Abstract
Helicases are biomolecular motors that unwind nucleic acids, and their regulation is essential for proper maintenance of genomic integrity. Escherichia coli Rep helicase, whose primary role is to help restart stalled replication, serves as a model for Superfamily I helicases. The activity of Rep-like helicases is regulated by two factors: their oligomeric state, and the conformation of the flexible subdomain 2B. However, the mechanism of control is not well understood. To understand the factors that regulate the active state of Rep, here we investigate the behavior of a 2B-deficient variant (RepΔ2B) in relation to wild-type Rep (wtRep). Using a single-molecule optical tweezers assay, we explore the effects of oligomeric state, DNA geometry, and duplex stability on wtRep and RepΔ2B unwinding activity. We find that monomeric RepΔ2B unwinds more processively and at a higher speed than the activated, dimeric form of wtRep. The unwinding processivity of RepΔ2B and wtRep is primarily limited by ‘strand-switching’—during which the helicases alternate between strands of the duplex—which does not require the 2B subdomain, contrary to a previous proposal. We provide a quantitative model of the factors that enhance unwinding processivity. Our work sheds light on the mechanisms of regulation of unwinding by Rep-like helicases.
INTRODUCTION
Helicases are a ubiquitous and diverse class of motor proteins that utilize the energy of NTP hydrolysis to translocate along single-stranded nucleic acids and unwind double-stranded nucleic acids (1–5). They serve a variety of important roles in the cell, in particular maintaining the genome (6,7). Since indiscriminate unwinding of nucleic acids would be detrimental to genomic integrity, helicase activity must be tightly regulated. In addition, many helicases have been shown to carry out multiple, distinct activities (1,8–11), but how these are controlled in the cell remains unclear. Thus, the mechanisms of regulation of helicase activity are critical to understanding their functions in the cell.
Rep, UvrD, and PcrA are structurally homologous proteins and prototype members of the large Superfamily I (SF1) of helicases (12), and serve as models for understanding the relationship between structure and behaviors such as translocation, unwinding, and regulation. Rep is involved in a variety of cellular processes such as DNA repair, replication, and replication restart (13–15) and shares a common structural organization with UvrD and PcrA, consisting of four subdomains (1A/2A/1B/2B; Figure 1A) (1,3,16–19). Two highly conserved RecA-like subdomains (1A/2A) comprise the canonical motor core that translocates in a 3′ to 5′ direction on single-stranded DNA (ssDNA) (3,12,20,22). An accessory subdomain (2B) can freely swivel about a hinge connected to the 2A subdomain populating multiple rotational conformations relative to the motor core, with the extreme states referred to as ‘open’ and ‘closed’ (8,16,23–26). Crystallographic studies (17,18) have indicated that the DNA duplex backbone interacts with a set of residues on the 2B subdomain of UvrD and PcrA monomers called the GIG motif, implicating 2B as an active player in duplex destabilization. The 2B subdomain of Rep has also been implicated in its role to displace proteins from DNA during replication (15). However, the 2B subdomain in Rep is auto-inhibitory for monomer helicase activity (20,27) and several lines of evidence point to 2B acting primarily as a switch-like regulator of unwinding activity (1,8,20,23,25–27).
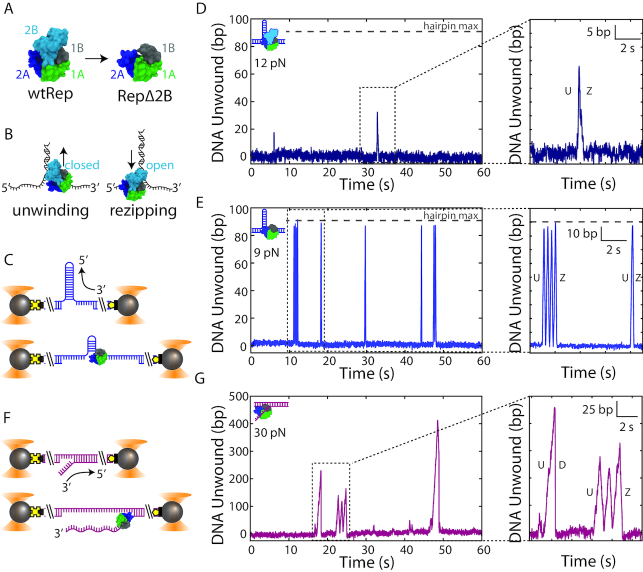
Figure 1.
Removal of the 2B subdomain activates Rep monomer unwinding. (A) wtRep structure in the ‘closed’ conformation (PDB 1uaa) showing its four subdomains (left), and a structural model of RepΔ2B with 2B replaced by three glycine residues (right). (B) Schematic of SF1 helicase (here, wtRep) monomer activity on DNA. 3′-to-5′ translocation on the 3′-strand of a DNA fork leads to unwinding of the duplex (left) with 2B in the closed state. 3′-to-5′ translocation on the 5′-strand leads to duplex rezipping behind the helicase (right) with 2B in the open state. (C) Schematic of dual-optical trap measurement of DNA hairpin unwinding. Two beads (grey spheres) held in traps (orange cones) stretch a DNA hairpin (blue) at constant tension via biotin-streptavidin (black square-yellow cross) and digoxigenin-anti-digoxigenin (yellow pentagon-black rectangle) linkages. Unwinding of a hairpin base pair releases two nucleotides, increasing DNA extension. (D) Representative data trace of DNA hairpin unwinding by wtRep dimer (schematic, top left corner). Close-up (inset) of one round of activity showing unwinding (U) followed by hairpin rezipping (Z). (E) Representative data trace of processive unwinding of DNA hairpin by RepΔ2B monomer (schematic, top left corner). The close-up (inset) of two rounds of activity shows hairpin unwinding (U) and rezipping (Z). The grey horizontal dashed line (D–E) indicates the limit expected for complete hairpin opening. (F) Schematic of dual-optical trap measurement of DNA fork (magenta) unwinding. Unwinding of a single base pair of the fork releases one nucleotide, changing DNA extension. (G) Representative data trace of processive DNA fork unwinding by RepΔ2B monomer (schematic, top left corner). The close-up (inset) shows two rounds of activity; protein unwinding (U) ends either by mid-fork dissociation (D) or dissociation at the base of the fork after three rounds of unwinding (U) and rezipping (Z).
The Rep protein is monomeric in solution and can bind DNA as a monomer (28–32). Multiple studies have shown that, in the absence of force, monomers of Rep, UvrD or PcrA alone are rapid and processive single-stranded DNA translocases, but show no DNA unwinding activity (20–23,26,27,32–36). However, DNA unwinding by monomers is possible through conformational control of the 2B subdomain. For example, removal of the 2B subdomain to form RepΔ2B activates the monomeric helicase (20,27). RepΔ2B monomers also translocate on single-stranded DNA faster than wtRep monomers (20,27). RepΔ2B appears functional in the cell; it can support replication of a phage that requires Rep function in vivo, although cells expressing RepΔ2B grow at a slower rate (27).
