Abstract
RNA-binding proteins (RBPs) play pivotal roles in directing RNA fate and function. Yet the current annotation of RBPs is largely limited to proteins carrying known RNA-binding domains. To systematically reveal dynamic RNA–protein interactions, we surveyed the human proteome by a protein array-based approach and identified 671 proteins with RNA-binding activity. Among these proteins, 525 lack annotated RNA-binding domains and are enriched in transcriptional and epigenetic regulators, metabolic enzymes, and small GTPases. Using an improved CLIP (crosslinking and immunoprecipitation) method, we performed genome-wide target profiling of isocitrate dehydrogenase 1 (IDH1), a novel RBP. IDH1 binds to thousands of RNA transcripts with enriched functions in transcription and chromatin regulation, cell cycle and RNA processing. Purified IDH1, but not an oncogenic mutant, binds directly to GA- or AU-rich RNA that are also enriched in IDH1 CLIP targets. Our study provides useful resources of unconventional RBPs and IDH1-bound transcriptome, and convincingly illustrates, for the first time, the in vivo and in vitro RNA targets and binding preferences of IDH1, revealing an unanticipated complexity of RNA regulation in diverse cellular processes.
INTRODUCTION
Pervasive transcription of mammalian genomes gives rise to thousands of long noncoding RNA (lncRNA) transcripts (1–4). LncRNAs are highly versatile molecules that carry out many regulatory functions at multiple levels in diverse cellular processes, and their dysregulation often contributes to human diseases (5–8). Considering that lncRNAs must enlist proteins to execute their regulatory roles, the revelation and characterization of lncRNA–protein interactions is a prerequisite for the mechanistic dissection of the regulatory processes governed by lncRNAs.
RNA-binding proteins (RBPs) are well-known for their roles in regulating RNA fate from synthesis to decay and participating in protein translation by assisting and/or directing RNAs (9). Tuschl's group previously assembled a repertoire of RBPs, which included all the proteins carrying annotated RNA-binding domains (RBDs) and those reside in well-characterized ribonucleoprotein (RNP) complexes (9). This RBP repertoire contains 1542 RBPs, comprising 7.5% of the ∼20 500 human protein-coding genes. These RBPs tend to be ubiquitously expressed, typically at higher levels than average cellular proteins and transcription factors (9). Interestingly, proteome-wide surveys of mRNA and newly transcribed RNA–protein interactomes using mass spectrometry-based approaches in human and mouse cells have revealed many RNA-binding proteins that were not included in Tuschl's RBP repertoire (10–19), suggesting that novel RBPs await recognition and further characterization. On the other hand, methods using immunoprecipitation against RBPs in the presence of RNase followed by deep sequencing [UV crosslinking and immunoprecipitation followed by sequencing (CLIP-seq) and derivative methods] have revealed that a single RBP can often bind to thousands of different RNA species at defined binding sites in cells (20,21). Thus, the landscape of RNA–protein interactions appears to be more diverse and vast than previously appreciated.
Methods using a reverse RNA immunoprecipitation methodology followed by mass spectrometry have been developed. For example, ChIRP-MS (chromatin isolation by RNA purification followed by mass spectrometry) and its derivatives, such as CHART and RAP, utilize complementary oligonucleotides as baits to capture the target RNA–protein and RNA-DNA complexes in cells (22–25). Other conventional methods utilizing RNA aptamer tagging and in vitro transcribed RNA are frequently used to capture interacting proteins in cells or cell lysates (25,26). When applied to lncRNAs, however, these methods often require large amounts of starting materials to ensure a sufficient detection level of mass spectrometry due to the paucity of target lncRNAs expressed in cells and limited pull-down efficiency. In addition, these approaches are heavily biased towards highly abundant RBPs, which harbor strong RNA binding activity toward hundreds or thousands of transcripts. Thus, high backgrounds caused by non-specific RBPs may obscure transient or weak RNA–protein interactions.
Protein microarrays have been used to detect a wide range of protein-ligand interactions and to identify substrates of a wide variety of enzymes involved in protein posttranslational modifications (25,27–30). Previously, Zhu et al. systematically profiled protein-DNA interactions using a protein microarray-based approach and unexpectedly identified > 300 unconventional DNA-binding proteins (uDBPs). In depth characterization of one such uDBP, a mitogen-activated kinase ERK2, revealed that ERK2 acts as a transcriptional repressor that regulates interferon gamma signaling pathway (31). This study suggested that there exist many moonlighting functions of well-annotated proteins yet to be discovered. Protein microarrays have been shown as a useful tool to identify RNA–protein interactions (32). For example, a previous study probed a small set of coding and noncoding RNAs to protein microarrays (i.e. ProtoArrays), comprised of ∼9,125 human proteins, and identified 137 RNA-binding proteins, most of which fall into the classical RBPs with common RBDs (33). Two studies using yeast protein microarrays revealed that several metabolic enzymes and vesicle trafficking proteins bind to mRNAs and total RNA from Saccharomyces cerevisiae as moonlighting functions (34,35). However, the confirmation and to what extend that such unconventional RNA-binding proteins exist in mammals are still not fully explored.
It has been proposed that cellular (intermediary) metabolism and the regulation of gene expression could be more closely networked than has been appreciated (36). Recent proteome-wide surveys of RNA interactomes also uncovered some metabolic enzymes, including isocitrate dehydrogenase 1 (IDH1), to interact with mRNA and nascent RNA transcripts in cells (10–13), although lack of further confirmation. IDH1 catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG). Interestingly, IDH1 is often mutated at codon Arginine 132 to Histidine (R132H) in multiple human cancers, including glioma, sarcoma, and acute myeloid leukemia (AML) (37–41). R132H of IDH1 is a gain-of-function mutation and confers an enzymatic activity that converts α-KG to the oncometabolite 2-hydroxyglutarate (2HG), which is a competitive inhibitor of α-KG-dependent DNA hydroxylases and histone demethylases, leading to globally elevated methylation and aberrant gene expression in tumor cells (41,42). RNA N6-methyladenosine (m6A) is the most prevalent RNA modification in higher eukaryotes and has been linked to the post-transcriptional regulation of gene expression (43–45). It was recently reported that IDH1/2 mutations promote m6A level of total RNA in AML (46). The idea that some metabolic enzymes, such as IDH1, moonlight as RNA-binding proteins has been postulated, but not tested. The relationship between IDH1 and RNA remains to be fully defined.
To identify lncRNA–protein interactomes that are highly dynamic and exist in low abundance, we profiled binding activities of 13 lncRNA transcripts on human proteome arrays (HuProt). Using a highly stringent cutoff value, we identified 671 RBPs, 525 of which lack any annotated classical RBDs. Interestingly, these unconventional RBPs are comprised of a large number of transcriptional and epigenetic modulators, metabolic enzymes, and small GTPases. To further characterize novel RNA-binding activities, we determined the RNA targets and binding preferences of IDH1 as a representative of unconventional RBPs. We demonstrated that IDH1 binds directly to RNA in vitro and in vivo. In ESCs, IDH1 binds to thousands of mature RNA transcripts, the protein products of which are enriched in functions related to transcription and chromatin regulation, cell cycle and RNA processing. Quantitative binding studies of purified IDH1 with RNAs of defined sequences show that IDH1 preferentially binds to GA- or AU-rich, but not GC-rich, sequences in single-stranded RNA, and showed little binding to double-stranded RNA, single/double-stranded DNA, or RNA/DNA hybrid. Intriguingly, the oncogenic IDH1 (R132H) mutant protein shows a decrease or loss of the RNA-binding activity in vivo and in vitro. Together, our work expands the current catalogue of protein families with novel RNA-binding activity, and also suggests an involvement of RNA in regulating diverse cellular processes to a much greater degree than was previously anticipated.
MATERIALS AND METHODS
Human protein microarray construction
Human ORF cloning using the Gateway recombinant cloning system (Invitrogen, CA, USA) and Human protein purification using a high-throughput protein purification protocol as described previously (30,31).
Cell culture
ESCs expressing 3 × FLAG and biotin tagged proteins were maintained in complete ESC culture medium: DMEM (Dulbecco's modified Eagle's medium) supplemented with 15% heat-inactivated FCS (fetal calf serum), 2 mM GlutaMax (100 × Life Technology), 1% Nucleoside mix (100 × stock, Millipore), penicillin–streptomycin solution (100 × stock, Life Technologies), 0.1 mM non-essential amino acids (NEAA), 0.1 mM β-mercaptoethanol and supplied with 1000 U/ml recombinant Leukemia Inhibitory Factor (LIF, Millipore). ESCs were cultured on plates which were pre-coated with 0.1% gelatin.
Protein expression and purification
Purification from yeast: plasmids expressing GST-tagged proteins were transformed into yeast (Y258 strain). The yeast strains were cultured in 5 ml SC-ura/glucose liquid medium at 30°C with shaking for >24 h to saturation. About 50 μl–1 ml of the saturated culture was transferred to 50 ml SC-ura/raffinose liquid medium and was incubated at 30°C with shaking overnight. When the OD600 reached 0.7–0.9, protein expression was induced by 2% galactose at 30°C with shaking for 4 h. Washed cell pellets were resuspended by cold Lysis Buffer [50 mM Tris–HCl pH 7.5, 100 mM NaCl, 1 mM EGTA, 10% glycerol, 0.1% Triton X-100, 0.1% β-mercaptoethanol, 1 mM PMSF and Protease inhibitor cocktail (Sigma)] and mixed with 500 μl of zirconia beads (0.5 mm diameter). Cells were lysed by vortex for 1 min for 12 times with 1 min intervals on ice. Meanwhile, glutathione beads (GE Healthcare, USA) were washed three times with cold lysis buffer without protease inhibitors. Then, the beads were mixed with cell lysate for incubation at 4°C for 1 h and washed with wash buffer I (50 mM Tris–HCl pH 7.5, 500 mM NaCl, 1 mM EGTA, 10% glycerol, 0.1% Triton X-100, 0.1% β-mercaptoethanol, 1 mM PMSF) for three times and Wash Buffer II (50 mM HEPES pH 7.5, 100 mM NaCl, 1 mM EGTA, 10% glycerol, 0.1% β-mercaptoethanol, 1 mM PMSF) for another three times. Proteins were eluted by elution buffer (50 mM HEPES pH 8.0, 100 mM NaCl, 30% glycerol, 40 mM reduced glutathione, 0.03% Triton X-100).
Purification from bacteria: plasmids expressing proteins of interest fused with N-terminal GST tag or 6× His tag were transformed into Escherichia coli BL21 (DE3) strain. Bacteria were cultured in 100 ml lysogeny broth (LB) media at 37°C with shaking for 5 h and 10 ml culture was inoculated into 2 l LB media for amplification. When OD600 reached 0.6, protein expression was induced by 0.5 mM isopropy-β-d-thiogalactoside (IPTG). After cultured at 16°C with shaking for 16–18 h, bacteria were harvested by centrifugation at 4°C, 4000 rpm for 15 min and pellets were resuspended by Cell Lysis Buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, supplied with 1 mM PMSF, 40 μg/ml lysozyme, 1 ng/ml DNase I, 1 mM MgCl2). Cells in lysis buffer were sonicated on ice using 50% amplification with 3 s on 7 s off for 30 min. Cell debris were removed by centrifugation at 4°C, 18 000 rpm for 1 h. For GST-tagged proteins, the supernatant was loaded onto glutathione column (Bio-Rad) and then washed by cell lysis buffer. The target protein was finally eluted by elution buffer (20 mM Tris–HCl pH 8.0, 20 mM reduced glutathione). Eluted proteins were added with 2 mM DTT and 2 mM EDTA and free glutathione was removed by ultracentrifugation using Amicon Ultra centrifugal filter (3 kD MWCO, Milipore). For His-tagged proteins, the supernatant was loaded onto Ni-NTA column (Bio-Rad) and then successively washed by cell lysis buffer, W2 Buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 20 mM imidazole) and W3 Buffer (20 mM Tris–HCl pH 8.0). The target protein was finally eluted by elution buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 300 mM imidazole).
Proteins were further purified by anion-exchange chromatography using Source Q column (GE® Healthcare) with Buffer A (20 mM Tris–HCl pH 8.0, 2 mM DTT) and Buffer B (20 mM Tris–HCl pH 8.0, 1 M NaCl, 2 mM DTT). Target proteins were concentrated and finally purified by size-exclusion chromatography using Superdex 200 10/300 GL column (GE® Healthcare) in SEC Buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM DTT). Purity of proteins were checked by SDS-PAGE and concentrations were measured by Nanodrop™2000 spectrophotometry (Thermo Scientific) according to the A280 absorbance and divided by extinction coefficient acquired from Expasy ProProm website (http://web.expasy.org/protparam/). Aliquots of proteins were stored in –80°C.
