Abstract
Background
Ovarian cancer (OV) is the most lethal gynecological cancer in women. We aim to develop a generalized, individualized immune prognostic signature that can stratify and predict overall survival for ovarian cancer.
Methods
The gene expression profiles of ovarian cancer tumor tissue samples were collected from 17 public cohorts, including 2777 cases totally. Single sample gene set enrichment (ssGSEA) analysis was used for the immune genes from ImmPort database to develop an immune-based prognostic score for OV (IPSOV). The signature was trained and validated in six independent datasets (n = 519, 409, 606, 634, 415, 194).
Findings
The IPSOV significantly stratified patients into low- and high-immune risk groups in the training set and in the 5 validation sets (HR range: 1.71 [95%CI: 1.32–2.19; P = 4.04 × 10−5] to 2.86 [95%CI: 1.72–4.74; P = 4.89 × 10−5]). Further, we compared IPSOV with nine reported ovarian cancer prognostic signatures as well as the clinical characteristics including stage, grade and debulking status. The IPSOV achieved the highest mean C-index (0.625) compared with the other signatures (0.516 to 0.602) and clinical characteristics (0.555 to 0.583). Further, we integrated IPSOV with stage, grade and debulking, which showed improved prognostic accuracy than clinical characteristics only.
Interpretation
The proposed clinical-immune signature is a promising biomarker for estimating overall survival in ovarian cancer. Prospective studies are needed to further validate its analytical accuracy and test the clinical utility.
Fund
This work was supported by National Key Research and Development Program of China, National Natural Science Foundation of China and Natural Science Foundation of the Jiangsu Higher Education Institutions of China.
Keywords: Ovarian cancer, Immune, Gene expression, Prognostic signature
List of Abbreviations
| OV | ovarian cancer |
| IPSOV | immune-based prognostic signature for OV |
| HR | hazard ratio |
| CI | confidence interval |
| GEO | Gene Expression Omnibus |
| TCGA | The Cancer Genome Atlas ovarian cancer |
| FIGO | International Federation of Gynecology and Obstetrics |
| ssGSEA | single sample gene set enrichment analysis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| GO | Gene Ontology |
| BP | biological process |
| MF | molecular function |
| CC | cellular component |
| FDR | False-Discovery Rate |
| C-index | concordance statistic |
| RMS | restricted mean survival |
1. Introduction
Ovarian cancer (OV) is the most lethal gynecological cancer among women, with >14,000 estimated new deaths in United States, 2018 [1]. Biomarkers especially gene expression in tumor tissues are reliably related to cancer prognosis and survival [[2], [3], [4]]. Thus, identification of the subset of patients with worse survival and higher mortality is needed for additional clinical therapy. The availability of large-scale public cohorts with gene expression data and the well-developed biological database bring the opportunity to identify a more generalized prognostic signature with biological background for ovarian cancer.
Immune system has been shown to be a determining factor during cancer initiation and progression [5,6]. Various evidence have proved that ovarian cancer are immunogenic tumors [[7], [8], [9]] and immunotherapy is strongly pursued through targeting on the immune checkpoints [10,11]. In addition, previous study has preliminary confirmed the prognostic value of immune system in ovarian cancer [12]. Thus, the immune based prognostic signature remains a potential to be applied in ovarian cancer.
In this study, we integrated multiple cohorts with gene expression which contained 2777 cases totally to develop and validate an individualized gene-set based prognostic signature for ovarian cancer from immune-related genes. With sufficient validation of 5 independent datasets, we proved the model stability and reliability. Further, to leverage the complementary value of clinical characteristics, we performed an integrated analysis to improve the predicted accuracy for overall survival.
2. Methods
2.1. Study population and eligibility criteria
We retrospectively collected the ovarian cancer gene expression profiles of frozen primary tumor tissue samples from 17 public microarray datasets, including 16 cohorts from the Gene Expression Omnibus (GEO) repository and 1 from The Cancer Genome Atlas ovarian cancer (TCGA-OV) cohort. All patients had received primary debulking surgery. Only patients with available follow-up time, status and gene expression data were included. The main outcome of our study was overall survival. Staging was assessed in accordance to the International Federation of Gynecology and Obstetrics (FIGO) stage system. Optimal debulking was defined as ≤1 cm of gross residual disease while sub-optimal debulking was defined as >1 cm of residual disease. We did not limit histology type, FIGO stage, grade, debulking and neoadjuvant chemotherapy status for sample collection. The sample quality control details were described in Table S1.
