Abstract
Basic helix-loop-helix family member e41 (BHLHE41) serves an important role in regulating cell differentiation, circadian rhythms and the response to hypoxia. However, the roles of BHLHE41 in clear cell renal cell carcinoma (ccRCC) remain unclear. The aim of the present study was to analyze the expression of BHLHE41 in ccRCC and investigate the effect of downregulated BHLHE41 on the growth and migration of ccRCC cells. The expression of BHLHE41 in ccRCC was demonstrated to be significantly increased in gene expression microarray datasets and RNA sequencing data. Reverse transcription-quantitative polymerase chain reaction and western blot analysis demonstrated that BHLHE41 expression in fresh ccRCC tissues was increased, compared with than their adjacent non-tumorous controls. BHLHE41 knockdown significantly reduced cell proliferation and migration of A498 and CAKI-1 cells. For the investigation of the molecules mediated by BHLHE41, immunoblotting analyses revealed that phosphorylation of p70S6K and protein levels of E-cadherin were reduced. Additionally, a lower frequency methylation was determined in the BHLHE41 3′-untranslated region through The Cancer Genome Atlas dataset analysis for the first time. These observations demonstrated that BHLHE41 could be a biomarker and an oncogene for ccRCC.
Keywords: clear cell renal cell carcinoma, basic helix-loop-helix family member e41, cell proliferation, DNA methylation
Introduction
Renal cell carcinoma (RCC) is one of the most common urologic cancer types. Its incidence rate is 2–3% of adult malignant tumor cases (1). Clear cell RCC (ccRCC) is the main subtype of RCC, and ~30% of patients with ccRCC have distal metastasis at the time of diagnosis (2,3). Additionally, 20–40% of patients will suffer from local recurrence or distant metastasis following resection (2,3). Therefore, investigating the molecular mechanism is important for this deadly disease. Identifying novel genes as diagnostic and therapeutic targets is beneficial for the diagnosis of the disease and provides more options for treatment.
Basic helix-loop-helix family member e41 (BHLHE41) is a member of the BHLH family proteins that are involved in differentiation, circadian rhythms and the response to hypoxia (4–6). Previous studies demonstrated that BHLHE41 serves a different role in tumors. BHLHE41 inhibits tumor proliferation and metastasis in pancreatic (7) and gastric cancer (8). However, BHLHE41 is associated with hypoxia inducible factor-1 (HIF-1) activation and promotes tumor invasion in osteosarcoma (9). BHLHE41 expression antagonizes paclitaxel-induced cell apoptosis in human prostate cancer cells (10).
The roles of BHLHE41 in ccRCC cell proliferation and migration have rarely been reported. In the present study, the expression of BHLHE41 in ccRCC tissues, the tumor-promoting effects of BHLHE41 in ccRCC cell lines A498 and CAKI-1, and the DNA methylation level of the gene were examined.
Materials and methods
Clinical specimens
A total of 50 ccRCC tissues and their matched normal adjacent tissues (>2 cm from the edge of the cancer tissue) were obtained from Fuzhou General Hospital (Fuzhou, Fujian) from November 2013 to November 2015. The separation and use of human tissues was approved by the Human Research Ethics Review Committee of Fuzhou General Hospital (approval no. 2013-017). Patients ranged in age from 28–77 years with the mean age of 55.5 years. All patients provided written informed consent. These patients were staged according to the Tumor-Node-Metastasis classification system of malignant tumors (7th) (11). Table I presents the patient's details.
Table I.
List of 50 clear cell renal cell carcinoma tissues.
| Characteristics | n (%) |
|---|---|
| Sex | |
| Male | 35 (70) |
| Female | 15 (30) |
| Age at surgery | |
| <60 | 30 (60) |
| ≥60 | 20 (40) |
| Tumor extenta | |
| T1 | 41 (82) |
| T2 | 7 (14) |
| T3 | 2 (4) |
| T4 | 0 (0) |
| Lymph node metastasisa | |
| N0 | 48 (96) |
| ≥N1 | 2 (4) |
| Distant metastasis a | |
| M0 | 48 (96) |
| M1 | 2 (4) |
| Tumor max diameter (cm)a | |
| <7 | 41 (82) |
| ≥7 | 9 (18) |
| Fuhrman gradeb | |
| G1 | 16 (32) |
| G2 | 33 (66) |
| G3 | 1 (2) |
| G4 | 0 (0) |
Cell lines
A498 cells were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China); CAKI-1 cells were obtained from Shanghai GeneChem Co., Ltd. (Shanghai, China). A498 cells are Von Hippel-Lindau tumor suppressor (VHL) mutant cells and CAKI cells are VHL wild-type cells (12). The cell lines used were all identified using short tandem repeat markers. A498 and CAKI-1 cells were maintained at 37°C and 5% CO2 in RPMI-1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific Inc.), and 1% penicillin/streptomycin (Thermo Fisher Scientific, Inc.).
Analysis of gene expression omnibus (GEO) and the cancer genome atlas (TCGA) data of ccRCC. Oncomine (http://www.oncomine.org) was used to select highly expressed genes in Beroukhim Renal_GSE14994 (13), Grumz Renal_GSE6344 (14), Jones Renal_GSE15641 (15) and Lenburg Renal_GSE781 (16). Gene expression microarray datasets (GSE53757) (17) were analyzed and normalized by GEODiver (http://www.geodiver.co.uk). The primary dataset was downloaded from the NCBI GEO database (http://ncbi.nlm.nig.gov/geo) (18). Kidney Renal Clear Cell Carcinoma (KIRC) mRNA expression and clinical data was downloaded from TCGA (http://cancergenome.nig.gov) (19). The transcriptome profiling data files for analysis were normalized and transferred into a txt file using a Perl script. Subsequently, the package edgeR of Bioconductor was used in RStudio (version 3.5.0; http://www.rstudio.com/) to screen out the differentially-expressed genes (DEGs) with a fold change >2, P< 0.01 and a false discovery rate value <0.01, which were considered statistically significant (20). Relative gene expression of BHLHE41 was reflected by the intensity of the probe signal and plotted using GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Kaplan-Meier survival curves were generated by Oncolnc (http://www.oncolnc.org/) for patients with ccRCC entered in TCGA database.
