Abstract
Many Clostridium species are found as commensal members of the intestinal microbiota. However, imbalances of the microbiota may lead to certain infections caused by these microorganisms, mainly Clostridium butyricum, Clostridium difficile, and Clostridium perfringens. In many cases, infection recurrence can occur after antibiotics, indicating the need for novel therapeutic options that act on the pathogens and also restore the microbiota. Herein, the in vitro antimicrobial activity and probiotic potential of clinical and reference strains of Bifidobacterium and Lactobacillus were investigated against Clostridium species. Antimicrobial activity was evaluated by the agar spot test and inhibition of gas production. Then, the probiotic potential of selected strains was assessed by analyzing their coaggregation ability, adhesive properties to host cells and mucin, tolerance to acidic pH and bile salts, and antimicrobial susceptibility profiles. Lactobacillus plantarum ATCC 8014 was the most promising strain based on its inhibitory activity against Clostridium spp. Also, this strain met criteria to be considered a probiotic based on its coaggregation ability, adhesive properties, and tolerance to harsh pH and bile acid salt conditions. The results indicate that among the studied strains, L. plantarum ATCC 8014 presents probiotic potential for controlling infections induced by the studied Clostridium species and should be further evaluated in in vivo animal models.
Keywords: probiotics, Lactobacillus plantarum, Clostridium, dysbiosis, antimicrobial activity
1. Introduction
In humans, the normal intestinal microbiota consists of a large number and high diversity of commensal microorganisms (about 1013 to 1014), mainly in the large intestine, where they establish a symbiotic relationship that influences the entire host organism [1,2,3]. Intestinal homeostasis is maintained through complex interactions between the host’s immune system and the microbiota [4,5]. However, this mutualistic relationship can be disrupted by a variety of factors, such as changes in diet, use of antibiotics, and immunomodulatory drugs, among others, leading to changes in both bacterial function and diversity [6,7].
An imbalance in the intestinal microbiota may cause or contribute to the establishment of infectious and inflammatory diseases such as inflammatory bowel disease [8], antibiotic-associated diarrhea (AAD) [9], irritable bowel syndrome [10], and necrotizing enterocolitis (NEC) [11]. In addition, there is growing evidence in the literature on the association of dysbiosis with other non-infectious diseases, including type 2 diabetes [12], asthma [13], non-alcoholic fatty liver disease [14], colorectal cancer [15], neurological conditions [16], and cardiovascular disease [17].
In some infectious disorders of the intestinal tract resulting from microbiota disruption, certain specific bacteria have been implicated as etiologic agents, especially potentially pathogenic Clostridium species. For example, Clostridium difficile is described as one of the leading causes of diarrhea and colitis associated with antibiotic use, with detection frequency ranging from 13% to 28% [9,18]. Mortality rates in patients with C. difficile infection may exceed 30%, especially in those individuals experiencing recurrence of infection within six months of initial treatment [19].
NEC is the most common and serious intestinal disorder among preterm infants and is diagnosed through radiological findings that include, among other manifestations, the presence of intestinal pneumatosis [20]. Its incidence may reach 12% in children that weigh less than 1 kg at birth [21] and mortality may range from 20% to 50% [22]. Risk factors for NEC include those that affect the normal microbiota, such as neonatal immaturity, enteral feeding, and intestinal colonization [23]. Its etiology is controversial and several causative organisms have been proposed, including viruses, Staphylococcus spp., various gram-negative bacilli, and Clostridium spp. [24]. Recent studies point to Clostridium butyricum as an important cause of NEC [25,26,27], although there are non-toxigenic strains that can be employed as probiotics [28].
Clostridium perfringens causes infection in both humans and animals [29]. Depending on the toxigenic type, C. perfringens may also cause other disorders in the intestinal tract, such as food poisoning and necrotic enteritis, in addition to tissue infections accompanied by myonecrosis, such as gas gangrene due to trauma [29]. In humans, C. perfringens has also been reported in cases of AAD with a lower prevalence [9,30].
Antibiotic therapy is the first line of treatment for these infectious intestinal disorders. However, recurrence is frequent, particularly in cases of AAD, since the microbiota remains unbalanced due to the use of broad-spectrum antibiotics [9,19]. Alternative interventions have been employed, such as narrow-spectrum antibiotics, dietary changes, fecal transplantation, and probiotics, which may attenuate the clinical symptoms, restore diversity of the intestinal microbiota, and improve host health [31,32,33].
Some studies have shown alleviation of AAD [34,35] and NEC [36,37] with probiotics. The efficacy of distinct probiotic strains, especially among Bifidobacterium and Lactobacillus species, suggests that they may have common properties that could positively impact patient health in such pathological states [31,38]. However, the basis of these properties is not yet fully understood [39] and there are no compelling explanations for the effects of probiotics in AAD or NEC. Several plausible mechanisms have been investigated and may contribute to the observed health benefits [39,40], but in terms of translational research, this is an evident shortcoming that hinders the development of improved therapies.
Therefore, the objective of this study was to carry out an in vitro screening of clinical and reference strains of Bifidobacterium and Lactobacillus with antimicrobial activity against C. butyricum, C. difficile, and C. perfringens. The most promising strain was subjected to analysis of criteria for consideration as a potential probiotic, including its ability to adhere to eukaryotic cells and mucus and its tolerance to acidic pH and bile salts.
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
The following Bifidobacterium and Lactobacillus reference strains were studied: Bifidobacterium longum subsp. longum ATCC 15707, Lactobacillus brevis ATCC 367, Lactobacillus delbrueckii subsp. delbrueckii ATCC 9649, Lactobacillus fermentum ATCC 23271, Lactobacillus paracasei subsp. paracasei ATCC 335, Lactobacillus plantarum ATCC 8014, and Lactobacillus rhamnosus ATCC 9595, which were obtained from the National Institute of Quality Control in Health (INCQS, FIOCRUZ, Rio de Janeiro, Brazil). Lactobacillus rhamnosus GG (LGG, ATCC 53103) was isolated from a commercial probiotic product (Floridral—Pharmaforce ApS, Copenhagen, Denmark) and used as a positive control. In addition, fecal isolates from newborn infants were evaluated and maintained in the Culture Collection Sector of Ceuma University, including Bifidobacterium longum 49.3, Bifidobacterium animalis subsp. lactis 56.1, Bifidobacterium bifidum 14.2, and Lactobacillus fermentum 54.2. All Bifidobacterium and Lactobacillus isolates were routinely cultured on agar or MRS broth (Man-Rogosa-Sharpe, Difco-BD, Detroit, MI, USA) with 0.25% l-cysteine and incubated at 37 °C for 24–48 h under anaerobic conditions. Cultures were stored in MRS broth with 20% glycerol at −80 °C.
C. butyricum ATCC 860, C. difficile ATCC 9689, and C. perfringens ATCC 12924 were obtained from INCQS (FIOCRUZ, Rio de Janeiro, Brazil). Clostridium strains were cultured in RCM (reinforced clostridial medium, Acumedia, Lansing, MI, USA) or thioglycolate medium (Acumedia) and incubated at 37 °C for 24–48 h in an anaerobic atmosphere. They were stored in RCM with 20% glycerol at −80 °C.
2.2. Antimicrobial Activity Screening
The ability of potential probiotics to inhibit Clostridium growth was evaluated in two distinct assays. All assays were performed in triplicate over three days.
