Abstract
Nucleic acid mimics of fluorescent proteins can be valuable tools to locate and image functional biomolecules in cells. Stacking between the internal G-quartet, formed in the mimics, and the exogenous fluorophore probes constitutes the basis for fluorescence emission. The precision of recognition depends upon probes selectively targeting the specific G-quadruplex in the mimics. However, the design of probes recognizing a G-quadruplex with high selectivity in vitro and in vivo remains a challenge. Through structure-based screening and optimization, we identified a light-up fluorescent probe, 9CI that selectively recognizes c-MYC Pu22 G-quadruplex both in vitro and ex vivo. Upon binding, the biocompatible probe emits both blue and green fluorescence with the excitation at 405 nm. With 9CI and c-MYC Pu22 G-quadruplex complex as the fluorescent response core, a DNA mimic of fluorescent proteins was constructed, which succeeded in locating a functional aptamer on the cellular periphery. The recognition mechanism analysis suggested the high selectivity and strong fluorescence response was attributed to the entire recognition process consisting of the kinetic match, dynamic interaction, and the final stacking. This study implies both the single stacking state and the dynamic recognition process are crucial for designing fluorescent probes or ligands with high selectivity for a specific G-quadruplex structure.
INTRODUCTION
G-quadruplex structures have been demonstrated to play important roles in mediating biological processes including functioning as diagnostic tools (1–7). However, the lack of ligands or fluorescent probes selectively targeting a specific G-quadruplex topology limits their unambiguous identification in live cells (8–10). Endogenic 4-hydroxybenzlidene imidazolinone (HBI) formed from three residues in the nascent green fluorescent proteins (GFP), work as essential visualizing tools that locates proteins and monitors biological process in living cells and organisms (11–13). As intrinsically fluorescent RNA is not known, an RNA aptamer ‘Spinach’ was identified as mimics of GFP with HBI derivatives as the exogenous fluorophores (14–16). Crystal structures demonstrated that it was the two-layer non-canonical G-quadruplex and an adjacent base-triplet in the RNA aptamer that served as a pocket to accommodate HBI derivatives, and restrain their vibration (17,18). Similar restraint was assumed to occur in DNA mimics of red fluorescent proteins and other RNA fluogenic light-up aptamers, such as Mango and Corn (19,20). The stacking between the G-quartet surface and the fluorophores constitutes the basis for the fluorescent property of these RNA\DNA mimics of FPs, which provide tools for genetic encoding of fluorescent nucleic acids and for tracking bio-molecules in cells (17,18,21,22). However, precision of these nucleic acid mimics of FPs on locating target biological molecules in cells is affected by multiple G-quadruplexes that can form physiologically (23,24). They share a common G-quartet surface, which has a potential to interact with the exogenous analogs of HBI (25–30). To improve precision, it is essential to develop methods for designing probes that recognize a specific G-quadruplex forming oligonucleotide with high selectivity and specificity in vitro and in vivo (9,10,31–34).
G-quadruplexes (G4) are four-stranded structures formed by guanine-rich DNA or RNA sequences, in which four guanines are assembled in a square co-planar arrangement by Hoogsteen hydrogen bonding to form a G-quartet (26–30). The G-quartets stack on top of one another to form a G-quadruplex. G-quadruplex-forming sequences are widely distributed in eukaryotic telomeres (35,36), and the promoter regions of genes such as c-MYC (37–39), c-KIT (40,41), k-RAS (42,43), BCL2 (44,45) and VEGF (46,47). G-quadruplex structures can be classified into various groups according to the orientation of the nucleic acid strands, such as parallel, antiparallel or hybrids thereof, and these structures share a common G-quartet. In the reported G-quadruplex–ligand complex structures in PDB, most of ligands interact with G-quadruplex via G-quartet stacking (30,48). Some compounds display light-up fluorescence property when interacting with G-quadruplex, and part of them exhibit selectivity to distinguish G-quadruplexes from the DNA duplex structure, which is derived from the π-π stacking between the planar aromatic core of the compounds and the common G-quartet surface (49,50). However, generic G-quartet stacking results in poor selectivity between G-quadruplex structures. Interaction with the flanking loops around the G-quartet enhances the binding of the stacked ligands (51). The adjacent grooves extend the binding pockets, and dynamic movement between G-quadruplex and the ligands explore more intermediate states (52,53). These could provide more diversified space for designing ligands with selectivity for a specific G-quadruplex structure.
The design of probes that recognizes a G-quadruplex topology with high selectivity and specificity in vitro and in vivo remains a challenge. In this study, we approach this issue by identifying a fluorescent probe that selectively recognizes c-MYC Pu22 G-quadruplex structure with the sequence (5′-TGAGGGTGGGTAGGGTGGGTAA-3′). c-MYC oncogene is overexpressed in some genetic aberrant solid tumors (54,55). The G-rich strand in its promoter region was assumed to mediate transcription through the formation of G-quadruplex structures. c-MYC Pu22 G-quadruplex forming sequence is modified from the wild-type sequence with two base mutations from G to T (Supplementary Table S1). This G-quadruplex adopts a propeller-type parallel conformation with 3′ and 5′ flanking sequences stacking on the top and the bottom end of the G-quadruplex structure, respectively (PDB code 1xav, Figure 2) (52). The complex structure between this G-quadruplex and quindoline (PDB code 2l7v) confirmed the stability of the G-quartet core and explored possible binding pockets for ligands or probes recognizing c-MYC Pu22 G-quadruplex (53). Generally, light-up fluorescent probes for biomolecules consist of a recognition group and a fluorescent signaling unit linked via rotatable bonds (32,56–58). The intra-molecular vibration of the fluorescent probes in their free state disperses their energy at the excited state. Stacking with biomolecules immobilizes the coplanar chromophore and ensures the dominance of fluorescence emission in excited state energy release (56). In this study, virtual screening based on the pockets was carried out to identify fluorescent probes of c-MYC Pu22 G-quadruplex. Commercially available compounds with fluorophores were docked into the ligand binding cavities in the structures to judge their fitness, and in vitro fluorescence assay was employed to measure their fluorescent response. Using identified hits as templates for further optimization, compound 9CI (6-(2-(anthracen-9-ylmethylene) hydrazinyl)-N2,N4-diphenyl-1,3,5-triazine-2,4-diamine; Scheme 1) was identified to selectively recognize c-MYC Pu22 G-quadruplex over other G-quadruplex structures in in vitro assays and ex vivo experiments. A DNA mimic of fluorescent proteins with this oligonucleotide and 9CI as the fluorescent response core located functional aptamer SL1 on the periphery of A549 human lung carcinoma cells. The interacting mode and the binding process analysis between 9CI and c-MYC Pu22 G-quadruplex highlighted that the entire recognition process might consist of a kinetic match, dynamic interaction, and the final stacking. This recognition mechanism rationalizes the high selectivity of 9CI, and has been further supported by the relationship between the structures of analogous compounds of 9CI and their measured fluorescent intensity.
Figure 2.
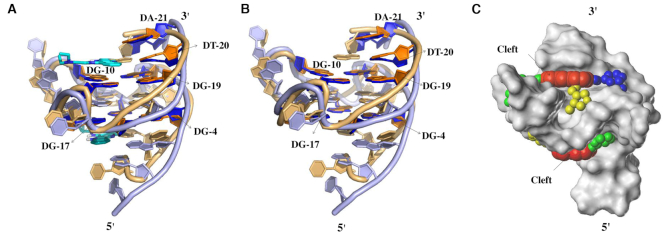
(A) Cartoon representation of superimposed c-MYC Pu22 G-quadruplex structures in apo state (brown, model 1 in the NMR structure, PDB 1xav) and with quindoline (purple, model 1 in the NMR structure, PDB 2l7v). The two quindoline molecules are shown in cyan. (B) Superimposed structures without ligands. (C) Connolly surface of c-MYC Pu22 G-quadruplex structure from PDB 2l7v. The red spheres highlight the central platform on top of 3′ end G-quartet and at the bottom of 5′ end G-quartet, while green, yellow and blue spheres represent the locations of grooves connecting to the central platform, respectively.
Scheme 1.
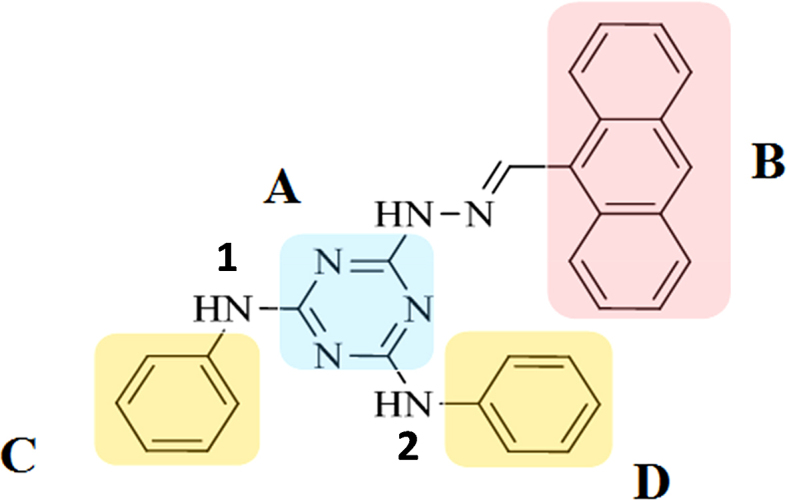
Structure of 6-(2-(anthracen-9-ylmethylene) hydrazinyl)-N2,N4-diphenyl-1,3,5-triazine-2,4-diamine (9CI) and its division into sub-structures. The triazine ring is labeled as A, and B represents the anthracene ring. C and D represent the phenyl rings. Nitrogen atoms connecting the triazine ring and the phenyl rings are labeled as 1 and 2, respectively.
