Abstract
There is growing evidence that most high-grade serous ovarian carcinomas likely arise from local dissemination of precursor lesions of the fallopian tube. Evolution of these lesions from early p53 signatures to latter-stage, serous tubal intraepithelial carcinomas (STICs) is characterized by cytologic atypia, accumulation of somatic mutations, and genomic instability, the etiologies of which remain unclear. Long interspersed element 1 (LINE-1) retrotransposon is expressed in many carcinomas, including high-grade serous ovarian carcinoma, where it contributes to genomic instability; however, the timing of LINE-1 activation during this evolution has yet to be elucidated. In this study, we assessed LINE-1 open reading frame 1 protein expression in 12 p53 signature lesions, 32 STICs, and 112 various types of ovarian cancers via immunohistochemical staining and examined LINE-1 promoter methylation in representative cases. We found that 78% and 57% of STICs, with and without concurrent ovarian carcinomas, respectively, exhibited intense LINE-1 immunoreactivity compared with adjacent, normal-appearing fallopian tube epithelium. Hypomethylation of the LINE-1 promoter was found in all STICs exhibiting overexpression. None of the 12 p53 signatures demonstrated significant LINE-1 expression. In ovarian cancer, 84 (75%) of 112 ovarian carcinomas overexpressed LINE-1. Our results indicate that LINE-1 retrotransposons often become deregulated during progression of ovarian cancer precursor lesions from the p53 signature to STIC stages and remain highly expressed in carcinoma.
Ovarian cancer is the fifth most common cancer in US women and the deadliest of all gynecologic malignancies. Although ovarian cancers were traditionally thought to originate from the ovarian surface epithelium, recent studies strongly suggest that most high-grade serous ovarian carcinomas (HGSOCs) likely originate from precursor lesions of the distal fallopian tube epithelium.1, 2, 3 These precursors have themselves been hypothesized to evolve in a stepwise manner from the so-called p53 signature lesions of normal-appearing, tubal epithelia that invariably harbor TP53 mutations to expansile serous tubal intraepithelial carcinomas (STICs), capable of seeding high-grade carcinomas to the ovary and peritoneum.3 Although there is considerable evidence to support this paradigm, there remains a limited understanding of the molecular and cellular events underlying this pathogenesis.
STIC lesions display many of the same genetic and histologic hallmarks that characterize HGSOCs, including TP53 mutations, copy number alterations, and highly similar gene expression and DNA methylation profiles,4, 5, 6 as well as a notable lack of genomic stability, as indicated by significant up-regulation of genes associated with DNA damage response mechanisms.7 Another commonly occurring phenomenon in carcinomas such as HGSOC is the up-regulated expression of long interspersed element 1 (LINE-1).8 LINE-1 sequences are a family of protein-coding, mobile genetic elements comprising approximately 17% of the human genome.9, 10 Although most (>99.8%) of LINE-1 elements in the human genome have been rendered inactive by 5′ truncation or internal mutations, there remain approximately 100 active loci—members of the L1PA1 or L1Hs subfamily that are fully functional and capable of propagating themselves in the genome through a process known as retrotransposition.11, 12 There is a growing body of evidence that expression of LINE-1 is not only a biomarker of carcinomas, but also results in activity of these mobile genetic elements in cancers.13, 14 De novo insertions of LINE-1 sequences are normally prevented by epigenetic silencing of these elements via heavy methylation of CpG dinucleotides within their promoter regions. However, during carcinogenesis, in a process that is currently poorly understood, LINE-1 promoter regions often become hypomethylated, leading to expression of LINE-1 proteins.
There are two proteins encoded by LINE-1, an RNA-binding protein [open reading frame 1 protein (ORF1p)] and a protein encoding reverse transcriptase and endonuclease activities (open reading frame 2 protein).15, 16 Both are required for retrotransposition, and somatically acquired LINE-1 insertions are commonplace in cancer genomes.17, 18 Whether LINE-1 plays causal roles in the development and progression of cancers or is epiphenomenal has not been well established, and the timing of its activation with respect to cellular transformation and disease progression has not been well characterized. LINE-1 protein expression and LINE-1 promoter epigenetic alterations have also yet to be evaluated in STICs.8, 19 In the present work, we investigate the time course and prevalence of LINE-1 expression in relation to the development of fallopian tube precursor lesions and HGSOC. Our results indicate that genomic LINE-1 copies typically become hypomethylated and LINE-1 ORF1p accumulates concurrent with STIC lesion formation, before disseminated progression to HGSOC, where LINE-1 expression persists.
Materials and Methods
Gynecologic Tissue Samples
This study was performed after approval by an institutional review board and conducted in accordance with the US Common Rule. All tissues were collected or retrieved from Johns Hopkins University gynecologic tissue bank with written consent given by all the participant subjects. No identifying information was associated with samples, and no medical record review was conducted for this study. Five tissue microarrays in total were constructed from archived paraffin tissue blocks of ovarian cancers: two from 33 high-grade serous carcinomas, two from 37 clear cell carcinomas, and one from 30 endometrioid carcinomas. Tissue cores were obtained at 3 mm in diameter from two, or at 1.5 mm in diameter from three, representative tumor-rich regions of each tumor block to minimize sampling error. A separate cohort of fallopian tube specimens were also obtained from salpingo-oophorectomies from 26 patients. Of these 26, 12 had pathologically confirmed HGSOC, from which 12 tumor tissue specimens, 18 STICs, and 3 p53 signatures were obtained. From the 14 specimens without concurrent HGSOC, 14 STICs and 9 p53 signatures were obtained. Adjacent, normal-appearing fallopian tubes (NFTs) were obtained from all 26 salpingo-oophorectomies. All the cases were carefully reviewed by three pathologists (S.A., S.-.F.L., and I.-.M.S.).