2.2.1. Agar Spot Test
The agar spot test procedure was performed as described previously [41], with modifications. Briefly, in a Petri dish containing 10 mL of MRS agar, 5 μL of each probiotic culture was spotted onto one quadrant of the culture medium, followed by incubation at 37 °C for 24 h under anaerobic conditions. After incubation, 10 mL of thioglycolate agar was overlaid onto the MRS agar containing the growth of Bifidobacterium and Lactobacillus strains. After solidification of the culture medium at room temperature (25–28 °C), Clostridium spp. suspensions (McFarland standard No. 0.5, 1.5 × 108 colony-forming units per milliliter (CFU/mL)) were spread with the aid of a swab. The plates were incubated at 37 °C for 24 h under anaerobic conditions. The formation of a clear halo around growth of the probiotics was indicative of antimicrobial activity. The diameter of the growth inhibition halo was measured and expressed in millimeters.
2.2.2. Inhibition of Gas Production
The ability of probiotics to inhibit the growth of Clostridium strains was also evaluated by assessing the inhibition of gas production due to the fermentative action of the pathogens, as described previously [42], with some modifications. Briefly, the assay was performed by inoculating 1 μL (~107 UFC) of Clostridium culture into the upper third of the RCM agar layer (supplemented with 1.5 g/100 mL bacteriological agar), composed of 3 mL per tube. Subsequently, 3 mL of MRS containing 0.7 g% bacteriological agar was melted, cooled to 50 °C, and inoculated with 30 μL (~108 UFC) of each probiotic culture. The contents were homogenized by vortexing and immediately poured over the RCM agar layer in tubes inoculated with the Clostridium strains. RCM agar with Clostridium and MRS agar without inoculated probiotics were used as negative controls. The tubes were incubated under anaerobic conditions at 37 °C for 24 h. The assays were performed in triplicate, with and without buffering of the MRS medium with K2HPO4 and KH2PO4 (100 mM). A positive assay for antimicrobial activity was characterized by the absence of gas production, that is, in the absence of bubbles in the culture media, or medium breakage.
2.3. Antimicrobial Activity after pH Adjustment
The strain with the highest antimicrobial activity was selected to verify if growth inhibition of Clostridium spp. was due to the acidic pH. Culture supernatants were tested after pH adjustment essentially as described by Gaspar et al. [43]. After cultivation of the microorganism in MRS broth (Difco-BD), 10 mL of the bacterial culture was heated at 70 °C for 30 min to inhibit protease activity, cooled at room temperature, and centrifuged (5,000 × g for 15 min at 4 °C). Hydrogen peroxide was eliminated by the addition of 5 mg/mL catalase from bovine liver (Sigma-Aldrich, St Louis, MO, USA) followed by filtration through a 0.2 μm pore-size cellulose acetate (Whatman®, Clifton, NJ, USA). Antimicrobial activity of the cell-free culture supernatants (CFCN) was evaluated with and without pH adjustment to 6.5 with 10 M NaOH solution. Antimicrobial activity was evaluated by the antagonist well-diffusion method. Briefly, 20 mL of MRS soft agar (0.7% agar) was inoculated with 200 μL of each Clostridium strains. Wells with 4 mm diameter were punched in agar plates and filled with 100 μL of CFCN. Phosphate-buffered saline (PBS, pH 7.4) was used as a negative control. Plates were incubated under anaerobic conditions at 37 °C for 24 °C in an upright position. The inhibition zone diameters were measured and expressed in millimeters.
2.4. Coaggregation Test
After cultivation of the probiotic strains and Clostridium species, aliquots of 1 mL of each culture were washed twice with phosphate-buffered saline (PBS, pH 7.2), centrifuged at 5000× g for 15 min, and resuspended in PBS. The optical density of each suspension was adjusted (OD620nm = 0.1), and 500-μL aliquots of the probiotic suspensions were mixed with 500 μL of each pathogen suspension in 24-well plates (Nunc, Roskilde, Denmark) and incubated at 37 °C for 4 h under constant stirring (100 rpm) on an orbital shaker. Plates were observed for macroscopically visible clumps and under inverted microscopy [44]. Glass slides were also prepared with 5 μL of each suspension and evaluated under the microscope for visualization of bacterial coaggregates after Gram staining. L. fermentum ATCC 23271 was used as a positive control in this assay, since high coaggregation scores were previously demonstrated [45]. Control assays were performed with individual bacterial samples to assess their ability to autoaggregate.
2.5. Mucin Binding Assay
The ability of selected probiotic strains to bind to mucin was evaluated essentially as described by Tallon et al. [46]. A volume of 100 μL of a 10 mg/mL mucin solution in PBS (pH 7.2) was added to the wells of polystyrene microtiter plates (Nunc) and incubated overnight at 4 °C. The wells were washed twice with 200 μL PBS and saturated with a 2% (w/v) bovine serum albumin (BSA) solution (Sigma-Aldrich, St. Louis, MO, USA) for 4 h at 4 °C. Finally, the wells were washed twice with 200 μL PBS. At least four replicates were used to estimate the adhesion of a given strain. Probiotic cultures in MRS broth were washed three times in PBS, and the final suspension was standardized by spectrophotometry (OD600nm = 0.1). Aliquots of 100 μL of the bacterial suspension were added to each well, and the microplates were incubated at 37 °C for 1 h. After this, the wells were washed 12 times with 1 mL PBS to remove non-adherent bacteria. The wells were treated with 200 μL of 0.5% Triton X-100 (Sigma-Aldrich), and the plates were then incubated for 2 h at room temperature under orbital shaking to release the adhered bacteria. Then, the wells were scraped with a sterile tip and the number of bacteria with binding ability to mucin was estimated by serial decimal dilutions in PBS and plating on MRS agar, followed by incubation at 37 °C for 24 h under anaerobic conditions. L. fermentum ATCC 23271 was used as a positive control [45]. Mucin-containing wells without bacteria were used as negative controls.
2.6. Adhesion to Eukaryotic Cells
Adhesion to HeLa was evaluated according to the method of Carmo et al. [45]. A 300-μL aliquot of each potential probiotic cultured in MRS broth was washed three times with PBS (pH 7.4, Sigma-Aldrich) and the bacterial pellet was resuspended in 300 μL Dulbecco’s modified Eagle’s medium (DMEM, Sigma-Aldrich). Monolayers of HeLa cells grown in 24-well microplates (Nunc) containing DMEM supplemented with 10% fetal bovine serum (Gibco, Gaithersburg, MD, USA), with or without glass coverslips, were inoculated with 50 μL (~2.3 × 107 CFU) of bacterial suspension and incubated at 37 °C under 5% CO2 for 3 h. Then, each well was washed three times with PBS to remove non-adherent bacteria. For quantification of the adherent bacteria, the HeLa cell monolayers in the wells without coverslips were treated with 1 mL of 0.1% Triton X-100 (Sigma-Aldrich) for 5 min and scraped with the aid of a tip. Thereafter, serial decimal dilutions were spread on MRS agar plates and incubated at 37 °C for 24 h. The total number of bacteria adhered to the cells was expressed as CFU per milliliter. Wells with HeLa cells incubated in the absence of bacteria were used as negative controls. Visualization of bacterial adherence to eukaryotic cells was performed after fixation with methanol (Amresco, Gymea, Australia) and staining with May–Grunwald and Giemsa (Amresco). Gram staining was also used to better visualize gram-positive Lactobacillus adhered to the cells. The coverslips were then mounted on glass slides and visualized by light microscopy under a 100× oil immersion objective.