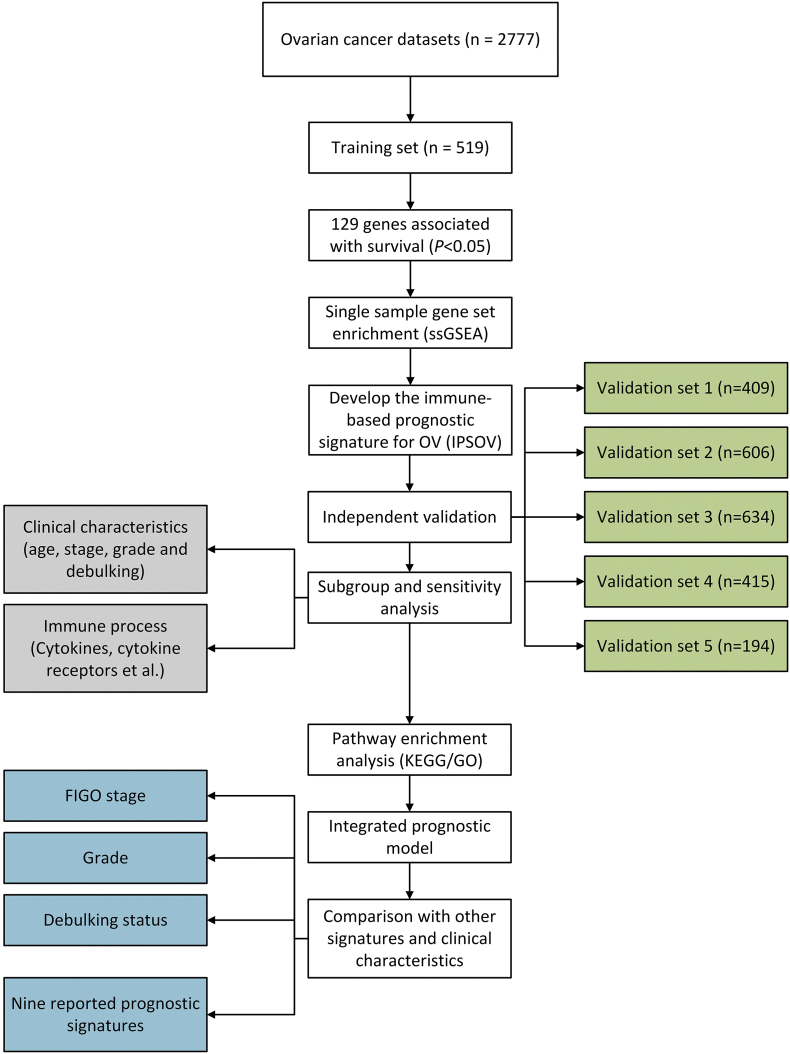
The study characteristics of the 17 public ovarian cancer datasets were described in Table S1. Finally, 2777 ovarian cancer cases were included in our study. The study design and workflow were provided in Fig. 1.
Fig. 1.
Flowchart of the study. 17 public ovarian cancer datasets containing 2777 cases were included and categorized into 6 independent datasets according to the microarray platform. We developed the IPSOV in the training set and validated in the other 5 datasets. Further, we integrated IPSOV with stage, grade and debulking status to improve the prognostic value.
2.2. Gene expression data preprocessing
For GEO datasets, all microarray data and clinical information were download from GEO repository (https://www.ncbi.nlm.nih.gov/geo/). To increase the statistical power and leverage cohorts with small sample size sufficiently, we combined the cohorts with the same microarray platform. Only the platforms with sample size ≥ 150 were included in the study.
For TCGA cohort, due to the samples with available RNA-Sequencing gene expression quantification were relatively small (n = 374), we collected Affymetrix Human Genome U133A Array microarray data instead (n = 519). All level-2 data were download from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/).
Finally, six independent datasets were included according to the different platforms from GEO and TCGA (Table 1). Entrez IDs were used to represent genes across different platform. In the independent validation phase, if multiple probe sets correspond to the same Entrez ID in the validation sets, the one with the highest mean signal was selected as the expression level of the corresponding gene [13].
Table 1.
Summary of the eight datasets included in the study.
| Dataset | Platform | Sample size | Included cohortsa |
|---|---|---|---|
| Training set | Affymetrix Human Genome U133A Array | 519 | TCGA |
| Validation set 1 | Affymetrix Human Genome U133A Array | 409 | GSE14764, GSE23554, GSE26712, GSE3149 |
| Validation set 2 | Affymetrix Human Genome U133 Plus 2.0 Array | 606 | GSE18520, GSE19829, GSE26193, GSE30161, GSE63885, GSE9891 |
| Validation set 3 | Agilent-014850 Whole Human Genome Microarray 4x44K G4112F | 634 | GSE17260, GSE32062, GSE53963, GSE73614 |
| Validation set 4 | Operon human v3 ~35 K 70-mer two-color oligonucleotide microarrays | 415 | GSE13876 |
| Validation set 5 | ABI Human Genome Survey Microarray Version 2 | 194 | GSE49997 |
We used ComBat to adjust the batch effects between different cohorts within the same platform. Gene expression values of all probes were adjusted in each dataset, respectively.
The batch effects between different datasets within the same platform were adjusted by ComBat method [14]. Gene expression values of all probes were adjusted in each dataset, respectively. We also adjusted the batch effects within the TCGA cohort according to the tissue source site code. All the gene expression levels had been logarithmic transformed before batch effects adjustment.
2.3. Immune-related genes definition
We constructed a prognostic signature from the immune-related genes, which were downloaded from the ImmPort database (http://www.immport.org) [15]. It includes 17 immune categories according to different molecular function, such as antimicrobials, cytokine, interleukins, T-cell receptor signaling pathway, B-cell receptor signaling pathway, TNF family receptors.
2.4. Development of the immune-based prognostic signature for OV (IPSOV)
We performed a three-stage strategy to develop the signature. First, we used the TCGA cohort as the training set to screen the survival related probes. 1905 microarray probes for 1109 immune genes were included totally. The Cox proportional-hazards model was used to assess their association with overall survival with adjustment for age, stage, grade and debulking status. We excluded the probes with adjusted P values larger than 0.05.
To estimate the population specific immune infiltration, we used single sample gene set enrichment analysis (ssGSEA) that define a enrichment score to represent the degree of absolute enrichment of a gene set in each sample within a given dataset [16]. Normalized enrichment scores could be calculated for each immune category. The ssGSEA analysis were performed in R package GSVA.