Immunohistochemistry
All tissue samples were fixed with 10% neutral-buffered formalin and embedded in paraffin. Serial paraffin sections (4 μm thick) were generated from each paraffin block (5 tissue microarray blocks and 36 blocks from 26 salpingo-oophorectomies in total). Hematoxylin and eosin staining was performed for all the tissue sections for histologic examination. For the whole-tissue sections from salpingo-oophorectomies, immunohistochemistry for p53 (clone DO-7; 1:600; Leica Biosystems, Wetzlar, Germany) and Ki-67 (clone D2H10; 1:400; Cell Signaling Technology, Inc., Dancers, MA) was also performed for further histologic classification of HGSOCs, STICs, p53 signatures, and NFTs. All tissue microarrays and whole-tissue sections were then subjected to LINE-1 immunohistochemistry. For LINE-1 immunostaining, the mouse monoclonal antibody against an epitope of LINE-1 ORF1p (clone 4H1; 1:3000), which was developed and validated in the previous studies,8, 20 was used (catalog number MABC1152; Millipore Sigma, Burlington, MA). Tissue sections were deparaffinized and hydrated before the antigen retrieval step. Heat-induced antigen retrieval was performed using Dako Target Retrieval Solution, Citrate pH 6 (Agilent, Santa Clara, CA) at 90°C for 20 minutes. The sections were then incubated with the primary antibody at 4°C overnight. Immunoreactivity was detected using the immunoenzyme polymer method (Dako Envision + System − HRP Polymer Anti-Mouse for p53 and LINE-1, Anti-Rabbit for Ki-67; Agilent) with 3,3′-diaminobenzidine as the chromogen (Dako Liquid DAB+ Substrate Chromogen System; Agilent).
Immunostained lesions were scored on a semiquantitative scale, whereby staining intensity grades of 2 (moderate) and 3 (strong) were considered as positive for LINE-1 expression, whereas lesions with that of 0 (no staining) and 1 (weak) were considered negative. The staining intensity grades (strong, moderate, and weak) correspond with LINE-1 (low, intermediate, and high, respectively), on the basis of representative images of cases of each intensity grade, as previously shown.8 In addition, LINE-1 immunoreactivity was also evaluated using the H-score system. The H-score, on the basis of the modification of McCarty et al,21 is calculated by adding the multiplication of the different staining intensities in four-grade semiquantitative scale (0, no staining; 1, weak; 2, moderate; and 3, strong) with each percentage of positive cells using the following formula: 1 × (% of weakly positive cells) + 2 × (% of moderately positive cells) + 3 × (% of strongly positive cells).
Finally, a score from 0 to 300 points is obtained. Herein, the H-scores between ovarian cancers, fallopian tubal lesions, and NFTs were compared. LINE-1 immunoreactivity was scored and reviewed twice by a single pathologist (S.A.).
LINE-1 staining H-scores were compared using methods based on the most reasonable assumptions according to the respective groups. A t-test was used to compare H-scores of HGSOCs versus STICs with and without HGSOC because these were confirmed to exhibit a normal distribution and equal variance according to a normal quantile plot and two-sided F-test. A Wilcoxon test (Mann-Whitney test) was used to compare HGSOCs, clear cell carcinomas, endometrioid carcinomas, STICs, p53 signatures, and NFTs because these did not meet the assumptions necessary for comparison using a parametric test.
LINE-1 Promoter Methylation
Seven STIC samples with adjacent NFTs and 12 NFT samples from women without malignant diseases were sectioned on MembraneSlide 1.0 PEN slides (Carl Zeiss Microscopy, LCC, Thornwood, NY) for laser-capture microdissection (LMD7000; Leica Biosystems). Genomic DNA from each sample was purified by using the QIAamp FFPE DNA tissue kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Bisulfite conversion of DNA extracted from fallopian and control tissues was performed using the EZ DNA Methylation-Lightning Kit (Zymo Research, Irvine, CA), according to the manufacturer's instructions, and eluted into 40 μL DNA Elution buffer (Zymo Research). Post-bisulfite treatment yields were quantified by MethyLight using primer and probe sequences for β-actin recognizing both methylated and unmethylated templates: forward primer, 5′-TAGGGAGTATATAGGTTGGGGAAGTT-3′; reverse primer, 5′-AACACACAATAACAAACACAAATTCAC-3′, spanning a 104-bp region (chromosome 7:5,532,169 to 5,532,271); and 100 nmol/L probe, 5′-∖56-FAM∖TGTGGGGTG∖ZEN∖GTGATGGAGGAGGTTTAG∖3IABkFQ∖-3′. MethyLight assays were performed using 10× Master Mix to yield a final volume of 25 μL and final working concentrations of 16.6 mmol/L (NH4)2SO4, 67 mmol/L tris pH 8.8, 6.7 mmol/L MgCl2, 10 mmol/L β-mercaptoethanol, 200 μmol/L of each dNTP, and 0.04 U/μL of Platinum Taq polymerase (ThermoFisher Scientific, Waltham, MA). Cycling conditions were 95°C for 5 minutes, followed by 50 cycles of 95°C for 5 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. Standards for quantification were generated by serial dilution of a 104-bp synthetic target equivalent to the bisulfite-converted locus. Assuming 5 pg DNA/genomic equivalent (two copies β-actin), approximate bisulfite-treated DNA concentrations ranged from 0.343 to 0.870 ng/μL, with an average of 0.583 ± 0.180 ng/μL.