2.7. Tolerance to Acidic pH and Bile Salts
Tolerance of selected bacteria to acidic pH (2 and 4) and bile salts (0.5% and 1%, Oxgall, Sigma-Aldrich) was evaluated as previously described, with minor modifications [47]. Briefly, 900 μL MRS, adjusted to pH 2 or 4, or non-adjusted (control), or supplemented with 0%, 0.5%, or 1.0% (w/v) Oxgall (Sigma-Aldrich), was inoculated with 100 μL of a 24-h culture, which had been previously washed three times with PBS and resuspended in the same volume of MRS broth. After incubation at 37 °C for 3 h under anaerobic conditions, the percentage of viable bacteria relative to that in the control was determined by plate counting on MRS agar.
2.8. Antibiotic Susceptibility Testing
Antibiotic susceptibility of probiotics was determined by a modification of the agar overlay diffusion method, as previously described [48]. Commercial discs (Oxoid) containing different antibiotics, including ciprofloxacin (5 μg), clindamycin (2 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), penicillin (10 μg), rifampicin (5 μg), co-trimoxazole (25 μg), vancomycin (30 μg), and tetracycline (30 μg), were placed on MRS agar plates inoculated with Lactobacillus (108 CFU/mL). The plates were incubated anaerobically at 37 °C for 24 h. Antibiotic susceptibility was evaluated based on the diameter (in millimeters) of the growth inhibition zone around the discs [48]. The reference strain Staphylococcus aureus ATCC 25923 was used for quality control of antibiotic discs and tested in Mueller–Hinton agar, as recommended [49].
2.9. Statistical Analyses
Statistical analyses were performed using NCSS 11 Statistical Software (2016; NCSS, Kaysville, UT, USA). Adherence to eukaryotic cells and to mucin was expressed as log CFU/mL (±SD). Tolerance to acidic pH and to bile salts was compared to growth in standard MRS medium. The Shapiro–Wilk test was carried out and confirmed that all variables were normally distributed. Thus, all comparisons were carried out by the Student t-test. Statistical significance was established at p < 0.05. All assays were performed in three independent experiments conducted on three different days.
3. Results
3.1. Selection of Strains with Antimicrobial Activity against Clostridium spp.
The antimicrobial effects of potential probiotics, as determined by the agar spot test, are shown in Table 1. C. butyricum was inhibited by 11 strains (91.7%), whereas C. difficile and C. perfringens were inhibited by 9 (75%) strains each. The diameter of the inhibition zone varied among the clinical and reference probiotic strains. Of the 12 species tested, 8 (66.7%) exhibited antimicrobial activity against all three Clostridium species. The largest inhibition zones were produced by L. plantarum ATCC 8014, mainly against C. butyricum (17 mm). Only B. animalis 56.1 showed no inhibitory activity against any Clostridium species in this assay.
Table 1.
Inhibitory activity of Bifidobacterium and Lactobacillus against Clostridium spp. based on the agar spot test.
| Potential Probiotics | Diameter of Inhibition Zones (mm ±SD) of: | ||
|---|---|---|---|
| Clostridium butyricum | Clostridium difficile | Clostridium perfringens | |
| Bifidobacterium animalis 56.1 | 0 | 0 | 0 |
| Bifidobacterium bifidum 14.2 | 12 (1.6) | 12 (0.4) | 11 (1.7) |
| Bifidobacterium longum ATCC 15707 | 11 (0.4) | 12 (0.2) | 11 (1.4) |
| Bifidobacterium longum 49.3 | 12 (0.4) | 11 (0.0) | 10 (0.7) |
| Lactobacillus brevis ATCC 367 | 9 (0.4) | 0 | 0 |
| Lactobacillus delbrueckii ATCC 9649 | 10 (1.5) | 11 (0.7) | 12 (2.4) |
| Lactobacillus fermentum ATCC 23271 | 10 (1.1) | 10 (0.5) | 10 (0.6) |
| Lactobacillus fermentum 54.2 | 13 (0.7) | 9 (0,4) | 10 (0.0) |
| Lactobacillus paracasei ATCC 335 | 11 (0.5) | 12 (0.3) | 11 (0.4) |
| Lactobacillus plantarum ATCC 8014 | 17 (0.8) | 13 (1.1) | 13 (0.6) |
| Lactobacillus rhamnosus ATCC 9595 | 11 (0.0) | 0 | 0 |
| Lactobacillus rhamnosus GG ATCC 53103 | 10 (0.9) | 0 | 0 |
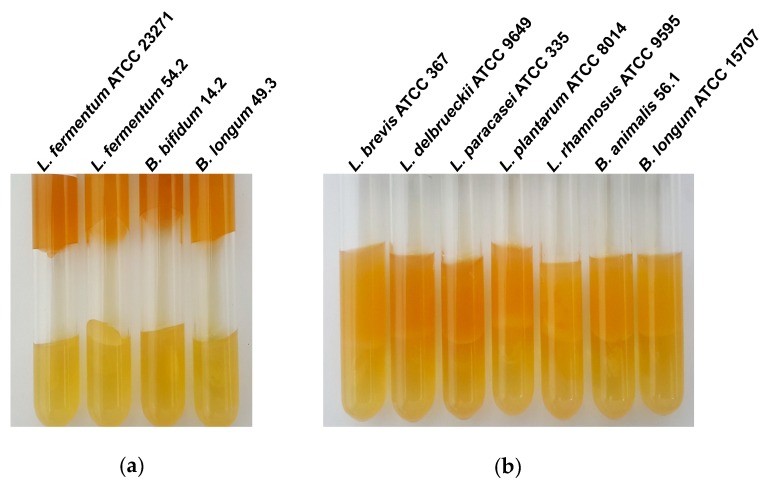
To assess whether or not the inhibitory activity on Clostridium spp. was due to acid produced by the probiotics, a preliminary attempt was made by supplementing MRS with phosphate buffer to evaluate the inhibition of gas production by C. butyricum ATCC 860 in culture medium. This test was also carried out in MRS medium without phosphate buffer. C. butyricum ATCC 860 was selected because this strain usually produces a large amount of gas resulting from its fermentative activity in thioglycolate medium or RCM. In the presence of buffer, the strains L. brevis ATCC 367, L. delbrueckii ATCC 9649, L. paracasei ATCC 335, L. plantarum ATCC 8014, L. rhamnosus ATCC 9595, B. animalis 56.1, and B. longum ATCC 15707 inhibited gas production by Clostridium spp., whereas the other strains did not (Figure 1). Among the strains that inhibited gas production in the absence of the phosphate buffer, only B. bifidum 14.2 gave a negative result when the assay was carried out in the of presence phosphate buffer in the MRS medium (Table 2).
Figure 1.
Inhibition of gas production assay. The lower layer corresponds to the RCM agar inoculated with Clostridium butyricum ATCC 860 and the upper layer is MRS medium with 0.7% agar and 100 mM phosphate buffer inoculated with Bifidobacterium and Lactobacillus strains; cultures were incubated at 37 °C for 24 h under anaerobic conditions. (a) Gas production by C. butyricum in buffered MRS medium, indicating the absence of inhibitory activity of Lactobacillus and Bifidobacterium strains. (b) Inhibitory activity of five Lactobacillus and two Bifidobacterium strains on gas production by Clostridium butyricum.