TCGA methylation analysis
Methylation data (Illumina Infinium Human Methylation 450 K) (21), which included 325 ccRCC tumor samples and 160 normal samples, were downloaded from TCGA data portal in April 2018. Samples with expression and methylation data were screened. Individual gene methylation was extracted from the fasta file using a custom Perl script. The correlation between DNA methylation/loci methylation level and BHLHE41 gene expression was investigated with Spearman's test.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA Kit II Kit (Omega Bio-Tek, Inc., Norcross, GA, USA) was used to extract RNA from tissues. Subsequently, 3 µg total RNA was reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). SYBR-Green select Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to qPCR. cDNA was subjected to RT-qPCR with BHLHE41 and β-actin primers. BHLHE41 upstream primer, 5′-AAGGAGCATGAAACGAGACGA-3′, and downstream primer, 5′-CTCGGTTAAGGCGGTTAAAGC-3′. β-actin upstream primer, 5′-TGACGTGGACATCCGCAAAG-3′, and downstream primer, 5′-CTGGAAGGTGGACAGCGAGG-3′. PCR was conducted for 40 cycles with 20 pmol primers under the following conditions: 94°C for 30 sec, 56°C for 30 sec and 72°C for 30 sec. Relative fold changes in mRNA expression were calculated by normalization to β-actin mRNA using the formula 2−∆∆Cq (22).
Western blot analysis
Tissue samples, and A498 and CAKI-1 cell extracts were prepared by a Protein Extraction reagent (cat. no. P0013; Beyotime Institute of Biotechnology, Haimen, China). A Bicinchoninic acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.) was used to quantify total protein. A total of 15 µg protein from each sample were separated on 10% SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membranes (Immobilon-P PVDF Membrane; EMD Millipore, Billerica, MA, USA). Membranes were blocked with 5% non-fat milk at room temperature for 1 h, and then incubated with primary antibodies (BHLHE41, p-AKT, AKT, p-p70S6K, p70S6K, β-catenin, E-cadherin and ACTIN) overnight at 4°C. Subsequently, membranes were incubated with HRP-conjugated secondary IgG antibodies (anti-rabbit IgG, cat. no. 7074; and anti-mouse IgG, cat. no. 7076) (both 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA) at room temperature for 1 h. Proteins were detected using a SuperSignal West Pico chemiluminescent kit (Pierce; Thermo Fisher Scientific, Inc.). BHLHE41 antibody (cat. no. ab190093; 1:1,000) was purchased from Abcam (Cambridge, UK). phospho (p)-AKT (cat. no. 9271; 1:1,000), AKT (cat. no. 9272; 1:1,000), p-p70S6K (cat. no. 9234; 1:1,000), p70S6K (cat. no. 2708; 1:1,000), β-catenin (cat. no. 8480; 1:1,000) and E-cadherin (cat. no. 3195; 1:1,000) antibodies were purchased from Cell Signaling Technology Inc. β-actin antibody (cat. no. A2228; 1:5,000) was purchased from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany).
Lentivirus production
BHLHE41 short hairpin RNA (shRNA; pLKO.1-BHLHE41-shRNA, TRCN0000016946) and non-targeting control SHCOO2 (shRNA-NC) vector were purchased from Sigma-Aldrich (Merck KGaA). For virus packaging, 3 µg non-targeting sequence control or BHLHE41 shRNA constructs were co-transfected with 1 µg pMD2.G and 2 µg psPax2 into 293T cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). A498 and CAKI-1 cells were infected with virus media. The media was changed after 48 h and RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin containing 1 µg/ml puromycin was added at 37°C for 10 days. Subsequently, the stably transfected cells were obtained. Inhibition efficiency of gene expression detected by RT-qPCR (data not shown) and verified with a western blot assay as previously described.
WST-1 for cell proliferation assay
Selected transgenic A498 and CAKI-1 cell proliferation was measured using a WST-1 assay kit (Roche Diagnostics GmbH, Mannheim, Germany). At 24, 48, 72 and 96 h after cell plating at 37°C, 10% WST-1 solution was added to adherent cells, and then incubated at 37°C for 45 min. The coloration of the solution was measured at 450 nm using a microplate reader.
BrdU staining
Selected transgenic A498 and CAKI-1 cells were seeded in a 24-well plate at 5×103 cells/cm2, and then cultured with RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C for 1 day, the cells were pulsed with 10 µM BrdU at 37°C for 1 h and fixed in 4% polyparaformaldehyde at room temperature for 15 min. Fixed cells were labeled with anti-BrdU antibody (cat. no. 5292; 1:100; Cell Signaling Technology, Inc.) at 4°C overnight, and then washed 3 times. Subsequently, the cells were treated with anti-mouse Alexa-488 (cat. no. 715-545-450; 1:100; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) at 37°C for 30 min, and counterstained with DAPI (Sigma-Aldrich; Merck KGaA) at room temperature for 5 min. Photomicrographs were captured using an epifluorescence microscope (X81; Olympus Corporation, Tokyo, Japan; ×100).
Transwell migration assay
Selected transgenic A498 and CAKI-1 cells were harvested for migration assays in 24-well plates with 8-µm pore size Transwell microporous membranes (EMD Millipore). Subsequently, 200 µl of the A498 and CAKI-1 cell suspension (containing 3×104 cells) was seeded in the upper chamber with serum-free RPMI-1640 medium, and the lower chamber was supplemented with 10% fetal bovine serum with 500 µl RPMI-1640 medium. After incubating for 36 h at 37°C in a 5% CO2 atmosphere, the cells were washed with PBS 3 times and fixed in 4% paraformaldehyde at room temperature for 15 min. The cells on the upper surface of the membrane were then removed with a cotton swab, and the migrating cells on the bottom surface were stained with 0.5% crystal violet at room temperature for 4 h. The number of cells in 5 random fields was counted under an epifluorescence microscope (X81; Olympus Corporation; ×100) and the mean was recorded.
