Graphical abstract

Abbreviations: CAMKK2, calcium/calmodulin-dependent protein kinase kinase 2; CDK, cyclin-dependent kinase; IC50, 50% inhibition concentration; PBD, Polo-box domain; PC, Polo-box cap; LK, Polo-like kinase
Keywords: Cell cycle, Natural products, Neoplasms, PyRx, Targeted chemotherapy, Virtual drug screening
Highlights
-
•
We were able to identify a novel anticancer molecule that targets PLK1.
-
•
The compound shows good activity in leukaemia-sensitive and -resistant cells.
-
•
Compound (1) induces mitotic arrest and interferes with normal spindle formation.
-
•
We showed the importance of virtual screening for facilitating drug discovery.
-
•
Compound (1) is a good scaffold for further chemical development.
Abstract
Polo-like kinase 1 (PLK1), a member of the Polo-like kinase family, plays an important regulatory role in mitosis and cell cycle progression. PLK1 overexpression is correlated with tumourigenesis and poor prognosis in cancer patients. Therefore, the identification of novel compounds that inhibit PLK1 would provide attractive therapeutic approaches. Although some PLK1 kinase inhibitors have been developed, their application has been limited by off-target effects. PLK1 contains a regulatory domain named the Polo-box domain (PBD), which is characteristic only for the Polo-like kinase family. This domain represents an alternative therapeutic target with higher selectivity for PLK1. In this study, we applied in silico virtual drug screening, fluorescence polarization and microscale thermophoresis to identify new scaffolds targeting the PBD of PLK1. One compound, 3-{[(1R,9S)-3-(naphthalen-2-yl)-6-oxo-7,11-diazatricyclo[7.3.1.02,7]trideca-2,4-dien-11-yl]methyl}benzonitrile (designated compound (1)), out of a total of 30,793 natural product derivatives, inhibited the PLK1 PBD with high selectivity (IC50: 17.9 ± 0.5 µM). This compound inhibited the growth of cultured leukaemia cells (CCRF-CEM and CEM/ADR5000) and arrested the cell cycle in the G2/M phase, which is characteristic for PLK1 inhibitors. Immunofluorescence analyses showed that treatment with compound (1) disrupted spindle formation due to the aberrant localization of PLK1 during the mitotic process, leading to G2/M arrest and ultimately cell death. In conclusion, compound (1) is a selective PLK1 inhibitor that inhibits cancer cell growth. It represents a chemical scaffold for the future synthesis of new selective PLK1 inhibitors for cancer therapy.
Introduction
Polo-like kinase 1 (PLK1), a member of the Polo-like kinase (PLK) family, is an enzyme mainly involved in cell cycle progression [1], [2]. PLKs consist of an N-terminal Ser/Thr kinase domain and a C-terminal regulatory Polo-box domain (PBD), which is characteristic for this family of kinases [3]. The PBD is crucial for the ligand binding and subcellular localization of kinases [4]. The PBD regulates the kinase domain by inhibiting its activity in the absence of its ligand [5].
The expression of PLK1 is increased during mitosis [6]. Activation occurs through the protein Bora, which binds to the PBD and induces a conformational change in PLK1 after the Aurora A kinase has phosphorylated the Thr210 of PLK1. PLK1 promotes spindle formation and centrosome maturation [7]. Activated PLK1 positively regulates the cyclin-dependent kinase (CDK1)/cyclin B1 complex [8], [9], [10]. PLK1 activates cell division cycle 25 (CDC25) phosphatase, which dephosphorylates CDK, thereby activating CDK. Activated CDKs and their attached cyclins promote mitotic entry.
The level of PLK1 in different tumours inversely correlates with patient survival [11]. Screening with small interfering RNA identified PLK1 as a potential target for cancer treatment [12], [13]. In contrast to PLK1, the kinases PLK2 and PLK3 act as tumour suppressors [14], [15]. The role of PLK1 as a tumour promoter inspired the search for specific PLK1 inhibitors. Two target sites of PLK1 may be feasible for small-molecule inhibitors: the ATP-binding site in the kinase domain and the substrate-binding site in the PBD. The classical target for kinase inhibition is the ATP-binding site. Several inhibitors of the ATP site of PLK1 have been identified, such as BI 2536, volasertib, GSK461364A, NMS-P937, HMN-214, and TKM-080301 [16]. Although these inhibitors have been efficient in treating cancer, the complete knowledge regarding their modes of action remains elusive. BI 2536 inhibits not only PLK1 but also death-associated kinase 2 (DAPK-2) and calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) [17], illustrating the general problem with inhibitors against the ATP-binding site of kinases. Due to the high degree of structural conservation among the ATP-binding pockets, the development of specific PLK1 inhibitors remains difficult. Because of the unique existence of Polo-box motifs in PLKs, the development of inhibitors against the PBD represents an alternative approach to solve problems of selectivity for other kinases [18].
The PBD of PLK1 is the regulatory domain for the function of the kinase and provides a potential target for drug discovery in cancer treatment. The PBD contains a Polo-box cap (PC) followed by characteristic Polo-box (PB) motifs [5]. Crystal structure analysis of the PBD revealed that it is composed of three α sheets and a 12 β-sandwich domain. Therefore, the PC is wrapped around the second Polo-box (PB2) and linked to the first Polo-box (PB1) (Fig. 1A). Binding of the ligand occurs at the cleft between both PBs [19], [20]. As shown in Fig. 1B, the amino acids involved in the interaction of ligands with the binding pocket are composed of a hydrophobic half, which includes Val411, Trp414, Leu490, and Leu491, and a positively charged half, which includes His538, Lys540, and Arg557 [21]. As there is a continuous need for new chemical identities for targeted cancer therapies to overcome the problem of drug resistance due to point mutations, we aimed to identify new compounds selectively targeting PLK1. Here, we report 3-{[(1R,9S)-3-(naphthalen-2-yl)-6-oxo-7,11-diazatricyclo[7.3.1.02,7]trideca-2,4-dien-11-yl]methyl}benzonitrile (designated compound (1)) as a novel PLK1 PBD inhibitor. This compound was identified by the in silico screening of a library of 30,793 natural product derivatives. The results were validated by fluorescence polarization assay and microscale thermophoresis. This novel PLK1 inhibitor may be used for the future pharmaceutical development of anticancer candidates.
Fig. 1.