Table 2.
Inhibition of gas production by C. butyricum induced by probiotics grown in MRS medium with and without phosphate buffer.
| Strains | Without Buffer | With Buffer |
|---|---|---|
| B. animalis 56.1 | + | + |
| B. bifidum 14.2 | + | − |
| B. longum ATCC 15707 | + | + |
| B. longum 49.3 | − | − |
| L. brevis ATCC 367 | + | + |
| L. delbrueckii ATCC 9649 | + | + |
| L. fermentum ATCC 23271 | − | − |
| L. fermentum 54.2 | − | − |
| L. paracasei ATCC 335 | + | + |
| L. plantarum ATCC 8014 | + | + |
| L. rhamnosus ATCC 9595 | + | + |
L. plantarum ATCC 8014, the strain with the highest antimicrobial activity in the agar spot test for the three Clostridium strains, was selected for investigation of the antimicrobial activity in cell-free culture supernatants (CFCN) with and without pH adjustment to 6.5 with NaOH 10N, after growth in MRS broth. CFCN at pH 4.3, the final pH after cultivation of L. plantarum ATCC 8014, showed growth inhibition zones ranging from 14.2 (±0.8) mm to 16.5 (±0.8) mm, whereas CFCN at pH 6.5 presented inhibition zones from 13.2 mm (±0.4) mm to 14.7 (±0.5) mm. However, the inhibition zones of the CFSN at pH 4.3 in comparison to CFSN at pH 6.5 only presented statistical significance against C. butyricum (p < 0.05, Table 3).
Table 3.
Antimicrobial activity of cell-free culture supernatants (CFSN) of L. plantarum ATCC 8014 at pH 4.3 and 6.5 against Clostridium spp. by the agar overlay diffusion.
| Clostridium spp. | Inhibition Zone Diameters of CFSN in mm (±SD) at: | t 1 | p Value 1 | |
|---|---|---|---|---|
| pH 4.3 | pH 6.5 | |||
| C. butyricum ATCC 860 | 16.5 (0.5) | 14.7 (0.5) | 5.97 | 0.002 |
| C. difficile ATCC 9689 | 14.2 (0.8) | 13.2 (0.4) | 2.24 | 0.076 |
| C. perfringens ATCC 12924 | 14.3 (0.8) | 13.7 (0.5) | 1.35 | 0.235 |
1 Comparative analysis was performed by paired Student’s t-test (p < 0.05).
3.2. L. plantarum ATCC 8014 Presents Probiotic Potential
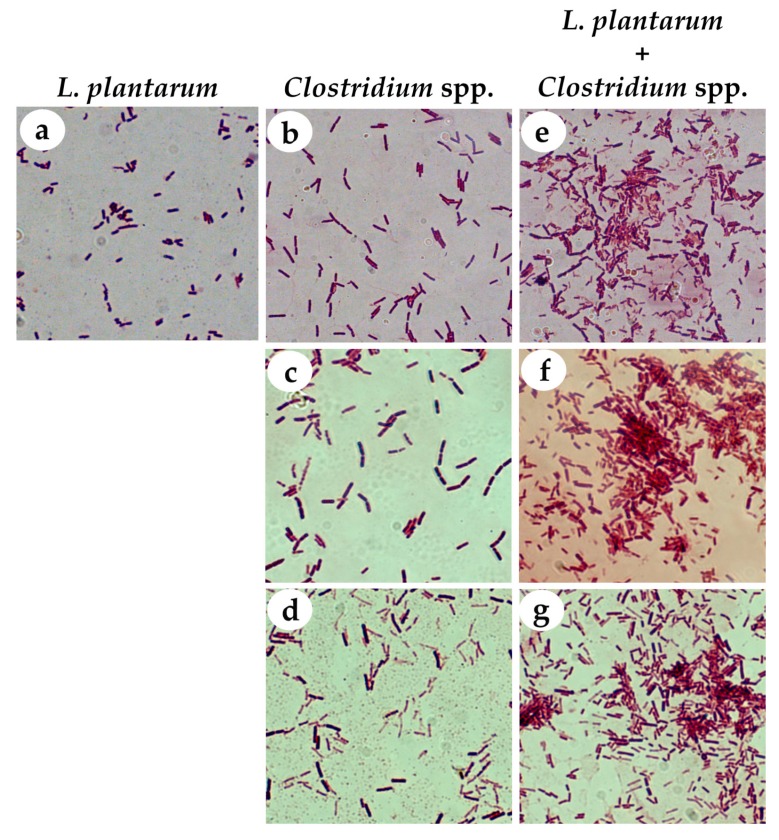
L. plantarum ATCC 8014 was selected for further analysis and characterization as a potential probiotic. In terms of its coaggregation capacity, we observed that L. plantarum ATCC 8014 interacted clearly with the pathogens, forming bacterial aggregates with the three species of Clostridium (Figure 2).
Figure 2.
Gram staining of bacterial isolates before (a–d) and after (e–g) the coaggregation assays of L. plantarum ATCC 8014 with the three species of Clostridium. (a) L. plantarum, (b) C. butyricum, (c) C. difficile, and (d) C. perfringens controls. Coaggregation of L. plantarum and (e) C. butyricum, (f) C. difficile, or (g) C. perfringens. Formation of bacterial aggregates were observed after mixing L. plantarum with each Clostridium strain. Gram stained slides were visualized by light microscopy under a 100× oil immersion objective.
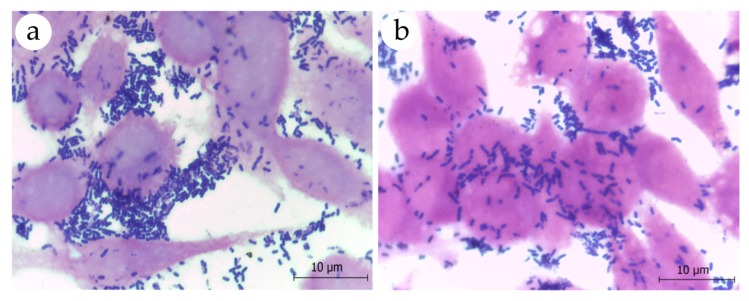
Investigation of the adhesion properties of L. plantarum ATCC 8014 revealed its ability to interact with eukaryotic cells, as observed in the assay with HeLa cells (Figure 3). In fact, adherence values for L. plantarum ATCC 8014 were higher than those observed for L. fermentum ATCC 23271 (Table 4, p = 0.0037). In contrast, L. fermentum ATCC 23271 exhibited a greater ability to bind mucin than L. plantarum ATCC 8014 (Table 4, p < 0.0001).
Figure 3.
Microscopic visualization of adhesion assays of (a) L. plantarum ATCC 8014 and (b) L. fermentum ATCC 23271 to HeLa cells after inoculation of approximately 107 CFU of bacterial suspensions. Cell monolayers grown on coverslips were stained by the Gram’s method and examined by light microscopy under a 100× oil immersion objective. Note the presence of numerous gram-positive bacilli adhered to HeLa cells.
Table 4.