Statistical processing
Data were collected from at least three technical replicates and expressed as the mean ± standard deviation. Statistical analyses of transcriptome profiling data results were performed in RStudio (version 3.5.0) software. Statistical differences between the groups were analyzed with the unpaired Student's t-test or one-way analysis of variance with the Bonferroni correction for multiple comparisons. All the data were graphically displayed using GraphPad Prism 6 software. P<0.05 was considered to indicate a statistically significant difference. Kaplan-Meier analysis of overall survival for patient's estimates based on all available data and compared with the log-rank test.
Results
BHLHE41 expression in GEO data
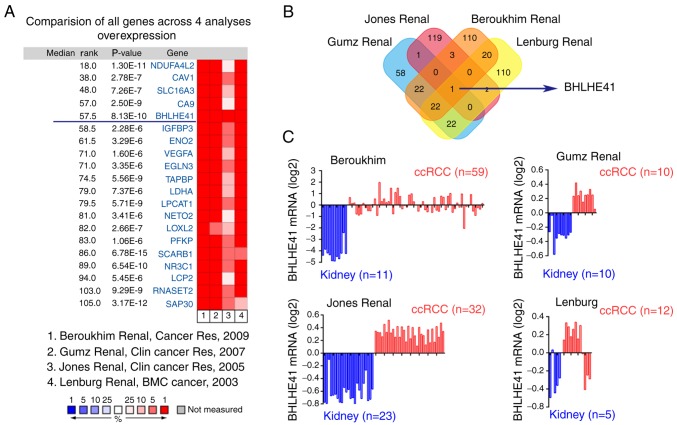
To search for ccRCC oncogenes, data mining was performed on the Oncomine database and 4 datasets were selected, which were analyzed using Affymetrix genome array (Human Genome U133A Array) (the detection method of the reference data). BHLHE41 was determined to be one of the highly DEGs (Fig. 1A). Fig. 1B depicts that BHLHE41 was the only overlapping gene among all of the 4 datasets. BHLHE41 is highly expressed in human ccRCC samples, compared with adjacent normal renal tissues (Fig. 1C) (Beroukhim Renal non-hereditary and hereditary ccRCC, n=59, fold change=19.414, P<0.001; Grumz Renal ccRCC, n=10, fold change=1.47, P<0.001; Jones Renal ccRCC, n=32, fold change=1.91, P<0.001; and Lenburg Renal ccRCC, n=12, fold change=1.32, P<0.001).
Figure 1.
BHLHE41 expression is upregulated in ccRCC based on Oncomine data mining. (A) Differential expression analysis was performed using Oncomine online analysis tools. (B) Venn diagram analysis of the upregulated top 1% (changes in expression) genes based on Beroukhim Renal (13), Gumz Renal (14), Jones Renal (15) and Lenburg Renal (16) data from the Oncomine database. (C) Bar charts with the BHLHE41 mRNA levels (log2) in the datasets used in (B). Blue represents low expression of BHLHE41 and red represents high expression of BHLHE41. ccRCC, clear cell renal cell carcinoma; BHLHE41, basic helix-loop-helix family member e41.
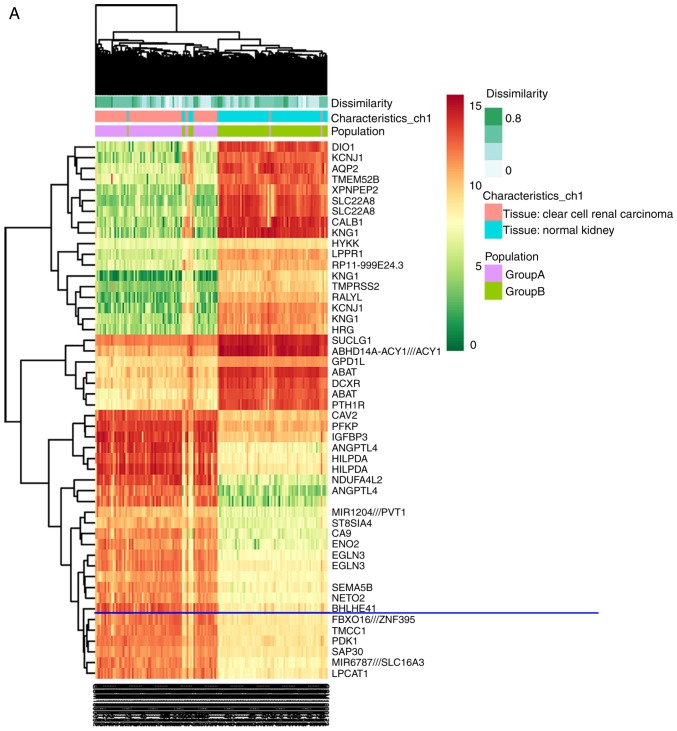
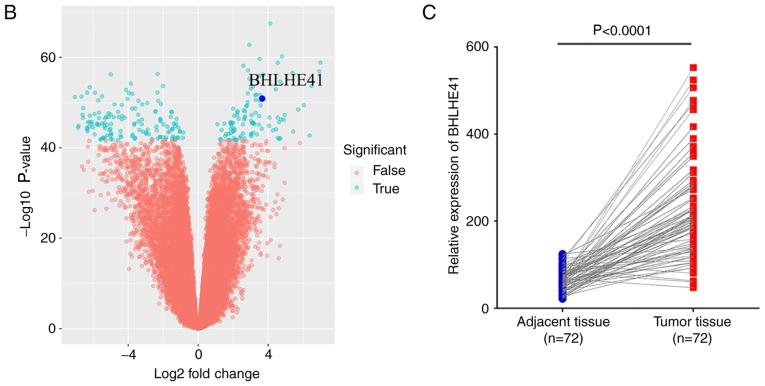
Subsequently, analysis of BHLHE41 expression from a large cohort ccRCC dataset (GSE53757, 72 pairs, HG-U133 Plus_2 array) was performed. BHLHE41 is one of the DEGs between ccRCC and their adjacent tissues (Fig. 2A and B). It was enhanced significantly in ccRCC tissues (P<0.0001), as demonstrated by paired Student's t-test analysis (Fig. 2C).