In the last step, we develop a prognostic signature named the immune-based prognostic signature for OV (IPSOV) to combine the effects of each immune category in the training set. The coefficients of each category were determined by multivariable Cox regression model: , where Si is the ssGSEA score for ith immune category.
2.5. Validation of the IPSOV
To get a uniform cutoff value to categorize the patients into low-risk and high-risk group, we performed a normalization for the gene expression values in each dataset with mean value = 0 and standard deviation (SD) = 1. After developing the signature, the prognostic score of IPSOV was further calculated in the 5 validation datasets, respectively. In the multivariable Cox regression, age, FIGO stage, grade, debulking status and study site were included as covariates.
2.6. Pathway enrichment analysis for the molecular function
To further understand the gene function in IPSOV, we performed a pathway enrichment analysis for the genes based on Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) database including biological process (BP), molecular function (MF) and cellular component (CC). The P values were adjusted by False-Discovery Rate (FDR) method for multiple comparison. All the analysis were performed by R package clusterProfiler [17].
2.7. Existing prognostic signatures for comparison
To evaluate the survival classification and prediction ability of IPSOV, we retrospectively collected 9 published prognostic signatures for comparison, which included from 7 genes to 300 genes (Table S7). Continuous prognostic scores were calculated for each signature. The P values of continuous scores in univariable Cox model and the overall concordance statistic (C-index) were compared in the six datasets.
2.8. Statistical analysis
Continuous variables were summarized as mean ± SD, and categorized variables were described by frequency (n) and proportion (%). In the survival analysis, associations between characteristics and overall survival were evaluated by Cox proportional hazard models. Kaplan-Meier survival curves were drawn and compared among subgroups using log-rank tests. The C-index and restricted mean survival (RMS) curve were estimated using R package survival and survRM2, while C-index was compared by R package compareC [18].
Statistical analyses were performed using R version 3.4.0 (The R Foundation). P values were two-sided, and P < 0.05 was considered statistically significant.
3. Results
3.1. Development and definition of the IPSOV
A total of 2777 patients with ovarian cancer (mean age ± SD, 59.7 ± 11.8; 12.8% early-stage [I, II], 87.2% late-stage [III, IV]; 43.5% optimal debulking, 56.5% sub-optimal debulking) from 6 independent datasets were included in the analysis (Table 2). In the training set, 156 probes of 129 genes from 15 immune categories were associated with overall survival among the 1905 immune-related microarray probes. Further, we used these probes to perform the ssGSEA analysis for each patient to calculate the immune enrichment score of each immune category. The IPSOV was defined as the combined effect of scores in different categories using the coefficients generated from multivariable Cox regression model (Table S3). The median score in the training set (−0.23) was used as the cutoff to stratify patients into low- and high-immune risk group across all the datasets.
Table 2.
Demographic and clinic characteristic descriptions for ovarian cancer patients in different datasets.
| Characteristicsa | Training set | Validation set 1 | Validation set 2 | Validation set 3 | Validation set 4 | Validation set 5 |
|---|---|---|---|---|---|---|
| Number of samples | 519 | 409 | 606 | 634 | 415 | 194 |
| Median survival time (month) (95% CI) | 44.5 (41–48.3) | 51 (45–65) | 41.9 (37.8–47.8) | 53 (49–62) | 24 (21−30) | >49b |
| Number of Death (%) | 284 (54.9) | 232 (56.7) | 346 (57.1) | 364 (57.1) | 302 (72.7) | 57 (29.3) |
| Age (years) | 59.8 ± 11.4 | – | 62.9 ± 10.6 | 62.6 ± 11.2 | 57.9 ± 12.2 | 57.6 ± 11.8 |
| Histology type (%) | ||||||
| Serous | 520 (100) | 96 (88.8) | 516 (85.5) | 534 (83.8) | 415 (100) | 171 (88.1) |
| Endometroid | 0 | 6 (5.5) | 41 (6.8) | 66 (10.3) | 0 | 0 |
| Clear cell | 0 | 2 (1.8) | 20 (3.3) | 37 (5.8) | 0 | 0 |
| Mucinous | 0 | 0 | 9 (1.4) | 0 | 0 | 0 |
| Other | 0 | 4 (3.7) | 17 (2.8) | 0 | 0 | 23 (11.8) |
| FIGO stage (%) | ||||||
| I | 15 (2.9) | 8 (4.1) | 41 (7.4) | 32 (5.0) | – | 0 |
| II | 25 (4.8) | 2 (1.0) | 111 (20.0) | 23 (3.6) | – | 9 (4.6) |
| III | 396 (76.7) | 164 (84.1) | 364 (65.8) | 462 (72.6) | – | 154 (79.3) |
| IV | 80 (15.5) | 21 (10.7) | 37 (6.6) | 119 (18.7) | – | 31 (15.9) |
| Grade (%) | ||||||
| 1 | 5 (0.9) | 9 (4.0) | 32 (5.8) | 26 (4.0) | – | 0 |
| 2 | 64 (12.5) | 86 (38.9) | 80 (14.6) | 204 (32.0) | – | 0 |
| 3 | 439 (86.2) | 125 (56.5) | 394 (72.0) | 324 (50.8) | – | 143 (100) |
| 4 | 1 (0.1) | 1 (0.2) | 41 (7.4) | 83 (13.0) | – | 0 |
| Debulking status (%) | ||||||
| Optimal | 337 (73.1) | 193 (54.0) | 209 (66.7) | 271 (51.4) | – | – |
| Sub-optimal | 124 (26.9) | 164 (45.9) | 104 (33.2) | 256 (48.5) | – | – |
Sum of frequency numbers may not equal to the total sample size due to missing values.
