Abstract
Beta-cryptoxanthin (β-cry) is a typical carotenoid found abundantly in fruit and vegetables such as the Japanese mandarin orange, persimmon, papaya, paprika, and carrot, and exerts various biological activities (e.g., antioxidant effects). We previously reported that β-cry suppressed lipopolysaccharide (LPS)-induced osteoclast differentiation via the inhibition of prostaglandin (PG) E2 production in gingival fibroblasts and restored the alveolar bone loss in a mouse model for periodontitis in vivo. In this study, we investigated the molecular mechanism underlying the inhibitory effects of β-cry on osteoclast differentiation. In mouse calvarial organ cultures, LPS-induced bone resorption was suppressed by β-cry. In osteoblasts, β-cry inhibited PGE2 production via the downregulation of the LPS-induced mRNA expression of cyclooxygenase (COX)-2 and membrane-bound PGE synthase (mPGES)-1, which are PGE synthesis-related enzymes, leading to the suppression of receptor activator of NF-κB ligand (RANKL) mRNA transcriptional activation. In an in vitro assay, β-cry directly suppressed the activity of the inhibitor of NF-κB kinase (IKK) β, and adding ATP canceled this IKKβ inhibition. Molecular docking simulation further suggested that β-cry binds to the ATP-binding pocket of IKKβ. In Raw264.7 cells, β-cry suppressed RANKL-mediated osteoclastogenesis. The molecular mechanism underlying the involvement of β-cry in LPS-induced bone resorption may involve the ATP-competing inhibition of IKK activity, resulting in the suppression of NF-κB signaling.
Keywords: beta-cryptoxanthin, bone resorption, lipopolysaccharide, periodontitis, osteoclast differentiation
1. Introduction
Bone homeostasis is precisely controlled by maintaining a balance between bone formation and bone resorption, and imbalances in bone remodeling result in various skeletal diseases, such as osteoporosis and periodontitis. Osteoclasts are multinucleated giant cells derived from hematopoietic stem cells that exhibit bone-resorbing activity, whereas osteoblasts are derived from mesenchymal stem cells and are responsible for bone formation. Receptor activator of NF-κB ligand (RANKL) is a key molecule involved in the differentiation of osteoclast precursors into multinucleated osteoclasts [1]. RANKL expressed on the cell surface of osteoblasts binds to RANK expressed on osteoclast precursor cells, and this RANKL–RANK interaction induces osteoclast differentiation and bone resorption via NF-κB signaling. The proinflammatory molecules, including lipopolysaccharide (LPS) and interleukin (IL)-1, promote prostaglandin (PG) E2 production and subsequently induce RANKL expression in osteoblasts, leading to osteoclast differentiation [2,3,4,5,6]. PGE2 is mainly produced by osteoblasts and fibroblasts under inflammatory conditions [7] and is synthesized by cyclooxygenase (COX)-2 and membrane-bound PGE synthase (mPGES)-1 in the arachidonic acid cascade. We previously reported that PGE2 bound to its receptor EP4 expressed on osteoblasts, leading to osteoclastic bone resorption associated with inflammation [3,4].
Periodontitis is a local bone disease associated with inflammation caused by the infection of Gram-negative bacteria, and the development of periodontitis results in alveolar bone resorption and tooth loss. LPS, an outer membrane component of Gram-negative bacteria, is a pathogen of periodontitis and is identified as a ligand for toll-like receptor (TLR) 4 [8]. Our previous study reported that LPS–TLR4 signaling stimulated PGE2 production through the upregulation of COX-2 and mPGES-1 mRNA expression in osteoblasts, resulting in osteoclastic bone resorption [5,9]. We also reported that the local injection of LPS into lower gingiva induced alveolar bone loss in mice; however, LPS injection failed to induce alveolar bone loss in mPGES1-deficient mice [5]. PGE2 is therefore an essential factor for LPS-induced inflammatory bone resorption.