IDH1 protein consists of 414 residues and the theoretical molecular weight is 49.9 kD. A single and symmetric peak with elution volume around 13 ml in Superdex 200 10/30 GL column indicated that the recombinant IDH1 forms a stable homodimer, free of nucleic acid contamination. And purified IDH1 is active as an isocitrate dehydrogenase revealed by kinetics assay.
HuProt arrays and data analysis
Purified human proteins were arrayed in a 384-well format and printed on FAST slides (Whatman, Germany) in duplicate. The protein chips were blocked with 3% BSA in hybridization buffer (25 mM HEPES pH 8.0, 150 mM NaCl, 8 mM magnesium acetate tetrahydrate, 3 mM DTT, 0.1% Triton X-100, 10% glycerol, 100 μg/ml yeast tRNA, 20 μg/ml heparin) at 4°C for 1.5 h. Then, the blocking buffer was drained and protein chips were immediately incubated with a Cy5-labeled lncRNA at final amount of 20 pmol in hybridization buffer with RNase inhibitor at 4°C for 1.5 h. The chips were washed three times by TBS-T buffer for 10 min each and followed by twice washes with ddH2O for 10 min every time. The slides were finally scanned at 635 nm with a GenePix 4000 scanner (MDS Analytical Technologies, CA, USA) and the binding signals were acquired using the GenePix software. Data quantification process using a protocol as previously described (31,47). Z-scores of representative RNA–protein interactions were visualized with a heatmap by TreeView. Gene Ontology analysis and domain analysis of RNA binding proteins was performed with DAVID (https://david.ncifcrf.gov/).
In vitro pull-down assay by RNAs
Biotin labeled RNA was generated by Biotin-16-rUTP (Roche, 11388908910) incorporation during in vitro transcription performed according to the manufacturer's protocol (Ambion, AM1334). Briefly, 1 μg linearized DNA template containing T7 promoter was mixed with NTPs (Biotin-16-rUTP/rUTP: 1:30) and enzyme mix in 1× transcription buffer. The reaction was incubated at 37°C overnight then DNA was digested by adding TURBO DNase for another 15 min at 37°C. Then, biotin-labeled RNA was extracted by TRIzol™ Reagent (Invitrogen) followed by ethanol precipitation. Concentration and quality of RNA were characterized by Nanodrop™2000 spectrophotometry and denatured agarose gel electrophoreses, respectively.
For pull-down assay, 2 μg biotin-labeled RNA was denatured at 65°C for 5 min, then cooled down to room temperature in structure buffer (10 mM Tris–HCl pH 7.0, 100 mM KCl, 10 mM MgCl2) for 25 min. Re-folded RNA was incubated with pre-blocked Streptavidin M280 beads (Invitrogen) in IPB (40 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 mM MgCl2, 20 μg/ml Heperin, 1 mM DTT, 0.01% NP40, 5% glycerol) buffer at 4°C for 2 h and 1 μg recombinant GST tagged protein was added into the mixture for another 6 h incubation at 4°C with rotation. The mixture was subjected to five wash cycles of 5 minutes each using 500 μl IPB buffer. After the last wash, the beads were resuspended in 25 μl Elution Buffer (150 mM NaCl, 10 mM Tris–HCl pH 8.0, 1 mM EDTA, 1% SDS) shaking for 1 h at 16°C, 12 000 rpm with 10 s on, 10 s off. Eluate was collected and mixed with 5 μl 6× SDS protein loading buffer, and the RNA-bound proteins were detected by western blot analysis using antibody against GST.
Tandem RNA immunoprecipitation followed by quantitative real-time PCR (FBioRIP-qPCR)
About 2 × 107 cells stably expressing FBioIDH1 or FBioEGFP were crosslinked at 254 nm UV light with 600 mJ/cm2 and lysed in ice-colded lysis buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 5% glycerol, supplemented with 1 mM DTT, 1 mM PMSF, 1:500 PI cocktail and 400 U/ml RNase Inhibitor). The cell lysates were treated with DNase I and 5% of the cell lysate was saved as input for RT-qPCR. Then, pre-equilibrated FLAG M2 resins (Sigma) in 2 × dilution buffer (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100) were added into the cell lysate and incubated at 4°C overnight. After three washes using IP200 Buffer (20 mM Tris–HCl pH7.4, 200 mM NaCl, 1 mM EDTA, 0.3% Triton X-100, 5% glycerol), the RNA/protein complex was eluted by 3× FLAG Elution Solution (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 150 ng/μl 3× FLAG peptides). The eluate was added into pre-equilibrated streptavidin (M-280, Invitrogen) dynabeads in NP40 lysis buffer (1× PBS, 0.5% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) and incubated at 4°C for 4 h or overnight. After twice washes with NP-40 lysis buffer and twice washes with the high salt buffer (5× PBS, 0.5% NP-40, 0.5% sodium deoxycholate, 0.5% SDS), the RNA/protein complex was eluted by Proteinase K digestion buffer (50 mM Tris–HCl pH7.4, 10 mM EDTA, 50 mM NaCl, 0.5% SDS, proteinase K was added freshly). The RNA was extracted by TRIzol™ Reagent (Invitrogen) according to the manufacturer's protocol. Reverse transcription was performed using Revert Aid First Strand cDNA Synthesis Kit (Fermentas, K1622) with random primers. Quantitative real-time PCR (qRT-PCR) was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, 1725121) on a Bio-Rad CFX384 RealTime System. Primers for RIP-qPCR are listed in Supplementary Table S6.
FBioCLIP-seq
About 3 × 107 cells expressing FBioIDH1 were crosslinked at 254 nm UV light with 600 mJ/cm2 and harvested in pre-chilled PBS. The cell pellets were resuspended in ice-colded lysis buffer used in RIP. Then treated the cell lysate with DNase I. The pre-equilibrated FLAG M2 resins (Sigma) were incubated with the lysate at 4°C overnight ad washed four times with RIP200 Buffer. The RNA/Protein complexes were eluted with 3 × FLAG elution buffer containing ∼200 ng/μl 3× FLAG peptides. Then, the eluate was incubated with pre-equilibrated streptavidin (M-280, Invitrogen) beads at 4°C overnight. After twice washes with NP-40 Lysis Buffer and twice with high salt buffer, the protein/RNA complex bound beads were washed four times with PNK buffer (50 mM Tris–HCl pH 7.4, 10 mM MgCl2, 0.5% NP-40) quickly. Then the RNAs were partially digested with Micrococcal nuclease (MNase) (NEB) at 37°C for 10 min (mixed with an Eppendorf Thermomixer at 1200 rpm for 5 s per 30 s). Here, we replaced the RNase A/T1 used in the original protocol with MNase (48,49), a relatively non-specific endo-exonuclease that can be inactivated by EGTA, in order to prevent continuous RNA trimming for better RNA recovery and to reduce cleavage biases.
The reaction was stopped by twice washes with ice-cold PNK-EGTA Buffer (PNK buffer with 2 mM EGTA). Then the beads were washed twice with ice-cold PNK buffer. To dephosphorylate the RNA, the fragmented RNA–protein complex bound beads were treated with CIP (NEB) for 10 min at 37°C (mixed with an Eppendorf Thermomixer at 1200 rpm for 5 s per 30 s). After two more washes with PNK Buffer, the pre-adenylated 3′ linker was ligated to the end of fragmented RNAs with T4 RNA ligase (NEB) at 16°C overnight (mixed with an Eppendorf Thermomixer at 1200 rpm for 5 s per 30 s). Then, after four times washes with PNK buffer, the bead-bound RNA fragments were phosphorylated by T4 PNK (NEB) for 10 min at 37°C and the reaction was stopped by washing three times with PNK-EGTA buffer. To elute the protein-bound RNA fragments, the samples were treated with Proteinase K (50 mM Tris–HCl pH 7.4, 150 mM NaCl, 0.5% SDS, and 20 μg Proteinase K) for 1 h at 55°C. The RNAs were purified by TRIzol™ Reagent and ligated with 5′ RNA adaptor. Then, the purified RNA fragments were reverse transcribed by Superscript III (Invitrogen) with RT primer and amplified for 20 cycles with 2× NEB HF PCR mix (NEB). Library index sequence was introduced by PCR with index primers for eight more cycles. Library was performed high-throughput sequencing by Illumina Hiseq 2500 sequencing platform. Analysis of FBioCLIP-seq was performed as described (50). Files in format of bedgraph were made for visualization in UCSC genome browser and IGV software.
Pre-adenylated 3′ linker: rAppAGATCGGAAGAGCACACGTCT-NH2, TAKARA.
5′ RNA adaptor: GUUCAGAGUUCUACAGUCCGACGAUC, TAKARA.
RT primer: AGACGTGTGCTCTTCCGATCT, TAKARA.
Amplification PCR primers:
Forward: GTTCAGAGTTCTACAGTCCGACGATC;
Reverse: AGACGTGTGCTCTTCCGATCT, TAKARA.
Index primers:
Forward (SR primer): AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGAC,
Reverse: CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT (the underlined hexamer indicates Illumina index sequence, TAKARA).
Immunofluorescence staining
ESCs were seeded on 18 × 18 mm glass coverslips. Pre-cold methanol was used to fix cells. Samples were blocked using 10% FBS with 0.5% saponin in PBS for 30 min at room temperature, and then incubated with primary antibodies for 1 h at 37°C followed by the secondary fluorescently labeled antibodies for 1 h at 37°C. Nuclei were counterstained with DAPI. Images were acquired using FV1200 confocal microscopy (Olympus IX83). Images were processed using software of ImageJ.
The following antibodies were used: IDH1 (Proteintech 12332-1-AP IF: 1:100, Cell Signaling Technology IDH1 (D2H1) Rabbit mAb #8137 WB: 1: 1000).
Electrophoretic mobility shift assay (EMSA)
RNA and DNA probes used in EMSA were chemical synthetized and purified by HPCL. Fluorescent probes were additionally labeled with Cy5 at their 5′ ends (Idobio Biological Technology). The sequences of probes are shown in Figure 6E and Supplementary Figures S7H and S8C.
Figure 6.
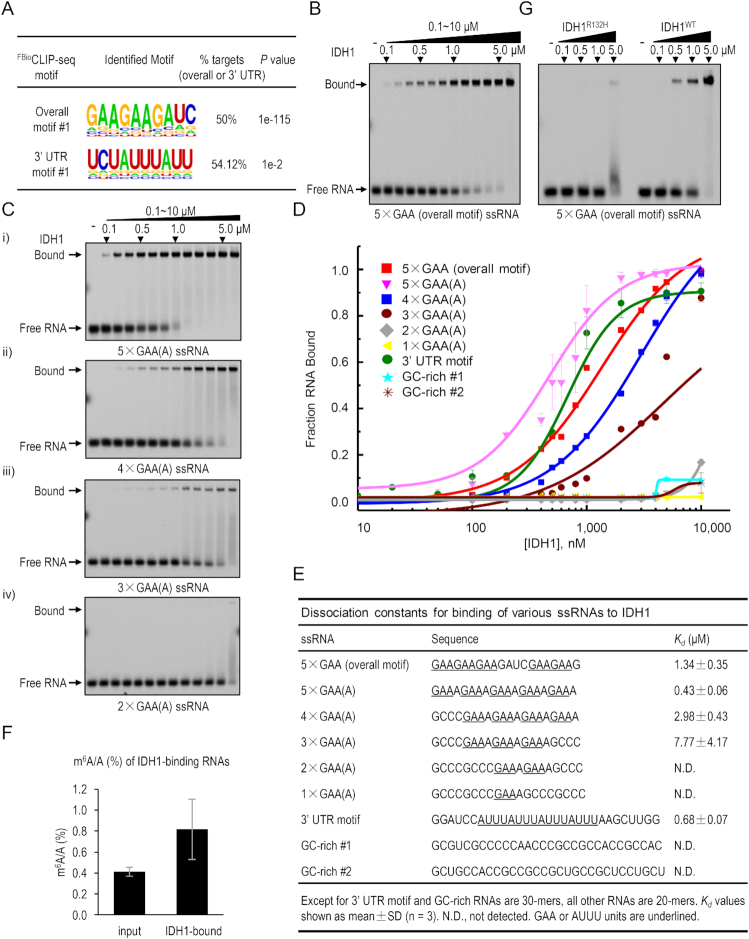
IDH1 binds to GA/AU-rich single-stranded RNA. (A) GA- and AU-rich RNA motifs are enriched in IDH1’s RNA targets in all sequences or in the 3′ UTR regions, respectively. (B–E) EMSA analysis of direct binding of purified wild-type to various fluorescence-labeled RNA/DNA probes as indicated. (B) EMSA with the 5 × GAA ssRNA probe containing the overall motif #1. (C) EMSA with ssRNA probes with 2–5 copies of GAA(A) nucleotides. (D) Binding isotherms for IDH1-RNA interactions. Each data point is shown as mean ± SD (n = 3 biological replicates). Error bars represent standard deviations (SD), some of which are too small to be visualized. (E) Summary of RNA-binding activities of IDH1 toward various ssRNA probes used above. Dissociation constants (Kd) are shown. N.D., not detected. GAA and AUUU units are underlined. (F) Relative m6A level of IDH1 RNA targets in ESCs was determined by HPLC-MS/MS following FBioRIP assays. Data are shown as mean ± SD, n = 3. (G) Comparison of RNA-binding activities of wild-type (WT) and oncogenic R132H mutant of IDH1 to the 5× GAA ssRNA probe as shown in the panel (B).