MATERIALS AND METHODS
Materials
All oligonucleotides used in this study were purchased from Sangon (Shanghai, China). G-quadruplex forming oligonucleotides were annealed in the buffer (10 mM K2HPO4/KH2PO4 pH 7.0, 100 mM KCl) by heating to 95°C for 5 min, followed by gradual cooling to room temperature. Most compounds in this study were purchased from the Chinese national compound library of Peking University (PKU-CNCL), and compounds 5 (9Cl) and 6 were synthesized in-house (Scheme S1). During the synthesis, all chemical reagents were commercially available and treated with standard methods before use unless otherwise noted. Silica gel column chromatography (CC) (silica gel 200–300 mesh) was ordered from Qingdao Makall Group Co., Ltd, Qingdao, China. 1H NMR and 13C NMR spectra were recorded in DMSO-d6 on a 600 MHz Varian VNMRS Spectrometer and resonances were given in ppm relative to tetramethylsilane (TMS). The following abbreviations were used to designate chemical shift multiplicities: s = singlet, d = doublet, t = triplet, m = multiplet, br = broad. High-resolution mass spectra (HRMS) were acquired in positive mode on a MALDI SYNAPT G2 high-definition mass spectrometer (Waters, Milford, MA, USA).
Synthesis of 9Cl (compound 5) and compound 6
Compounds I (1.0 equivalent) and compound II (1.0 equivalent) (Scheme S1) were synthesized according to the literature (59,60). Both of them were dissolved in 10 ml anhydrous EtOH, and two drops of acetic acid were added to the mixture. The reaction was kept at 80°C for 7 h; and then, the solvent was removed under reduced pressure, and the residue was purified by silica column chromatography to get 9Cl (compound 5) and 6 as yellow solid.
For compound 5 (9Cl): 73% yields. 1H NMR (600 MHz, DMSO-d6): 11.22 (s, 1H), 9.44 (s, 3H), 8.80 (s, 1H), 8.70 (d, J = 6.0 Hz, 2H), 8.17 (d, J = 6.0 Hz, 2H), 7.95–7.90 (m, 4H), 7.65–7.59 (m, 4H), 7.28 (s, 4H), 7.00 (s, 2H). 13C NMR (150 MHz, DMSO-d6): 164.87, 164.71, 142.27, 140.60, 131.46, 129.96, 129.41, 128.85, 127.42, 125.98, 125.57, 122.38, 120.60, 120.52. HRMS cacld for [M+H]+: 482.2088, found: 482.2104.
For compound 6: 80% yields. 1H NMR (600 MHz, DMSO-d6): 11.17 (s, 1H), 9.47 (s, 2H), 8.62 (s, 1H), 8.58 (s, 1H), 8.45 (s, 1H), 8.18–8.10 (m, 6H), 7.90 (s, 3H), 7.56–7.54 (m, 2H), 7.34 (s, 4H), 7.03 (t, J = 7.2 Hz, 2H). 13C NMR (150 MHz, DMSO-d6): 164.77, 164.33, 143.34, 140.60, 133.00, 132.17, 132.15, 131.73, 131.54, 129.15, 128.95, 128.84, 128.62, 128.60, 127.16, 126.66, 126.47, 122.46, 122.41, 120.68. HRMS cacld for [M+H]+: 482.2088, found: 482.2149.
Fluorescence measurement
Fluorescence spectra were scanned by Spectrofluorophotometer RF-5301PC (Shimadzu, Japan) at room temperature. For fluorescence intensity measurement, 0.5 μM annealed DNA samples were mixed with compounds at 0.5 μM in the buffer (10 mM K2HPO4/KH2PO4 pH 7.0, 100 mM KCl), and the mixture was incubated for 10 min at 30°C before the spectra collection. Full UV absorbance profiles were scanned for compounds and mixture with DNA and compounds. The peaks appeared in the profile were used as the initial excitation wavelengths to obtain the emission spectra, in which, the maximum emission wavelength was determined. With the fixed maximum emission wavelengths, the peak wavelength in the excitation spectra was chosen as the final excitation wavelengths. For the fluorescence change of 9CI with the titration of DNA samples, small aliquots of annealed DNA oligonucleotide were added into the solution with 0.5 μM 9Cl in the buffer (10 mM K2HPO4/KH2PO4 pH 7.0, 100 mM KCl). The final concentration of DNA samples varied from 0 to 2 μM. The mixture with DNA and 9CI was stirred and allowed to equilibrate for 10 min at 30°C before the measurement. The emission between 420 and 580 nm was recorded at the excitation wavelength of 405 nm with 5 nm excitation slit width and 5 nm emission slit width.
The fluorescence variation of 9CI with the titration of c-MYC Pu 22 G-quadruplex was used to derive Ka value according to the method of Benesi–Hildebrand (61), using the following equation: 1/(ΔF) = 1/(b[DNA][H]0Ka)+1/(b[H]0). ΔF represents the change in the fluorescence intensity (ΔF = F –F0); F denotes the fluorescence intensity in the presence of different concentration of c-MYC DNA; F0 is the maximum fluorescence intensity of the free compound 9Cl. [DNA] is the total added c-MYC Pu 22 G-quadruplex forming oligonucleotide concentration, and [H]0 is the total concentration of compound 9Cl.
Gel electrophoresis
The native gel electrophoresis was performed on a 20% polyacrylamide gel in 1× TBE running buffer containing 100 mM KCl at room temperature. 8 μl annealed DNA (10 μM) in 1× loading buffer were loaded into the different wells except for c-MYC Pu22 (20 μM) in lane 3. The electrophoresis ran at 100 V. SYBR Gold was used to stain one gel, after being diluted ten thousand times with TBE (Tris–borate–EDTA) buffer. The second gel was obtained with DNA samples premixed with 20 μM 9CI before the electrophoresis. Bands were detected by Tanon-1600 trans-UV light. The UV wavelength used for image acquisition is 302 nm.
Cell cytotoxicity assay
The cell growth effect of 9CI against A549 (human lung carcinoma cell) were tested via measuring the quantity of living cells using the Cell Counting Kit-8 (CCK8) (DOJINDO, Japan) according to the instruction. A549 cells were cultured in Dulbecco's Modified Eagle Medium (Gibco) with 1% glutamine and 10% fetal bovine serum (FBS) for 24 h at 37°C. These cells were treated with 9CI at the indicated concentrations (2.5, 5, 10, 20 μM) for 24 h. Then, 100 μl of 10% CCK-8 solution was added into wells, and the cells were incubated for another 1 h at 37°C. The cytotoxicity was evaluated basing on the percentage of cell survival in a dose-dependent manner regard to the control, and the optical density (OD) detected at 450 nm by automated microplate reader (Bio-Rad) was used to calculate the survival rate. All the experiments were performed in parallel and in triplicate.
Confocal imaging of co-localization
A549 cells were cultured in Dulbecco's modified Eagle's medium (Gibco) with 1% glutamine and 10% fetal bovine serum (FBS) for 24 h at 37°C. 40 pmol annealed c-MYC Pu22, TTA and c-KIT1 G-quadruplex forming oligonucleotides labelled with 5′-Cy5 was transfected into cells by using lipofectamine 2000 (Thermo Fisher) and incubated at 37°C for 5 h. Then 5 μM 9CI was added and incubated for another 2 h. The digital images were recorded using a confocal laser scanning microscopy (Olympus FV1000-MP). The location of 9CI in the cells was investigated by the fluorescence signal collected between 425–470 nm/500–545 nm at λex = 405 nm, and the location of G-quadruplex was checked by fluorescence signal collected between 655 and 755 nm with the excitation at 635 nm.
Bio-imaging for the functional aptamers in live cells
A549 cells was incubated with SL1-M (Table 1) for 30 min at room temperature, and washed by 1 ml DPBS twice. Then, the cells were incubated with 500 nM MFPs (c-MYC Pu22-MFP/ TTA-MFP, Table 1) and 5 μM 9CI for 10 min. Fluorescence was collected at 425–470 and 500–545 nm emission channels after the cells were washed with DPBS three times again.
Table 1.
Sequences of the oligonucleotides used in imaging SL1

|
Isothermal titration calorimetry
This experiment was carried out with MicroCal iTC200 (Malvern Instruments, Great Britain) at 25°C. 400 μl 9CI (20 μM in 10 mM K2HPO4/KH2PO4 buffer with 0.2% DMSO, pH 7.0) was loaded into the sample cell and 100 μl c-MYC Pu22 G-quadruplex oligonucleotide at 150 μM in the same buffer was prepared for titration. For a typical titration, 2 μl of DNA solution was injected into the sample cell with 9CI at 150 s interval with a stirring speed of 1000 rpm. Each injection lasted for 5 s, and 20 injections constituted the whole titration process. The heat output per injection was obtained through integration, and corrected by subtracting the dilution heat, which was determined by injecting the DNA solution into the buffer under the identical experimental conditions. The Kb value was obtained through fitting the binding isotherms by Origin.
Circular dichroism
CD spectra were collected with MOS-500 spectrophotometer (bio-logic, France) at room temperature by using a quartz cuvette with a 1 mm path length. 10 μM DNA oligonucleotides and 20 μM 9CI dissolved in buffer (10 mM K2HPO4/KH2PO4, 100 mM KCl, pH 7.0) were used in the assay. The spectrum signals between 230 and 320 nm were scanned with 1 nm bandwidth at 1 nm/s.
Job plots
The stoichiometry between c-MYC Pu22 G-quadruplex and 9CI was assessed by independent experiments at various concentrations of G-quadruplex and 9CI, while the sum of their concentrations was kept constant at 2 μM. Samples containing the suitable amount of 9CI were mixed with the correspondent amount of annealed G-quadruplex, and the fluorescence signal was measured after 10 min incubation. The fluorescence intensities of the solutions at the emission wavelength 472 nm were plotted as the function of the input mole fractions of 9CI. The break point in the resulting plot corresponded to the mole fraction of 9CI in the complex.