Figure 2.
mRNA expression of BHLHE41 in ccRCC based on GSE53757 data analysis. (A) Heat maps compiled from GSE53757 ccRCC samples, compared with adjacent normal tissues, demonstrated 50 differentially-expressed probe sets. Red and green indicate the upregulated and downregulated differentially-expressed genes, respectively. (B) Volcano plots revealed BHLHE41 is one of the most highly-expressed genes. (C) Paired Student's t-test indicated that BHLHE41 had significantly increased expression in GSE53757 ccRCC samples. ccRCC, clear cell renal cell carcinoma; BHLHE41, basic helix-loop-helix family member e41.
BHLHE41 expression in TCGA Data
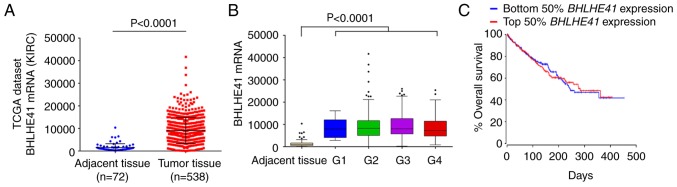
To further investigate the role of BHLHE41 in ccRCC, the expression of BHLHE41 was analyzed using TCGA's ccRCC (KIRC) RNA-seq data (21). The analysis results demonstrated that BHLHE41 was overexpressed in tumor tissues (P<0.0001; Fig. 3A). However, there were no significant differences among the various pathological grades (Fig. 3B) and high expression of BHLHE41 was not significantly associated with the overall survival rate in patients with ccRCC (Fig. 3C).
Figure 3.
mRNA expression of BHLHE41 in ccRCC based on TCGA data mining. (A) The relative mRNA expression of BHLHE41 in ccRCC tissues and normal tissues. KIRC: Kidney ccRCC, with 72 tumor adjacent tissues and 538 tumor tissues. (B) Box plot of BHLHE41 mRNA levels in non-tumorigenic tissues, Fuhrman tumor grade 1 (G1), 2 (G2), 3 (G3) and 4 (G4) of patients with ccRCC. Values shown are mean ± standard deviation. (C) Kaplan-Meier analysis of overall survival for patients with ccRCC relative to expression levels of BHLHE41. Patients were stratified as low and high expression of BHLHE41 mRNA (www.oncolnc.org). P=0.895 vs. the BHLHE41 low group. ccRCC, clear cell renal cell carcinoma; BHLHE41, basic helix-loop-helix family member e41; TCGA, The Cancer Genome Atlas.
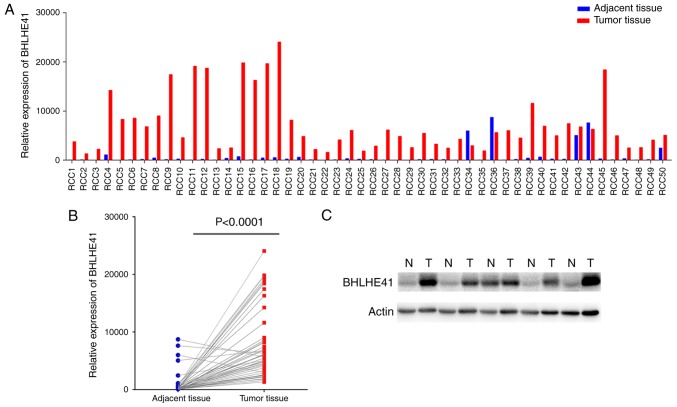
BHLHE41 expression in fresh ccRCC tissues
A total of 50 pairs of pathology confirmed and surgically removed ccRCC tissues, and their adjacent tissues, were collected at the Fuzhou General Hospital. The RT-qPCR data demonstrated that BHLHE41 was highly expressed in 94% of tumor tissues (Fig. 4A). Paired Student's t-test analysis revealed that BHLHE41 mRNA levels were significantly elevated in ccRCC tissues (P<0.0001; Fig. 4B). Subsequently, 5 pairs of samples were detected by western blot analysis. Fig. 4C indicates that the BHLHE41 protein level was increased in tumor tissues. For the samples collected, information on pathological Fuhrman grades (23), with the grades primarily being G1 and G2, was obtained, but there was no patient survival information. Therefore, an association between BHLHE41 expression and tumors was produced, but its association with tumor grade and patient survival was not analyzed.
Figure 4.
BHLHE41 is aberrantly upregulated in fresh human ccRCC samples. (A) BHLHE41 mRNA expression was detected in the 50 paired of ccRCC and matched adjacent non-tumorous tissues as determined by reverse transcription-quantitative polymerase chain reaction. (B) Relative levels of BHLHE41 expression in ccRCC and matched adjacent non-tumorous tissues were calculated by paired Student's t-test. (C) Western blot analysis demonstrated the BHLHE41 protein expression level in 5 paired ccRCC and matched adjacent non-tumorous tissues. N, non-tumor; T, tumor; ccRCC, clear cell renal cell carcinoma; BHLHE41, basic helix-loop-helix family member e41.