Activation of monomer unwinding is also possible by controlling 2B conformation. Single-molecule FRET (smFRET) and optical tweezers studies by Arslan et al. (25) demonstrate that intramolecular cross-linking of the 2B subdomain of Rep and PcrA into the closed state activates a monomer into a ‘superhelicase’ capable of unwinding double-stranded DNA (dsDNA) with very high processivity (>1 kb) compared to reported values for unmodified Rep-like helicases. Conversely, Rep cross-linked into an open state exhibits little unwinding activity. Interactions with accessory proteins (37) similarly regulate unwinding through conformational control of the 2B subdomain (25). Through simultaneous smFRET and optical tweezers measurement, Comstock et al. (23) observed that 2B conformational switching between closed and open states correlates with a switch in UvrD activity between DNA unwinding and DNA rezipping, respectively. A model was previously proposed in which UvrD can ‘strand-switch’, alternately translocating on one or the other of the two antiparallel DNA strands (11) (Figure 1B). To explain the correlation with 2B conformation, Comstock et al. proposed that 2B serves as an ‘orientational factor’, positioning the 1A/2A motor core on one strand, directing them into the ss-dsDNA fork junction (unwinding the duplex) or on the other strand, directing them away from the fork junction (allowing the duplex to rezip) (23).
Another mechanism to activate processive DNA unwinding of Rep-like helicases is through assembly into dimers. Rep can dimerize on DNA at high Rep/DNA ratios (20), and ensemble and single-molecule measurements have demonstrated that two molecules are needed for processive unwinding activity (20,32,34). Interestingly, the 2B subdomain has been implicated as a mediating factor for activation (20,27), and dimerization of UvrD has been shown to affect closing of the 2B subdomain (26). The 2B-deficient variant, RepΔ2B, neither dimerizes in solution nor upon binding to single-stranded DNA (20,27).
Together, these studies suggest that the 2B subdomain plays a primarily regulatory role, with the closed state activating Rep-like helicases for processive unwinding and the open state inhibiting unwinding activity. Moreover, they suggest that conformational switching between the two states may limit processivity by causing the helicase motor core to be redirected from the translocating strand to the opposing strand via ‘strand-switching’. However, our understanding of such mechanisms remains limited because the regulatory effects of 2B on Rep unwinding have not been well quantified. In the present study, we performed single-molecule optical trap measurements on full-length Rep helicase (wtRep; Figure 1A, left) and the 2B-deficient variant RepΔ2B (Figure 1A, right) (20,27) to quantify the effect of the 2B subdomain on Rep unwinding processivity and speed. By monitoring Rep activity on two different DNA substrates with unique geometries as a function of applied force, we show that a monomer of RepΔ2B is capable of unwinding hundreds of base pairs, making it much more processive than a dimer of wtRep, but not as processive as Rep with 2B cross-linked into the closed state. Our measurements confirm that the 2B subdomain is not essential for unwinding but plays a role in regulating the speed at which Rep can unwind DNA. We also show that 2B is not required for strand-switching, contrary to expectations. Furthermore, they reveal that Rep processivity is limited by the frequency of strand-switching events, which depends strongly on duplex stability. We place our findings in the context of Rep helicase's function in the cell and of the regulation mechanisms of SF1 helicases.
MATERIALS AND METHODS
Full-length Rep (here referred to as wtRep; Figure 1A left) and a 2B-deletion (RepΔ2B; Figure 1A, right) were expressed and purified as described (27,38). The ‘hairpin’ (Supplementary Figure S1A) and ‘fork’ (Supplementary Figure S1B) DNA constructs used in these experiments consisted of a variable ‘insert’ between two long double-stranded (ds)DNA ‘handles’ that were modified with biotin and digoxigenin to facilitate attachment to streptavidin and anti-digoxigenin antibody coated beads, respectively. The hairpin construct was made by ligating a left handle (LH, 1.5 kb) and a right handle (RHhairpin, 1.5 kb) to an 89-bp hairpin stem capped by a (dT)4 tetraloop (Supplementary Figure S1A). The fork construct was made by annealing and ligating four DNA fragments: a dsDNA left handle (LH, 1.5 kb) and right handle (RHfork 1.5 kb), a short ssDNA spacer, and a free 3′ poly-dT ssDNA tail for protein loading (Supplementary Figure S1B). All oligonucleotides for the synthesis of the constructs were purchased from Integrated DNA Technologies (IDT, Coralville, IA), and the sequences of the primers and inserts are listed in Supplementary Table S1.
All measurements were made using a custom-built dual-trap optical tweezers described previously (39–41). Data were collected at a rate of 100 Hz, using force feedback to maintain a constant tension in the tethered DNA. All data traces are plotted at 100 Hz. At this data acquisition rate, the noise was measured to be ≤2 bp in 97% of the DNA hairpin tethers formed and ≤4 bp in 96% of the DNA fork tethers formed (see SI Text). Events were scored as unwinding activity if their amplitude exceeded 1.5× the background noise and lasted more than 0.04 s. All single-molecule unwinding measurements were made in a custom flow chamber at 22°C, in a pH 8.0 buffer containing 2% glycerol, 20 mM NaCl, 35 mM TrisHCl, 5 mM MgCl2, conditions comparable to those in other in vitro studies of Rep and homologs (8,11,23,25,27,32,34). An oxygen scavenging system (1.2% glucose, 0.13 mg/mL catalase (EMD Millipore, Billerica, MA, USA), 0.29 mg/ml pyranose oxidase (Coriolus sp., Sigma-Aldrich, St. Louis, MO, USA)) was used to increase the lifetime of the DNA tethers (42,43). The ATP was at saturating concentrations ( 250 μM) for all experiments. The protein concentration was 6.8 or 13.6 nM for RepΔ2B and 44 nM for wtRep, unless otherwise specified. Detailed descriptions of the experimental and data analysis protocols are provided in SI Text.
250 μM) for all experiments. The protein concentration was 6.8 or 13.6 nM for RepΔ2B and 44 nM for wtRep, unless otherwise specified. Detailed descriptions of the experimental and data analysis protocols are provided in SI Text.
RESULTS
Deletion of Rep's 2B subdomain activates processive unwinding
To measure Rep helicase activity, we detected the unwinding of a DNA hairpin held under a constant force with dual-trap optical tweezers (39,40) (Figure 1C; see Materials and Methods). The hairpin consisted of an 89-bp stem capped with a (dT)4 tetraloop (Supplementary Figure S1A) and was held under a range of tensions (4–14 pN) below that necessary to mechanically unwind the hairpin (∼16 pN; see Supplementary Figure S1C). A poly-dT ssDNA loading site at the 3′-end of the hairpin (unless otherwise noted 10 nt in length) accommodated the 8-nt footprint of a single Rep monomer (16). The site allowed for monomeric protein in solution to bind and unwind the hairpin in the presence of saturating concentrations of ATP ( 250 μM). Each base pair unwound released 2 nt, and was detected as an increase in the end-to-end extension of the tethered DNA molecule stretched by the optical traps (Figure 1C). At the data collection rate of 100 Hz, unwinding events of >3 bp could typically be resolved with this assay (see Materials and Methods).