In order to generate double-stranded DNA or RNA and DNA/RNA hybrid probes, sense and antisense probes were mixed together with equal molar ratio in 1× SSC buffer (150 mM NaCl, 15 mM sodium citrate). And then annealed in PCR thermal cycler using the program: 90°C 2 min, 60°C 5 min, 20°C 30 min and then kept on ice until used.
RNAs were refolded by incubating at 95°C for 5 min, followed by snap cooling on ice for 5 min, then 1× refolding buffer (10 mM Tris–HCl pH 6.7, 50 mM KCl, 10 mM MgCl2) was added and let RNA gradually recover to room temperature for 30 min.
Purified recombinant IDH1 were thawed at room temperature and kept on ice. Proteins were serial diluted into 1× binding buffer (10 mM HEPES pH 7.4, 50 mM KCl, 1 mM EDTA, 0.05% Triton X-100, 5% glycerol, 0.01 mg/ml BSA, 1 mM DTT, 40 U/mL RNase inhibitor) and kept on ice until used. 20 nM RNA was mixed with increasing concentrations (0.1–10 μM) of proteins in 1× binding buffer. Samples were incubated at room temperature for 1 h and then resolved on a native polyacrylamide gel (6% acrylamide 37.5:1, 1× TBE, 0.08% APS, 0.1% TEMED) in 0.5× TBE running buffer and running for 2.5 h at 80 V at 4°C. Cy5 signals on the gel were scanned by Typhoon FLA 9500 fluorescence scanner (GE® Healthcare) and quantified by ImageJ software. Dissociation constants (Kd, μM) were determined by non-linear fitting of the binding curves using OriginPro 9 software. All the experiments are performed with three independent replicates.
Structural analysis
Protein structures of IDH1 (WT: 5YZH, 5YFM; R132H: 3MAR) (51,52) analyzed in this study were obtained from Research Collaboratory for Structural Bioinformatics (RCSB: www.rcsb.org) of World Wide Protein Databank (wwPDB: www.wwpdb.org) (53,54). The presented structures were analyzed and viewed in PyMOL software.
FBioRIP-HPLC–MS/MS
The FBioRIP procedure was described above. In brief, ESCs stably expressing FLAG-Biotin tagged IDH1 were subjected to FBioRIP procedure. Input and IDH1-bound RNA were extracted with TRIzol™ Reagent. Then, mRNA was purified using Dynabeads mRNA Purification Kit (Thermo Fisher # 61006). About 60 ng purified RNA was subjected to HPLC-MS/MS quantification of m6A level as previously reported (55).
Data availability
The accession numbers for LIN28A CLIP-seq data are GEO: GSM910955, GSM910956, GSM910957 (56). The accession number for mESCs RNA-seq is GSM1412826 (57). IDH1 FBioCLIP-seq data has been deposited in the GEO repository with the accession number GSE119798.
RESULTS
High-throughput profiling of lncRNA–protein interactions using HuProt arrays
To enable an unbiased, proteome-wide survey for dynamic RNA–protein interactions that are likely transient and/or less abundant in cells, we chose to profile the interactomes of 13 representative lncRNAs that show detectable expression in undifferentiated or differentiating embryonic stem cells (ESCs) (Supplementary Figure S1A and C). Six of them (i.e., Evx1as, Haunt, Lockd, Eprn, Apela and Platr14) were previously reported to play regulatory roles in modulating gene expression, lineage differentiation, and DNA damage response in ESCs or erythroid cells (57–62). For example, Haunt and its genomic locus play discrete and opposing roles in regulating the HOXA gene cluster located ∼40 kb downstream of Haunt during ESC differentiation (57). Evx1as promotes transcription of its adjacent gene EVX1 in cis to fine-tune mesendodermal lineage differentiation (58).
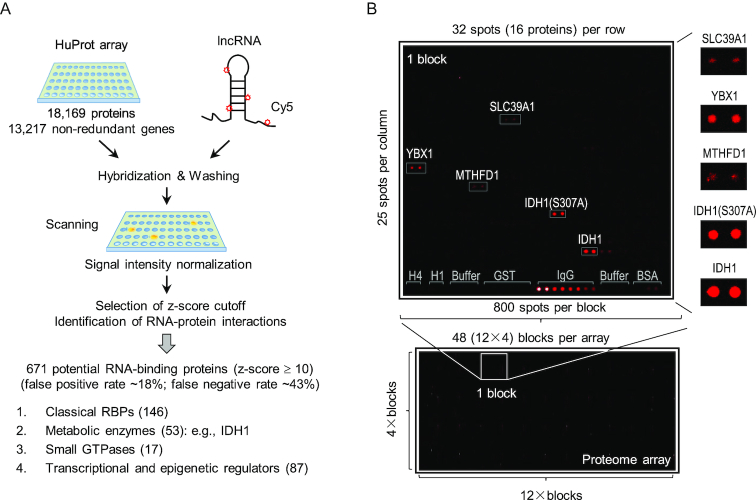
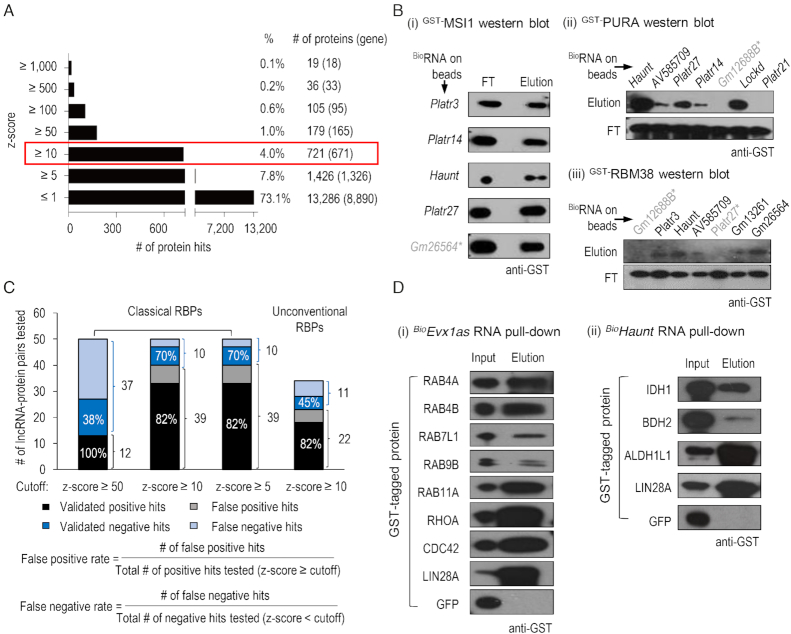
Using a previously established protocol (47), we hybridized 46 HuProt arrays with 19 Cy5-labeled RNA probes, comprised of 13 full-length lncRNAs and 6 truncated sequences of lncRNA candidates, in duplicate or triplicate (Figure 1A). Each HuProt array contained 18 169 full-length human proteins, representing 13 217 non-redundant gene products in duplicate. After stringent washes, the fluorescence signals of each protein spot on the HuProt arrays were acquired and normalized across all the binding assays to calculate the standard deviation values, based on which the signal intensity was represented as z-scores (Figure 1A and B). Overall, >73% of the proteins on HuProt arrays did not show any detectable fluorescent signals (z-scores ≤ 1) to any of the RNA probes tested, indicating that the background fluorescence was minimal on HuProt arrays (Figure 2A; Supplementary Table S1). Pearson correlation coefficients of replicates ranged mainly from 0.80 to 0.99, indicating a high reproducibility of the assays (Supplementary Figure S1B). After combining all of the binding data obtained with the 13 lncRNAs on 46 HuProt arrays, we identified 1326 (7.8%), 671 (4.0%) and 165 (1.0%) non-redundant proteins that bound to at least one RNA probe at a z-score value of ≥5, 10 and 50, respectively (Figure 2A).
Figure 1.
High-throughput profiling of lncRNA–protein interactions using HuProt arrays. (A) Scheme of HuProt array-based assay to reveal lncRNA-binding proteins in an unbiased manner. And the results of HuProt array screens are summarized and shown below. (B) A typical example of the results from HuProt arrays. Each microarray was divided into 48 blocks (bottom). Each block contains 800 protein spots. Controls, including negative controls (histone H1 and H4, buffer, GST and BSA) and the positive control IgG, are printed in the last row of each block as indicated, and tested proteins are printed in duplicate. In this example, the Cy5-labeled RNA probe selectively interacts with proteins such as YBX1, MTHFD1, SLC39A1, IDH1 and mutant IDH1 (S307A) (top).
Figure 2.
Validation of lncRNA–protein interactions in vitro. (A) Histogram showing the number of protein hits in different ranges of z-scores. The number of non-redundant genes is shown in brackets. (B) Typical results of in vitro pull-down assays to confirm lncRNA–protein interactions using z-score ≥ 10 as the cutoff. Various biotin-labeled lncRNA transcripts were immobilized on streptavidin beads, and incubated with GST-tagged recombinant MSI1 (i), PURA (ii) or RBM38 (IOH13305) (iii) proteins. After stringent washes, flow-through (FT) and eluted fractions were collected and subjected to anti-GST western blotting analysis. LncRNAs labeled in black: results of in vitro pull-down assays are consistent with that in HuProt arrays; otherwise, the lncRNAs are marked with ‘ * ’ in gray. (C) Summary of the success rates of lncRNA–protein interaction validations by in vitro pull-down assays when different cutoffs were used. The lncRNA–protein interactions chosen for validation have a wide range of z-scores from –1 to 2,161. When the z-score is larger than the chosen cutoff (50, 10 or 5), the interaction is regarded as a positive hit. On the contrary, it is regarded as a negative hit. The positive hits and negative hits that were confirmed by in vitro pull-down assays are labeled as ‘validated positive hits’ or ‘validated negative hits’, respectively. (D) Biotin-labeled Evx1as and Haunt lncRNAs capture a number of small GTPases and metabolic enzymes, respectively. Biotinylated RNA immobilized on streptavidin beads were incubated with various GST-tagged recombinant proteins. The input and eluted fractions were subjected to anti-GST western blotting. LIN28A and GFP are the positive or negative control, respectively.
Validation of lncRNA–protein interactions in vitro
To determine a reasonable cutoff value for identifying reliable interactions, we first chose 10 known RBPs such as LIN28A, MSI1, PURA and RBM38, which involved 49 pairs of lncRNA–protein interactions with a wide range of z-scores from –1 to 2161, for validation using an in vitro RNA pull-down assay (Supplementary Figure S2A; Supplementary Table S2). We expressed and purified these recombinant proteins in yeast or bacteria, transcribed the 13 lncRNAs in vitro, and labeled them with biotin (Supplementary Figure S2B and C). Gel electrophoresis and dot blot analysis indicated the successful production of biotinylated lncRNAs (Supplementary Figure S2B). GFP mRNA was also produced as a negative control. The biotinylated 13 lncRNAs and GFP mRNA were separately immobilized on streptavidin-coated beads and incubated with the corresponding protein partners. We optimized and utilized stringent incubation and wash conditions in presence of yeast transfer RNA (tRNA) as a non-specific competitor (Supplementary Figure S3A–C). Proteins captured by the immobilized lncRNA probes were then released and subjected to western blot analysis (Supplementary Figure S2A).