The median survival time is incalculable due to the mortality at the last follow-up time is <50%.
3.2. Validation of the IPSOV
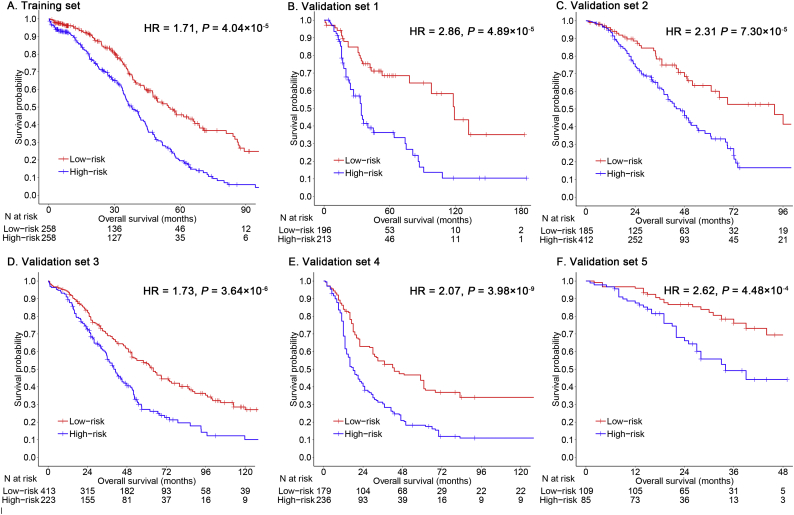
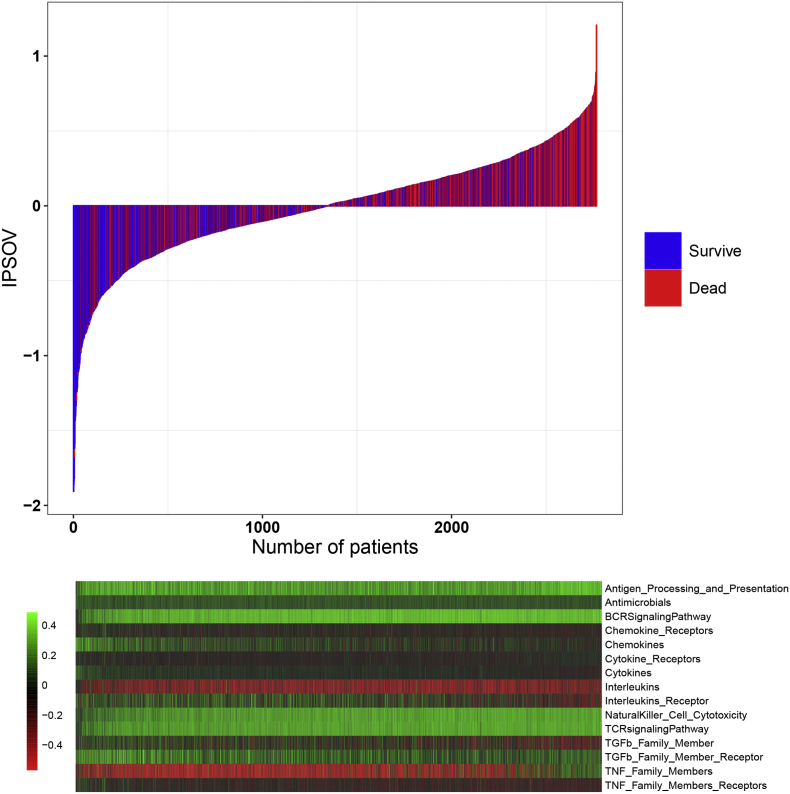
The IPSOV significantly stratified patients into low- and high-immune risk groups in the training set (hazard ratio [HR] = 1.82; 95% confidence interval [CI]: 1.43–2.30; P = 9.19 × 10−7) and in the 5 validation sets (HR range: 1.66 [95%CI: 1.27–2.15; P = 1.84 × 10−4] to 2.36 [95%CI: 1.38–4.00; P = 0.001]) (Table S4). In addition, it remained as an independent prognostic factor in the multivariable Cox model, after adjusting for age, stage, grade and debulking status (HR range: 1.71 [95%CI: 1.32–2.19; P = 4.04 × 10−5] to 2.86 [95%CI: 1.72–4.74; P = 4.89 × 10−5]) (Fig. 2). In the meta-analysis for all datasets, the high-immune risk group showed a 1.96-fold higher risk than the low-immune risk group (HR = 1.96; 95%CI: 1.73–2.23; P = 2.37 × 10−25) (Fig. S1). The IPSOV distribution with survival status in the combined dataset was shown in Fig. 3. Significant RMS time ratio (1.24 to 1.88) was observed in the 6 datasets, respectively (Table 3).
Fig. 2.