Carotenoids are abundantly contained in fruits and vegetables, and exert beneficial activity. Beta-cryptoxanthin (β-cry) is a typical carotenoid found abundantly in Japanese mandarin orange, persimmon, papaya, paprika, and carrot. β-cry has been reported to possess several beneficial functions, such as antioxidant, cancer-preventive effects, and anti-metabolic syndrome effects [10,11,12]. Park et al. [13] showed that the daily oral administration of β-cry prevented the progression of osteoarthritis and inhibited proinflammatory cytokines in mice. Ozaki et al. [14] reported that the daily intake of β-cry suppressed osteoclast differentiation via the repression of the NF-κB pathway, and ameliorated estrogen deficiency-induced bone loss in ovariectomized mice. Uchiyama et al. [15,16] showed that β-cry promoted osteoblastic bone formation and mineralization via the upregulated expression of insulin growth factor (IGF)-1, transforming growth factor (TGF)-β, collagen type 1 alpha 1 (Col1a1), runt-related transcription factor (RUNX) 2, and alkaline phosphatase in MC3T3-E1 cells. Regarding the molecular mechanisms, Yamaguchi et al. [17,18] reported that β-cry stimulated bone formation via the activation of TGF-β signaling, and β-cry bound to RXR or orphan receptors in the nucleus in order to regulate bone formation-related genes. Both mitogen-activated protein kinase (MAPK) and protein kinase (PK) C may also be target molecules of β-cry in osteoblastic bone formation [15]. Uchiyama and Yamaguchi [19,20,21] further reported that β-cry inhibited osteoclast differentiation and induced apoptosis in mature osteoclasts. We previously showed that β-cry suppressed PGE2 production in murine primary gingival fibroblasts and inhibited the LPS-induced bone loss of mandibular alveolar bone in a mouse model for periodontitis [22]; however, the target molecule of β-cry in bone resorption is unclear.
In the present study, we clarified the molecular mechanisms underlying the involvement of β-cry in osteoclast differentiation.
2. Materials and Methods
2.1. Animals and Reagents
Newborn, 6-week-old male mice of the ddY strain were obtained from Japan SLC Inc. (Shizuoka, Japan). All procedures were performed in accordance with the institutional guidelines for animal research. β-cry (purity: ≥97%) was obtained from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). LPS from Escherichia coli was provided by Sigma-Aldrich Co. LLC. (St. Louis, MO, USA). Recombinant human soluble RANK ligand (sRANKL) was purchased from Peprotech Co. Ltd. (Rocky Hill, NJ, USA).
2.2. Bone-Resorbing Activity in Organ Cultures of Mouse Calvariae
Mouse calvariae were collected from newborn mice and precultured for 24 h in BGJb medium supplemented with 1 mg/mL bovine serum albumin (BSA) at 37 °C under 5% CO2 in the air. Calvariae were treated with LPS (1 µg/mL) and β-cry after preculture and further cultured for 5 days. The concentration of Ca in the cultured medium was measured using o-cresolphthalein complexone (OCPC).
2.3. Cultures of Primary Mouse Osteoblastic Cells
Primary osteoblastic cells (POBs) were isolated from newborn mouse calvariae after digestion with 0.1% collagenase (Roche Diagnostics GmbH, Mannheim, Germany) and 0.2% dispase (Roche Applied Science, Mannheim, Germany). POBs were cultured in α-modified MEM (αMEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2 in the air, as reported previously [5].
2.4. Measurement of the PGE2 Content in the Cultured Medium
The concentration of PGE2 in conditioned medium in POB cultures was measured using an enzyme immunoassay system (EIA) (GE Healthcare UK Ltd., Little Chalfont, UK). The cross-reactivity of the antibody in the EIA was calculated as follows: PGE2: 100%, PGE1: 7.0%, 6-keto-PGF1α: 5.4%, PGF2α: 4.3%, and PGD2: 1.0%.