Bisulfite-converted DNA samples were assessed for LINE-1 promoter hypomethylation by methylation-sensitive PCR/MethyLight assay using the following: forward primer, 5′-GTTTTTAGTGTGAGTGATGTAGAAGAT-3′; reverse primer, 5′-CCTCACCCTACTTCAACTCACA-3′; and TaqMan probe with minor-groove binder (Applied Biosystems, Foster City, CA), 5′-∖6-FAM∖ACAATACACACACACACTAA∖MGB-NFQ∖-3′, spanning nucleotides 46 to 185 of the sense strand of bisulfite-converted (unmethylated) Ta-1d LINE-1 consensus sequence.11 MethyLight was performed with 300 nmol/L forward primer, 300 nmol/L reverse primer, and 100 nmol/L probe, using cycling conditions of 95°C for 5 minutes, followed by 50 cycles of 95°C for 30 seconds, 63.5°C for 30 seconds, and 72°C for 30 seconds using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA), and analyzed using the accompanying stock software, CFX Manager. A standard curve was generated using serial dilutions of bisulfite-converted Epitect unmethylated control DNA (Qiagen, Germantown, MD) mixed with bisulfite-converted CpG-Methylated HeLa Genomic DNA (New England BioLabs, Ipswich, MA). Percentage hypomethylation was calculated for each sample using the β-actin and LINE-1 standard curves, according to the following formula: % Hypomethylation = (number of copies hypomethylated LINE-1 in sample)/[(number of copies β-actin in sample) * (copies of LINE-1 per β-actin copy in unmethylated control)].
Hypomethylation between the three sample types (STIC, adjacent NFT, and healthy NFT without STIC or carcinoma) was compared using one-way analysis of variance, followed by Fisher's least significant difference test.
Results
LINE-1 Expression Is Common in Ovarian Cancer but Varies according to Histologic Subtype
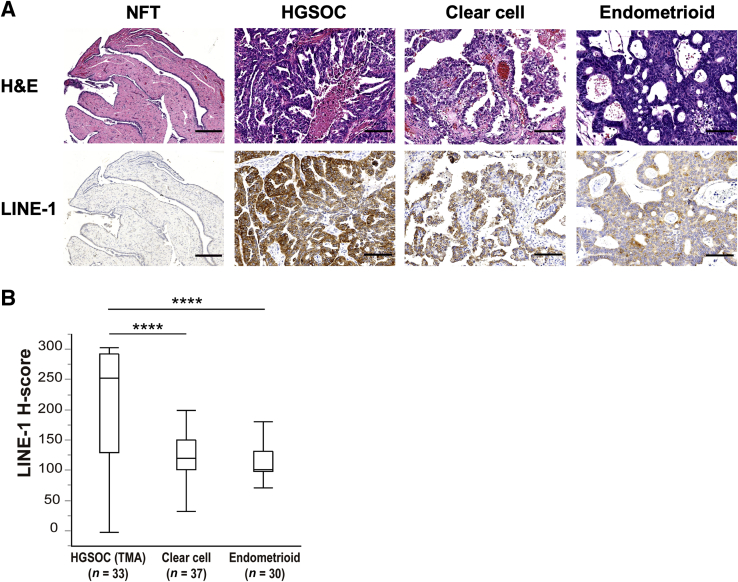
LINE-1 expression was assessed by performing immunohistochemical staining using a previously validated monoclonal antibody against LINE-1 ORF1p8, 20 and assigning an H-score21 on the basis of the resulting staining intensity and the overall percentage of positive cells. This method was used to survey the prevalence of LINE-1 ORF1p expression in five tissue microarrays, comprising 100 formalin-fixed, paraffin-embedded samples, of the three most common epithelial ovarian carcinoma subtypes: HGSOC, clear cell, and endometrioid (Figure 1A). As in previous studies, LINE-1 ORF1p was mainly observed in the cytoplasm of tumor cells with some heterogeneity throughout the tumor tissue.8, 20 Most ovarian cancers exhibited intense LINE-1 immunoreactivity, but prevalence was variable according to the histologic subtype (Table 1). Of the three, HGSOCs exhibited the highest proportion of samples with LINE-1 ORF1p expression (91% with the mean H-score of 212.4), followed by clear cell carcinomas (65% with the mean H-score of 124.7) and endometrioid carcinomas (60% with the mean H-score of 113.3). HGSOCs exhibited the strongest and most diffuse expression, whereas both of the clear cell carcinomas and endometrioid carcinomas tended to show weak-to-moderate LINE-1 expression. The observation of notably higher LINE-1 ORF1p H-scores in HGSOCs compared with the clear cell carcinomas (P < 0.0001) and endometrioid carcinomas (P < 0.0001) (Figure 1B) supports reports associating LINE-1 expression with TP53-deficient tumor types.8, 22
Figure 1.