Kaplan-Meier survival analyses of IPSOV. (a) Patients are stratified into low- (red) and high-immune (blue) risk groups with a cutoff of the median value in the training set. (b-f) Further, the prognostic signature IPSOV is validated in five independent validation sets. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
IPSOV distribution with survival status in the combined dataset. Upper half panel: IPSOV distribution with patient survival status. The X axis is sorted by IPSOV values. Red color indicates the patients are dead while blue color indicates survive. Lower half panel: Heatmap showing the corresponding 15 immune categories enrichment scores. The score of each immune category is normalized to mean = 0 and standard deviation = 1. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Restricted mean survival (RMS) time ratio between low- and high-IPSOV groups in different data sets.
| Dataset | NLow-IPSOV | NHigh-IPSOV | RMSLow-IPSOV (95%CI)a | RMSHigh-IPSOV (95%CI)a | RMS ratio (95%CI)b | P |
|---|---|---|---|---|---|---|
| Training set | 258 | 258 | 57.58 (52.61–62.55) | 42.73 (39.02–46.45) | 1.35 (1.19–1.52) | <0.001 |
| Validation set 1 | 196 | 213 | 84.51 (72.62–96.41) | 61.82 (52.53–71.11) | 1.37 (1.11–1.67) | 0.003 |
| Validation set 2 | 185 | 412 | 60.52 (54.77–66.28) | 46.40 (42.95–49.84) | 1.31 (1.15–1.47) | <0.001 |
| Validation set 3 | 413 | 223 | 92.42 (83.49–101.35) | 60.77 (51.99–69.56) | 1.52 (1.27–1.81) | <0.001 |
| Validation set 4 | 179 | 236 | 70.18 (60.61–79.75) | 37.17 (31.17–43.18) | 1.88 (1.52–2.33) | <0.001 |
| Validation set 5 | 109 | 85 | 40.50 (37.80–43.20) | 32.75 (29.00–36.51) | 1.24 (1.08–1.41) | 0.002 |
RMS time: months.
RMS ratio = RMSLow-IPSOV/ RMSHigh-IPSOV.
3.3. Subgroup and sensitivity analysis for IPSOV
To evaluate the model stability in different clinical subgroups, we performed a sensitivity analysis according to age, histology, stage, grade and debulking status. IPSOV was significant in most subgroups and had a relatively stable HRs except endometrioid and early-stage patients, which might due to the insufficient sample size (Fig. S2). This indicated IPSOV was potentially independent of clinical characteristics.
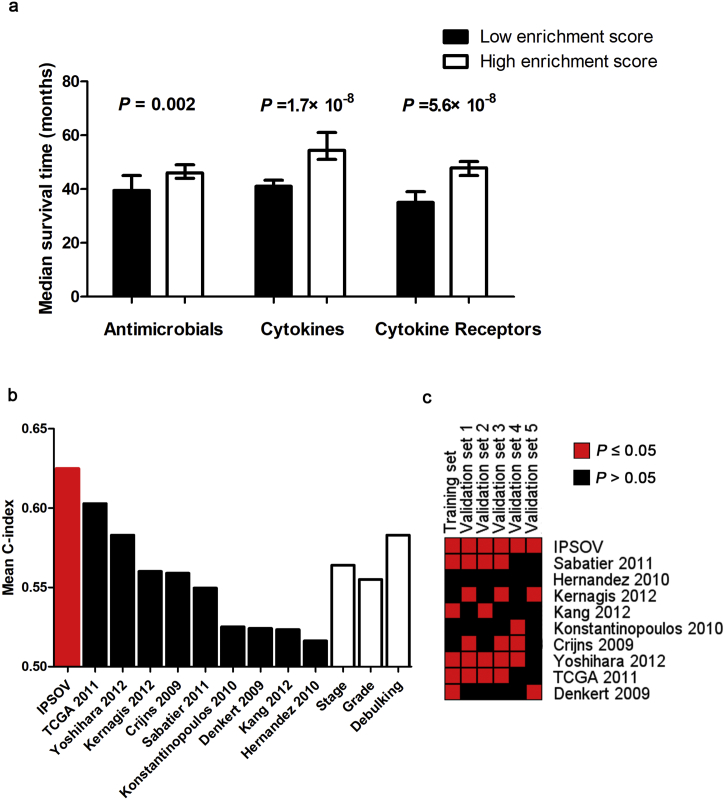
Further, we found the 129 genes were mostly involved in antimicrobials (33.3%), cytokine receptors (32.6%) and cytokines (29.5%) immune process (Table S2). We calculated the corresponding subgroup immune scores based on ssGSEA method in each immune process. The patients with low immune scores had a significant longer median survival time in each process (Fig. 4A).
Fig. 4.
(a) Immune scores are calculated based on the coefficients of IPSOV in antimicrobials, cytokines and cytokine receptors immune processes. We dichotomize the scores into low- and high-immune risk group by the median value. Median survival time is compared using log-rank test. (b) Mean C-index of IPSOV, stage, grade, debulking and 9 reported signatures. (c) P value comparison of IPSOV and 9 reported signatures. Red block indicates the model is significant (P ≤ 0.05) while black indicates unsignificant (P > 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To test the robustness of IPSOV, we randomly resampled 500 cases for 10,000 times from the combined dataset. Only 0.17% of P values failed to pass the 0.05 threshold (Fig. S3A). The median C-index was 0.624 with a SD of 0.017, which showed its stable predictive power (Fig. S3B).
3.4. Pathway enrichment analysis
Enrichment analysis for the 129 unique genes identified 85 significant KEGG pathways (Padjust < 0.05), such as Cytokine-cytokine receptor interaction, MAPK signaling pathway, Ras signaling pathway, B/T cell receptor signaling pathway and PI3K-Akt signaling pathway (Table S6). Further, pathway analysis based on Gene Ontology identified various significant pathways, including 1034 biological process (BP) pathways, 44 molecular function (MF) pathways and 77 cellular component (CC) pathways, which revealed the abundant biological function background.