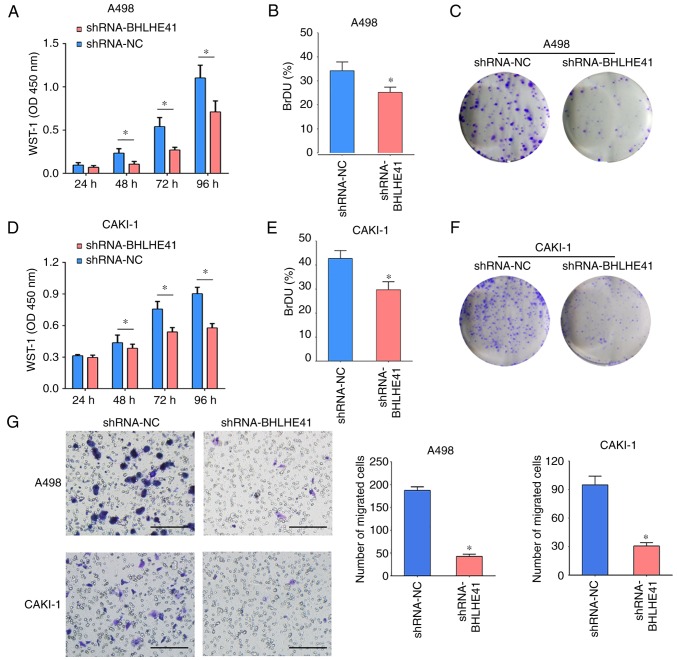
BHLHE41 knockdown impairs ccRCC cell proliferation and migration
BHLHE41 was stably knocked down in A498 and CAKI-1 cells using a BHLHE41 shRNA (shRNA-BHLHE41) or a scrambled control (shRNA-NC) to evaluate their proliferation and migration. Using WST-1 assays, it was observed that the knockdown of BHLHE41 in A498 and CAKI-1 cells significantly suppressed cell proliferation at 48, 72 and 96 h detection time points (Fig. 5A and B). The BrdU assay demonstrated that BHLHE41-knockdown cells proliferation rate was significantly reduced in both cell lines (Fig. 5C and D). BHLHE41-knockdown cell colonies were reduced, compared with shRNA-NC cells, in the ccRCC cell lines (Fig. 5E and F). Subsequently, a Transwell chamber assay was used to assess the effect of BHLHE41 on cell migration. Cell migration was significantly decreased in BHLHE41-knockdown A498 and CAKI-1 cells, compared with their control cells (Fig. 5G). These data indicate that BHLHE41 acts as a novel tumor-promoting molecule regulating ccRCC growth.
Figure 5.
BHLHE41 knockdown inhibits the cell proliferation and migration of ccRCC cells. (A) The inhibition effect of BHLHE41 knockdown on cell proliferation was examined by WST-1 in A498 cells. (B) The inhibition effect of BHLHE41 knockdown on cell proliferation was examined by WST-1 in CAKI-1 cells. (C) BrdU assay in A498 cells. (D) BrdU assay in CAKI-1 cells. (E) Colony formation assay in A498 cells. (F) Colony formation assay in CAKI-1 cells. (G) BHLHE41 knockdown also decreased the migration cell number in ccRCC cells. Scale bar, 100 µm. All the experiments were repeated 3 times, and the central horizontal line represented the mean. *P<0.05. ccRCC, clear cell renal cell carcinoma; BHLHE41, basic helix-loop-helix family member e41; NC, negative control; shRNA, short hairpin RNA; O.D., optical density; WST-1, water-soluble tetrazolium salt.
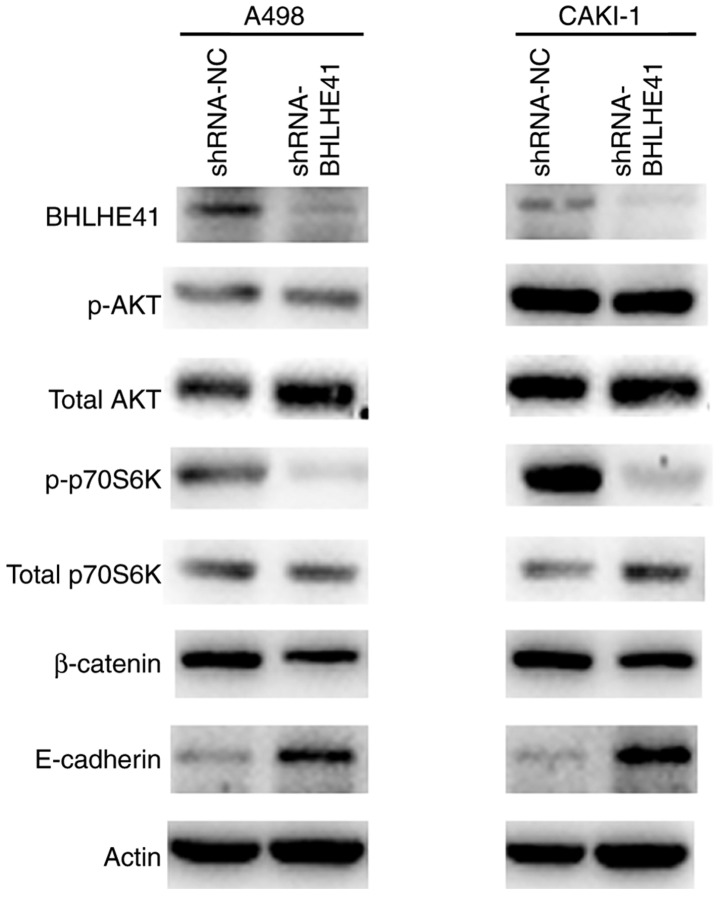
BHLHE41 ablation impairs signaling pathways associated with tumor progression in ccRCC cells
Since mechanistic target of rapamycin kinase (mTOR), AKT and epithelial-mesenchymal transition (EMT) signaling pathways are common tumorigenic pathways (24,25), to further determine the mechanism by which BHLHE41 drives the ccRCC tumorigenesis, western blot analyses was performed with a number of marker genes of mTOR, AKT and EMT signaling pathways. p70S6K activity was significantly reduced in BHLHE41 knockdown A498 and CAKI-1 cells. p-AKT levels were incomparable between the BHLHE41 knockdown and control cells. BHLHE41-knockdown cells and shRNA-NC cells exhibited relative high expression levels of E-cadherin protein in A498 and CAKI-1. There was no detected change in the amount of β-catenin (Fig. 6). These data indicated that the p-p70S6K and E-cadherin expression levels, which are involved in the mTOR and EMT pathways (26,27) respectively, were coordinated by BHLHE41.
Figure 6.
Representative western blot analysis of shRNA-BHLHE41 or control cells. The levels of BHLHE41, β-catenin, E-cadherin and AKT, p70S6K, p-AKT and p-p70S6K in A498 and CAKI-1 cells were measured. The result is representative of three separate experiments. BHLHE41, basic helix-loop-helix family member e41; NC, negative control; shRNA, short hairpin RNA; p-, phospho-; ACTIN: β-actin.