Adherence quantification of L. plantarum ATCC 8014 to HeLa cells and to mucin in comparison with the control.
| Assays 1 |
L. plantarum ATCC 8014 |
L. fermentum ATCC 23271 |
p Value 2 |
|---|---|---|---|
| Cell adhesion | 7.602 (±0.135) | 7.349 (±0.053) | 0.0037 |
| Mucin binding | 5.057 (±0.062) | 5.370 (±0.031) | <0.0001 |
1 Data are expressed as mean log10 CFU/mL (standard deviations) of triplicate experiments performed on three independent days. 2 Comparative analysis was performed by Student’s t-test (p < 0.05).
L. plantarum ATCC 8014 exhibited growth at pH 2 and 4 after 180 min of exposure, with growth values of 70.3% and 97.8%, respectively, relative to that observed in standard MRS medium (Table 4). Assays performed at bile salt concentrations of 0.5% and 1.0% resulted in growth values of 110.8% and 90.1%, respectively, relative to that observed in standard medium. No differences were observed relative to the growth of L. fermentum strain ATCC 23271 (Table 5).
Table 5.
Survival of L. plantarum ATCC 8014 and L. fermentum ATCC 23271 in the presence of acidic pH and bile salts.
| Conditions | % Survival (±SD) 1 | p Value 2 | |
|---|---|---|---|
| L. plantarum | L. fermentum | ||
| pH 2.0 | 70.3 (±4.46) | 64.7 (±6.49) | 0.1155 |
| pH 4.0 | 97.8 (±5.67) | 106.2 (±9.36) | 0.7606 |
| Bile salts 0.5% | 110.8 (±12.04) | 112.7 (±8.79) | 0.0912 |
| Bile salts 1.0% | 90.1 (±3.77) | 92.6 (±3.07) | 0.2419 |
1 Data represent survival percentage of microorganisms after 180 min of exposure to distinct conditions in comparison to bacterial growth of each under standard conditions. 2 Comparative analysis was performed by the Student’s t-test (p < 0.05).
The antibiotic susceptibility profile of L. plantarum ATCC 8014 was evaluated using the overlay diffusion method. L. plantarum ATCC 8014 showed resistance to ciprofloxacin and vancomycin and sensitivity to all other antibiotics tested (Table 6).
Table 6.
Antimicrobial susceptibility of L. plantarum ATCC 8014 by the agar overlay diffusion method.
| Antibiotics | Inhibition Zone Diameters mm (±SD) | Interpretation 1 |
|---|---|---|
| Clindamycin | 25.7 (1.1) | Susceptible |
| Chloramphenicol | 20.3 (1.5) | |
| Erythromycin | 30.0 (1.0) | |
| Gentamicin | 14.5 (0.5) | |
| Penicillin | 33.7 (1.2) | |
| Rifampicin | 23.3 (0.6) | |
| Tetracycline | 31.0 (1.0) | |
| Co-trimoxazole | 14.3 (0.6) | Moderately susceptible |
| Ciprofloxacin | 9.3 (0.6) | Resistant |
| Vancomycin | 0 |
1 The interpretive criteria for the diameters of inhibition zones were those described by Chateris et al. [48].
4. Discussion
Some probiotic strains have been successfully used in clinical studies for the treatment or prevention of AAD [34,35] and NEC [36,37]. Their mechanisms of action still remain unclear, perhaps because it seems that this is not important since some probiotics interventions have been effective in certain patients with some reliability [40,50]. On the other hand, controversial findings regarding AAD and NEC interventions have been reported in the literature, as a positive effect does not always occur when used in cases of dysbiosis [51]. A better understanding of the relevant “central probiotic properties” that contribute to their inhibitory effects on major etiological agents of these clinical syndromes could therefore aid in the design of a more rational strategy for selecting and producing more effective probiotics [31].
In this study, we showed that different clinical and reference strains of Bifidobacterium and Lactobacillus present different levels of antimicrobial efficacy against C. butyricum, C. difficile, and C. perfringens. Evaluation of the antimicrobial activity of these bacteria (12 potential probiotic bacteria) indicated that L. plantarum strain ATCC 8014 exhibited the greatest capacity to inhibit the growth of the three reference strains of Clostridium, based on the activity detected using screening methods. In addition to L. plantarum ATCC 8014, other species showed inhibitory activity against one or all Clostridium strains. However, their antimicrobial activities were evidenced by lower zones of inhibition or were variable in comparison to the inhibition test of gas production. This variability in performance has previously been reported and suggests that more than one method should be used to assess the antimicrobial activity of probiotics, given that the conditions of each methodology may interfere with the results [52].
Various species of Lactobacillus are able to produce compounds with antimicrobial activities, including organic (acetic and lactic) acids, low-molecular-weight compounds, antifungal peptides, and antibacterial peptides (bacteriocins) [53,54]. It appears, however, that the inhibition of Clostridium growth exhibited by the probiotic strains was not the result of the overproduction of acids, since the addition of buffer to the MRS media did not affect the inhibitory activity of the majority of these species. Furthermore, some species of Clostridium exhibit intense fermentative activity, resulting in the production of large quantities of organic acids, including acetic, lactic, formic, butyric, and propionic acid, among other substances; thus, they would likely already be habituated to these and would survive acidic pH conditions [54,55]. In addition, as Lactobacillus and Bifidobacterium can produce a large amount of organic acids [55] and the concentration of the phosphate buffer used in the assay might not be sufficient to neutralize them, we tested the culture supernatants of L. plantarum ATCC 8014 at pH 4.3 and 6.5, but the inhibitory effect continued to be evidenced even after pH adjustment, although there was a higher activity against L. butyricum at pH 4.3.
This evidence allows us to suggest that other molecules produced by the probiotic strain are expected to be involved in their inhibitory actions on Clostridium strains. Indeed, a recent study demonstrated that Lactobacillus metabolites isolated from vaginal smears presented with in vitro bacteriostatic effects against C. perfringens [56]. These have been associated with the production of bacteriocins, which are produced by several species of probiotics and have bactericidal or bacteriostatic actions. Bacteriocins may increase the permeability of the inner membrane of bacteria, thus contributing to their rupture and interfering with bacterial cell wall synthesis, resulting in pore formation by binding to the peptidoglycan precursor lipid [57]. Lacticin 3147, for example, produced by Lactococcus lactis, forms selective pores in the cell walls of some pathogenic gram-positive bacteria, including C. difficile, resulting in its death [58].
The nature of the compound(s) produced by L. plantarum ATCC 8014 and their potential bactericidal or bacteriostatic actions are not yet known. However, even if such compounds are bacteriostatic or capable of inducing only sporulation, such actions could be relevant to controlling the clinical manifestation of infections caused by Clostridium, neutralizing the metabolic activity of the pathogen and, consequently, the production of toxins and other virulence factors involved. In addition, it is worth mentioning that Clostridium spp. are gram-positive bacteria and bacteriocins have a more targeted inhibitory action against this type of microorganism rather than against gram-negative bacteria [52,54].
Several criteria are used to define microorganisms as probiotics, including the ability to: (1) coaggregate with microbial pathogens; (2) adhere to eukaryotic cells and mucus; and (3) tolerate conditions of acidic pH and bile salts, among other properties [59,60]. L. plantarum ATCC 8014 fulfilled all these criteria, as it tolerated acidic pH and bile salts under the conditions tested, coaggregated with Clostridium spp., and exhibited adhesive properties suggesting its capacity for in vivo colonization. Although this bacterium demonstrated lower mucin binding than the L. fermentum strain ATCC 23271, the results obtained were of great relevance.