2.5. Reverse Transcription-Quantitative PCR
Mouse POBs were cultured for 24 h in αMEM with 1% FBS with LPS (1 ng/mL) and β-cry (30 µM). Total RNA was isolated using ISOGEN (Nippon Gene Co., Ltd., Tokyo, Japan), and cDNA was prepared from RNA via reverse transcription. For real-time PCR, 5 µg of RNA was mixed with SsoAdvanced SYBR green supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and PCR primer pair, and real-time PCR was performed. The primer sequences for real-time PCR were as follows: mouse Rankl (NM_011613.3): 5′-aggctgggccaagatctcta-3′ (forward) and 5′-gtctgtaggtacg cttcccg-3′ (reverse), mouse Cox2 (NM_011198.4): 5′-gggagtctggaacattgtgaa-3′ (forward) and 5′-gtgcacatt gtaagtaggtggact-3′ (reverse), mouse mPges1 (NM_022415.3): 5′-gcacactgctggtcatcaag-3′ (forward) and 5′-acgtttcagcgcatcctc-3′ (reverse), mouse Ctsk (NM_007802.4): 5′-gcctagcgaacagattctcaa-3′ (forward) and 5′-cactgggtgtccagcattt-3′ (reverse), mouse β-actin (NM_007393.5): 5′-ccccattgaacatggcattg-3′ (forward) and 5′-acgaccagaggcatacagg-3′ (reverse). The results are shown as the relative fold expression normalized by β-actin compared with the control.
2.6. Dual-Luciferase Reporter Assay
Plasmid pNFκB-TA-Luc (0.4 µg) contained 4 tandem copies of the NF-κB consensus sequence with the firefly luciferase reporter gene (Clontech Laboratories, Inc., Mountain View, CA, USA), and the pGL4.74[hLuc/TK] plasmid (40 ng) contained the Renilla luciferase reporter gene (Promega Corp., Madison, WI, USA) as an internal control reporter vector. Both plasmids were transfected into mouse POBs using Lipofectamine 2000 (Thermo Fisher Scientific Inc., Waltham, MA, USA). The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corp.) with an ARVO MX multilabel/luminescence counter (Perkin Elmer Corp., Waltham, MA, USA).
2.7. Inhibitor of NF-κB Kinase (IKK) Activity Assay
The kinase activity of IKKβ was elucidated with or without β-cry (0.05–5 mM) using the Cyclex IKKα and β Assay/Inhibitor Screening Kit (CycLex Co. Ltd., Nagano, Japan) with IKKβ, IκBα, and anti-phospho-IκBα antibody.
2.8. Protein Structure Preparation
The three-dimensional X-ray crystal structure of IKKβ was obtained from a protein databank (PDB ID:4KIK, 2.83-Å resolution) [23]. For docking simulations, default parameters (H-atoms) were added into the protein structures using AutoDock Tools (Molecular Graphics Laboratory, La Jolla, CA, USA).
2.9. Ligand Structure Preparation
The chemical structure of β-cry was optimized using the online compound editor InDraw (http://in.indraw.integle.com; Integle Chemistry, Inc., Shanghai, China). All two-dimensional structures were converted into three-dimensional structures in the pdb format and saved in the mol format using Open Babel (http://www.openbabel.org/) [24].
2.10. Molecular Docking Studies
The protein–ligand molecular docking study was performed using AutoDock Vina [25]. Subsequently, AutoDock Vina was used to implement fast docking of the inhibitor ligand into the active pocket of both the IKKβ and kinase domains, which considered the flexibility and mobility of the ligand molecules and protein active-site residues, and used the Lamarckian genetic algorithm to fully explore the conformational space for the IKKβ inhibitor interactions. The rotational bonds of the protein were kept rigid, while those of the ligands were treated as flexible. The amino acids Leu21, Gly22, Tyr 23, Val29, Ala42, Lys44, Glu61, Val74, Met96, Glu97, Tyr98, Cys99, Gly102, Asp103, Glu149, Asn150, Val152, Ile165, Asp166, and the surrounding residues within a distance of 6.5 Å were defined as active sites.