250 μM). Each base pair unwound released 2 nt, and was detected as an increase in the end-to-end extension of the tethered DNA molecule stretched by the optical traps (Figure 1C). At the data collection rate of 100 Hz, unwinding events of >3 bp could typically be resolved with this assay (see Materials and Methods).
Figure 1D and E display a representative time trace of wtRep and RepΔ2B unwinding activity at a force of 12 and 9 pN, respectively. Each series of peaks represents individual rounds of hairpin unwinding followed by protein dissociation and eventual replacement by other protein. We routinely observed the deletion mutant RepΔ2B unwinding the hairpin DNA construct with a loading site accommodating a monomer (10 dT), across the force range assayed (4–14 pN). In contrast, wtRep unwinding was more difficult to detect and generally required an elongated protein loading site (38 dT)—allowing the binding of multiple monomers—and an increased protein concentration (44 nM, compared to 6.8 or 13.6 nM used for RepΔ2B). This observation is consistent with prior findings that Rep monomers are poor helicases and that activation requires at least a Rep dimer for processive unwinding (27,32,34). We thus attributed wtRep activity such as that observed in Figure 1D to that of a dimer. We detected rare cases of unwinding of a hairpin with a 10-dT loading site that could be attributed to monomeric wtRep only when high forces (15 pN), near those sufficient to unwind the hairpin mechanically (∼16 pN), were applied (Supplementary Figure S2). Presumably the latter events were observed because the base pairs of the hairpin were highly destabilized by the large force, facilitating its unwinding.
In many instances of wtRep dimer activity, unwinding was limited to a fraction of the 89-bp long hairpin stem. For example, in Figure 1D (inset) wtRep unwinds 35 bp before it reverses direction and the duplex rezips. In contrast, we observed numerous examples of RepΔ2B unwinding through the entire 89-bp hairpin stem past the (dT)4 tetraloop capping the hairpin stem, and translocating onto the opposing strand allowing the hairpin to rezip behind it (e.g. see Figure 1E, inset).
The low concentration of protein, short DNA loading site, long dwell times between rounds of activity compared to the average round duration, and prior findings that RepΔ2B is monomeric in solution (20) strongly suggest that the observed activity results from monomeric RepΔ2B, with multiple proteins simultaneously acting on DNA unlikely. To test this further, we carried out control experiments that ensured that only a single Rep monomer could bind to the DNA hairpin at a time. As shown in Supplementary Figure S3, we used a laminar flow chamber containing two streams of buffer flowing parallel to each other with minimal mixing to control the assembly of the RepΔ2B-DNA complex (see SI Text). One stream contained protein plus non-hydrolyzable ATP analog ATP-γS (250 μM) and the other ATP ( 250 μM) only (Supplementary Figure S3A). Positioning the optically trapped beads and tethered DNA hairpin in the protein + ATP-γS stream allowed a single RepΔ2B to bind to the 10-dT loading site but not unwind the DNA. Moving the protein-bound DNA tether into the ATP stream resulted in ATP binding and hairpin unwinding (Supplementary Figure S3B). Unwinding was followed by protein dissociation but no replacement, confirming a single protein turnover. Again, we observed the same high DNA unwinding processivity for monomeric RepΔ2B, with frequent examples of complete hairpin unwinding (Supplementary Figure S3B). In contrast, no unwinding activity was detected with wtRep under these single-round conditions. Thus, removal of the 2B subdomain dramatically increased unwinding activity relative to wtRep monomer or dimer.
250 μM) only (Supplementary Figure S3A). Positioning the optically trapped beads and tethered DNA hairpin in the protein + ATP-γS stream allowed a single RepΔ2B to bind to the 10-dT loading site but not unwind the DNA. Moving the protein-bound DNA tether into the ATP stream resulted in ATP binding and hairpin unwinding (Supplementary Figure S3B). Unwinding was followed by protein dissociation but no replacement, confirming a single protein turnover. Again, we observed the same high DNA unwinding processivity for monomeric RepΔ2B, with frequent examples of complete hairpin unwinding (Supplementary Figure S3B). In contrast, no unwinding activity was detected with wtRep under these single-round conditions. Thus, removal of the 2B subdomain dramatically increased unwinding activity relative to wtRep monomer or dimer.
Rep processivity is controlled by duplex stability
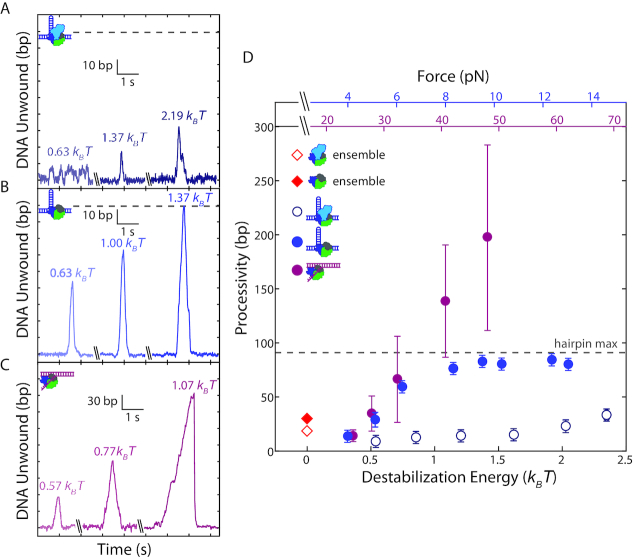
We next asked what aspects of Rep helicase activity are affected by 2B removal. We considered the processivity of Rep, which we define as the maximum number of base pairs unwound in one round of activity, and the effect of force. Representative time traces at different tensions show that wtRep dimer processivity increased marginally across the force range (Figure 2A) whereas that of RepΔ2B increased more dramatically (Figure 2B; distributions plotted in Supplementary Figure S5). (The processivities at each force were the same under single protein-turnover conditions or under conditions where protein could be replaced. Figure 2 combines both data sets.) Figure 2D summarizes this analysis by plotting the median processivity against force. We also provide in Figure 2C and D the corresponding decrease in base-pairing energy due to the destabilizing effect of force (see SI Text). At forces beyond ∼8 pN, the true RepΔ2B processivity exceeds 89 bp but is limited by the hairpin stem length.
Figure 2.
Effect of duplex stability on unwinding processivity of RepΔ2B and wtRep. (A–C) Representative traces of hairpin DNA unwinding by wtRep dimer (A, shades of dark blue), hairpin DNA unwinding by RepΔ2B monomer (B, shades of blue), and fork DNA unwinding by RepΔ2B monomer (C, shades of magenta) showing unwinding processivity with increasing force. Values in units of kBT correspond to destabilization energies (see SI Text). (D) Median processivity as a function of destabilization energy for wtRep hairpin (open dark blue circles, N = 37–67), RepΔ2B hairpin (filled blue circles, N = 34–770), and RepΔ2B fork (filled magenta circles, N = 37–180) unwinding activity. Maximum processivity at zero force determined from stopped-flow ensemble measurements for wtRep dimer (red open diamond) and RepΔ2B monomer (red filled diamond). The top axes show the forces applied to the hairpin (blue) or fork (magenta), and the bottom axis shows the corresponding destabilization energy. Error bars represent standard error of the median. The gray dashed lines in A–B, D indicate the extension expected for complete hairpin opening, placing a limit on the maximum attainable processivity.
