doi: 10.1021/pr300630k. [DOI] [PubMed] [Google Scholar]
- 40.Wang H., Zhang Q., Wen Q., Zheng Y., Lazarovici P., Jiang H. Proline-rich Akt substrate of 40kDa (PRAS40): a novel downstream target of PI3k/Akt signaling pathway. Cell Signal. 2012;24(1):17–24. doi: 10.1016/j.cellsig.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Cross D.A., Alessi D.R., Cohen P., Andjelkovich M., Hemmings B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 42.Weller M., Roth P., Sahm F., Burghardt I., Schuknecht B., Rushing E.J. Durable control of metastatic AKT1-mutant WHO grade 1 meningothelial meningioma by the AKT inhibitor, AZD5363. J Natl Cancer Inst. 2017;109(3):1–4. doi: 10.1093/jnci/djw320. [DOI] [PubMed] [Google Scholar]
- 43.James M.F., Han S., Polizzano C., Plotkin S.R., Manning B.D., Stemmer-Rachamimov A.O. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29(15):4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Martinez J.M., Moran J., Clarke R.G., Gray A., Cosulich S.C., Chresta C.M. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421(1):29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Berel D., Wang Y., Li P., Bhowmick N.A., Figlin R.A. A comparison of Ku0063794, a dual mTORC1 and mTORC2 inhibitor, and temsirolimus in preclinical renal cell carcinoma models. PLoS One. 2013;8(1):e54918. doi: 10.1371/journal.pone.0054918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mukhopadhyay A., Tabanor K., Chaguturu R., Aldrich J.V. Targeting inhibitor 2 of protein phosphatase 2A as a therapeutic strategy for prostate cancer treatment. Cancer Biol Ther. 2013;14(10):962–972. doi: 10.4161/cbt.25943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seo S.B., McNamara P., Heo S., Turner A., Lane W.S., Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104(1):119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 48.Beresford P.J., Zhang D., Oh D.Y., Fan Z., Greer E.L., Russo M.L. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276(46):43285–43293. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- 49.Au-Yeung N., Mandhana R., Horvath C.M. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2(3):e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Offenhauser N., Borgonovo A., Disanza A., Romano P., Ponzanelli I., Iannolo G. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol Biol Cell. 2004;15(1):91–98. doi: 10.1091/mbc.E03-06-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giampietro C., Disanza A., Bravi L., Barrios-Rodiles M., Corada M., Frittoli E. The actin-binding protein EPS8 binds VE-cadherin and modulates YAP localization and signaling. J Cell Biol. 2015;211(6):1177–1192. doi: 10.1083/jcb.201501089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lanzetti L., Rybin V., Malabarba M.G., Christoforidis S., Scita G., Zerial M. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408(6810):374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- 53.Hoffenberg S., Liu X., Nikolova L., Hall H.S., Dai W., Baughn R.E. A novel membrane-anchored Rab5 interacting protein required for homotypic endosome fusion. J Biol Chem. 2000;275(32):24661–24669. doi: 10.1074/jbc.M909600199. [DOI] [PubMed] [Google Scholar]
- 54.Matika C.A., Wasilewski M., Arnott J.A., Planey S.L. Antiproliferative factor regulates connective tissue growth factor (CTGF/CCN2) expression in T24 bladder carcinoma cells. Mol Biol Cell. 2012;23(10):1976–1985. doi: 10.1091/mbc.E11-08-0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maziveyi M., Alahari S.K. Cell matrix adhesions in cancer: the proteins that form the glue. Oncotarget. 2017;8(29):48471–48487. doi: 10.18632/oncotarget.17265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roig J., Mikhailov A., Belham C., Avruch J. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 2002;16(13):1640–1658. doi: 10.1101/gad.972202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan B.C., Lee S.C. Nek9, a novel FACT-associated protein, modulates interphase progression. J Biol Chem. 2004;279(10):9321–9330. doi: 10.1074/jbc.M311477200. [DOI] [PubMed] [Google Scholar]