Structural features of PLK1. (A) Schematic representation of the PLK1 domains. Shown are the kinase domain and the PBD, which consists of polo-box 1 (PB1, blue), polo-box 2 (PB2, yellow) and the polo-box cap (PC, green). (B) Crystal structure of the PLK1 PBD (PDB:4X9R) showing the amino acids involved in the ligand interaction between the two polo boxes of the PBD.
Material and methods
Cell lines
Drug-sensitive CCRF-CEM and multidrug-resistant P-glycoprotein-overexpressing CEM/ADR5000 leukaemia cells were kindly provided by Dr Axel Sauerbrey (University of Jena, Department of Paediatrics, Jena, Germany). The cells were cultured in RPMI 1640 medium (Life Technologies, Schwerte, Germany) supplemented with 10% foetal bovine serum (FBS), penicillin (100 U/mL) and streptomycin (100 µg/mL) (Invitrogen, Darmstadt, Germany) in a 5% CO2 atmosphere at 37 °C. For maintenance of the multidrug-resistance phenotype, the CEM/ADR5000 cells were treated every other week with 5 µg/ml doxorubicin (provided by the University Medical Center, Mainz, Germany). The multidrug-resistance phenotype of the CEM/ADR5000 cells has been previously reported [22], [23], [24], [25], [26].
Virtual screening
The ZINC natural derivative (Znd109) library contains 30,793 chemically modified natural products, the information of which was downloaded from the ZINC database (http://zinc.docking.org/). The crystal structure of the PLK1 PBD was obtained from the Protein Data Bank (http://www.rcsb.org/) with PDB entry 4X9R [27]. A structurally based virtual screening was then performed using PyRx software (http://pyrx.scripps.edu). The test compounds were energetically minimized, and seven amino acids of PLK1 (Trp414, Asp416, His489, Leu491, Arg516, His538, and Lys540) were used to create the grid box for defined docking [28], [29].
If screening results with a binding affinity less than or equal to − 10.0 kcal/mol were achieved, the corresponding compounds were used for subsequent molecular docking with AutoDock 4.2. Based on the virtual screening results, 25 identified candidate compounds were purchased from AnalytiCon Discovery GmbH (Potsdam, Germany) for in vitro verification of the in silico results.
Molecular docking with AutoDock
The test compounds obtained from the virtual screening results were further tested for their binding affinity for PLK1 (PBD:4X9R) and their selectivity with respect to the PLK2 PBD (PDB:4XB0) using molecular docking in AutoDock 4.2. All sdf files were converted to PDBQT (Protein Data Bank Partial Charge and Atom Type) files. The grid boxes were created around the amino acids of PLK1 (as described above in the virtual screening section) and PLK2 (Trp507, Asp509, Tyr578, Lys607, Tyr626, His629, Lys631, and Arg650) involved in ligand binding according to the literature [30].
Docking was performed using the Lamarckian Algorithm, and the parameters were set to 250 runs and 25,000,000 energy evaluations for each cycle, as previously described [31], [32]. The lowest binding, the mean binding affinity, and the predicted inhibition constant (pKi) were obtained from the docking log (dlg) file. The amino acids of the kinase involved in hydrogen bonding and hydrophobic interactions with the compound were analysed using AutoDockTools 1.5.7 (ADT). AutoDock 4.2 was used for docking visualization.
Molecular docking with FlexX
For molecular docking, we also used the FlexX module provided by LeadIT v.2.3.2 from BiosolveIT (BioSolveIT GmbH, Sankt Augustin, Germany). The PLK1 PBD 3D structure was retrieved from the Protein Data Bank (PBD:4X9R). The compounds were uploaded as mol2 files from the ZINC database. The programme uses the FlexX algorithm to dock the compound to the protein, and the binding site is determined according to a reference ligand by superimposition of the tested compounds. The top 10 poses of ligand binding to the protein binding site were used to estimate the binding energy.
Protein expression and purification
Human PLK1, PLK2 and PLK3 PBDs were expressed as previously described [33]. Gene sequences were amplified and cloned into a modified pET28a vector (PLK1) or into a modified pQE70 vector carrying a C-terminal 6× His tag and an N-terminal MBP tag (PLK2 and PLK3). Plasmids were transfected into Novagen Rosetta BL21DE3 cells (Merck, Darmstadt, Germany) for protein expression. Then, His•Bind® Resin (Merck) was used for protein purification and dialysis into buffer containing 50 mM Tris (pH 8.0) 200 mM NaCl (Plk1: 400 mM NaCl), 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 0.1% Nonidet P-40.
Fluorescence polarization assay
The fluorescence polarization assay conditions used have been previously described [34], [35]. Briefly, the peptide binding of the PLK PBDs was analysed using the following fluorescence-labelled peptides at a final concentration of 10 nM: PLK1, 5-carboxyfluorescein-GPMQSpTPLNG-OH; PLK2, 5-carboxyfluorescein-GPMQTSpTPKNG-OH; and PLK3, 5-carboxyfluorescein-GPLATSpTPKNG-NH2. Fluorescence polarization assays were performed in buffer containing 10 mM Tris/HCl, 50 mM NaCl, 1 mM EDTA, 0.1% NP-40 substitute, 2% DMSO and 1 mM DTT at pH 8. The final concentration of each PBD was as follows: PLK1, 20 nM; PLK2, 80 nM; and PLK3, 250 nM. Pipetting was carried out using a Biomek FXp robot (Beckman-Coulter, USA). Proteins were incubated with the compound for 1 h at room temperature before and after peptide addition. Fluorescence polarization was measured using an Infinite F500 plate reader (Tecan, Crailsheim, Germany). Fluorescence polarization measurements were converted into % inhibition by comparison with binding curve fits and controls in the absence of the compound using OriginPro software (OriginLab, USA).
Microscale thermophoresis (MST) analysis
MST experiments were performed using a Monolith NT1.115 instrument (NanoTemper Technologies, Munich, Germany) [36]. The procedure was performed as previously reported [37]. PLK1 PBD (500 nM) was labelled using a Monolith Protein Labelling Kit BLUE-NHS and was then mixed 1:1 with different concentrations of compound (1). All reactions were performed in buffer containing 50 mM Tris buffer pH 7.0, 150 mM NaCl, 10 mM MgCl2 and 0.05% Tween 20. Measurements were performed with 80% instrument MST power. Analysis was performed using NT Analysis 1.5.41 software, Monolith.