Automated docking
Virtual screening for fluorescent probes was carried out by the combined usage of AutoDock Vina (62) and Autodock 4.0 (63). Compounds with fluorophores from the database PKU-CNCL were clustered according to Tanimoto similarity analysis (64), and the representative compounds with similarity coefficient smaller than 0.7 were chosen as the candidates for virtual screening. Three-dimensional coordinates of c-MYC Pu22 DNA G-quadruplex (PDB codes: 1xav and 2l7v) (52,53) were retrieved from PDB (www.rscb.org). The ligand in the structure was removed, and two potassium ions were placed in the central electronegative channel. The final coordinate was converted into the PDBQT format using the graphical user interface AutoDock Tools (ADT) (http://autodock.scripps.edu/resources/adt/index_html). The Gasteiger–Marsili partial charges were assigned to the selected representative compounds. A search box centered at the quality center of compound quindoline in the complex structure was set up with size_x = 30, size_y = 25, size_z = 23 to encompass the cleft and three grooves in the c-MYC Pu22 G-quadruplex structure. Ten separate docking runs were performed for each selected compound by AutoDock Vina, using the default search parameters. Compounds with docking scores from AutoDock Vina in the top 30% were re-docked using Autodock 4.0 for more accurate docking. Grid points of 80 × 66 × 60 with a 0.375 Å spacing, centered on quindoline structure were calculated for all the ligand atom types using AutoGrid 4.2. Each docking calculation with Autodock 4.0 consisted of 25 million energy evaluations and a maximum of 270 000 generations using the Lamarckian genetic algorithm local search method. A population size of 300 and 300 rounds of Solis and Wets local search were applied with a probability of 0.06. A mutation rate of 0.02 and a crossover rate of 0.8 were used to generate new docking trials for subsequent generations, and the best individual from each generation was propagated to the next generation. One hundred binding poses were predicted for each compound by Autodock 4.0, and were clustered with root-mean-square deviation (RMSD) cutoff value 2 Å. The predicted binding poses with the frequency of occurrence higher than 10/100 were visually inspected for their fitness with the binding pocket.
Molecular dynamics simulations
The stability of 9CI interacting with c-MYC Pu22 G-quadruplex was further assessed by MD simulations. The docked 9CI - c-MYC Pu22 G-quadruplex complex was set up using the xleap module in AMBER16 molecular dynamics package (http://ambermd.org/). The ligand parameter was generated using the GAFF2 force field implemented in the Antechamber program. The simulation was run using parmbsc1 (65) version of Cornell et al. force field with OL15 modifications (66–68). Two potassium ions were placed at the center of the G-tetrads. The complex was further neutralized by adding K+ ions. The system was solvated using Amber-adapted Joung and Chetham (69) parameters specific for TIP3P waters. The solvated box whose boundary extended at least 10 Å from any solute atom was used as the starting configuration for our simulation. The complex was first minimized using 1000 steps of steepest descent, followed by 1000 steps of conjugate gradient minimization. A 300 ps MD equilibration was run in which the quadruplex was restrained, while the ions and water were allowed to equilibrate. The system was gently heated from 0 to 300 K with a time step of 0.5 ps followed by another round at constant pressure at 300 K for 1 ns. The constraints were gradually relaxed until no constraints were applied on the systems. The final production MD run was carried out for 1000 ns using ACEMD (70) molecular dynamics engine. The default parameters were used with 10 Å non-bonded cut-off, and 0.0001 Å tolerance was allowed for the SHAKE algorithm, an integration step of 2 fs, constant pressure of 1 ATM at 300 K temperature. The frames were collected every 20 ps and the analysis of the trajectory was performed using the CPPTRAJ (71) module of AMBER. VMD (72) was used to visualize the trajectories, and the figures were made in the ICM-Pro software (www.molsoft.com) and PyMOL (www.pymol.org).
RESULTS
Structure-based virtual screening for identifying fluorescent probes
In the quindoline–c-MYC Pu22 G-quadrupelx complex structure (PDB code 2l7v), both 3′ and 5′ flanking sequences exhibit ligand-induced reorientation in the binding pockets. Pockets in the complex structure and in apo state (Figure 2) were used as potential binding sites. Consensus docking using a combination of Autodock Vina (62) and Autodock 4.0 (63) was carried out to screen for fluorescent probes. Compounds with fluorophores from the Chinese National Compound Library of Peking University (PKU-CNCL) were clustered through structural similarity analysis using Open Babel (64). The representative compounds were selected for virtual screening. The binding modes of the representative compounds in the clefts were first predicted by AutoDock Vina, which achieves an approximately two orders of magnitude speed-up compared with AutoDock 4.0. Compounds with docking scores ranked in the top 30% from Autodock Vina were selected and re-docked using Autodock 4.0 to predict their binding poses with greater accuracy. Binding poses with a cluster population size >10 (out of 100) were visually inspected for their fitness within the binding pockets. Twenty representative compounds were purchased and tested for their fluorescence response to DNA oligonucleotides that adopt different secondary structures.
Fluorescence intensity measurement in solution and structure optimization
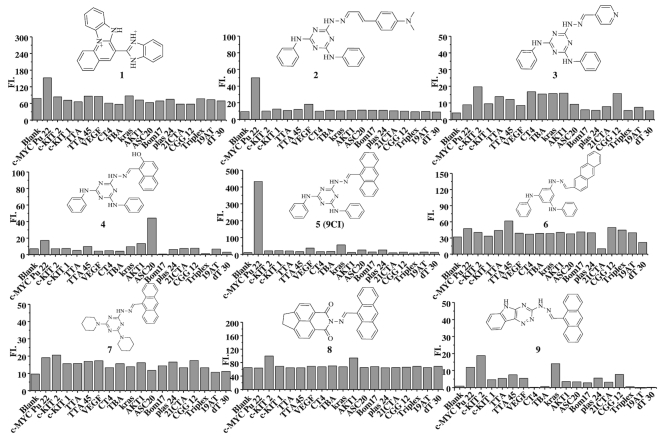
Eighteen DNA oligonucleotides (Supplementary Table S1) with the potential to form single strand, duplex, triplex, and G-quadruplexes were employed to check the fluorescence response of the selected compounds. Compound 1 and 2 exhibited enhanced fluorescence intensity at different exciting and emitting wavelengths (Figure 1, Supplementary Figure S3 and Table S2). In view of the selectivity of compound 2 for c-MYC Pu22 G-quadruplex, it was chosen as the template to further search for compounds with higher selectivity from the database. New ligands with groups common in compound 2 or with bioisosteres to them were purchased and synthesized (Figure 1, Supplementary Figure S1, Schemes S1 and Figures S4 and S5), and their fluorescence response to the eighteen oligonucleotides was measured (Figure 1 and Supplementary Figure S1).
Figure 1.
Structures of compounds 1–9 and their fluorescence response to eighteen DNA oligonucleotides. The excitation and emission wavelengths of these compounds are listed in Supplementary Table S2.
In vitro fluorescence selectivity of 9CI
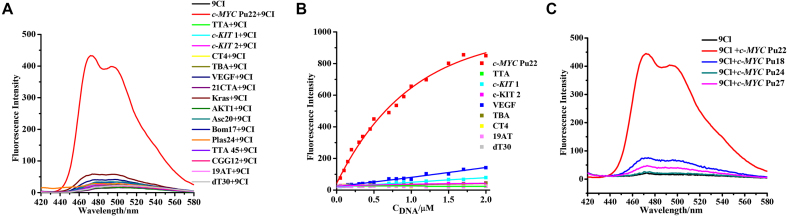
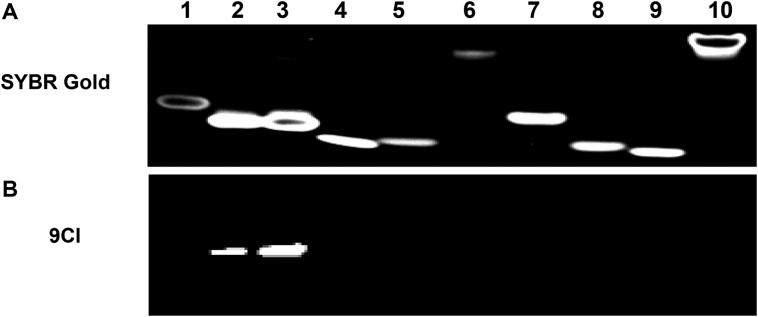
9CI (compound 5), with anthracene ring replacing 4-ethenyl-N,N-dimethylaniline in compound 2, exhibited high fluorescence response and selectivity against c-MYC Pu 22 G-quadruplex (Figure 3). The absorbance spectrum of 9CI displayed two vibronic bands centered at 263 and 405 nm (Supplementary Figure S6A). To avoid overlap with the nucleic acid absorbance, the fluorescence spectra were scanned with the excitation wavelength at 405 nm. Compound 9CI alone in buffer displayed extremely negligible emission at 472 nm. With gradual titration of c-MYC Pu22 G-quadruplex DNA (up to 2 μM), the fluorescence emission intensity at 472 and 503 nm exhibited a dramatic increase (Figure 3B), even without a distinct shift in absorption (Supplementary Figure S6B). In contrast, non-significant fluorescence enhancements were observed when 9CI was mixed or titrated with other DNA sequences, including antiparallel G-quadruplex (Bom17, Asc20, CT4, TBA, 21CTA), parallel G-quadruplex (c-KIT 1, c-KIT 2, Kras, VEGF, AKT1), mixed G-quadruplex (TTA, TTA 45, Plas24), trinucleotide (CGG12), duplex (19AT), single strand and triplex (Figure 1, Supplementary Table S1, Figure 3). 9CI did not yield significant fluorescent response towards the wild-type c-MYC G-quadruplex forming DNA oligonucleotides (Pu18, Pu24 and Pu27) (Supplementary Table S1, Figure 3C). Selectivity was further visualized by staining different DNA oligonucleotides with 9CI and SYBR Gold on a native gel (Figure 4). 0.5 and 1 equiv. of 9CI selectively stained c-MYC Pu22 G-quadruplex forming oligonucleotide and exhibited concentration dependence (lanes 2–3 in Figure 4). Meanwhile, the fluorescence intensity response of 9CI with c-MYC Pu22 G-quadruplex in solution could be observed by visualization under UV light radiation (Figure 5). The detection limit assay showed that 3 nM c-MYC Pu22 G-quadruplex forming DNA oligonucleotide was able to activate fluorescence emission of 9CI (Supplementary Figure S7). With the fluorescence response of quinine sulfate as the standard, the fluorescence quantum yields of 9Cl in the free and c-MYC Pu22-bound state were calculated as 0.017 and 0.221, respectively. The fluorescence quantum yield of 9Cl increased around 13 folds after the addition of c-MYC Pu22 DNA. No obvious fluorescence enhancement was observed for 9Cl with eight RNA G-quadruplex forming oligonucleotides (Supplementary Table S1, Figure S2).