BHLHE41 3′untranslated region (UTR) hypomethylation in patients with ccRCC in TCGA
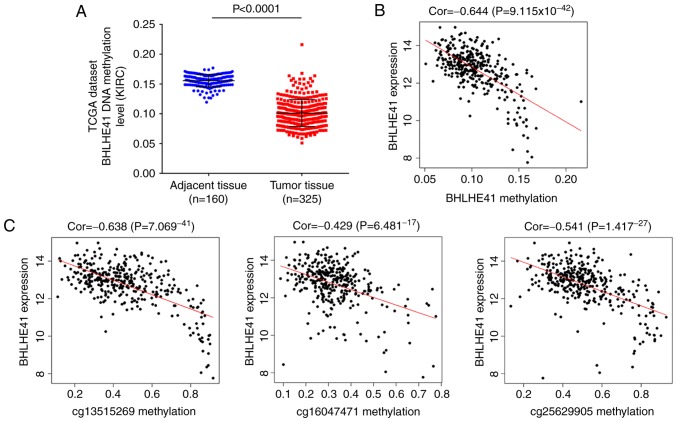
The expression of BHLHE41 has been reported to be regulated by HIF-1α (9). To further investigate potential mechanisms underlying the transcriptional enrichment of BHLHE41, DNA methylation levels and their association between copy number changes were analyzed in TCGA ccRCC samples in the present study. The analysis data demonstrated reduced DNA methylation of BHLHE41 in tumor samples (Fig. 7A; P<0.0001). Potential correlation between methylation and BHLHE41 gene expression was statistically significant (cor=−0.644, P=9.115×10−42; Fig. 7B). These results demonstrated that methylation changes are associated with BHLHE41 expression.
Figure 7.
Methylation and their correlation between methylation BHLHE41 expression. (A) The relative methylation level of BHLHE41 in ccRCC tissues and normal tissues. Values shown are mean ± standard deviation. (B) Scatter plot of BHLHE41 expression vs. methylation. (C) Scatter plot of BHLHE41 expression vs. methylation at probe cg13515269, cg16047471 and cg25629905. TCGA, The Cancer Genome Atlas; BHLHE41, basic helix-loop-helix family member e41.
A total of 17 probes of BHLHE41 were included in further analysis, and 5 loci were determined to be significantly reduced in tumor samples (Table II). It was observed that the correlations between expression and methylation of 3 loci (cg13515269, cg16047471 and cg25629905) within the 3′UTR region were statistically significant (cor=−0.638 and P=7.069 ×10−41; cor=−0.429 an P=6.481×10−17; and cor=−0.541 and P=1.417×10−27, respectively; Fig. 7C). These data indicated that increased BHLHE41 expression in ccRCC samples is associated with methylation level in the 3′UTR region.
Table II.
A total of 16 probes of BHLHE41 details in a dataset (21).
| Address ID | Strand | UCSC RefGene name | UCSC RefGene accession | UCSC RefGene group | Normal mean | Tumor mean | Fold change | P-value |
|---|---|---|---|---|---|---|---|---|
| cg02429656 | F | BHLHE41 | NM_030762 | Body | 0.012550 | 0.015537 | 1.237957 | 8.54×10−3 |
| cg03046445 | R | BHLHE41 | NM_030762 | 1stExon; 5′UTR | 0.056084 | 0.052514 | 0.936354 | 6.05×10−2 |
| cg05232853 | F | BHLHE41 | NM_030762 | Body | 0.075172 | 0.057177 | 0.760606 | 1.29×10−22 |
| cg05568797 | R | BHLHE41 | NM_030762 | Body | 0.035181 | 0.030349 | 0.862645 | 2.46×10−13 |
| cg08715914 | F | BHLHE41 | NM_030762 | TSS200 | 0.086382 | 0.091126 | 1.054917 | 6.15×10−05 |
| cg10049913 | R | BHLHE41 | NM_030762 | Body | 0.0228960 | 0.026840 | 1.172247 | 2.91×10−07 |
| cg10447982 | R | BHLHE41 | NM_030762 | TSS1500 | 0.013739 | 0.014931 | 1.086779 | 1.32×10−09 |
| cg13089947 | F | BHLHE41 | NM_030762 | 1stExon; 5′UTR | 0.035810 | 0.040915 | 1.142558 | 2.11×10−07 |
| cg13515269 | R | BHLHE41 | NM_030762 | 3′UTR | 0.818265 | 0.438801 | 0.536258 | 6.3×10−139 |
| cg15117548 | R | BHLHE41 | NM_030762 | TSS1500 | 0.022337 | 0.024664 | 1.104164 | 1.72×10−08 |
| cg15398617 | R | BHLHE41 | NM_030762 | 1stExon | 0.036305 | 0.036730 | 1.011723 | 8.08 ×10−1 |
| cg16047471 | R | BHLHE41 | NM_030762 | 3′UTR | 0.510720 | 0.323131 | 0.632697 | 2.65×10−47 |
| cg16739118 | R | BHLHE41 | NM_030762 | Body | 0.013970 | 0.015794 | 1.130527 | 6.49×10−16 |
| cg19243777 | F | BHLHE41 | NM_030762 | TSS1500 | 0.036049 | 0.037575 | 1.042343 | 9.37×10−2 |
| cg21335266 | R | BHLHE41 | NM_030762 | TSS1500 | 0.010229 | 0.013838 | 1.35275 | 4.12×10−2 |
| cg25629905 | F | BHLHE41 | NM_030762 | 3′UTR | 0.817583 | 0.490316 | 0.599714 | 2.90×10−146 |
BHLHE41, basic helix-loop-helix family member e41; UTR, untranslated region; F, forward; R, reverse.