Cytotoxicity assay
Cytotoxicity testing was performed using a resazurin reduction assay, as previously described [38], [39]. Exponentially growing CCRF-CEM and CEM/ADR5000 cells were seeded into 96-well plates (104 cells/well). The cells were then treated with different compound concentrations in a total volume of 200 µL and incubated for 72 h. Then, resazurin 0.01% w/v (Sigma Aldrich, Taufkirchen, Germany) was added in a total volume of 20 µl per well. The fluorescence intensity was measured using an Infinite M200 Pro plate reader (Tecan). Dose-response curves were formed by plotting the percent of viable cells against the applied concentrations of compound (1), and the 50% inhibition concentration (IC50) was calculated from three independent experiments, each with six parallel measurements, by linear regression analysis using Prism 7 GraphPad Software (La Jolla, CA, USA).
Cell cycle analysis
CCRF-CEM cells were seeded in 6-well plates (106 cells/well) and treated with 0.1% DMSO (v/v) as a negative control or various concentrations of compound (1) (10, 30 or 40 µM). The cells were incubated at 37 °C in a 5% CO2 atmosphere for 24 h. Then, the cells were washed twice with PBS (Life Technologies, Darmstadt, Germany), fixed with cold 70% ethanol and stored at 20 °C for 2 h. After the cells were washed with cold PBS and re-suspended in 500 µL of PBS, propidium iodide (PI) (Thermo Fisher Scientific, Dreieich, Germany) was added to a final concentration of 50 µg/ml and incubated with the cells for 15 min at 4 °C. PI fluorescence was measured using an LSR-Fortessa FACS analyser (Becton-Dickinson, Heidelberg, Germany). Experiments were independently performed three times, and standard deviations were calculated with Prism 7 GraphPad Software.
Immunofluorescence microscopy
CCRF-CEM cells were treated with 30 µM of compound (1) for 24 h. Then, the cells were washed with PBS and fixed with 3.7% paraformaldehyde for 30 min at room temperature. The cells were blocked for 1 h at room temperature using a blocking buffer (5% FBS and 0.3% Triton X-100 in PBS). Primary antibodies [rabbit α-tubulin antibody (Abcam, Cambridge, UK) and mouse PLK1 monoclonal antibody (Thermo Fisher Scientific)] were added and allowed to stand for 2 h at room temperature (dilution 1:1000). Then, the cells were washed with PBS three times. Secondary antibodies [goat anti-rabbit IgG H&L (Alexa Fluor® 488) (Abcam) or goat anti-mouse IgG H&L (Cy3®) preadsorbed (Abcam)] were subsequently added (dilution 1:1000). Then, the cells were washed with PBS, stained using 1 µg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Sigma Aldrich), and mounted using Fluoromount-G® (Southern Biotech, Birmingham, AL, USA). Fluorescence imaging was performed using an EVOS SL digital inverted microscope (Life Technologies).
Annexin V/PI staining
An annexin V/PI detection apoptosis kit (Life Technologies, Carlsbad, CA, USA) was used for the detection of early apoptosis and necrosis. CCRF-CEM cells (106 cells/well) were seeded in 6-well plates and then treated with 10 µM compound (1) for 24 h. The cells were washed with PBS and stained with annexin V/FITC for 15 min at room temperature. After the cells were washed and stained with PI for 15 min in the dark, analysis was carried out using a BD Accuri™ C6 system (Becton-Dickinson, Heidelberg, Germany).
Results
Virtual drug screening and molecular docking
As computer-based approaches have been evolving in the field of drug discovery, we performed virtual screening of the ZINC natural derivatives (Znd 109) library, which consists of 30793 compounds obtained from the ZINC main database, for binding to the PBD of PLK1. The results of screening the Znd 109 library with PyRx software revealed 29 compounds with predicted binding affinities of ≤−10 kcal/mol (Table 1). These compounds were further investigated using AutoDockTools for their predicted selectivity with respect to the PBD of PLK2. The results are also shown in Table 1. These 29 compounds showed predicted selective PLK1 PBD inhibition of variable potency, with the exception of three ligands, which were predicted to have unfavourable selectivity with respect to the PLK2 PBD: ZINC67899025, ZINC05415069, and ZINC05439871. These three ligands were excluded from further investigations.
Table 1.
PyRx and molecular docking results.
| Compound | PyRx Binding Affinity (kcal/mol) | PLK | Binding Affinity (kcal/mol) | Mean Binding Affinity (kcal/mol) | predKi (nM) | H Bonds | Number of Hydrophobic Interactions |