Figure 3.
(A) Fluorescence emission spectra of 0.5 μM 9CI in the presence of various oligonucleotides (0.5 μM), λex = 405 nm. Fluorescence emission of compound 9CI alone is shown in black. 0.5 μM oligonucleotide samples were in 10 mM K2HPO4/KH2PO4 buffer solution with 100 mM KCl at pH 7.0. (B) Fluorescence intensity enhancement of 0.5 μM 9CI against concentration of different DNA oligonucleotides. (C) Fluorescence intensity of 9Cl in the presence of c-MYC Pu18, c-MYC Pu22, c-MYC Pu24 and c-MYC Pu27. Both 9Cl and DNA are 0.5 μM. λex = 405 nm, λem = 472 nm.
Figure 4.
Electrophoresis of DNA oligonucleotides on 20% non-denaturing acrylamide gel. (A) Gel stained by SYBR Gold after electrophoresis; (B) DNA samples were premixed with 20 μM 9CI before the electrophoresis. SYBR Gold used for staining was diluted ten thousand times from original stocking solution with TBE (Tris-borate-EDTA) buffer. 80 pmol DNAs were loaded into the different wells except for c-MYC Pu22 (160 pmol) in lane 3. Lane 1: DNA marker with 22 nucleotides; lane 2: c-MYC Pu22 (80 pmol); lane 3: c-MYC Pu22 (160 pmol); lane 4: TTA; lane 5: c-KIT1; lane 6: c-KIT2; lane 7: VEGF; lane 8: TBA; lane 9: CT4; lane 10: 19AT.
Figure 5.
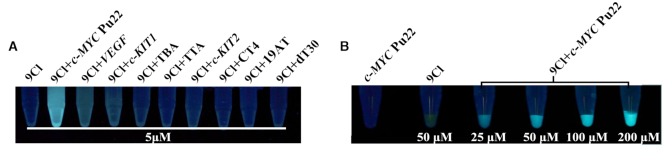
Visualization of 9CI mixed with DNA oligonucleotides under UV light. (A) 5 μM 9CI and 5 μM DNA samples in 10 mM KH2PO4/K2HPO4 buffer with 100 mM KCl at pH 7.0. (B) Lane 1: 200 μM c-MYC Pu22 DNA; lane 2: 50 μM 9CI; lane 3: 25 μM c-MYC Pu22 DNA + 25 μM 9CI; lane 4: 50 μM c-MYC Pu22 DNA + 50 μM 9CI; lane 5: 100 μM c-MYC Pu22 DNA + 100 μM 9CI; lane 6: 200 μM c-MYC Pu22 DNA + 200 μM 9CI.
Selective recognition of 9CI with c-MYC Pu22 G-quadruplex ex vivo
Co-localization of c-MYC Pu22 G-quadruplex and 9CI is an intuitive way to determine whether 9CI is able to recognize c-MYC Pu22 G-quadruplex in live cells. Negligible cytotoxic effect was observed for 9CI over 24 h incubation with cells (Supplementary Figure S8), and the G4-activated fluorescence of 9CI was barely influenced by the presence of cell lysates (Supplementary Figure S9). This implies its reliability and biocompatibility for bio-imaging in cells. Confocal laser scanning microscopy was employed to check their distribution in cells. 40 pmol c-MYC Pu22 DNA G-quadruplex-forming oligonucleotide labelled with 5′-Cy5 was transfected into A549 cells by using lipofectamine 2000. Negligible cell death was observed after 5 h incubation at 37°C, and the cells were further incubated with 5 μM 9CI for 2 h. Upon excitation of Cy5 at 635 nm, red fluorescence signals emitted from the cells explored the distribution of c-MYC Pu22 G-quadruplex in the cells (Figure 6A). Interaction with c-MYC Pu22 G-quadruplex enables 9CI to emit strong blue and green fluorescence signals in the cells at 405 nm (Figure 6B and C). Notably, most blue and green fluorescence signals (9CI) were co-localized with red fluorescence signals from the labelled c-MYC Pu22 G-quadruplex with Pearson correlation coefficient (PCC) 0.90, which indicated recognition between 9CI and c-MYC Pu22 G-quadruplex (Figure 6D). However, after 40 pmol labelled TTA (telomeric DNA sequence) or c-KIT1 G-quadruplex DNA was transfected into A549 cells with the addition of 9CI, almost no co-localization was observed between the TTA or c-KIT1 G-quadruplex structures and 9CI (Supplementary Figure S10 and S11). The recognition between 9CI and c-MYC Pu22 G-quadruplex was further proved by FRET (Förster resonance energy transfer) between 9CI and the Cy3 labelled c-MYC Pu22 G-quadruplex forming oligonucleotide in the cells (Supplementary Figure S12).
Figure 6.
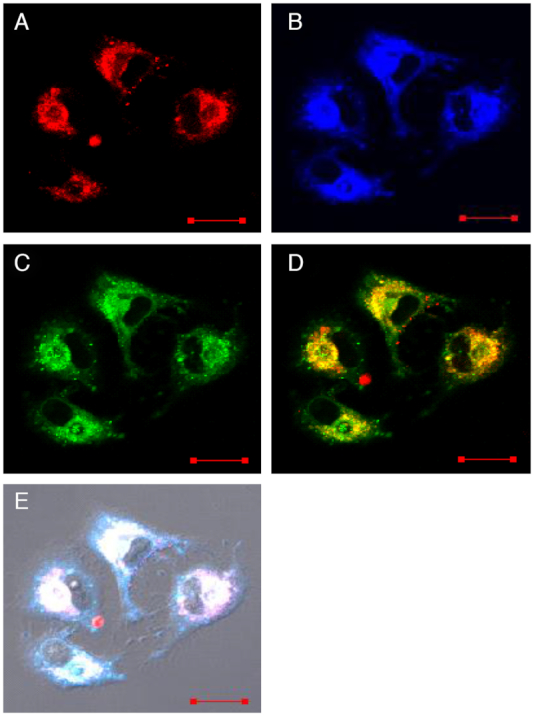
Fluorescence images of A549 cells transfected with 40 pmol c-MYC Pu22 DNA G-quadruplex forming oligonucleotide labelled with 5′-Cy5 using lipofectamine 2000 and then incubated with 5 μM 9Cl for 2 hours successively. (A) Fluorescence signal collected between 655–755 nm at λex = 635 nm, (B) fluorescence signal collected between 425–470 nm at λex = 405 nm, (C) fluorescence signal collected between 500 and 545 nm at λex = 405 nm, (D) merged image of (A) and (C), (E) merged images of (A), (B), (C) and the bright field. Scale bar, 20 μm.