2.11. Structural Visualization and Analyses
Pymol (http://apbs.sourceforge.net) was used for the analysis and visualization of protein–ligand interaction. Amino acids of active sites and the surrounding residues were set to be shown, while other parts were hidden.
2.12. RANKL-Induced Osteoclastogenesis in RAW264.7 Cells
RAW264.7 cells (2.5 × 104 cells) were cultured in αMEM containing 10% FBS for 5 days with sRANKL (100 ng/mL) in the presence of β-cry (5–10 µM). The formed osteoclastic cells were stained for tartrate-resistant acid phosphatase (TRAP). TRAP-positive multinucleated cells containing three or more nuclei per cell were defined as osteoclasts.
2.13. Statistical Analyses
Data were analyzed using a one-way analysis of variance, followed by Tukey’s test for the post hoc analysis. All data are presented as the means ± standard deviation (SD), and all statistical analyses were performed using the IBM SPSS Statistics Ver.23 software program (Armonk, NY, USA).
3. Results
3.1. β-Cry Inhibited LPS-Induced Bone Resorption in Calvarial Organ Culture
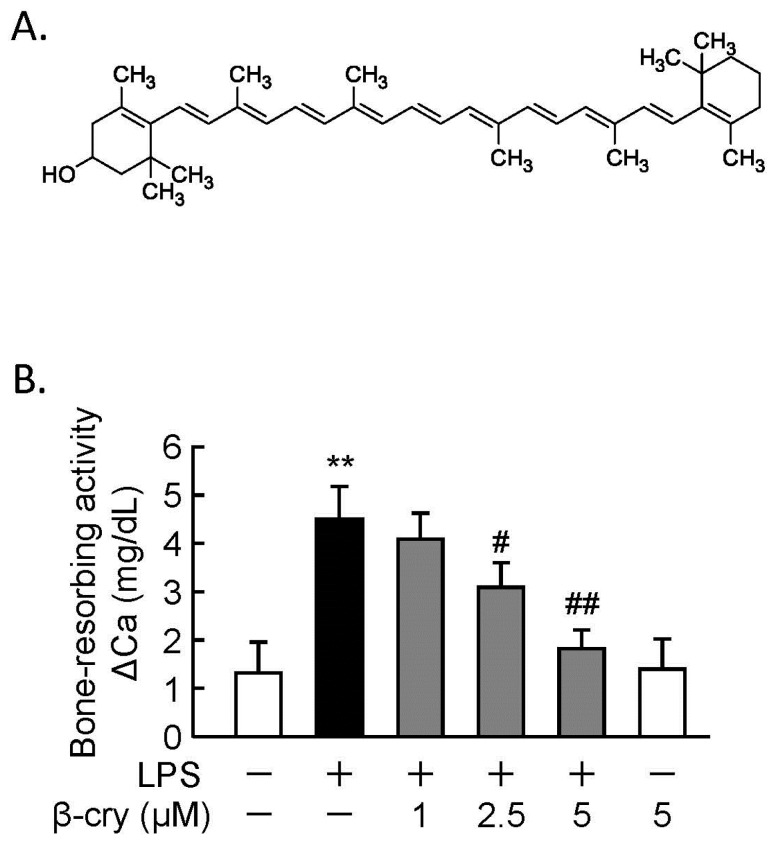
The chemical structure of β-cry is shown in Figure 1A. We previously showed that β-cry suppressed LPS-induced osteoclast differentiation in cocultures of mouse bone marrow cells and POBs [22]. To examine the effects of β-cry on bone resorption, calvariae from newborn mice were cultured with or without LPS (1 µg/mL) and β-cry (5 µM). β-cry suppressed the LPS-induced bone resorbing activity in a dose-dependent manner in calvarial organ cultures, as seen in Figure 1B.
Figure 1.