A worrying issue in the selection of probiotics to be used in foods or supplements is their potential to transmit genes involved in antibiotic resistance, especially if the microorganism in question carries plasmids [61,62]. Herein, L. plantarum ATCC 8014 showed resistance to ciprofloxacin and vancomycin. However, despite having plasmids [63], resistance to these antibiotics is considered intrinsic and non-transmissible in this species [64,65]. Of importance, vancomycin is among the antibiotic options to treat Clostridium infections [66,67]. Thus, we can assume that the development of a therapeutic strategy for intestinal disorders consisting of vancomycin administration in association with L. plantarum ATCC 8014 could be more effective than the single use of the antibiotic, if a synergistic effect is to be proven.
Currently, there is a large panel of probiotic strains in use. However, in most cases, probiotic action has been shown to be species-specific or even strain-specific [31,68,69,70]. Thus, the possibility of a potential probiotic with proven antimicrobial action against multiple Clostridium species, which are commonly associated with pathologies resulting from an imbalance in the microbiota, represents a significant advance for the reduction of morbidity and mortality rates arising from these clinical syndromes. In addition to being a more rational therapeutic approach, use of a probiotic would not have a negative impact on the intestinal microbiota and would not exert pressure for the selection of resistant bacteria, as with conventional antibiotic therapy [71].
Although the results presented herein are promising in terms of novel therapeutic strategies to treating intestinal infections caused by Clostridium spp., especially as L. plantarum ATCC 8014 is readily available to the scientific community as a culture collection strain, certain limitations of the study should be taken into consideration. Firstly, although there was an apparent cell monolayer integrity under microscopic examination of HeLa cells, its viability following incubation with the tested probiotic strain was not evaluated; thus, it is uncertain whether the adherence of L. plantarum ATCC 8014 was facilitated due to HeLa cell death. Also, it would be important to confirm the probiotic potential of L. plantarum ATCC 8014 in assays with clinical isolates of Clostridium, as not all reference strains tested in the present study are pathogenic, and to assure whether there is any variability in the spectrum of the antimicrobial activity against a larger panel of pathogenic isolates of C. butyricum, C. difficile, and C. perfringens. It is also important to highlight the need for addressing the safety of L. plantarum ATCC 8014 for oral administration in humans, especially in patients who have predisposing factors, such as preterm newborns or critically ill children [40]. Therefore, further studies in animal models and clinical trials would be essential to fill these gaps of knowledge and determine the in vivo benefits of L. plantarum ATCC 8014 against Clostridium spp.
5. Conclusions
Our findings allow us to conclude that L. plantarum strain ATCC 8014 has probiotic potential, with antimicrobial activity against C. butyricum ATCC 860, C. difficile ATCC 9689, and C. perfringens ATCC 12924. Additionally, this microorganism fulfills essential criteria to survive the harsh conditions of the gastrointestinal tract, as well as to colonize it.
Acknowledgments
The authors thank Hélio Euclides S. dos Santos for technical assistance.
Author Contributions
Conceptualization, C.R.A.V.M., M.S.d.C., E.S.F., and V.M.-N.; data curation, C.R.A.V.M. and S.G.M.; formal analysis, C.R.A.V.M., S.G.M., and V.M.-N.; funding acquisition, E.S.F. and V.M.-N.; investigation, C.R.A.V.M., M.S.d.C., B.O.M., M.S.A., C.I.d.S., M.R.Q.B., and E.S.F.; methodology, C.R.A.V.M., M.S.d.C., B.O.M., M.S.A., C.I.d.S., and M.R.Q.B.; project administration, V.M.-N.; resources, B.O.M., M.S.A., M.R.Q.B., E.S.F., and V.M.-N.; supervision, V.M.-N.; visualization, S.G.M.; writing—original draft, C.R.A.V.M. and V.M.-N.; writing—review and editing, E.S.F. and V.M.-N. All authors reviewed and approved the final manuscript.
Funding
This study was supported by grants from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001 to V.M.-N. and E.S.F.; Fundação de Amparo à Pesquisa e o Desenvolvimento Científico e Tecnológico do Maranhão (FAPEMA), Universal No.00995/17 and BEPP-02513/18 to V.M.-N.; and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPQ) grant No. 309046/2016-5 to E.S.F.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Perez-Lopez A., Behnsen J., Nuccio S.P., Raffatellu M. Mucosal immunity to pathogenic intestinal bacteria. Nat. Rev. Immunol. 2016;16:135–148. doi: 10.1038/nri.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.Reinoso Webb C., Koboziev I., Furr K.L., Grisham M.B. Protective and pro-inflammatory roles of intestinal bacteria. Pathophysiology. 2016;23:67–80. doi: 10.1016/j.pathophys.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonnenburg J.L., Bäckhed F. Diet–microbiota interactions as moderators of human metabolism. Nature. 2016;535:56. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Littman D.R., Pamer E.G. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conlon M.A., Bird A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donaldson G.P., Lee S.M., Mazmanian S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016;14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prosberg M., Bendtsen F., Vind I., Petersen A.M., Gluud L.L. The association between the gut microbiota and the inflammatory bowel disease activity: A systematic review and meta-analysis. Scand. J. Gastroenterol. 2016;51:1407–1415. doi: 10.1080/00365521.2016.1216587. [DOI] [PubMed] [Google Scholar]
- 9.McFarland L.V., Ozen M., Dinleyici E.C., Goh S. Comparison of pediatric and adult antibiotic-associated diarrhea and Clostridium difficile infections. World J. Gastroenterol. 2016;22:3078–3104. doi: 10.3748/wjg.v22.i11.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennet S.M., Ohman L., Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9:318–331. doi: 10.5009/gnl14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pammi M., Cope J., Tarr P.I., Warner B.B., Morrow A.L., Mai V., Gregory K.E., Kroll J.S., McMurtry V., Ferris M.J., et al. Intestinal dysbiosis in preterm infants preceding necrotizing enterocolitis: A systematic review and meta-analysis. Microbiome. 2017;5:31. doi: 10.1186/s40168-017-0248-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sircana A., Framarin L., Leone N., Berrutti M., Castellino F., Parente R., De Michieli F., Paschetta E., Musso G. Altered gut microbiota in type 2 diabetes: Just a coincidence? Curr. Diab. Rep. 2018;18:98. doi: 10.1007/s11892-018-1057-6. [DOI] [PubMed] [Google Scholar]
- 13.Durack J., Boushey H.A., Lynch S.V. Airway microbiota and the implications of dysbiosis in asthma. Curr. Allergy Asthma Rep. 2016;16:52. doi: 10.1007/s11882-016-0631-8. [DOI] [PubMed] [Google Scholar]