Bio-imaging for the functional aptamer in live cells
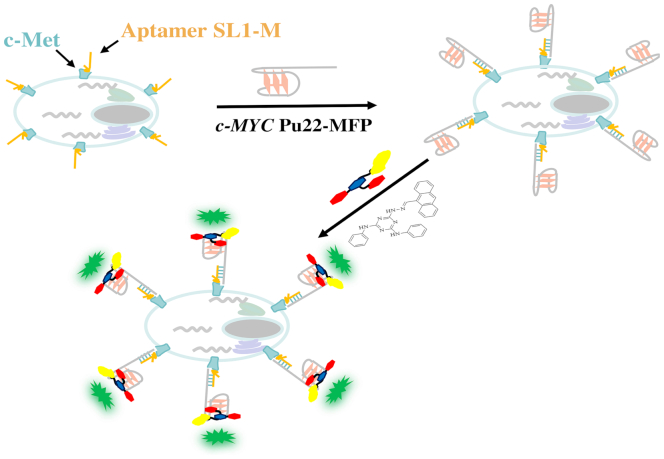
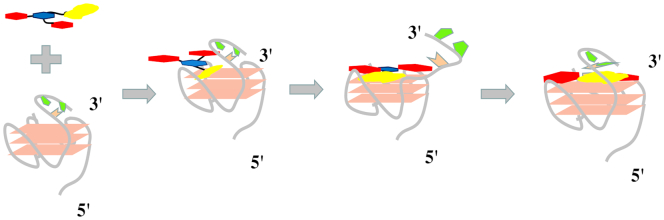
Compound 9CI displayed highly selective fluorescent recognition for c-MYC Pu22 G-quadruplex in vitro and in live cells. This encouraged us to exploit the 9CI - c-MYC Pu22 G-quadruplex complex as the fluorescent response core in a DNA mimic of fluorescent proteins to locate bio-functional molecules in cells, such as aptamers, and biomarkers. Aptamers are single-stranded DNA or RNA oligonucleotides with unique molecular recognition and binding capabilities specific to various targets, including small molecules, proteins, and live cells (73–75). The identification of their targeted cells and targets in live cell benefits the mechanism exploration and further optimization. Aptamer SL1 was found to affect the phosphorylation level in A549 and suppress cancer cell motility (76). To locate aptamer SL1 in cells, a fluorescent probe (c-MYC Pu22-MFP; Table 1) was constructed with two segments: a reporter strand bearing two repeated c-MYC Pu22 G-quadruplex sequences binding with 9CI and a matched recognition strand with the extended aptamer SL1-M (Table 1). The constructed fluorescent probe (c-MYC Pu22-MFP and 9CI) showed high fluorescent recognition for the extended SL1 (SL1-M) in vitro through hybridization (Supplementary Figure S13). At first, SL1-M was incubated with A549 cells in DPBS for 30 min at room temperature, following which c-MYC Pu22-MFP and 9CI were incubated to the cells sequentially (Figure 7). After the cells were washed with buffer three times, the bright blue ring and the green ring on the cellular periphery of A549 cells at 405 nm excitation suggested the aptamer SL1 might target the membrane receptors of A549 cells (Figure 8), whereas only weak fluorescence was observed in the control HepG2 cells. This is consistent with the reported mechanism that SL1 might target c-Met, a cell surface receptor for the hepatocyte growth factor in A549 cells (76). However, no signal was observed for SL1-M in control experiment carried out with a fluorescent probe (TTA2-MFP in Table 1) constructed with the TTA G-quadruplex forming oligonucleotide sequence replacing c-MYC Pu22 G-quadruplex forming oligonucleotide sequence (Figure 8). In order to highlight the application of locating aptamers in live cell, additional DNA oligonucleotides, based on AS1411, were constructed (Supplementary Table S3). Using the same protocol, a blue ring and a green ring was observed on the cellular periphery of A549 cells at 405 nm excitation (Supplementary Figure S14), suggesting aptamer AS1411 might target protein at the cell surface. This is consistent with the report that aptamer AS1411 could bind with nucleolin on the external surface of the tumor cells (77,78).
Figure 7.
Schematic illustration of locating aptamers in cells by DNA mimic of fluorescent proteins with the 9CI–c-MYC Pu22 G-quadruplex complex as the fluorescent response core.
Figure 8.
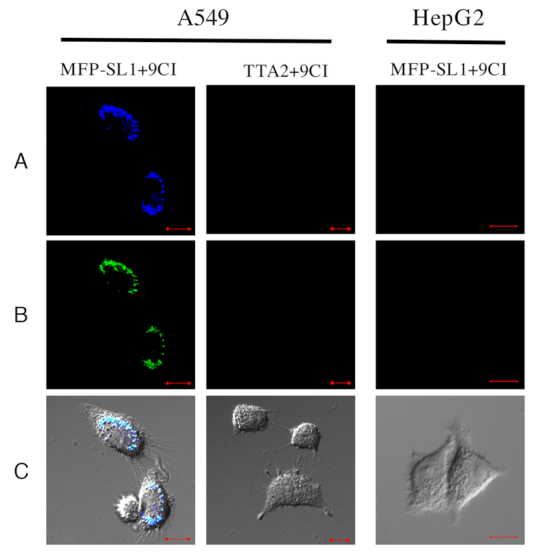
Confocal microscope images of the treated A549 cells and HepG2 cells at 405 nm excitation. A549 cells was incubated with SL1-M for 30 min at room temperature, and washed by 1 ml DPBS twice. Then, the cells were incubated with 500 nM MFP (c-MYC Pu22-MFP/ TTA-MFP) and 5 μM 9CI for 10 min. Fluorescence was collected after the cells were washed with DPBS three times again. (A) Fluorescence collected at 425–470 nm emission channel, (B) fluorescence collected at 500–545 nm emission channel, (C) merged image of (A) and (B) and bright field. Scale bar, 10 μm.
Binding site analysis
Compound 9CI exhibited high selectivity for c-MYC Pu22 G-quadruplex in in vitro and ex vivo and the constructed probe succeeded in locating aptamer SL1 at the periphery of A549 cells. These implied its potential on improving the precision of nucleic acid mimics of fluorescent proteins to track biomolecules in cells. Dissection of the recognition mechanism would provide important clues for designing ligands or fluorescent probes for a specific G-quadruplex structure. Job plot analysis showed that 9CI bound with c-MYC Pu22 G-quadruplex with 1:1 stoichiometry (Supplementary Figure S15). ITC (Isothermal titration calorimetry) assay and fluorimetric titration showed that the dissociation constants were on the order of magnitude of 10−5 M−1 (Supplementary Figures S16 and S17), suggesting a relative weak interaction between 9CI and c-MYC Pu22 DNA G-quadruplex. CD (Circular Dichroism) spectra showed that the addition of 9CI did not change the topology of the c-MYC Pu22 DNA G-quadruplex and other DNA structures (Supplementary Figure S18).
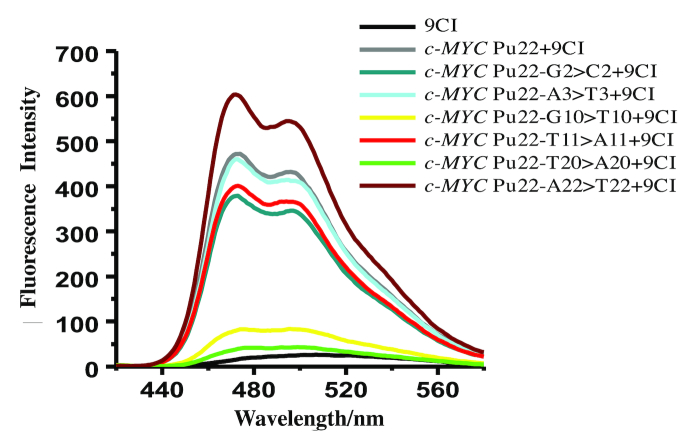
Fluorescence alterations upon recognition between 9CI and the mutant c-MYC Pu22 G-quadruplex sequences (Table 2) were analyzed to investigate the binding site of 9CI. The oligonucleotides mutated at the 5′ end loop (G2 > C2, A3 > T3), 3′ end loop (T20 > A20, A22 > T22), middle loop (T11 > A11), and 3′ end G-quartet (G10 > T10) were purchased from Sangon (Shanghai, China). The CD spectra showed that these mutated DNA oligonucleotides adopted the same topology as c-MYC Pu22 DNA G-quadruplex, and the addition of 9CI did not change their topology (Supplementary Figure S19). Upon the titration of 9CI, oligonucleotides with mutation at 5′ end (G2 > C2, A3 > T3) and the loop (T11 > A11) exhibited the same level of fluorescence response as the native oligonucleotide, suggesting that the binding site might be away from the 5′ end G-quartet and T11 loop (Figure 9). Surprisingly, the mutation of T for G10 in the 3′ end G-quartet did not change the G-quadruplex structure (Supplementary Figure S19). Instead, the weak fluorescence response (Figure 9) indicated that the mutation on G10 might prevent the interaction between 9CI and 3′ end G-quartet. The mutation from T20 to A20 resulted in negligible fluorescence response, and the oligonucleotide with mutation A22 > T22 exhibited the highest fluorescence enhancement to 9CI. Non-significant fluorescence change with mutation in the 5′ end loop and significant fluorescence variation with the mutation in the 3′ end G-quartet and the T20-A22 loop indicated that compound 9CI might be located between the T20-A22 loop and 3′ end G-quartet.
Table 2.
c-MYC Pu22 G-quadruplex sequence and the mutants in the binding site analysis
| Name | Sequence (5′-3′) |
|---|---|
| c-MYC Pu22 | TGAGGGTGGGTAGGGTGGGTAA |
| c-MYC Pu22-G2>C2 | TCAGGGTGGGTAGGGTGGGTAA |
| c-MYC Pu22-A3>T3 | TGTGGGTGGGTAGGGTGGGTAA |
| c-MYC Pu22-T11>A11 | TGAGGGTGGGAAGGGTGGGTAA |
| c-MYC Pu22-T20>A20 | TGAGGGTGGGTAGGGTGGGAAA |
| c-MYC Pu22-A22>T22 | TGAGGGTGGGTAGGGTGGGTAT |
| c-MYC Pu22-G10>T10 | TGAGGGTGGTTAGGGTGGGTAA |
Figure 9.
Fluorescence emission spectra of 0.5 μM 9CI with the addition of 0.5 μM c-MYC Pu22 DNA G-quadruplex oligonucleotide and the mutant oligonucleotides, λex = 405 nm. Fluorescence emission of 9CI alone is shown in black, and 0.5 μM oligonucleotides were prepared in 10 mM K2HPO4/KH2PO4, 100 mM KCl, pH 7.0.
Recognition mechanism explored by automated docking and molecular dynamics simulations.