- 58.Belham C., Roig J., Caldwell J.A., Aoyama Y., Kemp B.E., Comb M. A mitotic cascade of NIMA family kinases. Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J Biol Chem. 2003;278(37):34897–34909. doi: 10.1074/jbc.M303663200. [DOI] [PubMed] [Google Scholar]
- 59.Phadke M., Remsing Rix L.L., Smalley I., Bryant A.T., Luo Y., Lawrence H.R. Dabrafenib inhibits the growth of BRAF-WT cancers through CDK16 and NEK9 inhibition. Mol Oncol. 2018;12(1):74–88. doi: 10.1002/1878-0261.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li W., Wu C., Yao Y., Dong B., Wei Z., Lv X. MUC4 modulates human glioblastoma cell proliferation and invasion by upregulating EGFR expression. Neurosci Lett. 2014;566:82–87. doi: 10.1016/j.neulet.2014.02.033. [DOI] [PubMed] [Google Scholar]
- 61.Seshacharyulu P., Ponnusamy M.P., Rachagani S., Lakshmanan I., Haridas D., Yan Y. Targeting EGF-receptor(s) - STAT1 axis attenuates tumor growth and metastasis through downregulation of MUC4 mucin in human pancreatic cancer. Oncotarget. 2015;6(7):5164–5181. doi: 10.18632/oncotarget.3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anderson M., Marayati R., Moffitt R., Yeh J.J. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2016;8(34):56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolf A., Agnihotri S., Micallef J., Mukherjee J., Sabha N., Cairns R. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J Exp Med. 2011;208(2):313–326. doi: 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ko Y.H., Pedersen P.L., Geschwind J.F. Glucose catabolism in the rabbit VX2 tumor model for liver cancer: characterization and targeting hexokinase. Cancer Lett. 2001;173(1):83–91. doi: 10.1016/s0304-3835(01)00667-x. [DOI] [PubMed] [Google Scholar]
- 65.Lin H., Zeng J., Xie R., Schulz M.J., Tedesco R., Qu J. Discovery of a novel 2,6-disubstituted glucosamine series of potent and selective hexokinase 2 inhibitors. ACS Med Chem Lett. 2016;7(3):217–222. doi: 10.1021/acsmedchemlett.5b00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mercado-Pimentel M.E., Igarashi S., Dunn A.M., Behbahani M., Miller C., Read C.M. The novel small molecule inhibitor, OSU-T315, suppresses vestibular schwannoma and meningioma growth by inhibiting PDK2 function in the AKT pathway activation. Austin J Med Oncol. 2016;3(1) [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q., Fan S.Y., Qian J., Wang J.Y., Lu Y.C., Hu G.H. AKT2 expression in histopathologic grading and recurrence of meningiomas. Eur J Surg Oncol. 2014;40(9):1056–1061. doi: 10.1016/j.ejso.2014.02.243. [DOI] [PubMed] [Google Scholar]
- 68.Bhavanasi D., Speer K.F., Klein P.S. CKAP4 is identified as a receptor for Dickkopf in cancer cells. J Clin Invest. 2016;126(7):2419–2421. doi: 10.1172/JCI88620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karni R., de Stanchina E., Lowe S.W., Sinha R., Mu D., Krainer A.R. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14(3):185–193. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Proteins and phosphoproteins identified by LC-MS/MS in meningiomas and meninges.
Differentially expressed proteins and phosphoproteins of each meningioma grade vs. normal meningeal tissue.
Functional annotation enrichment analysis results from DAVID and IPA.
Statistical analysis of quantified proteins and phosphoproteins visualised in Volcano plots.
Molecular signature and GO enrichment analysis of grade II and III meningiomas.
Protein phosphorylation sites in meningiomas and functional annotation of phosphopeptides by DAVID and IPA.
Semi-quantitative scoring of immunohistochemical staining for validation of proteins and phosphoproteins.
Data Availability Statement
Proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository [30] with the following dataset identifiers and reviewer account details: global proteome PXD007073; phosphoproteome PXD007044; phosphopeptides PXD007125.