|---|---|---|---|---|---|---|---|
| ZINC08300249 | −10.5 | PLK1 | −9.31 | −9.20 | 150.27 | Lys540 Arg557 | 8 |
| PLK2 | −8.17 | −8.08 | 1020 | Trp507 | 10 | ||
| ZINC08856957 | −10.5 | PLK1 | −9.74 | −9.73 | 72.95 | Ser412 Trp414 Leu491 Asn533 | 6 |
| PLK2 | −9.36 | −9.36 | 138.03 | Met584 | 8 | ||
| ZINC67899025 | −10.4 | PLK1 | −10.56 | −10.20 | 18.26 | Ala495 Lys540 | 12 |
| PLK2 | −10.70 | −10.44 | 14.31 | Trp507 Asn624 | 7 | ||
| ZINC05415074 | −10.3 | PLK1 | −10.24 | −10.11 | 31.09 | – | 10 |
| PLK2 | −7.81 | −7.72 | 1870 | – | 9 | ||
| ZINC04237409 | −10.3 | PLK1 | −10.23 | −10.22 | 31.67 | Lys540 | 10 |
| PLK2 | −7.73 | −7.70 | 2160 | Asp509 | 6 | ||
| ZINC03840464 | −10.3 | PLK1 | −10.81 | −10.80 | 11.97 | – | 12 |
| PLK2 | −8.31 | −7.90 | 811.94 | Tyr578 Lys631 | 8 | ||
| ZINC03841379 | −10.3 | PLK1 | −11.57 | −11.01 | 3.28 | Asn533 Arg557 | 11 |
| PLK2 | −9.26 | −8.69 | 163.93 | Asp509 | 8 | ||
| ZINC04259479 | −10.3 | PLK1 | −12.66 | −11.48 | 0.52848 | Trp414 Lys492 | 11 |
| PLK2 | −9.17 | −8.66 | 189.5 | Trp507 Lys631 | 5 | ||
| ZINC20503376 | −10.3 | PLK1 | −9.65 | −9.44 | 84.54 | – | 11 |
| PLK2 | −9.61 | −9.38 | 89.6 | – | 10 | ||
| ZINC00990238 | −10.2 | PLK1 | −8.55 | −8.54 | 541.35 | Asn533 Lys540 | 8 |
| PLK2 | −7.72 | −7.72 | 2180 | Met584 Tyr626 | 5 | ||
| ZINC03841471 | −10.2 | PLK1 | −11.15 | −10.86 | 6.71 | Asn533 Arg557 | 11 |
| PLK2 | −8.98 | −8.17 | 260.89 | Met584 Arg650 | 8 | ||
| ZINC20503171 | −10.2 | PLK1 | −9.12 | −8.61 | 207.6 | – | 13 |
| PLK2 | −8.27 | −8.18 | 870.02 | – | 11 | ||
| ZINC20503194 | −10.2 | PLK1 | −9.30 | −8.97 | 151.61 | – | 14 |
| PLK2 | −8.24 | −8.20 | 919.64 | – | 11 | ||
| ZINC59676801 | −10.2 | PLK1 | −11.98 | −11.80 | 1.67 | Asp416 Arg557 | 10 |
| PLK2 | −8.12 | −7.96 | 1.12 | Asn582 Lys631 | 7 | ||
| ZINC04235865 | −10.1 | PLK1 | −8.44 | −8.37 | 649.98 | Trp414 | 11 |
| PLK2 | −8.00 | −7.92 | 1360 | Asp509 | 8 | ||
| ZINC03839453 | −10.1 | PLK1 | −9.49 | −9.25 | 110.32 | Gly494 | 11 |
| PLK2 | −7.41 | −7.31 | 3670 | – | 11 | ||
| ZINC05415069 | −10.1 | PLK1 | −9.56 | −9.44 | 98.57 | – | 14 |
| PLK2 | −9.72 | −9.64 | 74.82 | – | 11 | ||
| ZINC05439871 | −10.1 | PLK1 | −9.56 | −9.45 | 97.49 | – | 14 |
| PLK2 | −9.72 | −9.64 | 75.44 | – | 11 | ||
| ZINC03841381 | −10.1 | PLK1 | −11.27 | −10.71 | 5.53 | Asn533 Arg557 | 9 |
| PLK2 | −9.10 | −8.36 | 213.97 | Met584 Arg650 | 8 | ||
| ZINC59676822 | −10.1 | PLK1 | −12.14 | −11.90 | 1.26 | Arg557 | 11 |
| PLK2 | −8.33 | −8.27 | 783.73 | Met584 Lys607 | 5 | ||
| ZINC04235861 | −10.0 | PLK1 | −8.71 | −8.70 | 415.67 | – | 11 |
| PLK2 | −7.13 | −7.12 | 5990 | Trp507 | 6 | ||
| ZINC04235874 | −10.0 | PLK1 | −8.43 | −8.30 | 659.1 | Ser412 His538 Lys540 Arg557 | 7 |
| PLK2 | −7.75 | −7.67 | 2080 | Trp507 Asn624 Phe625 His629 | 6 | ||
| ZINC04235919 | −10.0 | PLK1 | −9.29 | −9.25 | 154.01 | Trp414 | 11 |
| PLK2 | −8.25 | −8.21 | 901.28 | Lys607 Lys631 | 6 | ||
| ZINC04236081 | −10.0 | PLK1 | −8.79 | −8.78 | 358.03 | Gly494 | 10 |
| PLK2 | −7.31 | −7.30 | 4400 | Lys607 His629 | 7 | ||
| ZINC05433649 | −10.0 | PLK1 | −11.27 | −9.95 | 5.44 | Lys540 Arg557 | 10 |
| PLK2 | −10.26 | −9.72 | 30.04 | – | 11 | ||
| ZINC08296353 | −10.0 | PLK1 | −11.26 | −10.99 | 5.61 | Leu491 Lys540 | 11 |
| PLK2 | −9.72 | −9.36 | 74.71 | Trp507 | 11 | ||
| ZINC08444225 | −10.0 | PLK1 | −10.72 | −10.57 | 13.82 | Leu491 | 11 |
| PLK2 | −8.60 | −8.59 | 496.82 | Trp507 | 8 | ||
| ZINC59676778 | −10.0 | PLK1 | −12.38 | −12.10 | 0.8435 | Asp416 Arg557 | 11 |
| PLK2 | −8.27 | −8.24 | 863.96 | Lys607 | 6 | ||
| ZINC59676860 | −10.0 | PLK1 | −11.30 | −10.70 | 5.19 | Lys540 | 11 |
| PLK2 | −9.66 | −8.75 | 82.5 | Lys631 | 11 |
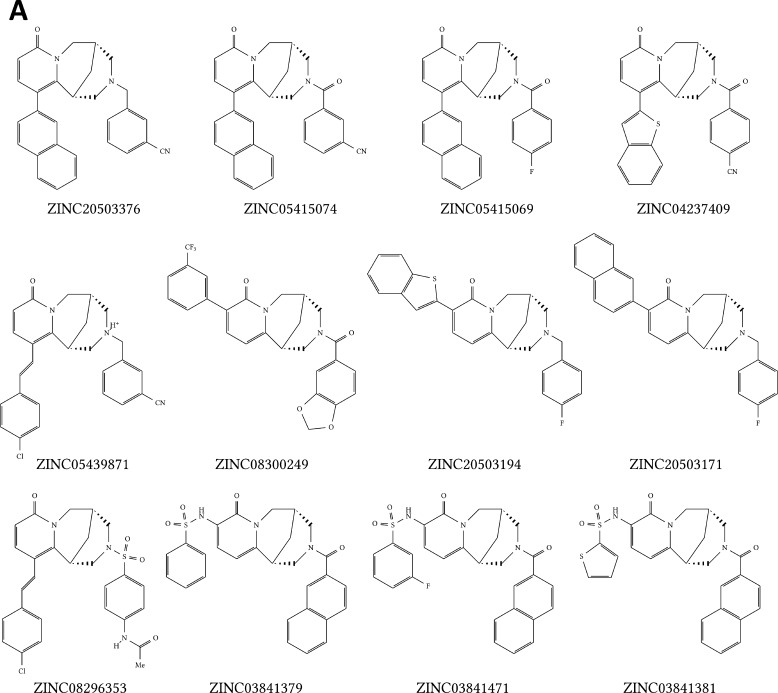
If the chemical structures of the candidate molecules were examined, two larger groups of compounds with the same basic structure could be formed. Twelve compounds shared a similar scaffold, suggesting a useful approach for the selective inhibition of the PLK1 PBD (Fig. 2A). Additionally, a second sub-group of eight compounds shared the same scaffold. This eight-member group may also lead to the discovery of a new compound (Fig. 2B) based on their structural consensus. Finally, the remaining compounds partly shared a similar basic structure (Fig. 2C). Due to the high similarity of the scaffolds, it was very likely for a new potential inhibitor to originate from the first or second group.