3.5. Comparison with other signatures and clinical characteristics
We compared model predictive accuracy of IPSOV with clinical characteristics including stage, grade and debulking as well as 9 reported ovarian cancer prognostic signatures. Continuous prognostic scores were calculated from each signature to compare among different datasets. Of the 13 survival predicted factors, IPSOV had a highest mean C-index (0.625) compared with stage (0.564), grade (0.555), debulking (0.583) and other signatures (0.516 to 0.602) (Table S5, Fig. 4B).
In addition, we also compared the P-values of continuous prognostic scores in the univariable Cox model. IPSOV was significant in all datasets, outperforming other signatures (from 0/6 to 5/6) (Fig. 4C).
3.6. Integrate IPSOV with clinical characteristics
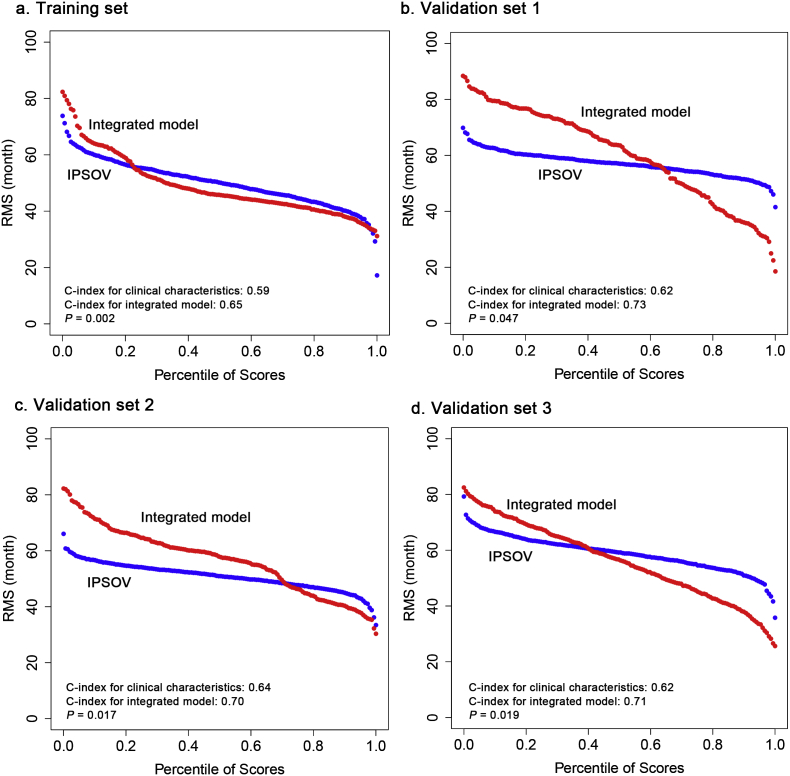
In addition to IPSOV, clinical characteristics including tumor stage, grade and debulking status were also independent prognostic factors, suggesting their complementary value. To further improve predict accuracy, we integrated IPSOV (continuous score) with them using the coefficients generated from the multivariable Cox regression model in the training set as: integrated model = 1.119 × IPSOV+0.335 × stage+0.368 × grade-0.501 × debulking. The integrated model was further validated in validation set 1, 2, 3 where full clinical information in the model was available. Significantly improved estimation of survival was achieved by the continuous score of integrated model relative to clinical characteristics only (C-index: 0.65 vs 0.59 in training set, P = 0.002; 0.73 vs 0.62 in validation set 1, P = 0.047; 0.70 vs 0.64 in validation set 2, P = 0.017; 0.71 vs 0.62 in validation set 3, P = 0.019) (Fig. 5).
Fig. 5.
Integration of IPSOV and clinical characteristics. (a) Training set. (b) Validation set 1. (c) Validation set 2. (d) Validation set 3. Restricted mean survival (RMS) curve for IPSOV and integrated model. In addition, we provided the C-index comparison of the model with clinical characteristics only and the integrated model. P-value represents the difference between the two models in term of C-index.
4. Discussion
Patients suffering from ovarian cancer often display a heterogeneous clinical outcome spanning <5 months to beyond 10 years [19]. In this respect, treatment options are lacking with all the patients being treated with similar drugs. Thus, a prognostic signature with a broad scope of application is needed to accurately identify those cases with refractory disease and worse survival. In this study, using multiple well-established public ovarian cancer cohorts, we developed a prognostic signature based on gene set enrichment and validated it in 5 independent validation datasets which covered most common microarray platforms. The IPSOV could further stratify clinically defined groups of patients (e.g. debulking status, FIGO stage and age) into subgroups with different survival outcomes.
To date, numerous studies have developed gene expression signatures to stratify ovarian cancer survival based on different cohorts. However, none of them has been incorporated into clinical practice might due to several limitations. First, the sample size was usually small that lacked sufficient validation to prove model stability. Second, most of them used specific genes to generate the prognostic score, which easily leaded to overfitting and ignored other casual genes [20]. In this study, we performed ssGESA analysis to determine the population specific immune infiltration based on immune pathway gene set level, which is more generic than the normal signature.