Due to the limited hairpin length (89 bp stem + 4 nt loop), we carried out additional measurements with an alternate DNA construct that provided a much longer track for DNA unwinding. We tethered between optically trapped beads a construct containing a fork junction with a free 3′-tail (10-dT unless otherwise noted) for protein loading followed by 1.5 kb of dsDNA (Figure 1F, see Materials and Methods). In this geometry, helicase unwinding of the duplex converted each broken dsDNA base pair into 1 nt of ssDNA (Supplementary Figure S1B, see SI Text), detected as an increase in the end-to-end extension of the tethered DNA over the range of forces applied (25–55 pN). Unwinding events >6 bp could be resolved at a data acquisition rate of 100 Hz with this assay (see Materials and Methods). As shown in Figure 1G, we consistently observed processive unwinding of the fork construct with RepΔ2B, with activity rounds reaching several hundred base pairs at the highest forces assayed. Again, wtRep activity was rarely detected, only when the 3′ tail was lengthened to 20 dT and the concentration of protein was high (13.6 or 35 nM) (Supplementary Figure S4). As above, we repeated measurements under single-round conditions ensuring monomer activity, and observed similarly processive RepΔ2B unwinding (Supplementary Figure S3C). As with the hairpin, processivity on fork DNA increased as a function of force (Figure 2B–D).
In order to compare our results on the two constructs, we considered how force would affect helicase activity, in particular processivity. To first order, we expect increased force to decrease base-pair stability (44), which may in turn facilitate unwinding. For the two construct geometries used, force is applied along different directions (perpendicular to the duplex strands for the hairpin, parallel in the fork) and thus contribute differently to duplex destabilization. By considering the mechanical work to stretch dsDNA and ssDNA to a force F, we calculated how force decreases the base-pairing energy stabilizing the duplex at each F (see SI Text). Despite detecting unwinding activity over different force ranges in the two constructs (4-14 pN for the hairpin, 25–55 pN for the fork), the corresponding destabilization energy ranges overlap (0.33-2.2 versus 0.38–1.7 kBT per base pair, respectively). Plotting the median processivities measured on hairpin and fork constructs along a common axis of force-mediated destabilization energy (Figure 2D), we observe the two data sets to match (P > 0.4, see SI Text) in the force range where unwinding is not limited by the hairpin stem length (0.3–0.7 kBT). The data sets diverge at higher destabilization energies (>0.8 kBT), only due to the increased number of events corresponding to complete hairpin unwinding. This strongly suggests that base-pair stability is a major determinant of processivity. Ensemble kinetic assays have also been used to estimate wtRep dimer and RepΔ2B monomer DNA unwinding processivity, yielding values of ∼18 and ∼30 bp, respectively, under similar buffer conditions as our measurements (50 mM NaCl versus 20 mM NaCl, see Materials and Methods) (20). These zero-force data points (Figure 2D, red open and filled diamond) match well with the trend with force observed in our data.
RepΔ2B and wtRep exhibit different unwinding speeds across the range of applied forces
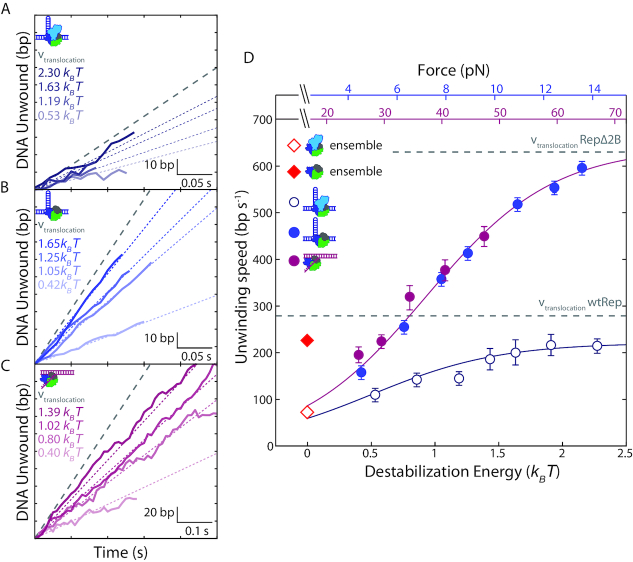
We next investigated the effect of 2B removal on DNA unwinding speed. Figure 3A and B shows sections of representative traces of DNA hairpin unwinding by wtRep and RepΔ2B, respectively. Figure 3D plots the average unwinding speed for both against force (and destabilization energy). (Speed distributions are shown in Supplementary Figure S6). At the lowest force assayed, ∼4 pN, the average unwinding speed of wtRep dimers on hairpin DNA was 110 ± 10 bp s−1, similar to that previously reported in ensemble stopped-flow experiments, 72 ± 20 bp s−1 (32) (Figure 3D, red open diamond). RepΔ2B monomer speed at the same force, 160 ± 4 bp s−1, was close to that of wtRep and similar to the previously reported value, 216 ± 22 bp s−1 (20) (Figure 3D, red filled diamond). Both unwinding speeds increased with force, to a greater extent for RepΔ2B. At the highest force assayed, ∼14 pN, the wtRep dimer unwinding speed approached the known wtRep monomer ssDNA translocation speed in the absence of force, 279 ± 2 nt s−1 (20) (Figure 3D, lower grey dashed line). RepΔ2B unwinding speed increased more dramatically to 600 ± 7 bp s−1, also comparable to its reported translocation speed, 630 ± 2 bp s−1 (20) (Figure 3D, upper grey dashed line). We expect the translocation speed on ssDNA to represent the fastest limiting speed of DNA unwinding that the protein can achieve.
Figure 3.
Effect of duplex destabilization on unwinding speed of RepΔ2B and wtRep. (A–C) Representative sections of hairpin DNA unwinding by wtRep dimer (A, shades of dark blue), hairpin DNA unwinding by RepΔ2B monomer (B, shades of blue), and fork DNA unwinding by RepΔ2B monomer (C, shades of magenta) showing unwinding speed with increasing destabilization energy. Color-coded dotted lines represent mean speed over all traces at the corresponding destabilization energy (see SI Text). The grey dashed line represents wtRep monomer (A) and RepΔ2B monomer (B, C) translocation speeds at zero force. For clarity, all selected traces start at the baseline (0 bp unwound) and extend to the first instance of a pause or a reversal in direction. (D) Mean unwinding speed as a function of destabilization energy for wtRep hairpin (open dark blue circles, N = 6–51), RepΔ2B hairpin (filled blue circles, N = 49–140), and RepΔ2B fork (filled magenta circles, N = 31–73) activity. The average unwinding speeds of wtRep dimer and RepΔ2B monomer at zero force are indicated by the red open and filled diamonds, respectively. The top axes show the forces applied to the hairpin (blue) or fork (magenta), and the bottom axis shows the corresponding destabilization energy. Error bars represent standard error of the mean. The grey dashed lines represent RepΔ2B and wtRep translocation speeds. The solid lines represent a model for the dependence of unwinding speed on destabilization energy for RepΔ2B (magenta) and wtRep (dark blue) (see main and SI text).