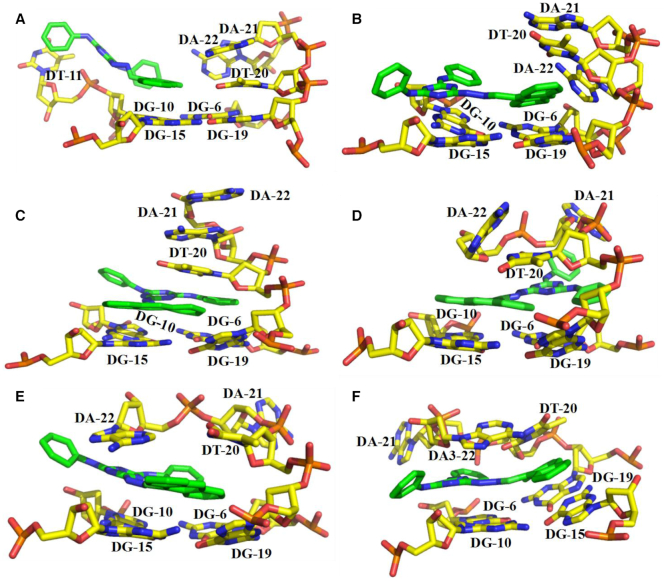
Fluorescence changes of 9CI with the mutant c-MYC Pu22 G-quadruplex oligonucleotides showed 3′ end cleft could be the recognition site. Exploring their recognition mode at the atomic level could provide detailed information for further structural modification. Automated docking coupled with molecular dynamics simulations were carried out to explore its recognition mechanism in the 3′ end cleft. 9CI was docked into the cleft at the 3′ end of c-MYC Pu22 G4-quadruplex structure (PDB ID: 2l7v) with 200 repeat runs using Autodock 4.0. The predicted binding conformation with the highest frequency of occurrence among 200 conformations was considered as the initial recognition mode. In this orientation (Supplementary Figure S20), the anthracene ring was found to stack on the top of G10 and G15 in 3′ end G-quartet surface; three nitrogen atoms in the triazine ring and nitrogen atoms N1 or N2 (Scheme 1) interacted with the phosphate oxygen atoms of T11 via electrostatic interaction and the phenyl ring stacked with the thymine of T11. In this case, the initial interaction between 9CI and the G-quadruplex structure constituted the basis for the dynamic recognition model.
One μsec molecular dynamics simulation (Supplementary Figure S21) was carried out to check the dynamic recognition between 9CI and c-MYC Pu22 G-quadruplex. In the initial contact, the triazine ring was attracted to the phosphate oxygen atoms through electrostatic interaction, while the anthracene ring of 9CI stacked on two guanines of the 3′ terminal end G-quartet (Figure 10A). As the anthracene ring approached the T20-A21-A22 loop, A22 moved away from the approaching ligand (Figure 10B). Further insertion of 9CI disrupted T20–A22 base pair, and forced the bases in loop T20–A21–A22 to stack (Figure 10C). In the following structural perturbations, 9CI and the T20–A21–A22 loop translated and rotated to obtain an optimal stacking mode. Finally, T20–A22 base pair reformed and the anthracene ring was sandwiched between T20–A22 base pair and G15–G19 on the 3′ end G-quartet (Figure 10F, Supplementary Figure S22). The triazine and the phenyl ring stacked on top of G6 (Figure 10F), while the bridge nitrogen atom interacted with the phosphate oxygen atom in T7 via electrostatic interaction (Supplementary Figure S22C). These stable interactions prevented intramolecular motions of 9CI, and ensured the dominance of emission pathway in excited state energy release. It has been confirmed by the remarkable fluorescence enhancement of 9Cl in the glycerol at different concentrations (Supplementary Figure S23).
Figure 10.
Dynamic recognition between 9CI and c-MYC Pu22 G-quadruplex explored by molecular dynamic simulations.
DISCUSSION
Selectivity for the cleft at the 3′ end
As a functional structural unit, G-quadruplex structures have been assumed to be anti-tumour or anti-viral targets (79,80). However, the design of ligands selectively targeting a specific G-quadruplex structure in cells still remains a challenge. Surprisingly, G-quadruplex structure was found to be a key component for the fluorescent response of exogenous fluorophores (HBI analogues) in some RNA/DNA mimics of fluorescent proteins. Due to the lack of selectivity for G-quadruplex structures in the mimics of FPs, the addition of the exogenous HBI analogues would result in enormous fluorescence noise by encountering hundreds of G-quadruplex in the genome. The noise can be reduced by identifying self-illuminating nucleic acid aptamers that generate endogenous fluorophores or an alternative approach is to discover probes that selectively recognize the specific G-quadruplex topology in the aptamer. However, majority of G-quadruplex ligands described thus far show little selectivity among different G-quadruplex structures. 9CI displays high selectivity for c-MYC Pu22 G-quadruplex structure over other G-quadruplex topologies. No obvious fluorescence enhancement with three wide-type c-MYC G-quadruplex forming DNA oligonucleotides (Pu18, Pu24 and Pu27) (Figure 3C) further suggested the potential of 9CI and c-MYC Pu22 G-quadruplex complex as the fluorescence response core on precisely locating target biological molecules in cells. CD spectra showed that addition of 9CI did not change the topologies of c-MYC Pu22 DNA G-quadruplex (Supplementary Figure S18). Fluorescence variance upon the recognition between 9CI and c-MYC Pu22 G-quadruplex forming sequences mutated in the loops at 3′ end, 5′ end or in the middle suggested that compound 9CI bound close to the T20-A22 loop and at the 3′-end G-quartet instead of 5′ end. The preference for only one terminal end G-quartet was further supported by Job plot analysis, which showed a 1:1 stoichiometry between 9CI and c-MYC Pu22 G-quadruplex.
Induced fit recognition mechanism supported by the relationship between the fluorescence response and the structures of 9CI analogues
Generally, the fluorescence response of G-quadruplex probes arises from the π stacking stabilization with G-quartet (32,81). The optimization of analogues was derived from the chemical skeleton of compound 2. Twenty three compounds with similar chemical groups in compound 2 or with bioisostere to them were measured for their fluorescence response to eighteen DNA oligonucleotides. The complicated relationship between these structures and their fluorescence response to c-MYC Pu22 G-quadruplex could not be explained by any single stacking state. However, some structure-fluorescence relationship could be rationalized.
Except for c-MYC Pu22 G-quadruplex, 9CI does not exhibit obvious fluorescence to other G-quadruplex structures (Figure 1). This suggests that stacking with the common G-quartet is not determinant for the fluorescence of 9CI.
Naphthalimides and benzoheterocycles groups stack well with the G-quartet (82,83). With these two fragments replacing the triazine ring and N-phenyl rings in compound 5 (9CI), compounds 8 and 9 were purchased and measured for their fluorescence response to different nucleic acid structures (Figure 1). Their negligible fluorescence response implies that the stacking between the anthracene ring in 9CI and the G-quartet is not crucial for the strong fluorescence response.
The anthracene ring in 9CI is assumed to be the fluorophore. Compounds 3–6 with different fluorophores were measured for their fluorescence response. Compared with 9CI, compounds 3 and 4 with small aromatic rings did not display obvious fluorescent signals with or without DNA oligonucleotides. This implies that fluorophores with big stacking surface are helpful. With the same structural components, compound 5 (9CI) and compound 6 with substitutions at different positions of the anthracene ring possess conjugated system at the similar size. However, completely different fluorescence response with c-MYC Pu22 G-quadruplex suggests that only big stacking surface with the G-quartet is not enough for the strong fluorescent response.
In spite of sharing >70% similarity with 9CI, compound 7 did not display fluorescence. This suggests that the N-phenyl ring in 9CI is important in the recognition process and cannot be replaced by the piperidine ring in compound 7.
Compounds 10–25 (Supplementary Figure S1) with random replacement for the N-phenyl, the triazine ring and the anthracene ring (parts A–D in Scheme 1) did not show obvious fluorescence response, suggesting that the cooperation of the rings play roles in the strong fluorescence response of 9CI.
Molecular dynamics simulations demonstrated that the recognition between 9CI and c-MYC G-quadruplex consisted of the kinetic match, dynamic recognition and the final stacking. The harmonious cooperation between the structural rearrangement of 3′ end loop and the systematic translocation of 9CI was critical for the induced fit (Figure 11). When N-phenyl is replaced by piperidine ring (compound 7), significant reduction in fluorescence could be due to the absence of the stacking between N-phenyl and T11 in the initial recognition. Weak fluorescence response of compounds 8 and 9 suggests the triazine ring and N-phenyl rings in 9CI play an important role in the initial fitting and the dynamic recognition before 9CI reaches the final stacked state. If compound 6 were able to reach the same final stacked state as 9CI, the anthracene ring could enable compound 6 to be a light-up fluorescent probe for G-quadruplex. However, insignificant fluorescence enhancement was observed for compound 6, suggesting that the relative orientation among the anthracene ring, the triazine ring and the phenyl rings might interfere with the initial contact and prevent further dynamic recognition. Finally, the stronger fluorescence response of 9CI for c-MYC Pu22 G-quadruplex over other G-quadruplexes highlights the importance of A20–T22 loop in the dynamic recognition and the sandwich interaction. The fluorescence intensity variance with different bioisoterisms of 9CI reflects the roles of different structural components play in the recognition process.
Figure 11.
Schematic illustration for the induced fitting between 9CI and c-MYC Pu22 G-quadruplex.