LINE-1 ORF1p expression in ovarian carcinomas. A: Representative images of hematoxylin and eosin (H&E) staining and LINE-1 immunostaining in the normal-appearing fallopian tube (NFT), high-grade serous ovarian carcinoma (HGSOC), clear cell carcinoma, and endometrioid carcinoma. HGSOCs exhibit the strongest and the most diffuse expression, whereas both clear cell carcinomas and endometrioid carcinomas tend to show only weak or moderate LINE-1 ORF1p expression. B: Comparison of LINE-1 ORF1p immunostaining H-scores between ovarian carcinomas. The H-scores of HGSOCs are significantly higher than endometrioid and clear cell carcinomas. ∗∗∗∗P < 0.0001 (Wilcoxon test). Scale bars: 250 μm (A, NFT); 100 μm (A, HGSOC, clear cell, and endometrioid). Original magnifications: ×5 (A, NFT); ×20 (A, HGSOC, clear cell, and endometrioid). TMA, tissue microarray.
Table 1.
Results of LINE-1 ORF1p Immunostaining in Ovarian Carcinomas and Fallopian Tubal Lesions
| Ovarian carcinomas and fallopian lesions | Total cases, n | Positive (strong or moderate), n (%) | Negative (weak or no staining), n (%) | Mean H-score |
|---|---|---|---|---|
| Tissue microarrays | ||||
| HGSOC | 33 | 30 (91) | 3 (9) | 212.4 |
| Clear cell carcinoma | 37 | 24 (65) | 13 (35) | 124.7 |
| Endometrioid carcinoma | 30 | 18 (60) | 12 (40) | 113.3 |
| Whole tissue sections | ||||
| HGSOC | 12 | 12 (100) | 0 (0) | 232.9 |
| STIC | 32 | 22 (69) | 10 (31) | 175.2 |
| With HGSOC | 18 | 14 (78) | 4 (22) | 201.9 |
| Without HGSOC | 14 | 8 (57) | 6 (43) | 140.7 |
| p53 signature | 12 | 0 (0) | 12 (100) | 15.0 |
| NFT | 26 | 0 (0) | 26 (100) | 33.1 |
HGSOC, high-grade serous ovarian carcinoma; NFT, normal-appearing fallopian tube; STIC, serous tubal intraepithelial carcinoma.
Initiation of LINE-1 Activity Occurs after TP53 Compromise in HGSOC Precursor Lesions
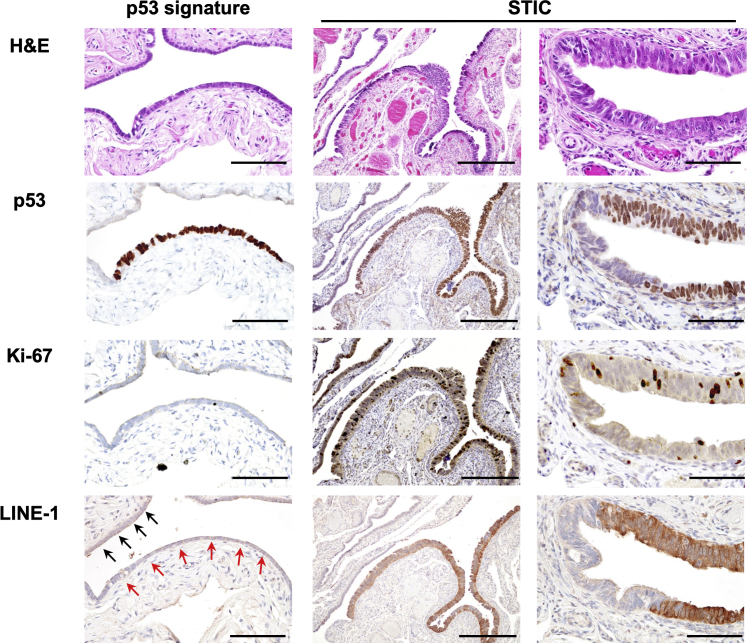
The dynamics of LINE-1 expression in relation to the development and progression of putative HGSOC precursor lesions of the fallopian tube were next studied. Individual lesions were initially identified by immunohistopathological assessment of formalin-fixed, paraffin-embedded fallopian tubes removed from women who underwent surgery for HGSOCs or benign conditions, including uterine leiomyoma and benign ovarian cysts. Individual lesions were stained for p53 and Ki-67 immunoreactivity for further classification as either a p53 signature or STIC, per previously established immunohistopathological criteria.23 In total, immunohistochemical staining was performed on 12 p53 signature lesions and 32 STICs (18 with HGSOC and 14 without HGSOC), as well as 12 HGSOCs and 26 adjacent NFTs (Figure 2).
Figure 2.
LINE-1 ORF1p expression in fallopian tubal lesions. Hematoxylin and eosin (H&E) staining indicates that, in contrast to the morphologically unremarkable p53 signature lesion, the serous tubal intraepithelial carcinoma (STIC) lesion exhibits nuclear stratification, enlargement, and prominent nucleoli. p53 Immunostaining shows diffuse positivity in both the p53 signature and STIC lesion. Ki-67 immunostaining shows a high positivity ratio in the STIC lesion (approximately 15% of the tumor cells), whereas only a single, very rare Ki-67–positive cell is seen in the p53 signature lesion. Similarly, strong LINE-1 expression is observed only in STIC lesions, in comparison to no or only focal and weak staining of LINE-1 in the p53 signature lesion (red arrows) and adjacent normal-appearing fallopian tube epithelium (black arrows). Scale bars: 100 μm (left column and right column); 250 μm (middle column). Original magnifications: ×20 (left column and right column); ×10 (middle column).