Individual traces for RepΔ2B unwinding of fork DNA display the same trend of speeds increasing with force over the range 25–55 pN (Figure 3C). Plotting RepΔ2B unwinding speed on fork DNA against force-mediated destabilization energy (Figure 3D), as done above for processivity, shows excellent agreement (P > 0.7) with the corresponding rates on hairpin DNA over their overlapping energy range (0.4–1.4 kBT). This indicates that, similarly to processivity, the destabilizing effect of force on the DNA duplex is a significant factor determining RepΔ2B unwinding speed. In contrast, wtRep dimer unwinding is slower than that of RepΔ2B monomer and exhibits a smaller force dependence.
RepΔ2B strand-switching limits processivity
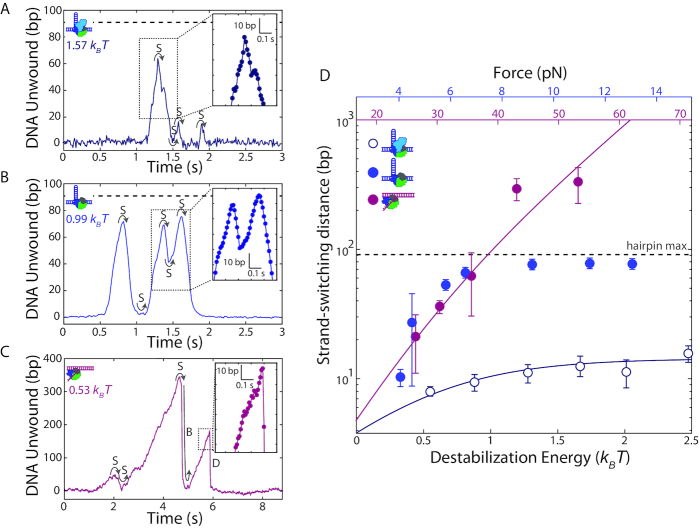
The similarity in the dependence of the unwinding processivity and speed on base pair destabilization energy suggests that the two are connected. In particular, it suggests that kinetic competition between unwinding and another process may limit processivity. Inspecting time traces of hairpin DNA unwinding by wtRep (Figure 4A) and RepΔ2B (Figure 4B) illuminates the primary process limiting processivity. In the great majority of unwinding events that did not reach the end of the hairpin (100% for wtRep and 94% for RepΔ2B over the destabilization energy range 0.5–0.8 kBT), unwinding ceased with a reversal in direction (e.g. see Figure 4A, t = 1.4, 1.6, and 1.9 s; Figure 4B, t = 0.9, 1.4, and 1.6 s) after which the hairpin gradually rezipped with rates comparable to DNA unwinding (Figure 4A and B, insets). In contrast to cases showing complete hairpin unwinding—in which Rep could translocate continuously on the same strand as it moved up the hairpin stem, past the tetraloop cap, and down the hairpin stem—we expect mid-hairpin reversals to involve Rep disengaging from one DNA strand and switching to the opposing strand. Strand-switching has been observed for UvrD (11) and correlated to 2B subdomain movement (23). Our results show that strand-switching of Rep does not require the 2B subdomain.
Figure 4.
Helicase processivity is limited by strand-switching. (A, B) Representative trace of wtRep dimer (A) and RepΔ2B monomer (B) activity on a hairpin DNA showing multiple rounds of unwinding and rezipping mediated by strand-switching (S). Close-up (insets) with individual data points (filled circles) shows that rezipping and unwinding have similar rates. The color-coded values in units of kBT correspond to the destabilization energies (see SI Text). (C) Representative trace of RepΔ2B monomer activity on fork DNA illustrating strand-switching (S), snap-back (B) and dissociation (D). Close-up (inset) shows that dissociation events are rapid and distinct from unwinding or rezipping (compare to insets in A, B). (D) Median strand-switching distance as a function of destabilization energy for wtRep hairpin (open dark blue circles, N = 50–120), RepΔ2B hairpin (filled blue circles, N = 11–66), and RepΔ2B fork (filled magenta circles, N = 12–44) activity. The top axes show the forces applied to the hairpin (blue) or fork (magenta), and the bottom axis shows the corresponding destabilization energy (see SI Text). The grey dashed lines in A–B, D indicate the limit set by the length of the DNA hairpin. The solid lines represent a model for the dependence of strand-switching distance on destabilization energy for RepΔ2B (magenta) and wtRep (dark blue) (see main and SI text). Error bars represent standard error of the median.
We observed strand-switching during transitions from hairpin unwinding to rezipping, but also from the reverse—rezipping to unwinding—often with the switch occurring at the base of the hairpin (e.g. see Figure 4A, at t = 1.5 s; Figure 4B, t = 1.1, 1.5 s). In about half of all bursts of activity, we observed multiple rounds of unwinding, strand-switching, and rezipping prior to protein dissociation. Strand-switching was similarly frequent (94% at 0.5-0.8 kBT) for RepΔ2B during unwinding of the fork construct (Figure 4C, t = 2 s).
Determining the mean distance unwound between strand-switching events—which we term strand-switching distance—from unwinding to rezipping, we detect a similar dependence on force as for processivity (Figure 4D). Again, plotting strand-switching distance against a common axis of force-mediated duplex destabilization energy allows for direct comparison of the data for hairpin and fork constructs. We observe excellent agreement of the RepΔ2B hairpin and fork data sets for data in which hairpin length is not limiting, emphasizing that strand-switching by RepΔ2B is not influenced by DNA geometry. Our analysis suggests that the primary factor limiting processive unwinding is strand-switching.
We also observed two distinct behaviors for RepΔ2B unwinding of the fork DNA construct. Figure 4C (at t = 6.3 s) shows a representative time trace during which the DNA extension rapidly dropped to the baseline, with no subsequent unwinding until the next protein was loaded. We interpret this signal as RepΔ2B dissociating followed by immediate rezipping of the DNA. In contrast, we observe RepΔ2B dissociation from the middle of the hairpin in only about 6% of the hairpin unwinding measurements; all of the mid-hairpin dissociation events happened at low forces (<6 pN). The most frequent (61%) dissociation events occurred with the helicase at the base of the hairpin (Figure 4A, t = 2 s, Figure 4B, t = 1.8 s). Furthermore, in a small fraction (2% over the range 0.5–0.8 kBT) of fork unwinding data we observed a pattern of sudden drops in extension back to the baseline followed by immediate restart of unwinding (Figure 4C, t = 5 s). Since our assay ensures a single round of unwinding for each loaded protein, we interpret such events as RepΔ2B ‘snapping back’ to the base of the fork, without dissociation. After spontaneous rezipping of the duplex, the same protein molecule unwinds the fork a second time. Interestingly, all snap-back events were preceded by strand-switching (Figure 4B, left inset). Similar snap-back behavior was first reported in single-molecule measurements of Rep translocation on ssDNA (8). Importantly, such behavior was proposed to be mediated by DNA looping, which would involve non-canonical interactions between Rep and the 3′ end of the DNA strand along which translocation occurs. Our observation of snap-back behavior on fork DNA is consistent with such a model. We did not observe snap-back behavior on hairpin DNA; in contrast to the 3′ tail of the fork, the 3′ end of the hairpin is under tension which presumably inhibits looping. Figure 5 summarizes the behaviors exhibited by wtRep and RepΔ2B on hairpin and fork DNA and their frequencies over the same range of destabilization energies, 0.5–0.8 kBT (see Discussion and SI text).