At a z-score ≥10, 32 of the 39 pairs (82%) tested were confirmed to show direct RNA–protein interactions, suggesting a false-positive rate of 18% (Figure 2C; Supplementary Figure S3D and E; Supplementary Table S2). No binding was detected for 7 of the tested 10 pairs with z-scores ranging from –1 to 9, suggesting a false-negative rate of 30% (Figure 2C; Supplementary Figure S3D and E; Supplementary Table S2). For example, four of five lncRNA–MSI1 interactions, six of seven lncRNA–PURA interactions and five of seven of lncRNA–RBM38 interactions were validated successfully by in vitro pull down (Figure 2B). At a cutoff of z-score ≥5, we observed similar false-negative rate (30%) and false-positive rate (18%) as the cutoff of z-score ≥10 (Figure 2C). With a cutoff of z-score ≥50, the false-negative rate increased to 62%, despite that all of the positive hits were validated (Figure 2C). To balance between the false-positive and false-negative rates, we decided to use z-score ≥10 as a reasonable cutoff, with which 671 were identified as candidate proteins with lncRNA-binding activity (Supplementary Table S3).
Next, we chose additional 16 proteins that do not encode any annotated RBDs for in vitro validation (Figure 2D; Supplementary Figure S3F and G; Supplementary Table S2). These proteins show a wide variety of RNA-unrelated biological functions, including seven small GTPases (RAB4A/4B/7L1/9B/11A, RHOA and CDC42), three metabolic enzymes (IDH1, BDH2 and ALDH1L1), four chromatin and transcription regulators (EED, TRIM24, NACA and PIR), and a protein kinase PAK2. RNA pull-down analysis validated 82% of the 22 RNA–protein interactions with z-scores ≥10 on HuProt arrays (Figure 2C). Thus, for unconventional RBPs, we estimated a false-positive rate of 18%, similar to that of classical RBPs (Figure 2C). Taken together, HuProt array-based approach recovers many expected binding activities but also reveals many unconventional RNA-binding proteins with a low false-discovery rate as estimated by in vitro validation.
FBioCLIP-seq validation of lncRNA–LIN28A interactions in vivo
We then sought to determine whether these in vitro interactions could be reproduced using in vivo assays. LIN28A (Lin-28 homolog A), a classical RBP, is known to bind to thousands of mRNAs to regulate microRNA biogenesis, splicing, translation, cell cycle and glucose metabolism (56,63–75). LIN28A has been implicated in cancer cell proliferation, stem cell pluripotency and reprogramming via its RNA-binding activity (66–71). On HuProt arrays, LIN28A bound to all 13 lncRNAs with high z-scores, ranging from 21 to 918 (Supplementary Table S3). Consistently, 6 of the 13 lncRNAs had LIN28A-binding signals in CLIP-seq data in ESCs that were reported previously by Kim et al. (Figure 3C; Supplementary Figure S4).
Figure 3.
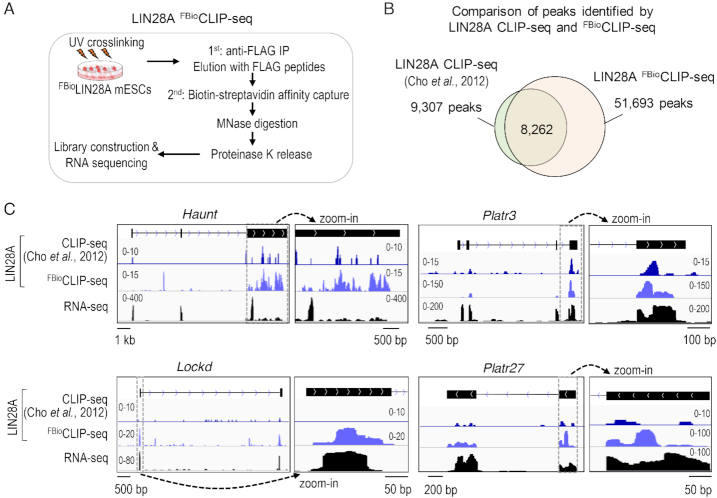
Validation of lncRNA–protein interactions in vivo. (A) Overview of the LIN28A FBioCLIP-seq method. (B) Venn diagram comparison of peaks identified by LIN28A FBioCLIP-seq and CLIP-seq (56). (C) Genome browser views of LIN28A FBioCLIP and CLIP signals at representative lncRNA loci.
To enable detection of interactions between proteins and lncRNAs of low abundance in cells, we employed a more sensitive and efficient CLIP method, dubbed as FBioCLIP-Seq (Crosslinking and Immunoprecipitation via a FLAG- and Biotin-double tags followed by sequencing). This method uses a stringent two-step, FLAG- and biotin-tags-mediated purification of RNA–protein complexes in the presence of Micrococcal Nuclease (MNase) (Figure 3A). We established an ESC line that stably expressed a FLAG- and biotin-tagged LIN28A protein. FBioCLIP-seq of LIN28A revealed ∼51 693 binding peaks corresponding to ∼8556 genes (Figure 3B), covering 89% (8262) of LIN28A CLIP-seq peaks (9307) as reported by Kim and colleagues (56). Notably, our method revealed significantly more CLIP peaks and target genes, demonstrating that LIN28A FBioCLIP-seq has a higher sensitivity and signal-to-noise ratio as compared with the traditional CLIP-seq. Consistent with LIN28A-binding profiles on HuProt arrays, all 12 lncRNAs that show detectable expression in ESCs also exhibited detectable signals of LIN28A FBioCLIP-seq (Figure 3C; Supplementary Figure S4). The lack of LIN28A binding on Evx1as transcripts in ESCs is consistent with the fact that Evx1as is not activated until ESCs differentiate into mesoendoderm cells. In vivo validation of LIN28A-lncRNA interactions indicated that HuProt array-based method is reliable and sensitive to identify novel RNA–protein interactions that are indeed present in cellular contexts.
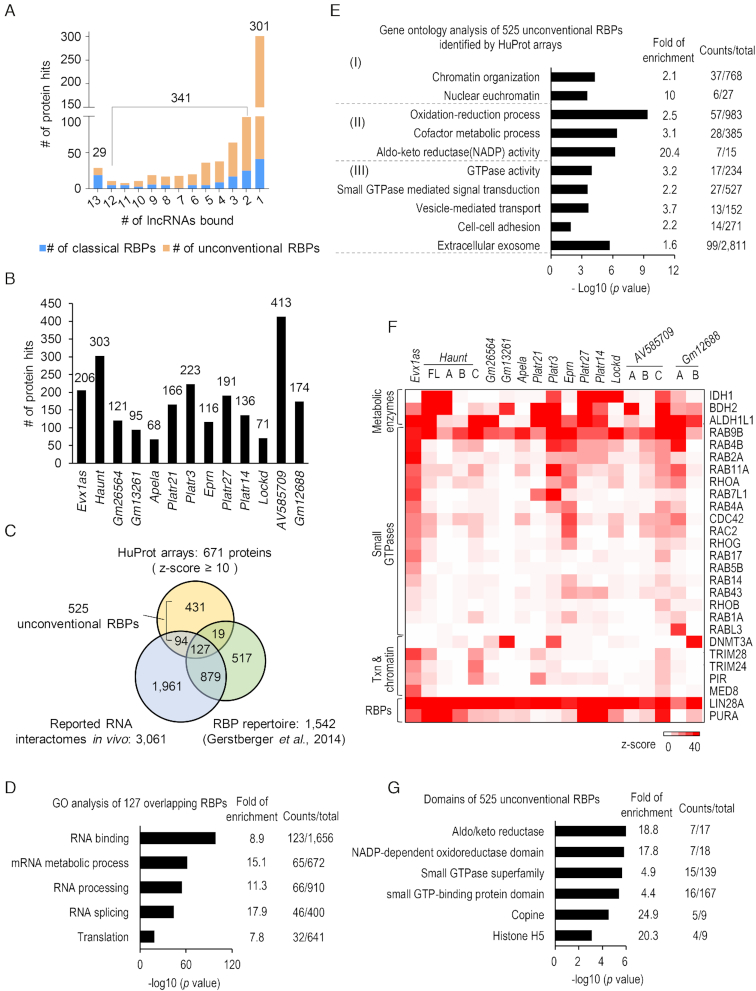
Revelation of unconventional RBPs with diverse functions
Among the 671 proteins with RNA-binding activity (z-score ≥ 10) identified in the HuProt assays, 301 (44.9%) show specific binding to only one lncRNA, 341 (50.8%) bind to 2–12 lncRNAs, and only 29 (4.3%) bind to all 13 lncRNAs (Figure 4A). On the other hand, each of the 13 lncRNAs bound to a large number of proteins, ranging from 68 to 413 (z-score ≥ 10) (Figure 4B). These observations suggest that these lncRNAs may execute pleiotropic functions by enlisting a large set of protein partners with diverse biological functions, and the majority of them exhibit different degrees of specificity towards the RNA sequences. For example, Haunt bound to 303 proteins on HuProt arrays, one of which was PURA (purine-rich element-binding protein A), a multifunctional, sequence-specific DNA- and RNA-binding protein that has been implicated in both transcriptional activation and repression (76). Haunt directly interacts with PURA as evidenced by our in vitro pull-down and in vivo RNA immunoprecipitation (RIP) assays (Figure 2B; Supplementary Figure S5). However, it is worth noting that PUR proteins were reported to be dispensable for the effect of Haunt on the HOXA cluster (57). Because Haunt transcripts associate with hundreds of proteins, we reasoned that inhibition of one or a few of the interacting proteins is unlikely to affect the function of a given lncRNA because it could be compensated by other interacting proteins.
Figure 4.
HuProt array profiling reveals hundreds of unconventional RBPs. (A) Histogram showing the number of proteins bound to different numbers of lncRNAs. Classical and unconventional RBPs are highlighted in blue and orange, respectively. (B) Histogram showing the number of proteins bound by each lncRNA. (C) Venn diagram for the comparison of RBPs identified by HuProt arrays with Tuschl's RBP repertoire (9) and the RNA interactomes we compiled (Supplementary Table S4) (10–17). (D) GO analysis of 127 overlapping RBPs shown in panel (C). Selected GO terms (fold enrichment ≥ 1.5, P value ≤ 2.3E–19) are shown. (E) Enriched GO terms of 525 unconventional RBPs shown in panel (C) (fold enrichment of a selected term ≥ 1.5, P value ≤ 1.9E–6). (F) Heatmap showing interactions among lncRNAs and representative proteins including metabolic enzymes, small GTPases, transcriptional and epigenetic regulators, and classical RBPs. (G) Enriched protein domains in 525 potential unconventional RBPs. Selected domains (fold enrichment ≥ 1.5, P value ≤ 7.9E–4) are shown.
We then compared the 671 newly identified lncRNA-binding proteins with Tuschl's RBP repertoire (1542) and a set of 3061 proteins reported in several studies of profiling protein interactomes of polyA or nascent RNA transcripts in human and mouse cells (9–17). Venn diagram analysis of the three datasets revealed an overlap of 127 RBPs, which are mainly involved in RNA processing, RNA splicing and translation (Figure 4C and D). In addition, 94 of the 671 proteins were also found in the in vivo RNA interactomes but not presented in Tuschl's RBP repertoire, suggesting that many newly discovered unconventional RBPs are likely to be physiologically relevant (10–17) (Figure 4C; Supplementary Table S3 and S4). Importantly, 431 were identified as potential RNA-binding proteins for the first time in our study, indicating that many novel RBPs are yet to be fully discovered (Figure 4C; Supplementary Table S3). In summary, a total of 525 (431 plus 94) proteins, identified in this study but not included in Tuschl's RBP repertoire, are potentially unconventional RBPs (Figure 4C).
Gene Ontology (GO) analysis indicated that the 525 potentially unconventional RBPs were significantly enriched in three functional categories (P value ≤ 1.9E–6), including (i) chromatin organization; (ii) proteins with GTPase activity implicated in vesicle transport, cell-cell adhesion, and extracellular exosome and (iii) metabolic and oxidation-reduction processes (Figure 4E and F; Table 1). In addition, protein domain analysis also revealed significant enrichments of small GTP-binding, aldo/keto reductase and NADP-dependent oxidoreductase domains, suggesting that these protein domains may possess moonlighting RNA-binding activities (Figure 4G).
Table 1.