CONCLUSION
As functional molecular structures, G-quadruplexes have attracted wide attention from different areas. However, it has been challenging to find a ligand or a fluorescent probe targeting a specific G-quadruplex structure. In this study, we identified compound 9CI exhibiting selective and strong light-up fluorescence response to c-MYC Pu22 G-quadruplex over other G-quadruplex structures. 9CI exhibited good biocompatibility without inhibition in some tumor cells (9CI at 20 μM), fungi (9CI at 50 ppm), and insects (9CI at 1000 ppm). The fluorescent probe constructed with 9CI and c-MYC Pu22 G-quadruplex forming oligonucleotides as the fluorescent response unit succeeded in locating functional aptamer SL1 on the cellular periphery of A549 cells. Experimental and computational analysis demonstrated that: (a) the initial conformation match led to the structural interaction; (b) the high selectivity came from the dynamic rearrangement and (c) the thermodynamic stability of the complex determined the strong fluorescence response. This was further supported by the structure-fluorescence intensity relationship of the 9CI analogues. This work demonstrates that: (i) it is possible to discover fluorescence probes or ligands to recognize a specific G-quadruplex structure; (ii) besides the single recognition state, the dynamic recognition process could provide clues for designing fluorescent probes or ligands with high selectivity for a specific G-quadruplex structure; (iii) complex consisting of a specific G-quadruplex and its unique light-up fluorescent probe could work as the fluorescent response core to improve their precision on biomedical imaging and clinical diagnosis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Stephen Neidle (UCL School of Pharmacy, University College London) for helpful comments and suggestions. We also thank Dr Likai Du (College of informatics, Huazhong Agricultural University) for the discussion about the theory of the fluorescence, Qi Wang (College of science, Huazhong Agricultural University) for the measurement of the binding affinity, and Shaoyong Ke (National Biopesticide Engineering Research Center, Hubei Academy of Agricultural Science) for the toxicity assays of 9CI.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [21502060, 31672558, 21732002]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2015RC013, 2662017PY113, 2662015PY208]; Open fund of The State Key Laboratory of Bio-organic and Natural Products Chemistry, CAS [SKLBNPC16343]; Open fund of Beijing National Laboratory For Molecular Sciences; Fundamental Research Funds for the Central Universities [2662016PY122]; Natural Science Foundation of Hubei Province of China [2016CFB230]. Funding for open access charge: Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2015RC013].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. [DOI] [PubMed] [Google Scholar]
- 2. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balasubramanian S., Hurley L.H., Neidle S.. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy. Nat. Rev. Drug Discov. 2011; 10:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Murat P., Zhong J., Lekieffre L., Cowieson N.P., Clancy J.L., Preiss T., Balasubramanian S., Khanna R., Tellam J.. G-quadruplexes regulate Epstein-Barr virus-encoded nuclear antigen 1 mRNA translation. Nat. Chem. Biol. 2014; 10:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Platella C., Riccardi C., Montesarchio D., Roviello G.N., Musumeci D.. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta. Gen. Subj. 2017; 1861:1429–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yuan Q., Wu Y., Wang J., Lu D., Zhao Z., Liu T., Zhang X., Tan W.. Targeted bioimaging and photodynamic therapy nanoplatform using an aptamer-guided G-quadruplex DNA carrier and near-infrared light. Angew. Chem. Int. Ed. Engl. 2013; 52:13965–13969. [DOI] [PubMed] [Google Scholar]
- 7. Zhu J., Zhang L., Li T., Dong S., Wang E.. Enzyme-free unlabeled DNA logic circuits based on toehold-mediated strand displacement and split G-quadruplex enhanced fluorescence. Adv. Mater. 2013; 25:2440–2444. [DOI] [PubMed] [Google Scholar]
- 8. Ohnmacht S.A., Neidle S.. Small-molecule quadruplex-targeted drug discovery. Bioorg. Med. Chem. Lett. 2014; 24:2602–2612. [DOI] [PubMed] [Google Scholar]
- 9. Gu Y., Lin D., Tang Y., Fei X., Wang C., Zhang B., Zhou J.. A light-up probe targeting for Bcl-2 2345 G-quadruplex DNA with carbazole TO. Spectrochim. Acta A: Mol. Biomol. Spectrosc. 2018; 191:180–188. [DOI] [PubMed] [Google Scholar]
- 10. Bag S.S., Pradhan M.K., Talukdar S.. Tetrazolylpyrene unnatural nucleoside as a human telomeric multimeric G-quadruplex selective switch-on fluorescent sensor. Org. Biomol. Chem. 2017; 15:10145–10150. [DOI] [PubMed] [Google Scholar]
- 11. Yang F., Moss L.G., Phillips G.N. Jr. The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996; 14:1246–1251. [DOI] [PubMed] [Google Scholar]
- 12. Ohashi T., Kiehart D.P., Erickson H.P.. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baker S.S., Vidican C.B., Cameron D.S., Greib H.G., Jarocki C.C., Setaputri A.W., Spicuzza C.H., Burr A.A., Waqas M.A., Tolbert D.A.. An epifluorescent attachment improves whole-plant digital photography of Arabidopsis thaliana expressing red-shifted green fluorescent protein. AoB Plants. 2012; 2012:pls003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paige J.S., Wu K.Y., Jaffrey S.R.. RNA mimics of green fluorescent protein. Science. 2011; 333:642–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ageely E.A., Kartje Z.J., Rohilla K.J., Barkau C.L., Gagnon K.T.. Quadruplex-flanking stem structures modulate the stability and metal ion preferences of RNA mimics of GFP. ACS Chem. Biol. 2016; 11:2398–2406. [DOI] [PubMed] [Google Scholar]
- 16. Okuda M., Fourmy D., Yoshizawa S.. Use of baby spinach and broccoli for imaging of structured cellular RNAs. Nucleic Acids Res. 2017; 45:1404–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang H., Suslov N.B., Li N.S., Shelke S.A., Evans M.E., Koldobskaya Y., Rice P.A., Piccirilli J.A.. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014; 10:686–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Warner K.D., Chen M.C., Song W., Strack R.L., Thorn A., Jaffrey S.R., Ferre-D’Amare A.R.. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol. 2014; 21:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng G., Luo C., Yi H., Yuan L., Lin B., Luo X., Hu X., Wang H., Lei C., Nie Z et al.. DNA mimics of red fluorescent proteins (RFP) based on G-quadruplex-confined synthetic RFP chromophores. Nucleic Acids Res. 2017; 45:10380–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neubacher S., Hennig S.. RNA structure and cellular applications of fluorogenic light-up aptamers. Angew. Chem. 2019; 58:1266–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trachman R.J. III, Abdolahzadeh A., Andreoni A., Cojocaru R., Knutson J.R., Ryckelynck M., Unrau P.J., Ferre-D’Amare A.R.. Crystal structures of the mango-II RNA aptamer reveal heterogeneous fluorophore binding and guide engineering of variants with improved selectivity and brightness. Biochemistry. 2018; 57:3544–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Warner K.D., Sjekloca L., Song W., Filonov G.S., Jaffrey S.R., Ferre-D’Amare A.R.. A homodimer interface without base pairs in an RNA mimic of red fluorescent protein. Nat. Chem. Biol. 2017; 13:1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kwok C.K., Marsico G., Sahakyan A.B., Chambers V.S., Balasubramanian S.. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 2016; 13:841–844. [DOI] [PubMed] [Google Scholar]
- 24. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 25. Neidle S., Parkinson G.N.. Quadruplex DNA crystal structures and drug design. Biochimie. 2008; 90:1184–1196. [DOI] [PubMed] [Google Scholar]
- 26. Parkinson G.N., Lee M.P., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 27. Haider S., Parkinson G.N., Neidle S.. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002; 320:189–200. [DOI] [PubMed] [Google Scholar]
- 28. Martadinata H., Phan A.T.. Structure of propeller-type parallel-stranded RNA G-quadruplexes, formed by human telomeric RNA sequences in K+ solution. J. Am. Chem. Soc. 2009; 131:2570–2578. [DOI] [PubMed] [Google Scholar]
- 29. Collie G.W., Haider S.M., Neidle S., Parkinson G.N.. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010; 38:5569–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Campbell N., Collie G.W., Neidle S.. Crystallography of DNA and RNA G-quadruplex nucleic acids and their ligand complexes. Curr.Protoc.Nucleic Acid Chem. 2012; doi:10.1002/0471142700.nc1706s50. [DOI] [PubMed] [Google Scholar]
- 31. Zhao D., Dong X., Jiang N., Zhang D., Liu C.. Selective recognition of parallel and anti-parallel thrombin-binding aptamer G-quadruplexes by different fluorescent dyes. Nucleic Acids Res. 2014; 42:11612–11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen S.-B., Wu W.-B., Hu M.-H., Ou T.-M., Gu L.-Q., Tan J.-H., Huang Z.-S.. Discovery of a new fluorescent light-up probe specific to parallel G-quadruplexes. Chem. Commun. 2014; 50:12173–12176. [DOI] [PubMed] [Google Scholar]
- 33. Laguerre A., Stefan L., Larrouy M., Genest D., Novotna J., Pirrotta M., Monchaud D.. A twice-as-smart synthetic G-quartet: PyroTASQ is both a smart quadruplex ligand and a smart fluorescent probe. J. Am. Chem. Soc. 2014; 136:12406–12414. [DOI] [PubMed] [Google Scholar]
- 34. Panda D., Saha P., Das T., Dash J.. Target guided synthesis using DNA nano-templates for selectively assembling a G-quadruplex binding c-MYC inhibitor. Nat. Commun. 2017; 8:16103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Neidle S., Parkinson G.N.. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003; 13:275–283. [DOI] [PubMed] [Google Scholar]