Figure 5.
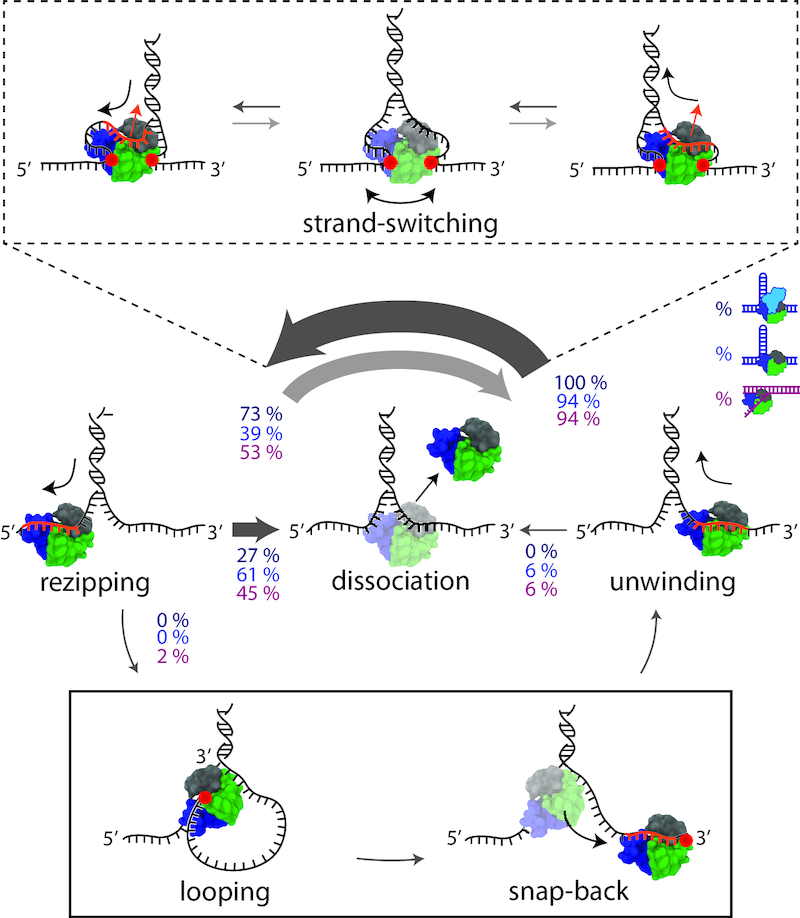
Schematic and model of RepΔ2B behaviors during DNA unwinding. Distinct protein behaviors, i.e. unwinding, rezipping, strand-switching, dissociation, and looping (as labelled) are observed during wtRep dimer and RepΔ2B monomer activity in the optical trap measurements. Percentages correspond to the frequency of a behavior possible at each junction as indicated by the direction of the arrow. Percentage values represent averages over a 0.5-0.8 kBT destabilization energy range for wtRep hairpin (dark blue, N = 15), RepΔ2B hairpin (blue, N = 305) and RepΔ2B fork (magenta, N = 169) activity (see SI Text for details). The thickness of the arrow represents the frequency of a behavior at each junction. During strand-switching (schematics in dashed-line box), ssDNA dissociates from the canonical binding site (orange portion of the DNA), but non-canonical protein-DNA interactions (red circles) allow Rep to remain bound to the junction. The opposing strand binds to the canonical site to complete strand-switching. During snap-back (schematics in solid-line box), ssDNA dissociates from the canonical binding site. A loop formed via non-canonical binding (red circle) allows Rep to snap back to the 3′ end without dissociating.
DISCUSSION
Our measurements show that the 2B subdomain of Rep plays a regulatory role and is not essential for DNA unwinding activity, consistent with previous reports (20,27). This rules out mechanisms for DNA unwinding by monomeric SF1 helicases (17,18) in which the interaction between the 2B subdomain and duplex DNA is proposed to be essential for DNA unwinding.
Our results show that RepΔ2B and wtRep exhibit very different DNA unwinding speeds under the influence of force (Figure 3), indicating a role for 2B in regulating speed. A quantitative model by Betterton and Jülicher (45) that describes the effect of duplex stability on helicase unwinding speed (see SI Text) fits both RepΔ2B and wtRep behaviors well (Figure 3, solid lines). In this model, both proteins unwind DNA by an active mechanism, contributing similarly to the destabilization of the duplex (see SI Text, Supplementary Table S2). The crucial difference between the two is the maximum limiting speed, 612 bp s−1 for RepΔ2B and 218 bp s−1 for wtRep, values close to their reported ssDNA translocation speeds, 630 nt s−1 and 279 nt s−1, respectively (20) (see Supplementary Table S2). The presence of the 2B subdomain thus limits Rep translocation speed, perhaps due to its interactions with DNA. Dimerization of wtRep is less likely to be a factor since we observe comparable unwinding speeds, 245 ± 70 bp s−1, in the rare examples of wtRep activity we attribute to monomers (Supplementary Figure S2).
Rep monomers lacking the 2B subdomain unwind DNA with a higher processivity than wtRep dimers, reaching up to several hundred base pairs with assisting force. Recent theoretical work has sought to model how helicase processivity depends on force (46). Our analysis of the force dependence of RepΔ2B processivity on the two different DNA construct geometries indicates that duplex stability is a key factor controlling processivity (Figure 2), similar to controlling unwinding speed (Figure 3). We further show (Figure 4) that strand-switching is the primary event limiting processivity (as opposed to protein dissociation or snap-back). These results together suggest a mechanism in which strand-switching and DNA unwinding are in kinetic competition. We devised a simple kinetic competition model (see SI Text) in which Rep enters the strand-switching pathway at a certain rate, while the unwinding speed determines the distance traveled between strand-switching events. For wtRep, the strand-switching distance is well fit by this simple model (Figure 4, solid dark blue line), assuming a constant rate of strand-switching (see Supplementary Table S2). For the corresponding RepΔ2B data, we found it necessary for the strand-switching rate also to depend on duplex destabilization energy to fit the results (see SI Text; Figure 4, solid magenta line). Thus RepΔ2B is more processive than wtRep as the duplex is destabilized in part because it unwinds faster but also because its rate of strand-switching decreases below that of wtRep. Duplex stability is thus a critical determinant of strand-switching frequency and processivity. This finding aligns well with prior observations that the locations on DNA at which the Rep homolog UvrD reverses direction correlate with more thermodynamically stable regions that would pose a barrier to unwinding (23).
This model of strand-switching would explain why such events occur, but not how they occur. Our observation of such DNA transactions in protein lacking the 2B subdomain do not support a proposed model for UvrD, in which strand-switching is mediated by 2B (23). In this model, interactions between the GIG motif in the 2B subdomain and the duplex above the fork junction allow the 1A–2A motor core to switch strands by providing an anchor point for UvrD to remain bound to DNA (23). However, it should be noted that the corresponding motif in Rep has a glutamic acid (E) substituting one glycine (G) (18), so the 2B subdomain interactions with dsDNA in Rep may be different. Nevertheless, our findings show that 2B-DNA interactions are not essential for strand-switching in Rep helicase.