A representative list of potential RBPs revealed by HuProt arrays. This list includes 53 oxidative-reductases, 17 small GTPases and 87 transcriptional and chromatin regulators. Proteins detected in previously reported RNA interactomes are labeled with ‘√’ or as indicated (10–17). Potential RBPs uniquely detected by HuProt arrays are indicated as ‘only in this study’ with maximal z-scores shown in brackets
| RNA interactomes reported in vivo | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mRNA interactomes by oligo (dT) capture | RBD mapping | aRICK-16 h | RICK-0.5 h, 1 h, 2 h | |||||||||||
| Proteins bound to RNA on HuProt arrays | Kwon et al., 2013 | Baltz et al., 2012 | Castello et al., 2012 | Bao et al., 2018 | Beckmann et al., 2015 | Castello et al., 2016 | Liao et al., 2016 | He et al., 2016 | Bao et al., 2018 | |||||
| Category | Protein hits | Max z-score | # of times identified | mESCs | HEK293 | HeLa | HeLa | HuH-7 | HeLa | HL-1 | mESCs | HeLa | HeLa | |
| Metabolic enzymes (53) | Reported previously | QDPR | 3352 | 1 | √ | |||||||||
| IDH1 | 1935 | 3 | √ | √ | √ | |||||||||
| MTHFD1 | 429 | 3 | √ | √ | √ | |||||||||
| BLVRB | 52 | 2 | √ | √ | ||||||||||
| ACOX1 | 41 | 1 | √ | |||||||||||
| IMPDH2 | 36 | 1 | √ | |||||||||||
| ENOX1 | 36 | 1 | √ | |||||||||||
| PDIA5 | 35 | 1 | √ | |||||||||||
| AKR1B1 | 33 | 1 | √ | |||||||||||
| COX4I1 | 24 | 2 | √ | √ | ||||||||||
| GBE1 | 20 | 1 | √ | |||||||||||
| PGK1 | 19 | 4 | √ | √ | √ | √ | ||||||||
| ENO1 | 19 | 6 | √ | √ | √ | √ | √ | √ | ||||||
| PRDX1 | 18 | 9 | √ | √ | √ | √ | √ | √ | √ | √ | √ | |||
| AKR1C3 | 16 | 1 | √ | |||||||||||
| DHCR7 | 15 | 1 | √ | |||||||||||
| GYS1 | 13 | 1 | √ | |||||||||||
| DLD | 12 | 4 | √ | √ | √ | √ | ||||||||
| G6PD | 11 | 3 | √ | √ | √ | |||||||||
| Only in this study | TP53I3 (3440) | BDH2 (1433) | ALDH1L1 (2603) | AKR1D1 (798) | COQ6 (87) | NDUFB11 (64) | CRYZ (60) | D2HGDH (43) | DECR2 (42) | CYB561 (38) | IMPDH1 (37) | ZADH2 (36) | ALDH1A1 (30) | |
| DCT (32) | CPOX (30) | HACL1 (29) | GLRX2 (29) | AKR7A2 (27) | PFKL (24) | MECR (24) | FOXRED2 (20) | DDO (19) | PIR (18) | ACAA1 (17) | CYBA (14) | CYP8B1 (13) | ||
| MLYCD (13) | HK3 (12) | LOXL3 (12) | SCCPDH (12) | GFOD1 (12) | AKR7A3 (11) | PTGIS (11) | CRAT (10) | |||||||
| Small GTPases (17) | Reported previously | RAB11A | 39 | 1 | √ | |||||||||
| CDC42 | 29 | 1 | √ | |||||||||||
| RAB14 | 11 | 1 | √ | |||||||||||
| Only in this study | RAB1A (11) | RAB2A (39) | RAB4A (22) | RAB4B (38) | RAB5B (15) | RAB7L1 (49) | RAB9B (109) | RAB17 (21) | RAB43 (14) | RHOA (27) | RHOB (10) | RHOG (17) | RAC2 (26) | |
| RABL3 (31) | ||||||||||||||
| Transcriptional & chromatin regulators (87) | Reported previously | TRIM28 (30) | TRIM24 (21) | DNMT3A (38) | SMARCE1 (44) | EED (18) | CBX1 (13) | HOXB6 (27) | CHD2 (11) | NACA (26) | ZNF326 (17) | HIST1H1C (29) | H1F0 (11) | H1FX (56) |
| GTF2I (39) | RBBP5 (18) | PAF1 (62) | SET (10) | SMARCC1 (11) | SMARCC2 (10) | RNF10 (10) | TRIP6 (28) | NAP1L1 (22) | HMGN1 (112) | NUCKS1 (13) | H2AFY2 (30) | ASCC1 (26) | ||
| CTNNB1 (14) | NDC80 (16) | PQBP1 (32) | SND1 (16) | G3BP1 (13) | ZCCHC9 (11) | CDC73 (12) | GTF3C2 (11) | ILF3 (24) | TARDBP (36) | UBE2N (42) | IFI16 (11) | |||
| Only in this study | MED8 (26) | DAXX (62) | PRDM15 (29) | POU4F2 (10) | FOXG1 (42) | APP (12) | ASXL2 (74) | BCOR (29) | CENPN (15) | EAF1 (14) | EYA4 (16) | HIST1H1A (35) | IKZF3 (17) | |
| RBBP8 (45) | TAF7 (15) | APBB1 (55) | MEF2C (16) | MNDA (12) | NEUROD1 (10) | NFYA (22) | NIF3L1 (19) | OVOL2 (16) | PCGF3 (17) | ING3 (17) | JDP2 (10) | PRMT8 (45) | ||
| SCMH1 (12) | SEH1L (12) | SIRT5 (23) | SSBP2 (30) | SUDS3 (16) | RORC (11) | TCEAL5 (305) | TCEAL6 (132) | TERF2IP (14) | TLX2 (10) | TP53 (11) | TTN (50) | VGLL4 (17) | ||
| WT1 (13) | ZNF226 (19) | ZNF274 (22) | ZNF350 (21) | ZNF485 (17) | ZNF502 (13) | ZNF528 (12) | ZNF791 (11) | ZXDC (34) | PIR (18) | |||||
aRICK (capture of the newly transcribed RNA interactome using click chemistry) (13).
Chromatin regulators, metabolic enzymes and small GTPases display RNA-binding activity
HuProt arrays revealed ∼87 proteins known to directly regulate transcription and chromatin functions, ∼53 metabolic enzymes involved in oxidation-reduction process, and 17 small GTPases (Table 1). This list included some well-known chromatin regulators (e.g., DNMT3A, MED8, and tripartite motif proteins TRIM28 and TRIM24), RAB and RHO family small GTPases (e.g., RAB11A and CDC42), and metabolic enzymes (e.g., IDH1, BDH2 and ALDH1L1) (Figure 4F; Supplementary Table S3). For example, IDH1 bound to Haunt with the highest z-score of 1935, as well as six other lncRNAs, including Lockd, with z-scores ranging from 11 to 155. DNMT3A, an essential DNA methyltransferase, was found to bind to three lncRNAs (Gm13261, Platr3, Gm12688) with z-scores ranging from 27 to 38 on HuProt arrays. MED8 specifically interacted with Evx1as (z-score = 26), consistent with a role of the Mediator complex in facilitating Evx1as function on nearby gene transcription (58). TRIM28 is known to interact with 7SK small nuclear ribonucleoprotein (snRNP) and to regulate the pausing release of RNA polymerase II (77). TRIM28 interacted with lncRNAs Evx1as, Haunt, and AV585709 (z-score ≥ 10) on HuProt arrays, suggesting other RNA-binding activities in addition to 7SK snRNP. TRIM24, an oncogenic transcriptional activator in prostate cancer (78), bound to Evx1as and Haunt (z-score ≥ 17) (Supplementary Table S3).
The RAS superfamily small GTPases, composed of ∼154 proteins in human, usher vesicles and membranes in the exocytic and endocytic pathways, and play a key role in cell-cell or cell-matrix adhesions (79,80). Among the 126 small GTPases available on HuProt arrays, seventeen (including eleven RAB and five RHO family members) were found to exhibit RNA-binding activities (Figure 4F; Table 1). Consistent with this finding, RAB1A and RAB11A (yeast homologues YPT1 and YPT32) were also detected with RNA-binding activity in previous studies using yeast protein microarrays (34,35).
As additional evidence to support our observations, previous studies of mapping RNA-binding regions in cells detected numerous RNA-crosslinked peptides from many proteins, such as IDH1, DNMT3A, TRIM24, RAB11A and CDC42, which were also identified in HuProt array screens (Table 1) (10–17). Notably, compared with the previously reported RNA interactomes (10–17), a set of 49 transcription and chromatin-related proteins (e.g., pirin or PIR), 34 metabolic enzymes (e.g., BDH2, ALDH1L1) and 14 small GTPases (e.g., RAB4A/4B/7L1/9B, RHOA) were identified for the first time to possess RNA-binding activity in our screens (Supplementary Table S3). For example, PIR, an iron-dependent redox sensor and regulator of NF-κB (81), was found to interact with three lncRNAs on HuProt arrays. The interaction between PIR and Haunt (z-score = 17) was validated using in vitro pull-down (Supplementary Figure S3F). Notably, the lncRNA Evx1as interacted with 13 small GTPases with z-scores ≥ 10. Of the seven small GTPases we chose for validation, all were confirmed to interact with Evx1as by in vitro pull-down, and 5 were shown for the first time to have RNA-binding activity (Figure 2D). In addition, Haunt RNA captured recombinant IDH1, BDH2 and ALDH1L1 proteins we confirmed in vitro (Figure 2D). Collectively, these results suggest that chromatin regulation, cellular metabolism and vesicular trafficking seem to be tightly coupled with the RNA-binding activities of key protein regulators or enzymes, expanding the catalog of RNA-binding proteins.
IDH1 FBioCLIP-seq reveals thousands of RNA targets in ESCs
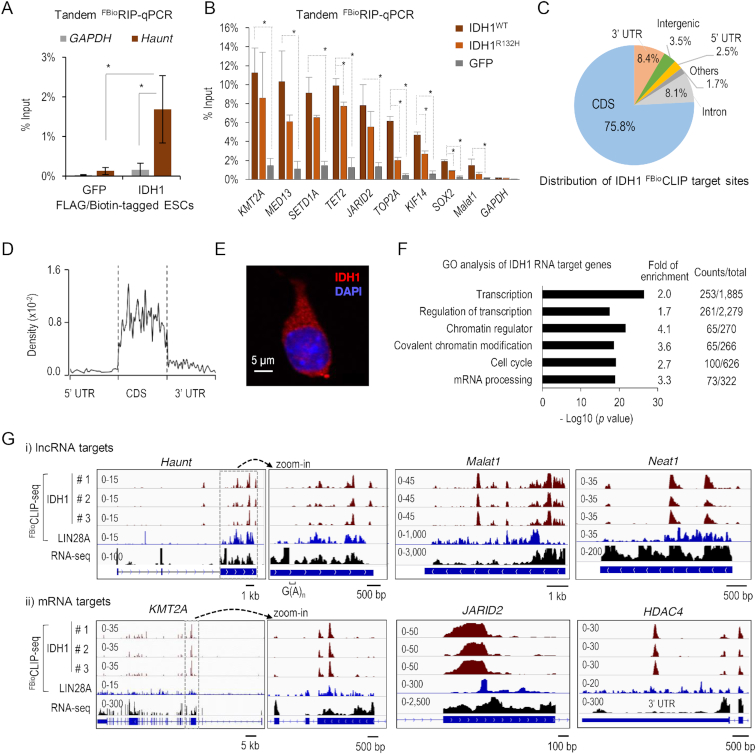
Subsequently, to further characterize the RNA-binding activity of novel RBPs, the metabolic enzyme IDH1 was chosen as a representative because of its strong affinity to RNA (z-score = 1935 to Haunt) and its well-characterized function in producing α-ketoglutarate, a key cofactor required for many histone and DNA demethylases. Having confirmed its in vitro RNA-binding activity, we sought to test whether IDH1 directly binds to RNA in cells. As commercially available antibodies against IDH1 are not suitable for immunoprecipitation, we established ESC lines that stably expressed the sub-endogenous level of FLAG- and biotin-tagged IDH1 (Supplementary Figure S6A). In addition, we also established a control ESC line that stably expressed FLAG- and biotin-tagged GFP. FLAG- and biotin-mediated tandem RNA immunoprecipitation followed by quantitative PCR analysis (tandem FBioRIP-qPCR) revealed that IDH1, but not GFP, robustly enriched for Haunt RNA (Figure 5A).