- 36. Bao H.-L., Ishizuka T., Sakamoto T., Fujimoto K., Uechi T., Kenmochi N., Xu Y.. Characterization of human telomere RNA G-quadruplex structures in vitro and in living cells using F-19 NMR spectroscopy. Nucleic Acids Res. 2017; 45:5501–5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siddiqui-Jain A., Grand C.L., Bearss D.J., Hurley L.H.. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boddupally P.V.L., Hahn S., Beman C., De B., Brooks T.A., Gokhale V., Hurley L.H.. Anticancer activity and cellular repression of c-MYC by the G-quadruplex-stabilizing 11-piperazinylquindoline is not dependent on direct targeting of the G-quadruplex in the c-MYC promoter. J. Med. Chem. 2012; 55:6076–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kaiser C.E., Gokhale V., Yang D., Hurley L.H.. Chaires JB, Graves D. Quadruplex Nucleic Acids. 2013; 330:1–21. [Google Scholar]
- 40. Rankin S., Reszka A.P., Huppert J., Zloh M., Parkinson G.N., Todd A.K., Ladame S., Balasubramanian S., Neidle S.. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005; 127:10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rigo R., Dean W.L., Gray R.D., Chaires J.B., Sissi C.. Conformational profiling of a G-rich sequence within the c-KIT promoter. Nucleic Acids Res. 2017; 45:13056–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cogoi S., Ferino A., Miglietta G., Pedersen E.B., Xodo L.E.. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018; 46:661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kaiser C.E., Van Ert N.A., Agrawal P., Chawla R., Yang D., Hurley L.H.. Insight into the complexity of the i-motif and G-quadruplex DNA structures formed in the KRAS promoter and subsequent drug induced gene repression. J. Am. Chem. Soc. 2017; 139:8522–8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onel B., Carver M., Wu G., Timonina D., Kalarn S., Larriva M., Yang D.. A new G-quadruplex with hairpin loop immediately upstream of the human BCL2 P1 promoter modulates transcription. J. Am. Chem. Soc. 2016; 138:2563–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Onyshchenko M.I., Gaynutdinov T.I., Englund E.A., Appella D.H., Neumann R.D., Panyutin I.G.. Quadruplex formation is necessary for stable PNA invasion into duplex DNA of BCL2 promoter region. Nucleic Acids Res. 2011; 39:7114–7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salvati E., Zizza P., Rizzo A., Iachettini S., Cingolani C., D’Angelo C., Porru M., Randazzo A., Pagano B., Novellino E et al.. Evidence for G-quadruplex in the promoter of vegfr-2 and its targeting to inhibit tumor angiogenesis. Nucleic Acids Res. 2014; 42:2945–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agrawal P., Hatzakis E., Guo K., Carver M., Yang D.. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013; 41:10584–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Haider S.M., Neidle S., Parkinson G.N.. A structural analysis of G-quadruplex/ligand interactions. Biochimie. 2011; 93:1239–1251. [DOI] [PubMed] [Google Scholar]
- 49. Kotar A., Wang B., Shivalingam A., Gonzalez-Garcia J., Vilar R., Plavec J.. NMR structure of a triangulenium-based long-lived fluorescence probe bound to a G-quadruplex. Angew. Chem. 2016; 55:12508–12511. [DOI] [PubMed] [Google Scholar]
- 50. Jin B., Zhang X., Zheng W., Liu X., Qi C., Wang F., Shangguan D.. Fluorescence light-up probe for parallel G-quadruplexes. Anal. Chem. 2014; 86:943–952. [DOI] [PubMed] [Google Scholar]
- 51. Dutta D., Debnath M., Mueller D., Paul R., Das T., Bessi I., Schwalbe H., Dash J.. Cell penetrating thiazole peptides inhibit c-MYC expression via site-specific targeting of c-MYC G-quadruplex. Nucleic Acids Res. 2018; 46:5355–5365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ambrus A., Chen D., Dai J.X., Jones R.A., Yang D.Z.. Solution structure of the biologically relevant G-quadruplex element in the human c-MYC promoter. implications for G-quadruplex stabilization. Biochemistry. 2005; 44:2048–2058. [DOI] [PubMed] [Google Scholar]
- 53. Dai J., Carver M., Hurley L.H., Yang D.. Solution structure of a 2:1 quindoline-c-MYC G-quadruplex: insights into G-quadruplex-interactive small molecule drug design. J. Am. Chem. Soc. 2011; 133:17673–17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W.. Identification of c-MYC as a target of the APC pathway. Science. 1998; 281:1509–1512. [DOI] [PubMed] [Google Scholar]
- 55. Baudino T.A., McKay C., Pendeville-Samain H., Nilsson J.A., Maclean K.H., White E.L., Davis A.C., Ihle J.N., Cleveland J.L.. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002; 16:2530–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu Y.-J., Yan S.-C., Chan F.-Y., Zou L., Chung W.-H., Wong W.-L., Qiu B., Sun N., Chan P.-H., Huang Z.-S et al.. Benzothiazole-substituted benzofuroquinolinium dye: a selective switch-on fluorescent probe for G-quadruplex. Chem. Commun. 2011; 47:4971–4973. [DOI] [PubMed] [Google Scholar]
- 57. Lin D., Fei X., Gu Y., Wang C., Tang Y., Li R., Zhou J.. A benzindole substituted carbazole cyanine dye: a novel targeting fluorescent probe for parallel c-myc G-quadruplexes. Analyst. 2015; 140:5772–5780. [DOI] [PubMed] [Google Scholar]
- 58. Xie X., Reznichenko O., Chaput L., Martin P., Teulade-Fichou M.-P., Granzhan A.. Topology-selective, fluorescent “Light-Up” probes for G-quadruplex DNA based on photoinduced electron transfer. Chem. Eur. J. 2018; 24:12638–12651. [DOI] [PubMed] [Google Scholar]
- 59. Dauselt J., Zhao J., Kind M., Binder R., Bashir A., Terfort A., Zharnikov M.. Compensation of the odd-even effects in araliphatic self-assembled monolayers by nonsymmetric attachment of the aromatic part. J. Phys. Chem. C. 2011; 115:2841–2854. [Google Scholar]
- 60. Dinari M., Gharahi F., Asadi P.. Synthesis, spectroscopic characterization, antimicrobial evaluation and molecular docking study of novel triazine-quinazolinone based hybrids. J. Mol. Struct. 2018; 1156:43–50. [Google Scholar]
- 61. Novikov E., Stobiecka A., Boens N.. Analysis of fluorometric titration curves. J. Phys. Chem. A. 2000; 104:5388–5395. [DOI] [PubMed] [Google Scholar]
- 62. Trott O., Olson A.J.. Software news and update AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010; 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J.. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998; 19:1639–1662. [Google Scholar]
- 64. O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R.. Open babel: an open chemical toolbox. J. Cheminform. 2011; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ivani I., Dans P.D., Noy A., Perez A., Faustino I., Hospital A., Walther J., Andrio P., Goni R., Balaceanu A et al.. Parmbsc1: a refined force field for DNA simulations. Nat. Methods. 2016; 13:55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Krepl M., Zgarbova M., Stadlbauer P., Otyepka M., Banas P., Koca J., Cheatham T.E. III, Jurecka P., Sponer J.. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA, and Z-DNA. J. Chem. Theory Comput. 2012; 8:2506–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zgarbova M., Javier Luque F., Sponer J., Cheatham T.E. III, Otyepka M., Jurecka P.. Toward improved description of DNA backbone: revisiting epsilon and zeta torsion force field parameters. J. Chem. Theory Comput. 2013; 9:2339–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zgarbova M., Sponer J., Otyepka M., Cheatham T.E. III, Galindo-Murillo R., Jurecka P.. Refinement of the sugar-phosphate backbone torsion beta for AMBER force fields improves the description of Z- and B-DNA. J. Chem. Theory Comput. 2015; 11:5723–5736. [DOI] [PubMed] [Google Scholar]
- 69. Joung I.S., Cheatham T.E. III.. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008; 112:9020–9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Harvey M.J., Giupponi G., De Fabritiis G.. ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 2009; 5:1632–1639. [DOI] [PubMed] [Google Scholar]
- 71. Roe D.R., Cheatham T.E. III.. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013; 9:3084–3095. [DOI] [PubMed] [Google Scholar]
- 72. Humphrey W., Dalke A., Schulten K.. VMD: visual molecular dynamics. J. Mol. Graph. 1996; 14:33–38. [DOI] [PubMed] [Google Scholar]
- 73. Zhou J., Rossi J.. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017; 16:181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ma H., Liu J., Ali M.M., Mahmood M.A.I., Labanieh L., Lu M., Iqbal S.M., Zhang Q., Zhao W., Wan Y.. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev. 2015; 44:1240–1256. [DOI] [PubMed] [Google Scholar]
- 75. Ozalp V.C., Kavruk M., Dilek O., Bayrac A.T.. Aptamers: molecular tools for medical diagnosis. Curr. Top. Med. Chem. 2015; 15:1125–1137. [DOI] [PubMed] [Google Scholar]
- 76. Ueki R., Sando S.. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun. 2014; 50:13131–13134. [DOI] [PubMed] [Google Scholar]
- 77. Soundararajan S., Chen W., Spicer E.K., Courtenay-Luck N., Fernandes D.J.. The nucleolin targeting aptamer AS1411 destabilizes bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008; 68:2358–2365. [DOI] [PubMed] [Google Scholar]
- 78. Park J.Y., Cho Y.L., Chae J.R., Moon S.H., Cho W.G., Choi Y.J., Lee S.J., Kang W.J.. Gemcitabine-Incorporated G-quadruplex aptamer for targeted drug delivery into pancreas cancer. Mol. Ther.-Nucleic Acids. 2018; 12:543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010; 277:1118–1125. [DOI] [PubMed] [Google Scholar]
- 80. Metifiot M., Amrane S., Litvak S., Andreola M.-L.. G-quadruplexes in viruses: function and potential therapeutic applications. Nucleic Acids Res. 2014; 42:12352–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hu M.-H., Chen S.-B., Guo R.-J., Ou T.-M., Huang Z.-S., Tan J.-H.. Development of a highly sensitive fluorescent light-up probe for G-quadruplexes. Analyst. 2015; 140:4616–4625. [DOI] [PubMed] [Google Scholar]
- 82. Yildiz U., Coban B.. Novel naphthalimide derivatives as selective G-quadruplex DNA binders. Appl. Biochem. Biotechnol. 2018; 186:547–562. [DOI] [PubMed] [Google Scholar]
- 83. Li Q., Xiang J.-F., Yang Q.-F., Sun H.-X., Guan A.-J., Tang Y.-L.. G4LDB: a database for discovering and studying G-quadruplex ligands. Nucleic Acids Res. 2013; 41:D1115–D1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.