Fig. 2.
Clustering of the candidate molecules. Virtual screening with PyRx resulted in 29 compounds with a predicted binding affinity of ≤−10 kcal/mol for PLK1. (A) Comparison of the structures of these 29 candidate molecules revealed a group of 12 molecules. (B) A group of 8 molecules each with the same basic structure. (C) The other 9 molecules partly share a similar framework.
Fluorescence polarization assays
In vitro experiments were performed to verify the predicted PLK1 PBD inhibition. Therefore, we performed competitive fluorescence polarization assays using the 25 selected compounds obtained via in silico screening. Among the tested candidates, (3-{[(1R,9S)-3-(naphthalen-2-yl)-6-oxo-7,11-diazatricyclo[7.3.1.02,7]trideca-2,4-dien-11-yl]methyl}benzonitrile) (ZINC20503376), designated as compound (1), inhibited the binding of the fluorescent peptide to the PLK1 PBD with an IC50 value of 17.9 ± 0.4 µM (Fig. 3A). Furthermore, compound (1) showed over 5-fold selectivity for the PLK1 PBD compared with the PLK2 PBD (IC50 value: 95.5 ± 16.4 µM) and the PLK3 PBD (IC50 value >100 µM), which suggests compound (1) as a novel PLK1 inhibitor.
Fig. 3.
Compound (1) binds selectively to the PLK1 PBD. (A) Chemical structure of compound (1). Dose-response curves of competitive fluorescence polarization assays. The curves show the effect of compound (1) on the binding of fluorescein-labelled phosphopeptides to the PLK1, PLK2, and PLK3 PBDs. (B) Binding affinity of compound (1) for the PLK1 PBD measured by MST. The curve shows the difference in the bound and unbound state of the PLK1 PBD in presence of compound (1).
MST
To confirm the in silico binding between compound (1) and the PLK1 PBD, we used MST. As shown in Fig. 3B, different concentrations of compound (1) were titrated against a labelled protein. Compound (1) bound with an equilibrium binding constant of 283 ± 27 nM, indicating that compound (1) also binds to the PLK1 PBD in vitro.
Binding of compound (1) to the PLK1 PBD binding pocket
Compound (1) was docked to the binding site of PLK1 PBD using AutoDock software. The lowest binding energy of the interaction was −9.7 kcal/mol, which reflected a good predicted affinity of compound (1) for the binding site. The predicted value of the inhibitory constant was obtained from the binding interaction calculation (Ki = 77.33 nM, pKi = 7.11). As shown in Fig. 4A, multiple amino acids of the PLK1 PBD binding sites were predicted to be involved in hydrophobic interactions with compound (1). These amino acids included Ser412, Lys413, Trp414, Val415, Asp416, Leu490, Leu491, Lys492, Ala493, Asn533, Lys540, and Arg557.
Fig. 4.
Molecular docking of compound (1). (A) Docking of compound (1) (green) to the PLK1 PBD binding site (PDB code 4X9R). Compound (1) interacted with the amino acids in the PBD binding pocket. AutoDock 4.2 software was used for visualization. (B) Pose-view visualization of the docking of compound (1) to the PLK1 PBD using LeadIt software. Hydrophobic interactions are shown in green, and hydrogen bonds are shown in red.
The interaction of compound (1) with the PBD binding pocket was also analysed by docking with LeadIT software. A high affinity was estimated, with a FlexX score of −24.1 kJ/mol. An interaction of compound (1) with the same amino acids of the PBD binding pocket was also observed in the second model, plus a hydrogen bond interaction with His538, as shown in Fig. 4B. Hence, a high affinity of compound (1) for PLK1 PBD was suggested.
Prediction of physicochemical properties of compound (1)
In the process of drug discovery, the determination of physiochemical properties plays an important role. We used DataWarrior software to calculate drug likeness-related parameters. The results are described in Table 2. Compound (1) showed a logP value less than 5, indicating high hydrophilicity and therefore predicting good absorption and permeation. These findings agree with the good solubility predicted by the logS value. The compound also showed a favourable drug likeness based on topological descriptors. Furthermore, compound (1) was not predicted to be carcinogenic by DataWarrior.
Table 2.
Physiochemical properties predicted by DataWarrior software.
| Properties | Values |
|---|---|
| clog P | 4,34 |
| cLogS | −5.6 |
| Total surface area | 334.4 |
| Relative polar surface area | 0.102 |
| Topological polar surface area | 47.34 |
| Drug likeness | −1.2 |
| Mutagenic | None |
| Tumourigenicity | None |
Cytotoxicity of compound (1) towards leukaemia cell lines
It is expected that PLK1 inhibition through the PBD induces cancer cell death. Therefore, we performed cytotoxicity assays with compound (1). Treatment of the sensitive CCRF-CEM leukaemia cell line and its multidrug-resistant P-glycoprotein-expressing CEM/ADR5000 subline with different concentrations of compound (1) induced growth inhibition, with an IC50 value of 11.4 ± 1.05 µM and 13.58 ± 1.13 µM for CCRF-CEM and CEM/ADR5000, respectively (Fig. 5). Hence, compound (1) revealed considerable cytotoxic effects. Moreover, the multidrug-resistant cells were not cross-resistant to compound (1).
Fig. 5.
Growth inhibition of drug-sensitive CCRF-CEM and multidrug-resistant CEM/ADR5000 cells by compound (1). The dose-response curves represent the mean ± SD of three independent experiments with six parallel measurements each.
Cell cycle analysis
To test the anti-proliferative effect of compound (1) and its role in cell cycle progression, we used flow cytometry. The analysis of the percentage of cells in the G0/G1, S and G2/M phases was performed for CCRF-CEM cells treated with various concentrations of compound (1) and DMSO as a control. As shown in Fig. 6A and B, there was a considerable increase in cells in the G2/M phase after treatment with 30 µM or 40 µM of compound (1). The increased accumulation of mitotic cells is a characteristic phenotype for PLK1 inhibitors.