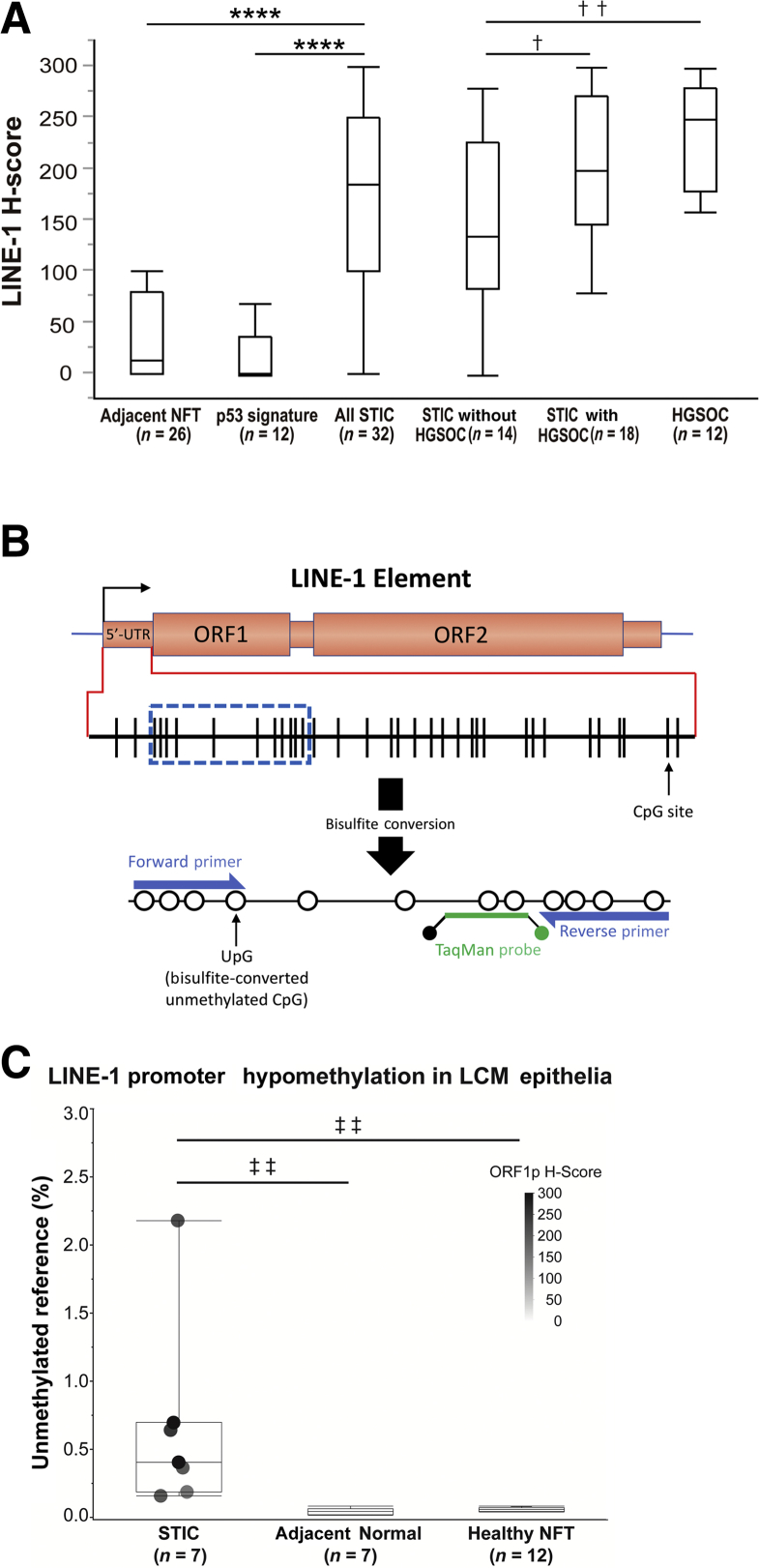
Immunohistochemistry staining for LINE-1 ORF1p revealed that 69% of STIC lesions exhibited strong LINE-1 ORF1p immunoreactivity (Table 1 and Figure 3A). In contrast, none of the p53 signatures or NFTs exhibited significant LINE-1 expression. LINE-1 H-scores in STICs were significantly higher than p53 signatures and adjacent NFTs (P < 0.0001 for both). There was also a higher proportion of STIC lesions with positive LINE-1 ORF1p staining in women diagnosed with HGSOC (78%), compared with HGSOC-free women (57%). LINE-1 H-scores in HGSOCs and STICs with HGSOC were significantly higher than STICs without HGSOC (P = 0.0031 and P = 0.0436, respectively), whereas there was no significant difference between HGSOCs and STICs with HGSOC (P = 0.4481). These results indicate that LINE-1 ORF1p overexpression is likely often established during the evolution of lesions from the p53 signature to the STIC stage. Moreover, LINE-1 overexpression increases in prevalence as lesions progress toward malignancy and seeding to the ovary, at which point the prevalence among HGSOC-concomitant STIC lesions (78%) approaches a value nearing that of the HGSOC tumors (>90%).
Figure 3.