Beta-cryptoxanthin (β-cry) suppressed the lipopolysaccharide (LPS)-induced bone-resorbing activity. (A) The chemical structure of β-cry. (B) Mouse calvariae were cultured with LPS (1 µg/mL) and β-cry (1–5 µM) for five days. The Ca concentration in the conditioned medium was measured by the o-cresolphthalein complexone (OCPC) method, and the increase in Ca was evaluated as the bone-resorbing activity. Data are expressed as the mean ± SD for three independent cultures. Significant differences are indicated, ** p < 0.01 vs. control; # p < 0.05, ## p < 0.01 vs. LPS.
3.2. Effects of β-Cry on the PGE2 Production and NF-κB Signaling in Osteoblasts
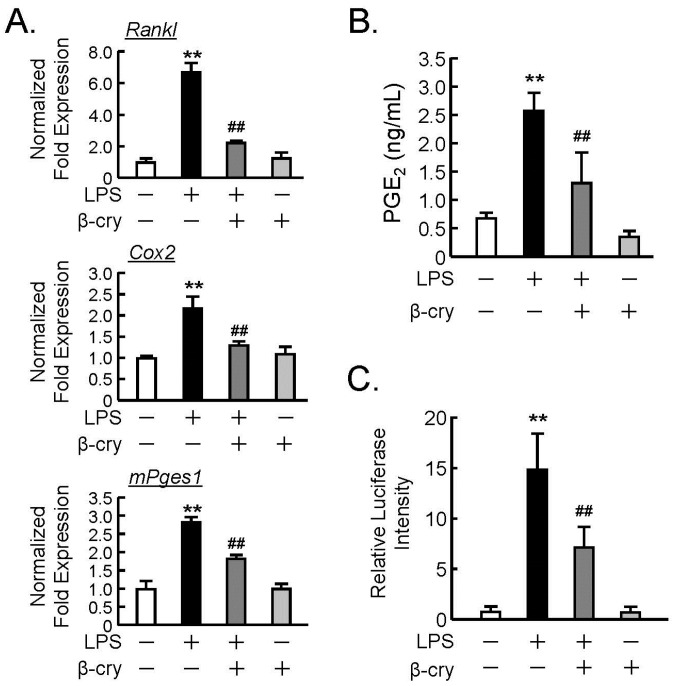
We previously reported that β-cry suppressed the PGE2 production induced by LPS in the murine fibroblast cell line NIH3T3 and primary gingival fibroblasts [22]. In order to assess the effects of β-cry on osteoblasts, the mRNA expression of PGE-related genes was analyzed by real-time PCR. LPS augmented the mRNA expression of COX-2, mPGES1, and RANKL, whereas β-cry significantly suppressed the LPS-induced mRNA expression of these genes, as seen in Figure 2A. In addition, β-cry clearly inhibited PGE2 production in the culture of osteoblasts, as seen in Figure 2B. We further analyzed the NF-κB transcriptional activity using a reporter gene assay and found that β-cry significantly attenuated the LPS-induced NF-κB activity, as seen in Figure 2C.
Figure 2.
The effects of β-cry on the LPS-induced expression of cyclooxygenase (COX)-2, membrane-bound PGE synthase (mPGES)-1, and receptor activator of NF-κB ligand (RANKL) mRNA on the production of prostaglandin (PG)E2 in osteoblasts. Primary osteoblastic cells (POBs) were pre-treated with β-cry (5 µM) for 24 h and further cultured for 24 h in the presence of LPS (1 µg/mL) and β-cry (5 µM). (A) The mRNA expression of RANKL, COX-2, and mPGES1 was analyzed by RT-qPCR. Data are expressed as the mean ± SD of three replicated wells in triplicate. (B) The PGE2 concentration was measured in the cultured medium of POBs. (C) The transcription activity of NF-κB was measured with or without β-cry (5 µM). Plasmid pNFkB-TA-Luc (0.4 µg) and the pGL4.74[hLuc/TK] plasmid (40 ng) were transfected into POBs, and the luciferase activity was measured with the Dual-Luciferase Reporter Assay System. Data are expressed as the mean ± SD for 3–4 independent wells. Significant differences are indicated, ** p < 0.01 vs. control; ## p < 0.01 vs. LPS.