Fig. 6.
Induction of G2/M arrest by compound (1). (A) CCRF-CEM cells treated with DMSO or different concentrations of compound (1) stained with PI and analysed for DNA content by flow cytometry. (B) Quantitative analysis of the cell cycle distribution. The results represent the mean ± SD of three independent experiments.
Immunofluorescence microscopy
The mitotic arrest described above might be due to the failure of PLK1 localization or of spindle formation. To investigate this hypothesis, CCRF-CEM cells treated with compound (1) were stained with antibodies specific for α-tubulin in mitotic spindles or PLK1. An increasing number of cells were arrested in prometaphase, as determined by chromosome morphology. The treated cells failed to form bipolar spindles; rather, mitotic cells with monopolar ‘Polo’ spindles accumulated (Fig. 7A). Additionally, positive staining for PLK1 was observed among the treated cells, confirming that the cells were arrested during early M phase. The staining of PLK1 revealed the mislocalization of PLK1 around the cell compartment rather than attached to mitotic spindles, as in the untreated control cells.
Fig. 7.
Compound (1) induced abnormal spindle formation. (A) Immunofluorescence staining of CCRF-CEM cells treated with 10 µM compound (1) for 24 h. The cells were stained with antibodies against α-tubulin (green) and PLK1 (red), and nuclei were counterstained with DAPI (blue). Increased numbers of cells with monopolar spindles were observed. (B) Fluorescence microscopy determination of nuclear integrity of CCRF-CEM cells as a parameter of apoptosis determined by DAPI staining. Cells with condensed or fragmented nuclei were not observed among either the untreated cells (left) or the cells treated with 10 µM compound (1) for 24 h (right). (C) Detection of CCRF-CEM cell apoptosis by flow cytometry and annexin V/PI staining.
To exclude artificial results due to dying and apoptotic cells, we treated cells with 10 µM compound (1) or left the cells untreated and performed staining with DAPI and annexin V FITC/PI (Fig. 7C). The DAPI staining did not show increased numbers of condensed or fragmented cell nuclei among the treated cells compared to the untreated cells (Fig. 7B). Similarly, the fraction of apoptotic cells shown by annexin V/PI staining was low (2.3% in untreated and 2.7% in treated cells, respectively) (Fig. 7C). These results provide evidence that the observed phenomenon of a high percentage of monopolar cells occurred in living cells and mostly occurred due to the failure of bipolar spindle formation as a result of PLK1 inhibition with compound (1).
Discussion
PLK1 represents an attractive target for cancer treatment, and continuous efforts focus on the development of targeted PLK1 inhibitors. PBD inhibitors are advantageous compared to other PLK1 inhibitors because of their improved selectivity [40]. In this study, we performed the virtual drug screening of > 30,000 natural product derivatives for their binding to a defined binding pocket in the PLK1 PBD. The screening yielded 25 molecules predicted to bind with high affinity (>-10 kcal/mol) to the active site. These results were further investigated by in silico molecular docking to the PLK1 PBD and PLK2 PBD to investigate whether the selected candidates selectively bound to PLK1 over PLK2, which has a tumour suppressive effect [41], [42], [43]. Clustering of the good candidates showed three main subgroups sharing a similar chemical scaffold. These results demonstrate the sensitivity of virtual screening, if applied to a defined binding site with recognized features.
To validate the screening results in vitro, we performed screening using competitive fluorescence polarization assays. Among the 25 tested candidates, compound (1) bound to the PLK1 PBD with 5-fold selectivity over the PLK2 and PLK3 PBDs. This result was further confirmed by MST binding assays with purified PLK1 PBD. As expected, the MST signals were different between the bound and unbound proteins, indicating that compound (1) indeed interacted with PLK1. Compound (1) belongs to a group of chemicals that are cytisine derivatives. Cytisine is a plant alkaloid that has long been used for facilitating smoking cessation [44].
Molecular docking suggested an interaction of compound (1) with vital amino acids in the PBD binding pocket, including His538, Trp414, Val415 and Leu490. These amino acids play an important role in the substrate recognition and biological activity of PLK1 [45], [46]. Understanding the binding mode of compound (1) will guide future drug development strategies for PLK1 inhibitors. The chemical characteristics of compound (1) are (i) the presence of aromatic rings that aid in hydrophobic interactions and (ii) the presence of at least two substitutions that are hydrogen bond acceptors. In the case of compound (1), this is represented by the cyano group (C—N triple bonds) and oxygen atoms. Additionally (iii), there is a positive ionizable area due to the presence of amino groups.
A resazurin reduction assay showed the cytotoxic activity of compound (1) towards CCRF-CEM leukaemia cells. Moreover, compound (1) inhibited P-glycoprotein-overexpressing CEM/ADR5000 leukaemia cells with similar efficacy as sensitive wild-type CCRF-CEM cells, indicating that compound (1) is not hampered by the multidrug-resistance phenotype, which represents a great obstacle in cancer therapy [47].
PLK1 is involved in cell division by regulating mitotic spindle formation, spindle maturation and activation of the cyclin B1-CDK1 complex [48], [49], [50]. Here, we report compound (1), which induced G2/M arrest in CCRF-CEM cells. The timely localization of PLK1 to the kinetochores is essential for the proper segregation of chromosomes [51]. This association is highly dynamic through different phases of mitosis, in which PLK1 is localized to spindles in metaphase [52]. PLK1 is fundamental for the generation of bipolar spindles, which is required to organize microtubule function. Loss of PLK activity leads to the formation of monopolar spindles in different organisms [8]. Upon treatment with compound (1), the immunofluorescence staining of mitotic spindles and PLK1 showed increased numbers of mitotic cells with monopolar spindles, which result in the failure of mitosis. The phenomenon of monopolar spindles has been previously reported in the context of PLK1 inhibitors [53], [54].
Conclusions
In the present investigation, we demonstrated that compound (1) represents a potent PLK1 inhibitor. Compound (1) inhibited the viability of human tumour cells and arrested them in the G2/M cell cycle phase. It led to an accumulation of cells with monopolar spindles, which is consistent with its PLK1-targeting activity. Compound (1) may serve as a starting point for medicinal chemistry projects aimed at the development of new anticancer drugs. Compound (1) was identified as a potential PLK1 PBD inhibitor via virtual drug screening. Our results further underscore the importance of in silico screening approaches for drug discovery.