Accumulating evidence indicates innate and adaptive immune systems make a crucial contribution to cancer initiation and development [[21], [22], [23]]. Moreover, cancer immunotherapy has made a great process and is moving fast that IPSOV may hold great promise for identifying novel molecular targets for it [24]. From the ImmPort database, we found most genes of IPSOV were involved in antimicrobials, cytokines and cytokine receptors processes. Recent pre-clinical and clinical trials demonstrate that adjuvant antimicrobial therapy is beneficial in cancer treatment. Antimicrobial agents can benefit cancer patients by killing oncogenic-related microorganisms and protecting from recurring immunosuppression-induced infection through their direct antiproliferative and cytotoxic effects [25]. Cytokines and cytokine receptors processes play important roles in inflammatory, angiogenesis and chemotaxis processes [26]. They can function to inhibit tumor development and progression [27] and also are proved to be effective in the treatment of cancer [28].
Further, the pathway enrichment analysis adds the evidence for its association with cancer and the clinical application potential. Ras-MAPK signaling pathway is considering playing an important role in multiple cancers and inhibitors for it have shown effectiveness in clinical trials [29]. Inhibitor of PI3K-Akt signaling pathway prevent the growth of ovarian cancer xenografts and potentiate the cytotoxic effects of paclitaxel and cisplatin [30]. In addition, other pathways such as kinase activity [31,32], growth factor [33,34] and ERK1/ERK2 cascade [35] were also involved in cancer cell growth, proliferation, invasion and metastasis and played important roles in ovarian cancer.
Our study has several strengths. First, the sample size is much larger than the studies before, which gives the signature sufficient validation and makes it more robust and reliable. Second, compared with the models screened from the whole genome that contain many genes with unknown function or a single candidate gene which shows less prognostic value than an aggregated model [36,37], the immune-related genes of IPSOV have a strong biological background, making it practicable to be applied in clinical adjuvant treatment. Third, IPSOV has a promising survival prediction ability through comparing with other signatures and clinical characteristics.
How high of a C-index is needed for a useful prognostic tool depend on the clinical context [38]. Although the absolute value of IPSOV C-index is not very high, it outperforms the traditional clinical factors and improves significantly in the integrated model. Thus, the utility of these tools could better estimate patient prognostic information and stratify into different subgroups to benefit from different treatment.
We acknowledge some limitations. First, we tried to include as many cohorts as possible to get a more sufficient validation of our biomarkers. However, gene expression values are subject to sampling bias due to different platforms. Second, the missing rate for the clinical characteristics was high, which decreased the statistical power in multivariable Cox regression analysis and the integrated prognostic model. Third, IPSOV is developed by numerous genes, where further biological experiments are warranted to validate their functions in ovarian cancer.
5. Conclusion
IPSOV is a promising prognostic biomarker in ovarian cancer which could be used to distinguish and predict patients' survival outcome. Prospective studies are needed to further validate its analytical accuracy for estimating prognoses and to test its clinical utility in individualized management of ovarian cancer.
Ethics approval and consent to participate
This study has been proved by the Nanjing Medical University institutional committee.
Availability of data and material
GEO datasets: https://www.ncbi.nlm.nih.gov/gds/.
TCGA dataset: https://portal.gdc.cancer.gov/.
The data analysis source code is provided in the github: https://github.com/fennudepenpen/IPSOV.
Consent for publication
All authors have reviewed the manuscript and consented for publication.
Conflicts of interest statements/Financial Disclosure statement
None.
Acknowledgments
Acknowledgements
We thank the patients and investigators who participated in TCGA and GEO for providing data.
Funding
This study was supported by National Key Research and Development Program of China (2016YFE0204900 to F.C.), National Natural Science Foundation of China (81530088 and 81473070 to F.C.), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (14KJA310002 to F.C.), Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). Y.W. and R.Z. were partially supported by the Outstanding Young Teachers Training Program of Nanjing Medical University.
Author contributions
SS, GW, and FC contributed to the study design. SS, GW contributed to data collection. SS, GW, RZ, YZ and YW performed statistical analysis and interpretation. SS and GW drafted the manuscript. All authors contributed to critical revision of the final manuscript and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.054.