Comparison of LINE-1 ORF1p immunostaining H-scores and correlation with LINE-1 promoter hypomethylation in fallopian tubal lesions. A: Boxes represent the interquartile range, with the upper whisker indicating the 75th percentile and the lower whisker indicating the 25th percentile. The median values are indicated by a horizontal line. LINE-1 ORF1p immunostaining H-scores in serous tubal intraepithelial carcinomas (STICs) are significantly higher than p53 signatures and adjacent normal-appearing fallopian tubes (NFTs). LINE-1 H-scores in high-grade serous ovarian carcinomas (HGSOCs) and STICs with HGSOCs are significantly higher than STICs without HGSOCs, whereas there is no significant difference between HGSOCs and STICs with HGSOCs. B: Design of LINE-1 MethyLight assay targeting unmethylated promoters within the 5′-untranslated region (UTR) of potentially active LINE-1 elements. C: LINE-1 hypomethylation of laser capture microdissection (LCM) epithelia from STIC versus adjacent NFTs and versus NFTs from healthy women (without STIC or carcinoma). Hypomethylation values are normalized to fully unmethylated control DNA. ∗∗∗∗P < 0.0001 (Wilcoxon test); †P < 0.05, ††P < 0.01 (t-test); ‡‡P < 0.01 (one-way analysis of variance using Fisher least significant difference test).
LINE-1 ORF1p Expression and Promoter Hypomethylation Are Correlated in Precursor Lesions
Hypomethylation of LINE-1 elements is a well-known cancer-associated phenomenon and normally assumed to be essential for expression of LINE-1 proteins to occur.24 LINE-1 hypomethylation is most typically quantified by assessing the average methylation status of LINE-1 elements throughout the entire genome. Several recent studies have studied individual transpositionally active LINE-1 loci and found that these typically exhibit complete or near-complete hypomethylation of their respective promoter.14, 25, 26 On the basis of this observation, we designed a unique methylation-specific PCR MethyLight assay27 (Figure 3B) highly specific to fully unmethylated copies of the promoter region of the consensus sequence of retrotransposition-competent LINE-1 elements.11 This assay was then employed to determine the relative proportion of potentially active, unmethylated LINE-1 elements with respect to LINE-1 ORF1p expression. Laser capture microdissection was performed for 7 select STIC lesions and respective adjacent NFT epithelia, as well as 12 samples of NFT epithelia from healthy women. The seven selected STIC lesions exhibited LINE-1 ORF1p H-scores ranging from 160 to 300, yielding an average staining score of 233. DNA was extracted from each of the microdissected epithelia samples and underwent bisulfite conversion before assessment for LINE-1 hypomethylation by MethyLight assay. Figure 3C shows the percentage of hypomethylated LINE-1 elements within each sample with respect to an equivalent amount of fully unmethylated control DNA. Overall, the STIC samples exhibited significantly elevated LINE-1 hypomethylation (average, 0.66%) with respect to both the adjacent NFT (average, 0.05%; P = 0.0037) and healthy NFT epithelia controls (average, 0.06%; P = 0.0017). Relatively speaking, all seven STIC samples were hypomethylated with respect to all 19 control samples, and the four STIC samples that stained most intensely for LINE-1 ORF1p (H-score > 200) also exhibited the highest levels of LINE-1 hypomethylation. Overall, these results confirm that expression of LINE-1 ORF1p is strongly associated with, and likely potentiated by, the relative fraction of fully unmethylated LINE-1 promoter elements within the respective tissue.
Discussion
Aberrant expression of LINE-1 is a commonly observed feature of carcinomas and a known contributor to genomic instability. Herein, we evaluated expression of LINE-1 ORF1p in type I and type II ovarian carcinomas28 and found it to be prevalent in all histologic subtypes, especially HGSOCs (type II carcinomas), the most common and aggressive type of ovarian cancer. Furthermore, in addition to expressing LINE-1 at a higher prevalence, HGSOC specimens also did so to a greater degree, as evidenced by a significantly higher average LINE-1 H-score than the other subtypes tested. These differences most likely reflect the fact that, in comparison to type I carcinomas such as clear cell and endometrioid subtypes, virtually all HGSOCs harbor TP53 mutations,29 a key regulator of transpositional activity,22 and consequently exhibit a higher prevalence of genome rearrangements, including somatically acquired LINE-1 insertions.18
Further investigation into the chronological course of LINE-1 up-regulation in precursors of HGSOC revealed that LINE-1 expression is also frequently present in STIC lesions, the putative precursors of many HGSOCs,2, 30, 31 indicating that LINE-1 expression is an early event in HGSOC carcinogenesis that likely precedes the development of ovarian cancer itself. More specifically, we pinpoint the start of LINE-1 ORF1p expression to when lesions of the fallopian tube evolve beyond the p53 signature stage. The lack of detectable LINE-1 expression in the p53 signatures implies a separation between TP53 compromise and LINE-1 activation, such that although TP53 loss may permit LINE-1 expression, additional molecular mechanisms may be responsible for orchestrating LINE-1 promoter hypomethylation. The absence of LINE-1 expression in p53 signatures, presumably because of epigenetic repression, is also notable in that it provides tangential support for recent studies reporting that epigenetic mechanisms likely work in concert with driver mutations in the development of HGSOC32 and the emergence of STIC lesions6 in particular. Interestingly, such a paradigm has also been shown to occur in the initiation of melanoma, where epigenetic mechanisms precipitate the onset of melanoma from a p53-deficient precancerized field.33 The concordance between the increasing prevalence of LINE-1 ORF1p expression among precursor lesions as they ostensibly progress toward malignancy, a process that is estimated to occur over a few years,3 suggests a close relationship between the two. However, whether and to what extent LINE-1 expression works in coordination with other molecular alterations, such as cyclin E1 (CCNE1) amplification/overexpression,30 telomere abnormalities,34, and up-regulation of HGSOC-associated proteins,35 to effect the characteristic phenotypic traits of HGSOC remains a conspicuously open question. Further studies will be required to elucidate these mechanisms.