Conflict of interest
The authors have declared no conflict of interest.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (INST 268/281-1 FUGG) and the German Academic Exchange Service (DAAD). We are grateful for the support of the Institute of Molecular Biology Core Facility, especially that of the kind flow cytometry staff (IMB, Mainz, Germany).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2018.10.002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.van Vugt M.A., Medema R.H. Getting in and out of mitosis with Polo-like kinase-1. Oncogene. 2005;24:2844–2859. doi: 10.1038/sj.onc.1208617. [DOI] [PubMed] [Google Scholar]
- 2.Moshe Y., Boulaire J., Pagano M., Hershko A. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc Natl Acad Sci USA. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lowery D.M., Lim D., Yaffe M.B. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 4.Liu X. Targeting polo-like kinases: A promising therapeutic approach for cancer treatment. Transl Oncol. 2015;8:185–195. doi: 10.1016/j.tranon.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elia A.E.H., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 6.Seki A., Coppinger J., Jang C. Bora and aurora a cooperatively activate Plk1 and control the entry into mitosis. Science. 2008;320:1655–1658. doi: 10.1126/science.1157425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nigg E.A. Polo-like kinases: positive regulators of cell division from start to finish. Curr Opin Cell Biol. 1998;10:776–783. doi: 10.1016/s0955-0674(98)80121-x. [DOI] [PubMed] [Google Scholar]
- 8.Ohkura H., Hagan I.M., Glover D.M. The conserved Schizosaccharomyces pombe kinase plol, required to form a bipolar. Genes Dev. 1995:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 9.Lane H.A., Nigg E.A. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descombes P., Nigg E.A. The polo-like kinase Plx1 is required for M phase exit and destruction of mitotic regulators in Xenopus egg extracts. EMBO J. 1998;17:1328–1335. doi: 10.1093/emboj/17.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weichert W., Ullrich A., Schmidt M., Gekeler V., Noske A., Niesporek S. Expression patterns of polo-like kinase 1 in human gastric cancer. Cancer Sci. 2006;97:271–276. doi: 10.1111/j.1349-7006.2006.00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu K., Law J.H., Fotovati A., Dunn S.E. Small interfering RNA library screen identified polo-like kinase-1 (PLK1) as a potential therapeutic target for breast cancer that uniquely eliminates tumor-initiating cells. Breast Cancer Res. 2012;14:R22. doi: 10.1186/bcr3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seth S., Matsui Y., Fosnaugh K., Liu Y., Vaish N., Adami R. RNAi-based therapeutics targeting survivin and PLK1 for treatment of bladder cancer. Mol Ther. 2011;19:928–935. doi: 10.1038/mt.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syed N., Smith P., Sullivan A., Spender L.C., Dyer M., Karran L. Transcriptional silencing of Polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood. 2006;107:250–256. doi: 10.1182/blood-2005-03-1194. [DOI] [PubMed] [Google Scholar]
- 15.Yang Y., Bai J., Shen R., Brown S.A.N., Komissarova E., Huang Y. Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1?? under hypoxic conditions. Cancer Res. 2008;68:4077–4085. doi: 10.1158/0008-5472.CAN-07-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strebhardt K., Becker S., Matthess Y. Thoughts on the current assessment of Polo-like kinase inhibitor drug discovery. Expert Opin Drug Discov. 2014;441:1–8. doi: 10.1517/17460441.2015.962510. [DOI] [PubMed] [Google Scholar]
- 17.Raab M., Pachl F., Kramer A., Kurunci-Csacsko E., Dotsch C., Knecht R. Quantitative chemical proteomics reveals a Plk1 inhibitor-compromised cell death pathway in human cells. Cell Res. 2014;24:1141–1145. doi: 10.1038/cr.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shan H.M., Wang T., Quan J.M. Crystal structure of the polo-box domain of polo-like kinase 2. Biochem Biophys Res Commun. 2015;456:780–784. doi: 10.1016/j.bbrc.2014.11.125. [DOI] [PubMed] [Google Scholar]
- 20.Elia A.E.H. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z., Song Y., Rehse P.H. Thymoquinone blocks pSer/pThr recognition by plk1 polo-box domain as a phosohate mimic. ACS Chem Biol. 2013;8:303–308. doi: 10.1021/cb3004379. [DOI] [PubMed] [Google Scholar]
- 22.Kimmig A., Gekeler V., Neumann M., Frese G., Handgretinger R., Kardos G. Susceptibility of multidrug-resistant human leukemia cell lines to human interleukin 2-activated killer cells. Cancer Res. 1990;50:6793–6799. [PubMed] [Google Scholar]
- 23.Efferth T., Sauerbrey A., Olbrich A., Gebhart E., Rauch P., Weber H.O. Molecular modes of action of artesunate in tumor cell lines. Mol Pharmacol. 2003;64:382–394. doi: 10.1124/mol.64.2.382. [DOI] [PubMed] [Google Scholar]
- 24.Gillet J.-P., Efferth T., Remacle J. Chemotherapy-induced resistance by ATP-binding cassette transporter genes. Biochim Biophys Acta. 2007;1775:237–262. doi: 10.1016/j.bbcan.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Efferth T., Konkimalla V.B., Wang Y.-F., Sauerbrey A., Meinhardt S., Zintl F. Prediction of broad spectrum resistance of tumors towards anticancer drugs. Clin Cancer Res. 2008;14:2405–2412. doi: 10.1158/1078-0432.CCR-07-4525. [DOI] [PubMed] [Google Scholar]
- 26.Kadioglu O., Cao J., Kosyakova N., Mrasek K., Liehr T., Efferth T. Genomic and transcriptomic profiling of resistant CEM/ADR-5000 and sensitive CCRF-CEM leukaemia cells for unravelling the full complexity of multi-factorial multidrug resistance. Sci Rep. 2016;6:1–18. doi: 10.1038/srep36754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian W.J., Park J.E., Grant R., Lai C.C., Kelley J.A., Yaffe M.B. Neighbor-directed histidine N (τ)-alkylation: A route to imidazolium-containing phosphopeptide macrocycles. Biopolymers. 2015;104:663–673. doi: 10.1002/bip.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakkiah S., Senese S., Yang Q., Lee K.W., Torres J.Z. Dynamic and multi-pharmacophore modeling for designing Polo-box Domain inhibitors. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scharow A., Raab M., Saxena K., Sreeramulu S., Kudlinzki D., Gande S. Optimized Plk1 PBD inhibitors based on poloxin induce mitotic arrest and apoptosis in tumor cells. ACS Chem Biol. 2015;10:2570–2579. doi: 10.1021/acschembio.5b00565. [DOI] [PubMed] [Google Scholar]