Appendix A. Supplementary data
Supplementary material
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Calon A., Lonardo E., Berenguer-Llergo A., Espinet E., Hernando-Momblona X., Iglesias M. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47(4):320. doi: 10.1038/ng.3225. [DOI] [PubMed] [Google Scholar]
- 3.Li B., Cui Y., Diehn M., Li R. Development and validation of an individualized immune prognostic signature in early-stage nonsquamous non–small cell lung cancer. JAMA Oncol. 2017;3(11):1529–1537. doi: 10.1001/jamaoncol.2017.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng S.W., Mitchell A., Kennedy J.A., Chen W.C., McLeod J., Ibrahimova N. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433. doi: 10.1038/nature20598. [DOI] [PubMed] [Google Scholar]
- 5.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21(8):938. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angell H., Galon J. From the immune contexture to the immunoscore: the role of prognostic and predictive immune markers in cancer. Curr Opin Immunol. 2013;25(2):261–267. doi: 10.1016/j.coi.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Kandalaft L.E., Powell D.J., Jr., Singh N., Coukos G. Immunotherapy for ovarian cancer: what's next? J Clin Oncol. 2010;29(7):925–933. doi: 10.1200/JCO.2009.27.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kershaw M.H., Westwood J.A., Parker L.L., Wang G., Eshhar Z., Mavroukakis S.A. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin Cancer Res. 2006;12(20):6106–6115. doi: 10.1158/1078-0432.CCR-06-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodell V., Salazar L.G., Urban N., Drescher C.W., Gray H., Swensen R.E. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. J Clin Oncol. 2006;24(5):762–768. doi: 10.1200/JCO.2005.03.2813. [DOI] [PubMed] [Google Scholar]
- 10.Nagarsheth N., Wicha M.S., Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Topalian S.L., Drake C.G., Pardoll D.M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosuke Y., Tatsuhiko T., Daichi S. High-risk ovarian cancer based on 126-gene expression signature is uniquely characterized by downregulation of antigen presentation pathway. Clin Cancer Res. 2012;18(5):1374–1385. doi: 10.1158/1078-0432.CCR-11-2725. [DOI] [PubMed] [Google Scholar]
- 13.Miller J.A., Cai C., Langfelder P., Geschwind D.H., Kurian S.M., Salomon D.R. Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinformatics. 2011;12(1):322. doi: 10.1186/1471-2105-12-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya S., Andorf S., Gomes L., Dunn P., Schaefer H., Pontius J. ImmPort: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2–3):234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 16.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang L., Chen W., Petrick N.A., Gallas B.D. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34(4):685–703. doi: 10.1002/sim.6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mok S.C., Bonome T., Vathipadiekal V., Bell A., Johnson M.E., Park D.-C. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: microfibril-associated glycoprotein 2. Cancer Cell. 2009;16(6):521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian J., Simon R. Response: Re: gene expression–based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(21):1678–1679. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zitvogel L., Apetoh L., Ghiringhelli F., Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8(1):59. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- 22.Yu H., Kortylewski M., Pardoll D. Tumour immunology: Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7(1):41. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 23.Ogino S., Galon J., Fuchs C.S., Dranoff G. Cancer immunology—analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8(12):711. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg S.A., Yang J.C., Restifo N.P. Cancer immunotherapy: moving beyond current vaccines. Nat Med. 2004;10(9):909. doi: 10.1038/nm1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alibek K., Bekmurzayeva A., Mussabekova A., Sultankulov B. Using antimicrobial adjuvant therapy in cancer treatment: a review. Infect. Agents Cancer. 2012;7(1):33. doi: 10.1186/1750-9378-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin W.-W., Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117(5):1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dranoff G. Cytokines in cancer pathogenesis and cancer therapy. Nat Rev Cancer. 2004;4(1):11. doi: 10.1038/nrc1252. [DOI] [PubMed] [Google Scholar]
- 28.Lee S., Margolin K. Cytokines in cancer immunotherapy. Cancer. 2011;3(4):3856–3893. doi: 10.3390/cancers3043856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santarpia L., Lippman S.M., El-Naggar A.K. Targeting the MAPK–RAS–RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi: 10.1517/14728222.2011.645805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu L., Hofmann J., Lu Y., Mills G.B., Jaffe R.B. Inhibition of phosphatidylinositol 3′-kinase increases efficacy of paclitaxel in in vitro and in vivo ovarian cancer models. Cancer Res. 2002;62(4):1087–1092. [PubMed] [Google Scholar]
- 31.Siu M.K., Chan H.Y., Kong D.S., Wong E.S., Wong O.G., Ngan H.Y. p21-activated kinase 4 regulates ovarian cancer cell proliferation, migration, and invasion and contributes to poor prognosis in patients. Proc Natl Acad Sci. 2010;107(43):18622–18627. doi: 10.1073/pnas.0907481107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang M.K., Zhou H.Y., Yam J.W., Wong A.S. c-Met overexpression contributes to the acquired apoptotic resistance of nonadherent ovarian cancer cells through a cross talk mediated by phosphatidylinositol 3-kinase and extracellular signal-regulated kinase 1/2. Neoplasia. 2010;12(2):128–IN5. doi: 10.1593/neo.91438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moghaddam S.M., Amini A., Morris D.L., Pourgholami M.H. Significance of vascular endothelial growth factor in growth and peritoneal dissemination of ovarian cancer. Cancer Metastasis Rev. 2012;31(1–2):143–162. doi: 10.1007/s10555-011-9337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rattan R., Graham R.P., Maguire J.L., Giri S., Shridhar V. Metformin suppresses ovarian cancer growth and metastasis with enhancement of cisplatin cytotoxicity in vivo. Neoplasia. 2011;13(5):483–491. doi: 10.1593/neo.11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friday B.B., Adjei A.A. Advances in targeting the Ras/Raf/MEK/Erk mitogen-activated protein kinase cascade with MEK inhibitors for cancer therapy. Clin Cancer Res. 2008;14(2):342–346. doi: 10.1158/1078-0432.CCR-07-4790. [DOI] [PubMed] [Google Scholar]
- 36.Shen S., Bai J., Wei Y., Wang G., Li Q., Zhang R. A seven-gene prognostic signature for rapid determination of head and neck squamous cell carcinoma survival. Oncol Rep. 2017;38(6):3403–3411. doi: 10.3892/or.2017.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen S., Wang G., Shi Q., Zhang R., Zhao Y., Wei Y. Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenetics. 2017;9(1):88. doi: 10.1186/s13148-017-0392-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldron L., Haibe-Kains B., Culhane A.C., Riester M., Ding J., Wang X.V. Comparative meta-analysis of prognostic gene signatures for late-stage ovarian cancer. JNCI. 2014;106(5) doi: 10.1093/jnci/dju049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
GEO datasets: https://www.ncbi.nlm.nih.gov/gds/.
TCGA dataset: https://portal.gdc.cancer.gov/.
The data analysis source code is provided in the github: https://github.com/fennudepenpen/IPSOV.