- 14.Boursier J., Mueller O., Barret M., Machado M., Fizanne L., Araujo-Perez F., Guy C.D., Seed P.C., Rawls J.F., David L.A. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilg H., Adolph T.E., Gerner R.R., Moschen A.R. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Tremlett H., Bauer K.C., Appel-Cresswell S., Finlay B.B., Waubant E. The gut microbiome in human neurological disease: A review. Ann. Neurol. 2017;81:369–382. doi: 10.1002/ana.24901. [DOI] [PubMed] [Google Scholar]
- 17.Lezutekong J.N., Nikhanj A., Oudit G.Y. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in cardiovascular disease. Clin. Sci. 2018;132:901–904. doi: 10.1042/CS20180172. [DOI] [PubMed] [Google Scholar]
- 18.Rupnik M., Wilcox M.H., Gerding D.N. Clostridium difficile infection: New developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 2009;7:526. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 19.Olsen M.A., Yan Y., Reske K.A., Zilberberg M.D., Dubberke E.R. Recurrent Clostridium difficile infection is associated with increased mortality. Clin. Microbiol. Infect. 2015;21:164–170. doi: 10.1016/j.cmi.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 20.Coursey C.A., Hollingsworth C.L., Wriston C., Beam C., Rice H., Bisset G., 3rd Radiographic predictors of disease severity in neonates and infants with necrotizing enterocolitis. AJR Am. J. Roentgenol. 2009;193:1408–1413. doi: 10.2214/AJR.08.2306. [DOI] [PubMed] [Google Scholar]
- 21.Shah T.A., Meinzen-Derr J., Gratton T., Steichen J., Donovan E.F., Yolton K., Alexander B., Narendran V., Schibler K.R. Hospital and neurodevelopmental outcomes of extremely low-birth-weight infants with necrotizing enterocolitis and spontaneous intestinal perforation. J. Perinatol. 2012;32:552–558. doi: 10.1038/jp.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fitzgibbons S.C., Ching Y., Yu D., Carpenter J., Kenny M., Weldon C., Lillehei C., Valim C., Horbar J.D., Jaksic T. Mortality of necrotizing enterocolitis expressed by birth weight categories. J. Pediatr. Surg. 2009;44:1072–1075. doi: 10.1016/j.jpedsurg.2009.02.013. discussion 1075–1076. [DOI] [PubMed] [Google Scholar]
- 23.Neu J., Walker W.A. Necrotizing enterocolitis. N. Engl. J. Med. 2011;364:255–264. doi: 10.1056/NEJMra1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coggins S.A., Wynn J.L., Weitkamp J.-H. Infectious causes of necrotizing enterocolitis. Clin. Perinatol. 2015;42:133–154. doi: 10.1016/j.clp.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cassir N., Benamar S., Khalil J.B., Croce O., Saint-Faust M., Jacquot A., Million M., Azza S., Armstrong N., Henry M. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin. Infect. Dis. 2015;61:1107–1115. doi: 10.1093/cid/civ468. [DOI] [PubMed] [Google Scholar]
- 26.Sato Y., Kujirai D., Emoto K., Yagami T., Yamada T., Izumi M., Ano M., Kase K., Kobayashi K. Necrotizing enterocolitis associated with Clostridium butyricum in a Japanese man. Acute Med. Surg. 2018;5:194–198. doi: 10.1002/ams2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schonherr-Hellec S., Klein G.L., Delannoy J., Ferraris L., Roze J.C., Butel M.J., Aires J. Clostridial strain-specific characteristics associated with necrotizing enterocolitis. Appl. Environ. Microbiol. 2018;84:e02428-17. doi: 10.1128/AEM.02428-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cassir N., Benamar S., La Scola B. Clostridium butyricum: From beneficial to a new emerging pathogen. Clin. Microbiol. Infect. 2016;22:37–45. doi: 10.1016/j.cmi.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Uzal F.A., Freedman J.C., Shrestha A., Theoret J.R., Garcia J., Awad M.M., Adams V., Moore R.J., Rood J.I., McClane B.A. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future Microbiol. 2014;9:361–377. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asha N., Tompkins D., Wilcox M. Comparative analysis of prevalence, risk factors, and molecular epidemiology of antibiotic-associated diarrhea due to Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus. J. Clin. Microbiol. 2006;44:2785–2791. doi: 10.1128/JCM.00165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 32.Kelly C.R., de Leon L., Jasutkar N. Fecal microbiota transplantation for relapsing Clostridium difficile infection in 26 patients: Methodology and results. J. Clin. Gastroenterol. 2012;46:145–149. doi: 10.1097/MCG.0b013e318234570b. [DOI] [PubMed] [Google Scholar]
- 33.Walker A.W., Lawley T.D. Therapeutic modulation of intestinal dysbiosis. Pharmacol. Res. 2013;69:75–86. doi: 10.1016/j.phrs.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Hempel S., Newberry S.J., Maher A.R., Wang Z., Miles J.N., Shanman R., Johnsen B., Shekelle P.G. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: A systematic review and meta-analysis. JAMA. 2012;307:1959–1969. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- 35.Lau C.S., Chamberlain R.S. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: A systematic review and meta-analysis. Int. J. Gen. Med. 2016;9:27. doi: 10.2147/IJGM.S98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AlFaleh K., Anabrees J. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Evid. Based Child Health. 2014;9:584–671. doi: 10.1002/ebch.1976. [DOI] [PubMed] [Google Scholar]
- 37.Underwood M.A. Impact of probiotics on necrotizing enterocolitis. Semin. Perinatol. 2017;41:41–51. doi: 10.1053/j.semperi.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanders M.E., Benson A., Lebeer S., Merenstein D.J., Klaenhammer T.R. Shared mechanisms among probiotic taxa: Implications for general probiotic claims. Curr. Opin. Biotechnol. 2018;49:207–216. doi: 10.1016/j.copbio.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Lebeer S., Bron P.A., Marco M.L., Van Pijkeren J.P., O’Connell Motherway M., Hill C., Pot B., Roos S., Klaenhammer T. Identification of probiotic effector molecules: Present state and future perspectives. Curr. Opin. Biotechnol. 2018;49:217–223. doi: 10.1016/j.copbio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 40.Singhi S.C., Kumar S. Probiotics in critically ill children. F1000Res. 2016;5 doi: 10.12688/f1000research.7630.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anas M., Eddine H.J., Mebrouk K. Antimicrobial activity of Lactobacillus species isolated from Algerian raw goat’s milk against Staphylococcus aureus. World J. Dairy Food Sci. 2008;3:39–49. [Google Scholar]
- 42.Golic N., Veljovic K., Popovic N., Djokic J., Strahinic I., Mrvaljevic I., Terzic-Vidojevic A. In vitro and in vivo antagonistic activity of new probiotic culture against Clostridium difficile and Clostridium perfringens. BMC Microbiol. 2017;17:108. doi: 10.1186/s12866-017-1015-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaspar C., Donders G.G., Palmeira-de-Oliveira R., Queiroz J.A., Tomaz C., Martinez-de-Oliveira J., Palmeira-de-Oliveira A. Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Express. 2018;8:153. doi: 10.1186/s13568-018-0679-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reid G., McGroarty J.A., Domingue P.G., Chow A.W., Bruce A.W., Eisen A., Costerton J.W. Coaggregation of urogenital bacteria in vitro and in vivo. Curr. Microbiol. 1990;20:47–52. doi: 10.1007/BF02094024. [DOI] [Google Scholar]