Discussion
BHLHE41, a member of circadian clock genes, is involved in the regulation of biological circadian rhythms (28). Disruption of circadian rhythms results in sleep disturbance and the progression of cancer (29,30). Therefore, the biological effects of BHLHE41 are not limited to the regulation of biological rhythms. BHLHE41 is involved in regulating the cellular immune (31) and hypoxia responses (32,33). In the present study, it was determined that BHLHE41 expression was elevated in the GSE53757 dataset and in TCGA ccRCC (KIRC). Despite the lack of analysis of the association between BHLHE41 expression and clinicopathological parameters in a large number of samples, an elevated gene expression was demonstrated in 50 pairs of fresh ccRCC tissues. BHLHE41 was knocked down in ccRCC cell lines (A498 and CAKI-1) through shRNA lentivirus infection. WST-1 and BrdU assays indicated that downregulation of BHLHE41 inhibited A498 and CAKI-1 cell proliferation. The colony formation ability of shRNA-BHLHE41 cells was reduced significantly in these cell lines. The number of migrated cells in BHLHE41-knockdown group was decreased in Transwell migration assays. BHLHE41-knockdown cells exhibited a significant reduction in p-p70S6K and protein production of E-cadherin, indicating that BHLHE41 serves a critical role in regulating tumor progression-associated signaling proteins in ccRCC cells. In order to investigate the role of BHLHE41, a gene array or more signaling pathways gene protein levels between BHLHE41 expressed and control cells should be analyzed in future studies. These results indicated that BHLHE41 may be a biomarker and present a novel treatment option for ccRCC. Additionally, through bioinformatics analysis, a decrease in the methylation frequency of the 3′UTR region of BHLHE41 in ccRCC was reported. This proposed a novel regulatory model for BHLHE41 gene transcription.
Bigot et al (34) reported an association for a single-nucleotide polymorphism rs7132434 and the risk for ccRCC. The rs7132434 polymorphism influences BHLHE41 expression. Overexpressing BHLHE41 produce a larger mouse xenograft tumor, but no significant differences in ACHN cell growth rate or cell migration were observed in in vitro experiments. In the present study, BHLHE41 knockdown was determined to significantly inhibit cell proliferation and migration in A498 and CAKI-1 ccRCC cells. However, Bigot et al did not determine an association between BHLHE41 expression and survival, as indicated in the present study. This could be explained by BHLHE41 not contributing to RCC disease progression robustly.
A previous study demonstrated an association between BHLHE41 expression and the hypoxic adaptation (35). BHLHE41 genetic variants are selected during adaptation to an anoxic environment (35). It is well known that the VHL/HIF pathway is involved in the pathogenesis of ccRCC (36,37). The present data demonstrated that the activity of the mTOR signaling pathway gene, p70S6K, was attenuated by BHLHE41 downregulation in ccRCC cells, indicating that BHLHE41 activates the mTOR pathway. Future studies must determine whether BHLHE41 regulates the VHL/HIF signaling pathways-induced mTOR activation and mTOR inhibitors should be examined, as to whether they affect the BHLHE41-induced proliferation, migration and clonogenic activities of ccRCC cell-lines. Additionally, the present study demonstrated that BHLHE41 regulated the expression of E-cadherin. In a future study, to further clarify the impact of BHLHE41 on EMT signaling pathways, further molecules, including TWIST and SNAILS, need to be detected.
Studies on DNA methylation primarily concentrate on the promoter regions (38,39). Other regulatory sequences, such as the 3′UTR, and other non-coding and coding regions remain poorly understood. However, a number of studies have reported the role of DNA methylation in the 3′UTR on the regulation of gene expression (40,41). 3′UTR hypermethylation interferes p15INK4b gene expression in primary lymphomas (40). The 3′UTR hypermethylation levels of pancreatic and duodenal homeobox 1 and orthodenticle homeobox 1 genes were observed in colorectal cancer cases, with a decrease in gene expression (41). These studies demonstrated that the methylation of the 3′UTR can affect the occurrence of tumors by regulating the level of RNA and/or protein. Therefore, regulation of the BHLHE41 gene expression may come from combinatorial mechanisms. The specific function and regulated mechanism will also be required to be verified through cell experiments in vitro.
In summary, BHLHE41 expression is significantly upregulated in ccRCC tissues, compared with normal renal tissues. BHLHE41 knockdown reduced cell proliferation and migration of ccRCC cells. Additionally, 3′UTR hypomethylation could be a potential mechanism that affects BHLHE41 expression.
Acknowledgements
The authors would like to thank TCGA and GEO projects for providing high-quality clinical data on clear cell renal carcinoma.
Funding
The present study was supported by The National Natural Science Foundation of China (grant no. 81570748) and Natural Science Foundation of Fujian Province (grant nos. 2018J01345 and 2017XQ1194).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
JL and ZS designed the experiments. ZS, LZ, CZ and XC performed the experiments. JL provided the patient samples. JL, LZ, CZ and XC analyzed the data. JL wrote the manuscript. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
The use of human tissues was approved by the Fuzhou General Hospital IRB (Fuzhou, China) (approval no. 2013-017). All the tissue detections were conducted following the receipt of informed written consent from the patients.