Acknowledgments
We thank Daniel Ardeljan for helpful discussions.
T.R.P., K.H.B., T-.L.W., and I-.M.S. conceived and designed the study; T.R.P., S.A., I-.M.S., and K.H.B. designed methods; T.R.P., S.A., S-.F.L., H.S., and A.B-.T. acquired data; T.R.P. and S.A. analyzed data; T.R.P., S.A., K.H.B., T-.L.W., and I-.M.S. wrote the manuscript; K.H.B., T-.L.W., and I-.M.S. supervised the study; T-.T.Y., A.B-.T., and T-.H.W. provided administrative, technical, and/or material support.
Footnotes
Supported by The Department of Defense Congressionally Directed Medical Research Programs grant W81XWH-11-2-0230; NIH/National Cancer Institute grants EDRN U01CA200469, R01CA215483, R01GM124531, P50CA228991, and P50GM107632; the Tina's Wish Foundation; the Ovarian Cancer Research Alliance; a TEAL award; the Mathers Foundation; the Richard W. TeLinde Endowment; and the Johns Hopkins Discovery Award.
T.R.P. and S.A. contributed equally to this work.
Disclosures: None declared.
References
- 1.Crum C.P., Drapkin R., Miron A., Ince T.A., Muto M., Kindelberger D.W., Lee Y. The distal fallopian tube: a new model for pelvic serous carcinogenesis. Curr Opin Obstet Gynecol. 2007;19:3–9. doi: 10.1097/GCO.0b013e328011a21f. [DOI] [PubMed] [Google Scholar]
- 2.Kurman R.J., Shih IeM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186:733–747. doi: 10.1016/j.ajpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu R.C., Wang P., Lin S.F., Zhang M., Song Q., Chu T., Wang B.G., Kurman R.J., Vang R., Kinzler K., Tomasetti C., Jian Y., Shih I.M., Wang T.L. Genomic landscape and evolutionary trajectories of ovarian cancer early precursor lesions. J Pathol. 2018 doi: 10.1002/path.5219. [Epub ahead of print] doi: 10.1002/path.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuhn E., Kurman R.J., Vang R., Sehdev A.S., Han G., Soslow R., Wang T.L., Shih I.M. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma: evidence supporting the clonal relationship of the two lesions. J Pathol. 2012;226:421–426. doi: 10.1002/path.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kindelberger D.W., Lee Y., Miron A., Hirsch M.S., Feltmate C., Medeiros F., Callahan M.J., Garner E.O., Gordon R.W., Birch C., Berkowitz R.S., Muto M.G., Crum C.P. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: evidence for a causal relationship. Am J Surg Pathol. 2007;31:161–169. doi: 10.1097/01.pas.0000213335.40358.47. [DOI] [PubMed] [Google Scholar]
- 6.Pisanic T.R., Cope L., Lin S.-F., Yen T.-T., Athamanolap P., Asaka R., Nakayama K., Fader A.N., Wang T.-H., Shih I.-M., Wang T.-L. Methylomic analysis of ovarian cancers identifies tumor-specific alterations readily detectable in early precursor lesions. Clin Cancer Res. 2018;24:6536–6547. doi: 10.1158/1078-0432.CCR-18-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chene G., Ouellet V., Rahimi K., Barres V., Caceres K., Meunier L., Cyr L., De Ladurantaye M., Provencher D., Masson A.M.M. DNA damage signaling and apoptosis in preinvasive tubal lesions of ovarian carcinoma. Int J Gynecol Cancer. 2015;25:761–769. doi: 10.1097/IGC.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 8.Rodić N., Sharma R., Sharma R., Zampella J., Dai L., Taylor M.S., Hruban R.H., Iacobuzio-Donahue C.A., Maitra A., Torbenson M.S., Goggins M., Shih I.-M., Duffield A.S., Montgomery E.A., Gabrielson E., Netto G.J., Lotan T.L., De Marzo A.M., Westra W., Binder Z.A., Orr B.A., Gallia G.L., Eberhart C.G., Boeke J.D., Harris C.R., Burns K.H. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol. 2014;184:1280–1286. doi: 10.1016/j.ajpath.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 10.Burns Kathleen H., Boeke Jef D. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns K.H. Transposable elements in cancer. Nat Rev Cancer. 2017;17:415–424. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- 14.Tubio J.M., Li Y., Ju Y.S., Martincorena I., Cooke S.L., Tojo M. Mobile DNA in cancer: extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kines K.J., Sokolowski M., deHaro D.L., Christian C.M., Belancio V.P. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. 2014;42:10488–10502. doi: 10.1093/nar/gku687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasior S.L., Wakeman T.P., Xu B., Deininger P.L. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J., 3rd, Lohr J.G., Harris C.C., Ding L., Wilson R.K., Wheeler D.A., Gibbs R.A., Kucherlapati R., Lee C., Kharchenko P.V., Park P.J., Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang Z., Steranka J.P., Ma S., Grivainis M., Rodic N., Huang C.R., Shih I.M., Wang T.L., Boeke J.D., Fenyo D., Burns K.H. Human transposon insertion profiling: analysis, visualization and identification of somatic LINE-1 insertions in ovarian cancer. Proc Natl Acad Sci U S A. 2017;114:E733–E740. doi: 10.1073/pnas.1619797114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardeljan D., Taylor M.S., Ting D.T., Burns K.H. The human long interspersed element-1 retrotransposon: an emerging biomarker of neoplasia. Clin Chem. 2017;63:816–822. doi: 10.1373/clinchem.2016.