- 45.Do Carmo M.S., Noronha F.M., Arruda M.O., Costa E.P., Bomfim M.R., Monteiro A.S., Ferro T.A., Fernandes E.S., Giron J.A., Monteiro-Neto V. Lactobacillus fermentum ATCC 23271 displays in vitro inhibitory activities against Candida spp. Front. Microbiol. 2016;7:1722. doi: 10.3389/fmicb.2016.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tallon R., Arias S., Bressollier P., Urdaci M. Strain-and matrix-dependent adhesion of Lactobacillus plantarum is mediated by proteinaceous bacterial compounds. J. Appl. Microbiol. 2007;102:442–451. doi: 10.1111/j.1365-2672.2006.03086.x. [DOI] [PubMed] [Google Scholar]
- 47.Munoz-Quezada S., Chenoll E., Vieites J.M., Genoves S., Maldonado J., Bermudez-Brito M., Gomez-Llorente C., Matencio E., Bernal M.J., Romero F., et al. Isolation, identification and characterisation of three novel probiotic strains (Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036) from the faeces of exclusively breast-fed infants. Br. J. Nutr. 2013;109(Suppl. 2):S51–S62. doi: 10.1017/S0007114512005211. [DOI] [PubMed] [Google Scholar]
- 48.Charteris W.P., Kelly P.M., Morelli L., Collins J.K. Antibiotic susceptibility of potentially probiotic Lactobacillus species. J. Food Prot. 1998;61:1636–1643. doi: 10.4315/0362-028X-61.12.1636. [DOI] [PubMed] [Google Scholar]
- 49.CLSI . Perfomance Standards for Antimicrobial Susceptibility Testing (Supplement M100) 27th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2017. p. 250. [Google Scholar]
- 50.Iannitti T., Palmieri B. Therapeutical use of probiotic formulations in clinical practice. Clin. Nutr. 2010;29:701–725. doi: 10.1016/j.clnu.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aceti A., Gori D., Barone G., Callegari M.L., Di Mauro A., Fantini M.P., Indrio F., Maggio L., Meneghin F., Morelli L., et al. Probiotics for prevention of necrotizing enterocolitis in preterm infants: Systematic review and meta-analysis. Ital. J. Pediatr. 2015;41:89. doi: 10.1186/s13052-015-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coman M., Verdenelli M., Cecchini C., Silvi S., Orpianesi C., Boyko N., Cresci A. In vitro evaluation of antimicrobial activity of Lactobacillus rhamnosus IMC 501®, Lactobacillus paracasei IMC 502® and SYNBIO® against pathogens. J. Appl. Microbiol. 2014;117:518–527. doi: 10.1111/jam.12544. [DOI] [PubMed] [Google Scholar]
- 53.Do Carmo M.S., Santos C.I.D., Araujo M.C., Giron J.A., Fernandes E.S., Monteiro-Neto V. Probiotics, mechanisms of action, and clinical perspectives for diarrhea management in children. Food Funct. 2018;9:5074–5095. doi: 10.1039/C8FO00376A. [DOI] [PubMed] [Google Scholar]
- 54.Nes I.F., Kjos M., Diep D.B. Antimicrobial Components of Lactic Acid Bacteria. In: Lahtinen A., Ouwehand A.C., Salminen S., Wright A., editors. Lactic Acid Bacteria: Microbiological and Functional Aspects. 4th ed. CRC Press; Boca Raton, FL, USA: 2012. pp. 285–330. [Google Scholar]
- 55.Lee Y.K., Salminen S. Handbook of Probiotics and Prebiotics. John Wiley & Sons; Hoboken, NJ, USA: 2009. [Google Scholar]
- 56.Amin M., Moradi Choghakabodi P., Alhassan Hamidi M., Najafian M., Farajzadeh Sheikh A. In vitro antimicrobial activities of metabolites from vaginal Lactobacillus strains against Clostridium perfringens isolated from a woman’s vagina. J. Chin. Med. Assoc. 2017;80:29–33. doi: 10.1016/j.jcma.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Wiedemann I., Bottiger T., Bonelli R.R., Schneider T., Sahl H.G., Martinez B. Lipid II-based antimicrobial activity of the lantibiotic plantaricin C. Appl. Environ. Microbiol. 2006;72:2809–2814. doi: 10.1128/AEM.72.4.2809-2814.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rea M.C., Clayton E., O’Connor P.M., Shanahan F., Kiely B., Ross R.P., Hill C. Antimicrobial activity of lacticin 3,147 against clinical Clostridium difficile strains. J. Med. Microbiol. 2007;56:940–946. doi: 10.1099/jmm.0.47085-0. [DOI] [PubMed] [Google Scholar]
- 59.Kos B., Šušković J., Vuković S., Šimpraga M., Frece J., Matošić S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J. Appl. Microbiol. 2003;94:981–987. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 60.Van Tassell M.L., Miller M.J. Lactobacillus adhesion to mucus. Nutrients. 2011;3:613–636. doi: 10.3390/nu3050613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devirgiliis C., Zinno P., Perozzi G. Update on antibiotic resistance in foodborne Lactobacillus and Lactococcus species. Front. Microbiol. 2013;4:301. doi: 10.3389/fmicb.2013.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FAO/WHO . Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. FAO/WHO; Rome, Italy: 2001. pp. 1–4. [Google Scholar]
- 63.Posno M., Leer R.J., van Luijk N., van Giezen M.J., Heuvelmans P.T., Lokman B.C., Pouwels P.H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fraqueza M.J. Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int. J. Food Microbiol. 2015;212:76–88. doi: 10.1016/j.ijfoodmicro.2015.04.035. [DOI] [PubMed] [Google Scholar]
- 65.Shao Y., Zhang W., Guo H., Pan L., Zhang H., Sun T. Comparative studies on antibiotic resistance in Lactobacillus casei and Lactobacillus plantarum. Food Control. 2015;50:250–258. doi: 10.1016/j.foodcont.2014.09.003. [DOI] [Google Scholar]
- 66.Brook I. Clostridial Infections in Children: Spectrum and Management. Curr. Infect. Dis. Rep. 2015;17:47. doi: 10.1007/s11908-015-0503-8. [DOI] [PubMed] [Google Scholar]
- 67.Debast S., Bauer M., Kuijper E., Committee European Society of Clinical Microbiology and Infectious Diseases: Update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2014;20:1–26. doi: 10.1111/1469-0691.12418. [DOI] [PubMed] [Google Scholar]
- 68.Agustina R., Kok F.J., van de Rest O., Fahmida U., Firmansyah A., Lukito W., Feskens E.J., van den Heuvel E.G., Albers R., Bovee-Oudenhoven I.M. Randomized trial of probiotics and calcium on diarrhea and respiratory tract infections in Indonesian children. Pediatrics. 2012;129:e1155–e1164. doi: 10.1542/peds.2011-1379. [DOI] [PubMed] [Google Scholar]
- 69.Bron P.A., Tomita S., Mercenier A., Kleerebezem M. Cell surface-associated compounds of probiotic lactobacilli sustain the strain-specificity dogma. Curr. Opin. Microbiol. 2013;16:262–269. doi: 10.1016/j.mib.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 70.Snel J., Vissers Y.M., Smit B.A., Jongen J.M., van der Meulen E.T., Zwijsen R., Ruinemans-Koerts J., Jansen A.P., Kleerebezem M., Savelkoul H.F. Strain-specific immunomodulatory effects of Lactobacillus plantarum strains on birch-pollen-allergic subjects out of season. Clin. Exp. Allergy. 2011;41:232–242. doi: 10.1111/j.1365-2222.2010.03650.x. [DOI] [PubMed] [Google Scholar]
- 71.Hughes D., Andersson D.I. Evolutionary Trajectories to Antibiotic Resistance. Annu. Rev. Microbiol. 2017;71:579–596. doi: 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]