The observation of snap-back behavior on fork DNA (Figure 4C) may point to a potential mechanism. During snap-backs, the RepΔ2B motor core must disengage with its DNA strand, causing the duplex to rezip, yet the protein remains attached as evidenced by another round of unwinding. This behavior indicates that non-canonical ssDNA-helicase interactions must exist. This interpretation is compatible with smFRET measurements of wtRep monomer translocation on ssDNA. Myong et al. (8) showed that an encounter with a ds-ssDNA junction causes Rep to snap back to the 3′-end of the translocating strand. FRET between the labeled Rep and 3′ ssDNA end provides direct evidence that snap-backs occur through the formation of a ssDNA loop via Rep interaction with the 3′-end. Figure 5 illustrates (solid line box) how looping through non-canonical interactions with the DNA tail (red circle) could mediate snap-back events in our experiments. Similarly, Figure 5 shows (dashed-line box) how non-canonical interactions with either (or both) strands of the fork junction (red circles) could plausibly provide RepΔ2B additional attachment points to the DNA to facilitate strand-switching without dissociation.
Although we find that the 2B subdomain is not essential for strand-switching, we do not believe this contradicts the model that this subdomain can redirect the helicase motor core at a fork junction by rotating (23). The 2B subdomain rotational conformation is an important factor in Rep unwinding activity, as demonstrated by the fact that Rep cross-linked into a closed 2B conformation (‘Rep-X’) unwinds processively while Rep cross-linked into an open state (‘Rep-Y’) does not (25). Interestingly, Rep-X displays a much higher processivity than either wtRep dimers or RepΔ2B monomers, exceeding 6 kb in optical tweezers measurements with applied forces providing less duplex destabilization than in the present study (25). Importantly, no evidence for strand-switching was observed. We believe this is either because constraining 2B in a closed state prevents the motor core from being redirected to the other strand or because the topological enclosure of one DNA strand by the cross-link prevents strand exchange or dissociation. In contrast, strand-switching by RepΔ2B or wtRep is not impeded.
Our measurements also show that monomers of wtRep exhibit very little unwinding activity (Figure 2A and D), consistent with prior reports (8,32,34). We observe wtRep unwinding across a range of forces only when multiple proteins can bind, promoting dimerization. Dimerization of Rep is known to facilitate processive unwinding, and dimerization of the Rep homolog UvrD was recently shown to shift the 2B subdomain toward a more closed state (26), the conformation known to promote unwinding (25). However, assuming that this conformational shift occurs for Rep, dimerization cannot simply sequester the 2B subdomain into the closed state since dimeric wtRep is much less processive than Rep-X. We thus strongly suspect that 2B in a wtRep dimer may still exhibit conformational flexibility. Our results along with the findings mentioned above point to a mechanism in which conformational switching of the 2B subdomain does not simply mediate strand-switching, as shown for UvrD (23), but can promote it. This could explain why the rate of wtRep strand-switching is consistently larger than that of RepΔ2B (see SI Text, Supplementary Table S2). Suppression of wtRep helicase processivity may thus be the result of 2B conformational flexibility (i.e. 2B rotation), which is absent in both RepΔ2B and Rep-X.
SF1 helicases such as Rep have multiple activities and functions that need to be regulated. Rep not only functions as a helicase to unwind DNA, but can also displace proteins from the DNA that might interfere with replication (15). Interestingly, Rep's 2B subdomain plays a role in regulating both its ability to unwind DNA and displace proteins from DNA. As a monomer, Rep is a rapid and processive ssDNA translocase, but must be activated via some mechanism to exhibit helicase activity. As shown in this study and previously (20), the 2B subdomain of Rep is not essential for its DNA unwinding activity, but rather plays a role in inhibiting monomeric helicase activity. However, Rep also functions to displace DNA-bound proteins that would interfere with replication and the 2B subdomain, not just its translocase activity, is needed for Rep to carry out protein displacement (15). Hence it is possible that as a monomer, Rep functions as a ssDNA translocase using its 2B subdomain to displace proteins from DNA. However, since the 2B subdomain is inhibitory for Rep monomer helicase activity, Rep needs to oligomerize or interact with accessory proteins to remove the helicase-inhibitory function of the 2B subdomain. A possible interaction that lifts 2B autoinhibition of Rep monomer unwinding activity may occur in replication restart. During this process Rep is tasked with unwinding ∼100 bp of the nascent lagging strand at a stalled replication fork, allowing fork reversal to occur (47). This value is larger than the mean wtRep dimer processivity we measure at any force. In this capacity, Rep may thus require the aid of protein partners to be activated. For instance, Rep interaction with loading protein PriC at a stalled replication fork is known to stimulate its unwinding activity 6-fold (13).
DNA transactions such as strand-switching may also play a role in Rep's cellular function. While Rep must unwind lagging strand DNA, studies report that Rep can track on the leading strand template (48,49), where 3′-5′ translocation would direct it into the replication fork. Strand-switching would provide a mechanism for Rep to bypass the fork rather than unwind the parental dsDNA. Thus, such DNA transactions may play an important role in regulating Rep unwinding of lagging strand DNA during replication fork reversal.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Chemla and Lohman laboratories for scientific discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Work in the Lohman lab is support by National Institutes of Health (NIH) [R01 GM045948]; Work in the Chemla laboratory is supported by National Institutes of Health [R01 GM120353] National Science Foundation Physics Frontier Center (PFC) ‘Center for the Physics of Living Cells’ (CPLC) [PHY-1430124]. Funding for open access charge: NIH [R01 GM120353] and NIH [R01 GM045948].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lohman T.M., Tomko E.J., Wu C.G.. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 2008; 9:391–401. [DOI] [PubMed] [Google Scholar]
- 2. Lohman T.M., Bjornson K.P.. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996; 65:169–214. [DOI] [PubMed] [Google Scholar]
- 3. Singleton M.R., Dillingham M.S., Wigley D.B.. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007; 76:23–50. [DOI] [PubMed] [Google Scholar]
- 4. Delagoutte E., von Hippel P.H.. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part I: Structures and properties of isolated helicases. Q. Rev. Biophys. 2002; 35:431–478. [DOI] [PubMed] [Google Scholar]
- 5. Delagoutte E., von Hippel P.H.. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003; 36:1–69. [DOI] [PubMed] [Google Scholar]
- 6. Brabant A.J. Van, Stan R., Ellis N.A.. DNA helicases, genomic instability and human genetic disease. Annu. Rev. Genomics Hum. Genet. 2000; 1:409–459. [DOI] [PubMed] [Google Scholar]
- 7. Chu W.K., Hickson I.D.. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009; 9:644–654. [DOI] [PubMed] [Google Scholar]
- 8. Myong S., Rasnik I., Joo C., Lohman T.M., Ha T.. Repetitive shuttling of a motor protein on DNA. Nature. 2005; 437:1321–1325. [DOI] [PubMed] [Google Scholar]