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma R., Rodić N., Burns K.H., Taylor M.S. In: Immunodetection of human LINE-1 expression in cultured cells and human tissues. Transposons and Retrotransposons: Methods and Protocols. Garcia-Pérez J.L., editor. Springer New York; New York, NY: 2016. pp. 261–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarty K.S., Szabo E., Flowers J.L., Cox E.B., Leight G.S., Miller L., Konrath J., Soper J.T., Budwit D.A., Creasman W.T., Seigler H.F., McCarty K.S. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986;46:4244s–4248s. [PubMed] [Google Scholar]
- 22.Wylie A., Jones A.E., D'Brot A., Lu W.-J., Kurtz P., Moran J.V., Rakheja D., Chen K.S., Hammer R.E., Comerford S.A., Amatruda J.F., Abrams J.M. p53 Genes function to restrain mobile elements. Genes Dev. 2016;30:64–77. doi: 10.1101/gad.266098.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vang R., Visvanathan K., Gross A., Maambo E., Gupta M., Kuhn E., Li R.F., Ronnett B.M., Seidman J.D., Yemelyanova A., Shih I.-M., Shaw P.A., Soslow R.A., Kurman R.J. Validation of an algorithm for the diagnosis of serous tubal intraepithelial carcinoma. Int J Gynecol Pathol. 2012;31:243–253. doi: 10.1097/PGP.0b013e31823b8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miousse I.R., Koturbash I. The fine LINE: methylation drawing the cancer landscape. Biomed Res Int. 2015;2015:8. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wissing S., Muñoz-Lopez M., Macia A., Yang Z., Montano M., Collins W., Garcia-Perez J.L., Moran J.V., Greene W.C. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott E.C., Gardner E.J., Masood A., Chuang N.T., Vertino P.M., Devine S.E. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26:745–755. doi: 10.1101/gr.201814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eads C.A., Danenberg K.D., Kawakami K., Saltz L.B., Blake C., Shibata D., Danenberg P.V., Laird P.W. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurman R.J., Shih I.-M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vang R., Levine D.A., Soslow R.A., Zaloudek C., Shih I.-M., Kurman R.J. Molecular alterations of TP53 are a defining feature of ovarian high-grade serous carcinoma: a rereview of cases lacking TP53 mutations in The Cancer Genome Atlas Ovarian Study. Int J Gynecol Pathol. 2016;35:48–55. doi: 10.1097/PGP.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn E., Wang T.-L., Doberstein K., Bahadirli-Talbott A., Ayhan A., Sehdev A.S., Drapkin R., Kurman R.J., Shih I.-M. CCNE1 amplification and centrosome number abnormality in serous tubal intraepithelial carcinoma: further evidence supporting its role as a precursor of ovarian high-grade serous carcinoma. Mod Pathol. 2016;29:1254–1261. doi: 10.1038/modpathol.2016.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurman R.J., Shih I.-M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer: shifting the paradigm. Hum Pathol. 2011;42:918–931. doi: 10.1016/j.humpath.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bartlett T.E., Chindera K., McDermott J., Breeze C.E., Cooke W.R., Jones A., Reisel D., Karegodar S.T., Arora R., Beck S., Menon U., Dubeau L., Widschwendter M. Epigenetic reprogramming of fallopian tube fimbriae in BRCA mutation carriers defines early ovarian cancer evolution. Nat Commun. 2016;7:11620. doi: 10.1038/ncomms11620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman C.K., Mosimann C., Fan Z.P., Yang S., Thomas A.J., Ablain J., Tan J.L., Fogley R.D., van Rooijen E., Hagedorn E.J., Ciarlo C., White R.M., Matos D.A., Puller A.-C., Santoriello C., Liao E.C., Young R.A., Zon L.I. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science. 2016;351:aad2197. doi: 10.1126/science.aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn E., Meeker A., Wang T.-L., Sehdev A.S., Kurman R.J., Shih I.-M. Shortened telomeres in serous tubal intraepithelial carcinoma: an early event in ovarian high-grade serous carcinogenesis. Am J Surg Pathol. 2010;34:829–836. doi: 10.1097/PAS.0b013e3181dcede7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehdev A.S., Kurman R.J., Kuhn E., Shih I.-M. Serous tubal intraepithelial carcinoma upregulates markers associated with high-grade serous carcinomas including Rsf-1 (HBXAP), cyclin E and fatty acid synthase. Mod Pathol. 2010;23:844–855. doi: 10.1038/modpathol.2010.60. [DOI] [PMC free article] [PubMed] [Google Scholar]