- 30.Kim J.H., Ku B., Lee K.S., Kim S.J. Structural analysis of the polo-box domain of human Polo-like kinase 2. Proteins. 2015:1201–1208. doi: 10.1002/prot.24804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kadioglu O., Jacob S., Bohnert S., Naß J., Saeed M.E.M., Khalid H. Evaluating ancient Egyptian prescriptions today: Anti-inflammatory activity of Ziziphus spina-christi. Phytomedicine. 2016;23:293–306. doi: 10.1016/j.phymed.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Wu C.F., Seo E.J., Klauck S.M., Efferth T. Cryptotanshinone deregulates unfolded protein response and eukaryotic initiation factor signaling in acute lymphoblastic leukemia cells. Phytomedicine. 2016;23:174–180. doi: 10.1016/j.phymed.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 33.Reindl W., Yuan J., Krämer A., Strebhardt K., Berg T. Inhibition of polo-like kinase 1 by blocking polo-box domain-dependent protein-protein interactions. Chem Biol. 2008;15:459–466. doi: 10.1016/j.chembiol.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 34.Reindl W., Strebhardt K., Berg T. A high-throughput assay based on fluorescence polarization for inhibitors of the polo-box domain of polo-like kinase 1. Anal Biochem. 2008;383:205–209. doi: 10.1016/j.ab.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 35.Reindl W., Gräber M., Strebhardt K., Berg T. Development of high-throughput assays based on fluorescence polarization for inhibitors of the polo-box domains of polo-like kinases 2 and 3. Anal Biochem. 2009;395:189–194. doi: 10.1016/j.ab.2009.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Jerabek-Willemsen M., Wienken C.J., Braun D., Baaske P., Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9:342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo E.-J., Efferth T. Interaction of antihistaminic drugs with human translationally controlled tumor protein (TCTP) as novel approach for differentiation therapy. Oncotarget. 2016;7:16818–16839. doi: 10.18632/oncotarget.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuete V., Mbaveng A.T., Nono E.C.N., Simo C.C., Zeino M., Nkengfack A.E. Cytotoxicity of seven naturally occurring phenolic compounds towards multi-factorial drug-resistant cancer cells. Phytomedicine. 2016;23:856–863. doi: 10.1016/j.phymed.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Kuete V., Mbaveng A.T., Sandjo L.P., Zeino M., Efferth T. Cytotoxicity and mode of action of a naturally occurring naphthoquinone, 2-acetyl-7-methoxynaphtho[2,3-b]furan-4,9-quinone towards multi-factorial drug-resistant cancer cells. Phytomedicine. 2017;33:62–68. doi: 10.1016/j.phymed.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Raab M., Sanhaji M., Pietsch L., Béquignon I., Herbrand A.K., Süß E. Modulation of the allosteric communication between the polo-box domain and the catalytic domain in Plk1 by small compounds. ACS Chem Biol. 2018;13:1921–1931. doi: 10.1021/acschembio.7b01078. [DOI] [PubMed] [Google Scholar]
- 41.Matthew E.M., Yang Z., Peri S., Andrake M., Dunbrack R., Ross E. Plk2 loss commonly occurs in colorectal carcinomas but not adenomas: Relationship to mTOR signaling. Neoplasia. 2018;20:244–255. doi: 10.1016/j.neo.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu F., Zhang S., Zhao Z., Mao X., Huang J., Wu Z. MicroRNA-27b up-regulated by human papillomavirus 16 E7 promotes proliferation and suppresses apoptosis by targeting polo-like kinase2 in cervical cancer. Oncotarget. 2016;7:19666–19679. doi: 10.18632/oncotarget.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staib F., Robles A.I., Varticovski L., Wang X.W., Zeeberg B.R., Zhurkin V.B. NIH Public Access. 2006;65:10255–10264. doi: 10.1158/0008-5472.CAN-05-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prochaska J.J., Benowitz N.L. The past, present, and future of nicotine addiction therapy. Annu Rev Med. 2016;67:467–486. doi: 10.1146/annurev-med-111314-033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun S.-M., Moulaei T., Lim D., Bang J.K., Park J.-E., Shenoy S.R. Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1. Nat Struct Mol Biol. 2009;16:876–882. doi: 10.1038/nsmb.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng K.Y., Lowe E.D., Sinclair J., Nigg E.A., Johnson L.N. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hamdoun S., Fleischer E., Klinger A., Efferth T. Lawsone derivatives target the Wnt/β-catenin signaling pathway in multidrug-resistant acute lymphoblastic leukemia cells. Biochem Pharmacol. 2017;146:63–73. doi: 10.1016/j.bcp.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Sunkel C.E., Glover D.M. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89(Pt 1):25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 49.Kumagai A., Dunphy W.G. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 1996;273:1377–1380. doi: 10.1126/science.273.5280.1377. [DOI] [PubMed] [Google Scholar]
- 50.Glover D.M., Ohkura H., Tavares Á. Polo kinase: The choreographer of the mitotic stage? J Cell Biol. 1996;135:1681–1684. doi: 10.1083/jcb.135.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kang Y.H., Park J.E., Yu L.R., Soung N.K., Yun S.M., Bang J.K. Self-regulated Plk1 recruitment to kinetochores by the Plk1-PBIP1 interaction is critical for proper chromosome segregation. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Golsteyn R.M., Golsteyn R.M., Mundt K.E., Mundt K.E., Fry A.M., Fry A.M. Cell-cycle regulation of the activity and subcellular-localization of Plk1, a human protein-kinase implicated in mitotic spindle function. J Cell Biol. 1995;129:1617–1628. doi: 10.1083/jcb.129.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudolph D., Steegmaier M., Hoffmann M., Grauert M., Baum A., Quant J. BI 6727, a polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 54.Choi M., Kim W., Cheon M.G., Lee C.W., Kim J.E. Polo-like kinase 1 inhibitor BI2536 causes mitotic catastrophe following activation of the spindle assembly checkpoint in non-small cell lung cancer cells. Cancer Lett. 2015;357:591–601. doi: 10.1016/j.canlet.2014.12.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.