Figure 5.
A metabolic enzyme IDH1 emerges as an unconventional RBP. (A) FLAG- and biotin-mediated tandem FBioRIP of IDH1 and GFP followed by qPCR analysis of the Haunt and GAPDH transcripts in ESCs. Data are shown as mean ± SD (standard deviation, n = 4 biological replicates). *P < 0.01. (B) Tandem FBioRIP-qPCR of wild-type IDH1WT, IDH1 mutant (IDH1R132H) and GFP in ESCs. Data are shown as mean ± SD (n ≥ 3 biological replicates). *P < 0.01. (C) Distribution of IDH1 RNA-binding peaks. (D) Metagene analysis of IDH1 FBioCLIP read densities along the mRNA transcripts. (E) Immunofluorescence of endogenous IDH1 in ESCs. IDH1: red, DAPI: blue, n = 20 cells. (F) GO analysis of IDH1-binding transcripts. Selected GO terms (fold enrichment ≥ 1.5, P value ≤ 4.10E–15) are shown. (G) Genome browser views of IDH1 FBioCLIP-seq reads at representative loci of lncRNA (top) and mRNA (bottom), respectively. The tracks shown on the right are zoom-in views of the regions boxed by dotted lines. The region of G(A)n repeat (2 ≤ n ≤ 6) in the last exon of Haunt is indicated by a bracket on the top. Sequencing data processing removed ambiguous reads that can mapped to multiple genomic loci so that no FBioCLIP-seq signals were detected in the G(A)n repeat region. The zoom-out views of JARID2 and HDAC4 loci are shown in Supplementary Figure S7E.
Next, to obtain a genome-wide view of IDH1’s RNA targets, we performed IDH1 FBioCLIP-seq. The results from three biological replicates were highly correlated (Pearson coefficients > 0.9) (Supplementary Figure S6B). In total, we identified ∼2300 high-confidence overlapping peaks in three replicates, corresponding to 1341 genes (Supplementary Table S5). While the majority (∼98%) of IDH1 target genes are protein-coding transcripts (Supplementary Table S5), IDH1 also binds to a number of lncRNA transcripts, including Haunt, Malat1, Neat1 and B230354K17Rik (Figure 5G; Supplementary Figure S6E). Tandem FBioRIP-qPCR validated significant enrichment of IDH1 on a number of FBioCLIP RNA targets compared to the GFP control (Figure 5B).
IDH1 FBioCLIP-seq signals were found enriched in the coding sequences (CDS, 75.8%) and the 3′ untranslated regions (3′ UTRs, 8.4%), as compared with the expected percentage of these regions in the transcriptome (Figure 5C; Supplementary Figure S6C). Metagene analysis showed that IDH1 RNA reads are evenly distributed within the CDS (Figure 5D). Only 8.1% of IDH1 FBioCLIP-seq reads fall into intronic regions, indicating that IDH1 largely interacts with sequences within mature mRNA transcripts. This result is consistent with a dominant localization of IDH1 protein in the cytoplasm (Figure 5E). GO analysis showed that IDH1 target transcripts mainly encode proteins involved in transcription and chromatin regulations, cell cycle and mRNA processing (Figure 5F). In addition, the lack of correlation between RNA signal densities of IDH1 and gene expression implies that IDH1 specifically binds to a subset of RNA transcripts, arguing against a bias toward abundant transcripts (Supplementary Figure S6D).
GA- and AU-rich RNA motifs are enriched in IDH1 targets
Motif analysis of all target sequences of IDH1 FBioCLIP-seq revealed two GA-rich consensus sequences (Overall motif #1: GAAGAAGAUC; and #2: AGAAGGAGGAGA; P < 1e−112), which accounted for more than half of IDH1 target sites (Figure 6A; Supplementary Figure S7A). Interestingly, we noticed a 212-nt sequence mainly composed of G(A)n simple repeats (2 ≤ n ≤ 6) in the last (third) exon of Haunt RNA (Supplementary Figure S7D, bottom). Secondary structure prediction suggested that this G(A)n repeat region is likely to form a big extended and unstructured loop within the folded Haunt RNA (Supplementary Figure S7D, top). Interestingly, FBioCLIP-seq showed strong IDH1-binding signals in the last exon of Haunt (Figure 5G).
To dissect RNA elements that are required for Haunt binding to IDH1, we generated three truncated probes surrounding the G(A)n repeat region and analyzed their binding affinity to IDH1 on HuProt arrays (Supplementary Figure S6F). Both Haunt-A and C probes comprise the 5′ sequences of Haunt, but only the probe A, but not C, contains the G(A)n repeat region. The Haunt-B probe comprises the 3′ sequence but lacks the stretch of G(A)n repeats of Haunt. Interestingly, the Haunt-A RNA probe bound to IDH1 albeit with a reduced affinity compared to the full-length Haunt (z-scores 216 versus 1,935, respectively). However, both Haunt-B and C failed to be captured by IDH1 (z-scores 1 and 0). Thus, the G(A)n repeat region of Haunt appears to be required for IDH1 binding, while the 3′ sequence of Haunt in the Haunt-B probe is necessary but not sufficient for IDH1 binding. Together, these results demonstrate that IDH1 directly binds to Haunt RNA in vitro, probably via interaction with the GA-rich motif in the last exon.
IDH1 FBioCLIP-seq signals appeared to be slightly enriched in the 3′ UTR compared to the 5′ UTR and introns. Motif analysis of IDH1 target RNA signals that specifically fall into the 3′ UTR regions revealed distinct AU-rich sequences (3′ UTR motif #1: UCUAUUUAUU; and #2: UAAAAUCCAU; P < 1e−2), although they represent a minor portion of overall IDH1 RNA targets (Figure 6A; Supplementary Figure S7A).
IDH1 binds strongly to GA- or AU-rich single-stranded RNA in vitro
To determine whether IDH1 binds directly to the identified RNA sequences in vitro, we performed native gel electrophoretic mobility shift assay (EMSA) using purified IDH1 and Cy5 fluorescence-labeled, single-stranded RNA (ssRNA) probes of 20- to 30-nt in length. Recombinant IDH1 proteins were purified free of nucleic acid contamination using size exclusion chromatography. Purified IDH1 formed homodimers which represent the active conformation of IDH1, and were enzymatically active when tested (Supplementary Figure S7G and data not shown). Because IDH1 showed strong interaction with the GA-rich RNA motifs enriched in its FBioCLIP targets, we first tested a 20-nt ssRNA probe, named as ‘5 × GAA (overall motif)’, which contains the overall motif #1 (GAAGAAGAUC) flanked with GAA sequences, resulted in a total of five copies of GAA tri-nucleotides. Addition of recombinant IDH1, but not negative control proteins GFP and GST, caused super-shifted bands of the RNA probe, with a dissociation constant (Kd) of 1.34 ± 0.35 μM (Figure 6B–E; Supplementary Figure S7E).
To further quantify the effect of GAA repeat numbers on IDH1 binding, we designed a set of five fluorescence-labeled ssRNA probes carrying 1 to 5 copies of the GAA(A) sequence (frequently observed in the 212-nt G(A)n repeats of Haunt). We gradually replaced the GAA(A) with a GCC(C) sequence for probes with fewer copies of GAA(A) repeats (Figure 6C; Supplementary Figure S7B). Interestingly, IDH1 showed gradually enhanced RNA-binding activity along with increasing numbers of GAA(A) repeats (Figure 6C–E; Supplementary Figure S7B). The binding affinity to the 5 × GAA(A) probe (Kd = 0.43 ± 0.06 μM) is ∼7- and 18-fold higher than to the 4 × GAA(A) (Kd = 2.98 ± 0.43 μM) and 3 × GAA(A) probes (Kd = 7.77 ± 4.17 μM), respectively. In contrast, 2 × GAA(A) and 1 × GAA(A) RNA probes failed to super-shift with IDH1 (Figure 6C–E; Supplementary Figure S7B), indicating that at least three copies of GAA(A) are required for the binding of IDH1.
Motif analysis suggested that IDH1 could bind to a portion of target RNAs enriched in AU-rich motifs detected in 3′ UTR regions. We examined the binding of IDH1 to an AU-rich ssRNA probe which is 30-nt in length and contains four copies of AUUU unit with the core sequence of 3′ UTR motif #1. IDH1 super-shifted with the AU-rich ssRNA with a Kd of 0.68 ± 0.07 μM (Figure 6D and E; Supplementary Figure S7C). We then tested several AU-rich probes of the same sequence but in various nucleic acidic forms, such as double-stranded RNA (dsRNA), single- or double-stranded DNA (ssDNA or dsDNA), and DNA/RNA hybrid. In contrast, none of them super-shifted with IDH1 (Supplementary Figure S7F and H). To further test the binding specificity of IDH1, we employed two additional GC-rich ssRNA probes with 30-nt in length and found that both failed to super-shift with IDH1 unless IDH1 was added at extremely high concentrations (e.g., 10 μM) (Supplementary Figure S7E). Taken together, these results demonstrate that IDH1 preferentially recognizes ssRNA in a sequence-specific manner in vitro.
A functional link between IDH1/2 mutations and RNA m6A modification was recently reported in human cancer cells (46). We found that recombinant IDH1 has similar binding affinities to RNA probes with or without m6A modification shown by EMSA (Supplementary Figure S8A–C). Interestingly, RNA transcripts captured by IDH1 tandem RIP in ESCs exhibited ∼1-fold higher levels of m6A compared to the input RNA (Figure 6F). Thus, m6A modification does not appear to affect IDH1 binding to RNA in vitro. We posit that IDH1 binding may modulate m6A levels of its target RNA, which consequently influence the fate of RNA in processing, translation and decay.
R132H mutation in IDH1 reduces its RNA-binding activity in vitro and in vivo
The oncogenic R132H mutant of IDH1 has been reported for its role in influencing global DNA and histone methylation and promoting tumorigenesis via the oncometabolite 2-hydroxyglutarate (40–42). To explore a potential connection between the newly discovered RNA-binding activity and IDH1’s known function, we sought to test whether the RNA-binding activity of IDH1 is affected by the R132H mutation. First, by comparing ESCs that were stably expressing either the wild-type IDH1WT or R132H mutant (IDH1R132H) construct, we found that IDH1R132H exhibited decreased binding activity to a panel of IDH1WT -targeted transcripts, such as TET2, TOP2A, KLF14 and SOX2, as shown by tandem FBioRIP-qPCR (Figure 5B). Next, we tested the direct RNA-binding activity of purified IDH1R132H using EMSA. IDH1R132H failed to super-shift with the 5× GAA (overall motif) ssRNA unless it was added at a high concentration (i.e. 5 μM); whereas the wild-type IDH1 robustly super-shifted the same ssRNA probe at a much lower concentration (i.e. 0.5 μM) (Figure 6G). These results indicate that the R132H mutation attenuates the ssRNA-binding activity of IDH1 both in vitro and in vivo, suggesting a potential link of RNA binding with the biological function of IDH1.
Next, we sought to ask how R132H mutation disrupts the RNA-binding activity of IDH1. Intriguingly, addition of excess amounts of the substrate isocitrate at concentrations up to 1000 μM (∼50 000-fold higher than RNA) failed to alter the binding of 5× GAA(A) RNA probes (20 nM) to IDH1 (1 μM) (Supplementary Figure S9B). This observation suggests that the RNA-binding and catalytic activities of IDH1 may reside in different sites so that they do not compete with each other. Analysis of the crystal structure of an asymmetric homodimer of IDH1 (52,82) reveals that the large and small domains of IDH1 form a large, deep cleft as the catalytic site. At the bottom of the cleft, the substrate isocitrate contacts the R132 residue, which is known to form an extensive interaction network via salt bridges and hydrogen bonds with residues around it to stabilize IDH1 structure (52). Above the catalytic site, we identified a potential RNA-binding surface, which is covered by four positively charged amino acids (R249, K260, R314 and R317) and appears to be positioned far away from the R132 residue (Supplementary Figure S9A). To test the role of these four positively charged amino acids in IDH1-RNA binding, we mutated them individually or in combination to negatively charged aspartic acid (D) and purified recombinant mutant proteins for EMSA using 5× GAA(A) RNA probes. Interestingly, all these mutations attenuated the RNA-binding activity of IDH1 at different levels (Supplementary Figure S9C–E). For example, the mutant protein of IDH1 with R314D and R317D double mutations exhibited the most severe loss of RNA-binding activity with a Kd of 10.3 ± 1.5 μM, while R249D or K260D single mutant had a larger Kd of 7.5 ± 0.8 or 3.6 ± 0.2 μM, respectively, compared to a Kd of 0.43 ± 0.06 μM for the wild-type IDH1.