Patient consent for publication
All patients agree to the publication of the article.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J, Nathan P, Staehler M, de Souza P, Merchan JR, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med. 2013;369:722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 3.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 4.Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identificat ion of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277:47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 5.Gorski JP, Price JL. Bone muscle crosstalk targets muscle regeneration pathway regulated by core circadian transcriptional repressors DEC1 and DEC2. Bonekey Rep. 2016;5:850. doi: 10.1038/bonekey.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sato F, Bhawal UK, Yoshimura T, Muragaki Y. DEC1 and DEC2 crosstalk between circadian rhythm and tumor progression. J Cancer. 2016;7:153–159. doi: 10.7150/jca.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato F, Kawamura H, Wu Y, Sato H, Jin D, Bhawal UK, Kawamoto T, Fujimoto K, Noshiro M, Seino H, et al. The basic helix-loop-helix transcription factor DEC2 inhibits TGF-β-induced tumor progression in human pancreatic cancer BxPC-3 cells. Int J Mol Med. 2012;30:495–501. doi: 10.3892/ijmm.2012.1037. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Jia YF, Ma XL, Zheng Y, Kong Y, Zhang Y, Zong S, Chen ZT, Wang YS. DEC2 suppresses tumor proliferation and metastasis by regulating ERK/NF-kappaB pathway in gastric cancer. Am J Cancer Res. 2016;6:1741–1757. doi: 10.1158/1538-7445.AM2016-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu T, He N, Yang Y, Yin C, Sang N, Yang Q. DEC2 expression is positively correlated with HIF-1 activation and the invasiveness of human osteosarcomas. J Exp Clin Cancer Res. 2015;34:22. doi: 10.1186/s13046-015-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Wu Y, Yoshizawa T, Yan X, Morohashi S, Seino H, Kato Y, Kijima H. Basic helix-loop-helix transcription factor DEC2 functions as an anti-apoptotic factor during paclitaxel-induced apoptosis in human prostate cancer cells. Int J Mol Med. 2016;38:1727–1733. doi: 10.3892/ijmm.2016.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobin L Gosparowicz MK, Wittekind C. 7th edition; New York, Wiley: 2009. TNM classification of malignant tumours. [Google Scholar]
- 12.Feng C, Sun Y, Ding G, Wu Z, Jiang H, Wang L, Ding Q, Wen H. PI3Kβ inhibitor TGX221 selectively inhibits renal cell carcinoma cells with both VHL and SETD2 mutations and links multiple pathways. Sci Rep. 2015;5:9465. doi: 10.1038/srep09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beroukhim R, Brunet JP, Di Napoli A, Mertz KD, Seeley A, Pires MM, Linhart D, Worrell RA, Moch H, Rubin MA, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69:4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gumz ML, Zou H, Kreinest PA, Childs AC, Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et al. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin Cancer Res. 2007;13:4740–4749. doi: 10.1158/1078-0432.CCR-07-0143. [DOI] [PubMed] [Google Scholar]
- 15.Jones J, Otu H, Spentzos D, Kolia S, Inan M, Beecken WD, Fellbaum C, Gu X, Joseph M, Pantuck AJ, et al. Gene signatures of progression and metastasis in renal cell cancer. Clin Cancer Res. 2005;11:5730–5739. doi: 10.1158/1078-0432.CCR-04-2225. [DOI] [PubMed] [Google Scholar]
- 16.Lenburg ME, Liou LS, Gerry NP, Frampton GM, Cohen HT, Christman MF. Previously unidentified changes in renal cell carcinoma gene expression identified by parametric analysis of microarray data. BMC Cancer. 2003;3:31. doi: 10.1186/1471-2407-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Roemeling CA, Radisky DC, Marlow LA, Cooper SJ, Grebe SK, Anastasiadis PZ, Tun HW, Copland JA. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. 2014;74:4796–4810. doi: 10.1158/0008-5472.CAN-14-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: Archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomczak K, Czerwinska P, Wiznerowicz M. The cancer genome atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research Network, corp-author. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 24.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Ozes ON, Akca H, Mayo LD, Gustin JA, Maehama T, Dixon JE, Donner DB. A phosphatidylinositol 3-kinase/Akt/mTOR pathway mediates and PTEN antagonizes tumor necrosis factor inhibition of insulin signaling through insulin receptor substrate-1. Proc Natl Acad Sci USA. 2001;98:4640–4645. doi: 10.1073/pnas.051042298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;1:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 28.He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL, Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gery S, Koeffler HP. Circadian rhythms and cancer. Cell Cycle. 2010;9:1097–1103. doi: 10.4161/cc.9.6.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innominato PF, Palesh O, Dhabhar FS, Levi F, Spiegel D. Regulation of circadian rhythms and hypothalamic-pituitary-adrenal axis: An overlooked interaction in cancer. Lancet Oncol. 2010;11:816–817. doi: 10.1016/S1470-2045(10)70171-6. [DOI] [PubMed] [Google Scholar]
- 31.Olkkonen J, Kouri VP, Hynninen J, Konttinen YT, Mandelin J. Differentially expressed in chondrocytes 2 (DEC2) increases the expression of IL-1β and is abundantly present in synovial membrane in rheumatoid arthritis. PLoS One. 2015;10:e0145279. doi: 10.1371/journal.pone.0145279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura H, Tanimoto K, Hiyama K, Yunokawa M, Kawamoto T, Kato Y, Yoshiga K, Poellinger L, Hiyama E, Nishiyama M. Human mismatch repair gene, MLH1, is transcriptionally repressed by the hypoxia-inducible transcription factors. DEC1 and DEC2. Oncogene. 2008;27:4200–4209. doi: 10.1038/onc.2008.58. [DOI] [PubMed] [Google Scholar]
- 33.Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380–384. doi: 10.1038/nature11207. [DOI] [PubMed] [Google Scholar]
- 34.Bigot P, Colli LM, Machiela MJ, Jessop L, Myers TA, Carrouget J, Wagner S, Roberson D, Eymerit C, Henrion D, et al. Functional characterization of the 12p12.1 renal cancer-susceptibility locus implicates BHLHE41. Nat Commun. 2016;7:12098. doi: 10.1038/ncomms12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huerta-Sanchez E, Degiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, Cardona A, Montgomery HE, Cavalleri GL, Robbins PA, et al. Genetic signatures reveal high-altitude adaptation in a set of ethiopian populations. Mol Biol Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gossage L, Eisen T. Alterations in VHL as potential biomarkers in renal-cell carcinoma. Nat Rev Clin Oncol. 2010;7:277–288. doi: 10.1038/nrclinonc.2010.42. [DOI] [PubMed] [Google Scholar]
- 37.Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, Reuter VE, Hsieh JJ, Scandura JM, Massagué J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19:50–56. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 39.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 40.Malumbres M, Perez de Castro I, Santos J, Fernandez Piqueras J, Pellicer A. Hypermethylation of the cell cycle inhibitor p15INK4b 3′-untranslated region interferes with its transcriptional regulation in primary lymphomas. Oncogene. 1999;18:385–396. doi: 10.1038/sj.onc.1202299. [DOI] [PubMed] [Google Scholar]
- 41.Smith JF, Mahmood S, Song F, Morrow A, Smiraglia D, Zhang X, Rajput A, Higgins MJ, Krumm A, Petrelli NJ, et al. Identification of DNA methylation in 3′ genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–172. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.