- 9. Veaute X., Delmas S., Selva M., Jeusset J., Le Cam E., Matic I., Fabre F., Petit M.-A.. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 2005; 24:180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin C.-T., Tritschler F., Lee K.S., Gu M., Rice C.M., Ha T.. Single-molecule imaging reveals the translocation and DNA looping dynamics of hepatitis C virus NS3 helicase. Protein Sci. 2017; 26:1391–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dessinges M.-N., Lionnet T., Xi X.G., Bensimon D., Croquette V.. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:6439–6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heller R.C., Marians K.J.. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J. Biol. Chem. 2005; 280:34143–34151. [DOI] [PubMed] [Google Scholar]
- 14. Guy C.P., Atkinson J., Gupta M.K., Mahdi A.A., Gwynn E.J., Rudolph C.J., Moon P.B., van Knippenberg I.C., Cadman C.J., Dillingham M.S. et al.. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol. Cell. 2009; 36:654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brüning J.-G., Howard J.A.L., Myka K.K., Dillingham M.S., McGlynn P.. The 2B subdomain of Rep helicase links translocation along DNA with protein displacement. Nucleic Acids Res. 2018; 1:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Korolev S., Hsieh J., Gauss G.H., Lohman T.M., Waksman G.. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997; 90:635–647. [DOI] [PubMed] [Google Scholar]
- 17. Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B.. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999; 97:75–84. [DOI] [PubMed] [Google Scholar]
- 18. Lee J.Y., Yang W.. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006; 127:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dillingham M.S. Superfamily I helicases as modular components of DNA-processing machines. Biochem. Soc. Trans. 2011; 39:413–423. [DOI] [PubMed] [Google Scholar]
- 20. Brendza K.M., Cheng W., Fischer C.J., Chesnik M.A., Niedziela-Majka A., Lohman T.M.. Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:10076–10081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fischer C.J., Maluf N.K., Lohman T.M.. Mechanism of ATP-dependent translocation of E. coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 2004; 344:1287–1309. [DOI] [PubMed] [Google Scholar]
- 22. Niedziela-Majka A., Chesnik M.A., Tomko E.J., Lohman T.M.. Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 2007; 282:27076–27085. [DOI] [PubMed] [Google Scholar]
- 23. Comstock M.J., Whitley K.D., Jia H., Sokoloski J., Lohman T.M., Ha T., Chemla Y.R.. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science. 2015; 348:352–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jia H., Korolev S., Niedziela-Majka A., Maluf N.K., Gauss G.H., Myong S., Ha T., Waksman G., Lohman T.M.. Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J. Mol. Biol. 2011; 411:633–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Arslan S., Khafizov R., Thomas C.D., Chemla Y.R., Ha T.. Engineering of a superhelicase through conformational control. Science. 2015; 348:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nguyen B., Ordabayev Y., Sokoloski J.E., Weiland E., Lohman T.M.. Large domain movements upon UvrD dimerization and helicase activation. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12178–12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng W., Brendza K.M., Gauss G.H., Korolev S., Waksman G., Lohman T.M.. The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:16006–16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chao K.L., Lohman T.M.. DNA-induced dimerization of the Escherichia coli Rep helicase. J. Biol. Chem. 1991; 221:1165–1181. [DOI] [PubMed] [Google Scholar]
- 29. Wong I., Chao K.L., Bujalowski W., Lohman T.M.. DNA-induced dimerization of the Escherichia coli Rep helicase. Allosteric effects of single-stranded and duplex DNA. J. Biol. Chem. 1992; 267:7596–7610. [PubMed] [Google Scholar]
- 30. Wong I., Lohman T.M.. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase & DNA binding. Science. 1992; 256:350–355. [DOI] [PubMed] [Google Scholar]
- 31. Wong I., Moore K.J.M., Bjornson K.P., Hsieh J., Lohman T.M.. ATPase activity of Escherichia coli Rep helicase is dramatically dependent on DNA ligation and protein oligomeric states. Biochemistry. 1996; 35:5726–5734. [DOI] [PubMed] [Google Scholar]
- 32. Cheng W., Hsieh J., Brendza K.M., Lohman T.M.. E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 2001; 310:327–350. [DOI] [PubMed] [Google Scholar]
- 33. Yokota H., Chujo Y.A., Harada Y.. Single-molecule imaging of the oligomer formation of the nonhexameric escherichia coli UvrD helicase. Biophys. J. 2013; 104:924–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ha T., Rasnik I., Cheng W., Babcock H.P., Gauss G.H., Lohman T.M., Chu S.. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002; 419:638–641. [DOI] [PubMed] [Google Scholar]
- 35. Maluf N.K., Fischer C.J., Lohman T.M.. A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 2003; 325:913–935. [DOI] [PubMed] [Google Scholar]
- 36. Lee K.S., Balci H., Jia H., Lohman T.M., Ha T.. Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat. Commun. 2013; 4:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ordabayev Y.A., Nguyen B., Niedziela-Majka A., Lohman T.M.. Regulation of UvrD helicase activity by MutL. J. Mol. Biol. 2018; 430:4260–4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lohman T.M., Chaos K., Green J.M., Sage S., Runyon G.T.. Large-scale purification and characterization of the Escherichia coli rep gene product. J. Biol. Chem. 1989; 264:10139–10147. [PubMed] [Google Scholar]
- 39. Comstock M.J., Ha T., Chemla Y.R.. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011; 8:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Whitley K.D., Comstock M.J., Chemla Y.R.. High-resolution optical tweezers combined with single-molecule confocal microscopy. Methods in Enzymology. 2017; 582:Elsevier Inc; 137–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moffitt J.R., Chemla Y.R., Izhaky D., Bustamante C.. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9006–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landry M.P., McCall P.M., Qi Z., Chemla Y.R.. Characterization of photoactivated singlet oxygen damage in single-molecule optical trap experiments. Biophys. J. 2009; 97:2128–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swoboda M., Henig J., Cheng H.M., Brugger D., Haltrich D., Plumeré N., Schlierf M.. Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano. 2012; 6:6364–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Woodside M.T., Behnke-Parks W.M., Larizadeh K., Travers K., Herschlag D., Block S.M.. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:6190–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Betterton M.D., Jülicher F.. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E - Stat. NonlinearSoft Matter Phys. 2005; 71:1–11. [DOI] [PubMed] [Google Scholar]
- 46. Pincus D.L., Chakrabarti S., Thirumalai D.. Helicase processivity and not the unwinding velocity exhibits universal increase with force. Biophys. J. 2015; 109:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McGlynn P., Lloyd R.G.. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002; 3:859–870. [DOI] [PubMed] [Google Scholar]
- 48. Courcelle C.T., Landstrom A.J., Anderson B., Courcelle J.. Cellular characterization of the primosome and Rep helicase in processing and restoration of replication following arrest by UV-induced DNA damage in Escherichia coli. J. Bacteriol. 2012; 194:3977–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scott J.F., Eisenberg S., Bertsch L.L., Kornberg A.. A mechanism of duplex DNA replication revealed by enzymatic studies of phage phi X174: catalytic strand separation in advance of replication. Proc. Natl. Acad. Sci. U.S.A. 1977; 74:193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.