Intriguingly, we found that R132H mutation dramatically alters the positions of the above four positively charged residues as shown in the crystal structure of IDH1R132H mutant protein (Supplementary Figure S9G). In addition, we performed IDH1 truncation and in vitro RNA pull-down to map its RNA-binding domain (Supplementary figure S9F). The minimal segment of IDH1 that harbors the RNA-binding activity resides in amino acid residues 136–285. Coincidently, this segment comprises the two key flexible regions (residues 132–141 and residues 271–286), which exhibit large structural changes with greater flexibility for being mostly disordered as shown in the crystal structure of IDH1R132H mutant (52). Taken together, we propose that IDH1 has distinct RNA-binding and catalytic sites, and the residue R132 interacts with the substrate isocitrate, but not RNA, in the catalytic cleft. Structural changes caused by R132H mutation may disrupt the potential RNA-binding surface, consequently leading to its loss of RNA-binding activity. Yet, firm confirmation of the RNA-binding sites of IDH1 will have to await structural analysis of the IDH1–RNA complex.
DISCUSSION
Here, we described a rapid and unbiased approach that utilizes HuProt arrays to comprehensively interrogate proteins with RNA-binding activity, and identified 671 proteins that interact with 13 lncRNA transcripts (z-score ≥ 10). In vitro pull-down validated 82 pairs of randomly selected RNA–protein interactions that comprise the classical and unconventional RBPs, with a false positive rate of 18% and a false negative rate of 43% in our screens. The HuProt approach appears superior in terms of unbiased identification of transient or weak interactions of RNA and proteins, whose expression is not limited to a certain cell type. In comparison, the in vivo approaches often bias towards abundant proteins and/or those with strong RNA-binding activities. As the HuProt lacks protein complexes and cellular contexts, our screens may underestimate the scope of in vivo RNA–protein interactions. Nevertheless, FBioCLIP-seq profiling of LIN28A and IDH1 in ESCs demonstrated that our approach is reliable to identify physiologically relevant protein-RNA interactions. Interestingly, 525 of the 671 identified proteins do not carry any conventional RBDs and are enriched in functions that are not directly related to RNA; and 431 were identified as potential RNA-binding proteins for the first time in our study, thus expanding the current catalogue of RBPs. The interplay between RNA regulation and diverse cellular processes is of a much greater degree than previously anticipated.
About 87 proteins are directly involved in transcription and chromatin functions, serving as a bridge that connects lncRNAs to global transcriptional regulation. Several of them, such as EED, DNMT3A and TARDBP (TDP-43), have been identified as RBPs via candidate-based approaches (83–86), supporting a proposed role of RNA–protein interactions in mediating epigenetic and transcriptional regulation. Unexpectedly, we found that 17 small GTPases known to function in vesicle transport may harbor the RNA-binding activity, among 126 of ∼154 annotated small GTPases spotted on the HuProt array. Compared with other lncRNAs tested, lncRNA Evx1as showed a higher preference to small GTPases. In addition to the reported cis-regulatory role of Evx1as on chromatin (58), we speculate a possible involvement of Evx1as in cell-cell communication through its association with RAB proteins. Several lncRNAs, including lincROR, lncARSR and lncPARTICLE, have been implicated in regulating intercellular communication and drug sensitivity via exosome-mediated transmission in cancer cells (87–89). Signal recognition particle (SRP) RNA could enhance the GTPase activity of the SRP-receptor complex in co-translational protein targeting to cell membranes (90). Intriguingly, it was reported that ∼1.6% of lncRNAs associate with lipid directly or indirectly (91,92). Together, these observations suggest a potential role of small GTPases in linking RNA biology and vesicle transport. Small GTPases, particularly those in the RAB family, might be regulated by RNA or have RNA-related functions in which RNA transcripts act as cargo.
Moreover, this study revealed many more metabolic enzymes that moonlight as RNA-interacting proteins. About 53 newly identified unconventional RBPs are well characterized metabolic enzymes known to regulate oxidation-reduction processes and cover much of the landscape of intermediary metabolism (93). This finding lends support to the hypothesis that the RNA-binding activity of metabolic enzymes serves as a regulatory link to mediate cross-talks between gene expression and intermediary metabolism (94,95). Importantly, we provided complementary lines of evidence to illustrate, for the first time, the RNA targets and binding preferences of IDH1 in vitro and in vivo.FBioCLIP-seq in ESCs revealed that IDH1 targets to a large set of ∼1,341 mRNA transcripts enriched in GA- or AU-rich motifs. Direct interactions between IDH1 and ssRNA containing GA- or AU-rich but not GC-rich sequences revealed by native EMSA, and mapping potential residues and minimal sequences required for IDH1 binding to RNA, demonstrate that the ssRNA-binding activity of IDH1 is direct and sequence-dependent, corroborating the role of IDH1 as a novel RBP.
Interestingly, the mRNA targets of IDH1 are specifically enriched in functions related to transcriptional and chromatin regulation, cell cycle and RNA processing. It was reported that the subset of genes encoding transcription factors, chromatin modifying enzymes and cell-cycle-specific regulators are more likely to produce unstable mRNAs and proteins based on global quantification of the abundance and turnover of mammalian transcriptome and proteome (96). In addition, the GA-rich RNA motifs identified in more than half of IDH1 targeting sites in ESCs have been widely recognized by many protein regulators of splicing, translation, lineage development and innate immune response (56,75,97–100). Moreover, AU-rich motifs, enriched in IDH1 target RNAs in the 3′ UTR regions, are similar to the regulatory AU-rich element (ARE) within 3′ UTR, which is reportedly to be bound by metabolic enzymes such as GAPDH to regulate the translation and stability of target mRNAs (101–103). The fact that IDH1 preferentially binds to specific RNA elements on a specific set of target transcripts with distinct functional enrichments implies a potentially functional link of IDH1’s RNA-binding activity to particular cellular processes. Intriguingly, the RNA targets of IDH1 exhibit higher m6A levels, and the oncogenic R132H mutant of IDH1 shows impaired RNA-binding activity. Future studies to define the in vivo function of IDH1 binding to RNA should provide physiological and functional insights into IDH1-RNA interactions.
In summary, our work reports valuable resources of unconventional RBPs and novel RNA-binding activities of IDH1, providing a useful launch pad to study the biological roles and mechanisms of RNA-mediated regulations in unexpected layers of cellular regulatory networks. Elaborating RNA–protein interactions may contribute to intertwine regulatory networks of diverse cellular processes. Revelation of the functional and physiological significance of novel RNA–protein interactions and the underlying mechanisms will certainly bring future surprises of RNA-centered regulation in biology.
DATA AVAILABILITY
IDH1 FBioCLIP-seq data has been deposited in the GEO repository with the accession number GSE119798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119798). The accession numbers for LIN28A CLIP-seq data are GEO: GSM910955, GSM910956, GSM910957 (56). The accession number for mESCs RNA-seq is GSM1412826 (57).
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge Drs Junbiao Dai and Haitao Li for technical help, discussion and suggestions. We acknowledge the Protein Preparation and Characterization Core Facility of Tsinghua University Branch of China National Center for Protein Sciences Beijing for providing the facility support. We acknowledge members of Shen and Zhu Laboratories for insightful discussion and suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31471219, 31630095 to X.S.] (in part); National Basic Research Program of China [2017YFA0504204, 2018YFA0107604 to X.S.]; Center for Life Sciences (CLS) at Tsinghua University (to X.S.); National Institute of Healthy, U.S.A. [1R01GM111514 to H.Z.]. Funding for open access charge: National Natural Science Foundation of China, National Basic Research Program of China and the Center for Life Sciences (CLS) at Tsinghua University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al.. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 2. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al.. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. et al.. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017; 543:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S. et al.. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rinn J.L., Chang H.Y.. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012; 81:145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Batista P.J., Chang H.Y.. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013; 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011; 12:861–874. [DOI] [PubMed] [Google Scholar]
- 9. Gerstberger S., Hafner M., Tuschl T.. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014; 15:829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B.M., Strein C., Davey N.E., Humphreys D.T., Preiss T., Steinmetz L.M. et al.. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012; 149:1393–1406. [DOI] [PubMed] [Google Scholar]
- 11. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.M., Foehr S., Curk T., Krijgsveld J., Hentze M.W.. Comprehensive identification of RNA-Binding domains in human cells. Mol. Cell. 2016; 63:696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liao Y., Castello A., Fischer B., Leicht S., Foehr S., Frese C.K., Ragan C., Kurscheid S., Pagler E., Yang H. et al.. The cardiomyocyte RNA-Binding proteome: Links to intermediary metabolism and heart disease. Cell Rep. 2016; 16:1456–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bao X., Guo X., Yin M., Tariq M., Lai Y., Kanwal S., Zhou J., Li N., Lv Y., Pulido-Quetglas C. et al.. Capturing the interactome of newly transcribed RNA. Nat. Methods. 2018; 15:213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He C., Sidoli S., Warneford-Thomson R., Tatomer D.C., Wilusz J.E., Garcia B.A., Bonasio R.. High-Resolution mapping of RNA-Binding regions in the nuclear proteome of embryonic stem cells. Mol. Cell. 2016; 64:416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. et al.. The mRNA-Bound proteome and its global occupancy profile on Protein-Coding transcripts. Mol. Cell. 2012; 46:674–690. [DOI] [PubMed] [Google Scholar]
- 16. Kwon S.C., Yi H., Eichelbaum K., Fohr S., Fischer B., You K.T., Castello A., Krijgsveld J., Hentze M.W., Kim V.N.. The RNA-binding protein repertoire of embryonic stem cells. Nat. Struct. Mol. Biol. 2013; 20:1122–1130. [DOI] [PubMed] [Google Scholar]
- 17. Beckmann B.M., Horos R., Fischer B., Castello A., Eichelbaum K., Alleaume A.M., Schwarzl T., Curk T., Foehr S., Huber W. et al.. The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat. Commun. 2015; 6:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang R., Han M., Meng L., Chen X.. Transcriptome-wide discovery of coding and noncoding RNA-binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E3879–E3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitchell S.F., Jain S., She M., Parker R.. Global analysis of yeast mRNPs. Nat. Struct. Mol. Biol. 2013; 20:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Riley K.J., Steitz J.A.. The “Observer Effect” in genome-wide surveys of protein-RNA interactions. Mol. Cell. 2013; 49:601–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee F.C.Y., Ule J.. Advances in CLIP technologies for studies of Protein-RNA interactions. Mol. Cell. 2018; 69:354–369. [DOI] [PubMed] [Google Scholar]
- 22. Chu C., Zhang Q.C., da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y.. Systematic discovery of Xist RNA binding proteins. Cell. 2015; 161:404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simon M.D., Wang C.I., Kharchenko P.V., West J.A., Chapman B.A., Alekseyenko A.A., Borowsky M.L., Kuroda M.I., Kingston R.E.. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:20497–20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Engreitz J.M., Pandya-Jones A., McDonel P., Shishkin A., Sirokman K., Surka C., Kadri S., Xing J., Goren A., Lander E.S. et al.. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013; 341:1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chu C., Spitale R.C., Chang H.Y.. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015; 22:29–35. [DOI] [PubMed] [Google Scholar]
- 26. Yoon J.H., Srikantan S., Gorospe M.. MS2-TRAP (MS2-tagged RNA affinity purification): tagging RNA to identify associated miRNAs. Methods. 2012; 58:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bertone P., Snyder M.. Advances in functional protein microarray technology. FEBS J. 2005; 272:5400–5411. [DOI] [PubMed] [Google Scholar]
- 28. Hu S., Xie Z., Qian J., Blackshaw S., Zhu H.. Functional protein microarray technology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011; 3:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hall D.A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M.. Regulation of gene expression by a metabolic enzyme. Science. 2004; 306:482–484. [DOI] [PubMed] [Google Scholar]
- 30. Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T. et al.. Global analysis of protein activities using proteome chips. Science. 2001; 293:2101–2105. [DOI] [PubMed] [Google Scholar]
- 31. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H.S., Woodard C., Wang H., Jeong J.S. et al.. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009; 139:610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhu J., Gopinath K., Murali A., Yi G., Hayward S.D., Zhu H., Kao C.. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:3129–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Siprashvili Z., Webster D.E., Kretz M., Johnston D., Rinn J.L., Chang H.Y., Khavari P.A.. Identification of proteins binding coding and non-coding human RNAs using protein microarrays. BMC Genomics. 2012; 13:633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scherrer T., Mittal N., Janga S.C., Gerber A.P.. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS One. 2010; 5:e15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsvetanova N.G., Klass D.M., Salzman J., Brown P.O.. Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One. 2010; 5:e12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Castello A., Hentze M.W., Preiss T.. Metabolic Enzymes Enjoying New Partnerships as RNA-Binding Proteins. Trends Endocrinol. Metab. 2015; 26:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dang L., White D.W., Gross S., Bennett B.D., Bittinger M.A., Driggers E.M., Fantin V.R., Jang H.G., Jin S., Keenan M.C. et al.. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010; 465:966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L. et al.. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008; 321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y. et al.. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009; 324:261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Waitkus M.S., Diplas B.H., Yan H.. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 2018; 34:186–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F. et al.. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010; 18:553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H., Ito S., Yang C., Wang P., Xiao M.T. et al.. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011; 19:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gilbert W.V., Bell T.A., Schaening C.. Messenger RNA modifications: Form, distribution, and function. Science. 2016; 352:1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yue Y., Liu J., He C.. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015; 29:1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao B.S., He C.. Fate by RNA methylation: m6A steers stem cell pluripotency. Genome Biol. 2015; 16:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Elkashef S.M., Lin A.P., Myers J., Sill H., Jiang D., Dahia P.L.M., Aguiar R.C.T.. IDH mutation, competitive inhibition of FTO, and RNA methylation. Cancer Cell. 2017; 31:619–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rapicavoli N.A., Poth E.M., Zhu H., Blackshaw S.. The long noncoding RNA Six3OS acts in trans to regulate retinal development by modulating Six3 activity. Neural Dev. 2011; 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., Gage F.H.. An RNA code for the FOX2 splicing regulator revealed by mapping RNA–protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009; 16:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Darnell R.B. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip. Rev. RNA. 2010; 1:266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Moore M.J., Zhang C., Gantman E.C., Mele A., Darnell J.C., Darnell R.B.. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat. Protoc. 2014; 9:263–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cho H.J., Cho H.Y., Park J.W., Kwon O.S., Lee H.S., Huh T.L., Kang B.S.. NADP(+)-dependent cytosolic isocitrate dehydrogenase provides NADPH in the presence of cadmium due to the moderate chelating effect of glutathione. J. Biol. Inorg. Chem. 2018; 23:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang B., Zhong C., Peng Y., Lai Z., Ding J.. Molecular mechanisms of “off-on switch” of activities of human IDH1 by tumor-associated mutation R132H. Cell Res. 2010; 20:1188–1200. [DOI] [PubMed] [Google Scholar]
- 53. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Berman H., Henrick K., Nakamura H.. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003; 10:980. [DOI] [PubMed] [Google Scholar]
- 55. Xiao W., Adhikari S., Dahal U., Chen Y.S., Hao Y.J., Sun B.F., Sun H.Y., Li A., Ping X.L., Lai W.Y. et al.. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016; 61:507–519. [DOI] [PubMed] [Google Scholar]
- 56. Cho J., Chang H., Kwon S.C., Kim B., Kim Y., Choe J., Ha M., Kim Y.K., Kim V.N.. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012; 151:765–777. [DOI] [PubMed] [Google Scholar]
- 57. Yin Y., Yan P., Lu J., Song G., Zhu Y., Li Z., Zhao Y., Shen B., Huang X., Zhu H. et al.. Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell. 2015; 16:504–516. [DOI] [PubMed] [Google Scholar]
- 58. Luo S., Lu J.Y., Liu L., Yin Y., Chen C., Han X., Wu B., Xu R., Liu W., Yan P. et al.. Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell. 2016; 18:637–652. [DOI] [PubMed] [Google Scholar]
- 59. Li M., Gou H., Tripathi B.K., Huang J., Jiang S., Dubois W., Waybright T., Lei M., Shi J., Zhou M. et al.. An apela RNA-containing negative feedback loop regulates p53-Mediated apoptosis in embryonic stem cells. Cell Stem Cell. 2015; 16:669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paralkar V.R., Taborda C.C., Huang P., Yao Y., Kossenkov A.V., Prasad R., Luan J., Davies J.O., Hughes J.R., Hardison R.C. et al.. Unlinking an lncRNA from its associated cis element. Mol. Cell. 2016; 62:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li M.A., Amaral P.P., Cheung P., Bergmann J.H., Kinoshita M., Kalkan T., Ralser M., Robson S., von Meyenn F., Paramor M. et al.. A lncRNA fine tunes the dynamics of a cell state transition involving Lin28, let-7 and de novo DNA methylation. Elife. 2017; 6:e23468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bergmann J.H., Li J., Eckersley-Maslin M.A., Rigo F., Freier S.M., Spector D.L.. Regulation of the ESC transcriptome by nuclear long noncoding RNAs. Genome Res. 2015; 25:1336–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Moss E.G., Lee R.C., Ambros V.. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997; 88:637–646. [DOI] [PubMed] [Google Scholar]
- 64. Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N.. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008; 32:276–284. [DOI] [PubMed] [Google Scholar]
- 65. Viswanathan S.R., Daley G.Q., Gregory R.I.. Selective blockade of microRNA processing by Lin28. Science. 2008; 320:97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Xu B., Zhang K., Huang Y.. Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA. 2009; 15:357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shyh-Chang N., Daley G.Q.. Lin28: primal regulator of growth and metabolism in stem cells. Cell Stem Cell. 2013; 12:395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M.T., Tsanov K.M., Nguyen L., Asara J.M., Cantley L.C., Daley G.Q.. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013; 155:778–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang J., Ratanasirintrawoot S., Chandrasekaran S., Wu Z., Ficarro S.B., Yu C., Ross C.A., Cacchiarelli D., Xia Q., Seligson M. et al.. LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell. 2016; 19:66–80. [DOI] [PubMed] [Google Scholar]
- 70. Li N., Zhong X., Lin X., Guo J., Zou L., Tanyi J.L., Shao Z., Liang S., Wang L.P., Hwang W.T. et al.. Lin-28 homologue A (LIN28A) promotes cell cycle progression via regulation of cyclin-dependent kinase 2 (CDK2), cyclin D1 (CCND1), and cell division cycle 25 homolog A (CDC25A) expression in cancer. J. Biol. Chem. 2012; 287:17386–17397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ma X., Li C., Sun L., Huang D., Li T., He X., Wu G., Yang Z., Zhong X., Song L. et al.. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014; 5:5212. [DOI] [PubMed] [Google Scholar]
- 72. Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S., Takeuchi A., Engreitz J.M., Hagan J.P., Kharas M.G. et al.. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011; 147:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G.. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008; 10:987–993. [DOI] [PubMed] [Google Scholar]
- 74. Polesskaya A., Cuvellier S., Naguibneva I., Duquet A., Moss E.G., Harel-Bellan A.. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007; 21:1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wilbert M.L., Huelga S.C., Kapeli K., Stark T.J., Liang T.Y., Chen S.X., Yan B.Y., Nathanson J.L., Hutt K.R., Lovci M.T. et al.. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol. Cell. 2012; 48:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gallia G.L., Johnson E.M., Khalili K.. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000; 28:3197–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. McNamara R.P., Reeder J.E., McMillan E.A., Bacon C.W., McCann J.L., D’Orso I.. KAP1 recruitment of the 7SK snRNP complex to promoters enables transcription elongation by RNA polymerase II. Mol. Cell. 2016; 61:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Groner A.C., Cato L., de Tribolet-Hardy J., Bernasocchi T., Janouskova H., Melchers D., Houtman R., Cato A.C.B., Tschopp P., Gu L. et al.. TRIM24 Is an Oncogenic Transcriptional Activator in Prostate Cancer. Cancer Cell. 2016; 29:846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wennerberg K., Rossman K.L., Der C.J.. The Ras superfamily at a glance. J. Cell Sci. 2005; 118:843–846. [DOI] [PubMed] [Google Scholar]
- 80. Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009; 10:513–525. [DOI] [PubMed] [Google Scholar]
- 81. Liu F., Rehmani I., Esaki S., Fu R., Chen L., de Serrano V., Liu A.. Pirin is an iron-dependent redox regulator of NF-kappaB. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:9722–9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Xu X., Zhao J., Xu Z., Peng B., Huang Q., Arnold E., Ding J.. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004; 279:33946–33957. [DOI] [PubMed] [Google Scholar]
- 83. Denisenko O., Shnyreva M., Suzuki H., Bomsztyk K.. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol. Cell Biol. 1998; 18:5634–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Holz-Schietinger C., Reich N.O.. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 2012; 40:8550–8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wang L., Zhao Y., Bao X., Zhu X., Kwok Y.K., Sun K., Chen X., Huang Y., Jauch R., Esteban M.A. et al.. LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015; 25:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bhardwaj A., Myers M.P., Buratti E., Baralle F.E.. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013; 41:5062–5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Takahashi K., Yan I.K., Haga H., Patel T.. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014; 127:1585–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. et al.. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016; 29:653–668. [DOI] [PubMed] [Google Scholar]
- 89. O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N.. PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep. 2015; 11:474–485. [DOI] [PubMed] [Google Scholar]
- 90. Siu F.Y., Spanggord R.J., Doudna J.A.. SRP RNA provides the physiologically essential GTPase activation function in cotranslational protein targeting. RNA. 2007; 13:240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Lin C., Yang L.. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018; 28:287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lin A., Hu Q., Li C., Xing Z., Ma G., Wang C., Li J., Ye Y., Yao J., Liang K. et al.. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017; 19:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. McDonald A.G., Tipton K.F.. Elucidation of metabolic pathways from enzyme classification data. Methods Mol. Biol. 2014; 1083:173–186. [DOI] [PubMed] [Google Scholar]
- 94. Hentze M.W., Preiss T.. The REM phase of gene regulation. Trends Biochem. Sci. 2010; 35:423–426. [DOI] [PubMed] [Google Scholar]
- 95. Hentze M.W., Muckenthaler M.U., Galy B., Camaschella C.. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010; 142:24–38. [DOI] [PubMed] [Google Scholar]
- 96. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M.. Global quantification of mammalian gene expression control. Nature. 2011; 473:337–342. [DOI] [PubMed] [Google Scholar]
- 97. Caputi M., Zahler A.M.. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H'/F/2H9 family. J. Biol. Chem. 2001; 276:43850–43859. [DOI] [PubMed] [Google Scholar]
- 98. Jiang M., Zhang S., Yang Z., Lin H., Zhu J., Liu L., Wang W., Liu S., Liu W., Ma Y. et al.. Self-recognition of an inducible host lncRNA by RIG-I feedback restricts innate immune response. Cell. 2018; 173:906–919. [DOI] [PubMed] [Google Scholar]
- 99. Xue Z., Hennelly S., Doyle B., Gulati A.A., Novikova I.V., Sanbonmatsu K.Y., Boyer L.A.. A G-rich motif in the lncRNA braveheart interacts with a zinc-finger transcription factor to specify the cardiovascular lineage. Mol. Cell. 2016; 64:37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wang X., Goodrich K.J., Gooding A.R., Naeem H., Archer S., Paucek R.D., Youmans D.T., Cech T.R., Davidovich C.. Targeting of polycomb repressive complex 2 to RNA by short repeats of consecutive guanines. Mol. Cell. 2017; 65:1056–1067. [DOI] [PubMed] [Google Scholar]
- 101. Barreau C., Paillard L., Osborne H.B.. AU-rich elements and associated factors: are there unifying principles. Nucleic Acids Res. 2005; 33:7138–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nagy E., Rigby W.F.. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold). J. Biol. Chem. 1995; 270:2755–2763. [DOI] [PubMed] [Google Scholar]
- 103. Chang C.H., Curtis J.D., Maggi L.B. Jr, Faubert B., Villarino A.V., O'Sullivan D., Huang S.C., van der Windt G.J., Blagih J., Qiu J. et al.. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013; 153:1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for LIN28A CLIP-seq data are GEO: GSM910955, GSM910956, GSM910957 (56). The accession number for mESCs RNA-seq is GSM1412826 (57). IDH1 FBioCLIP-seq data has been deposited in the GEO repository with the accession number GSE119798.
IDH1 FBioCLIP-seq data has been deposited in the GEO repository with the accession number GSE119798 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE119798). The accession numbers for LIN28A CLIP-seq data are GEO: GSM910955, GSM910956, GSM910957 (56). The accession number for mESCs RNA-seq is GSM1412826 (57).