3.3. β-Cry Inhibited IKKβ Activity Via ATP-Competitive Binding
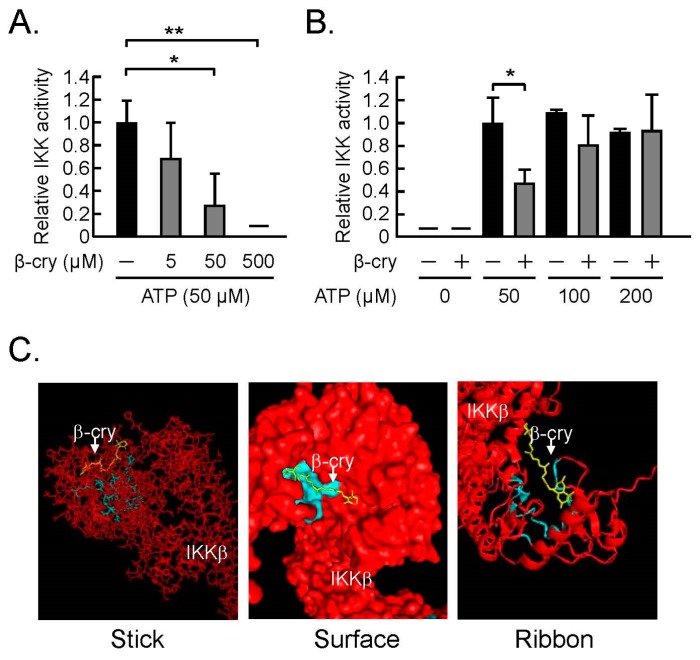
Kim et al. [26] suggested that flavonoid binds to the ATP-binding pocket of IKKβ to suppress its kinase activity and attenuate NF-κB activation. Our data similarly showed that β-cry inhibited the recombinant IKK activity in a dose-dependent manner, as seen in Figure 3A, and this effect of β-cry was also canceled by ATP in a dose-dependent manner, as seen in Figure 3B. In order to examine the possible binding model of β-cry to IKKβ, β-cry was subjected to molecular docking using a homology model of IKKβ. In Figure 3C, the 3D images of the ligand binding model are shown as three models. The colors in the three images were indicated as follows: yellow (β-cry), light blue (active site), and red (IKKβ). The amino acids Leu21, Gly22, Tyr 23, Val29, Ala42, Lys44, Glu61, Val74, Met96, Glu97, Tyr98, Cys99, Gly102, Asp103, Glu149, Asn150, Val152, Ile165, Asp166, and the surrounding residues within a distance of 6.5 Å were defined as active sites. β-cry was positioned in the catalytic center, a pocket-like structure of the assumed binding site (Figure 3C). These data suggest that β-cry binds to the ATP-binding pocket of IKKβ to suppress its kinase activity, leading to attenuation of the NFκB pathway.
Figure 3.
The direct action of β-cry on inhibitor of NF-κB kinase (IKK) activity in an in vitro assay. The IKK activity was elucidated by the IKK activity assay kit using IKKβ, IκBα, and anti-phospho-IκBα antibody. (A) The effects of β-cry on the IKK activity. (B) ATP was added to the IKK assay with β-cry, and the IKK activity was expressed as the percentage of the control without β-cry. Data are expressed as the mean ± SD for three independent wells. Significant differences are indicated, * p < 0.05, ** p < 0.01. (C) Predicted binding model of β-cry docked into a homology of IKKβ. The 3D images of the ligand binding model are shown as three models. The colors in the three images were indicated as follows: yellow (β-cry), red chain (IKKβ), and light blue (active site in IKKβ).
3.4. β-Cry Directly Suppressed sRANKL-Induced Osteoclast Differentiation
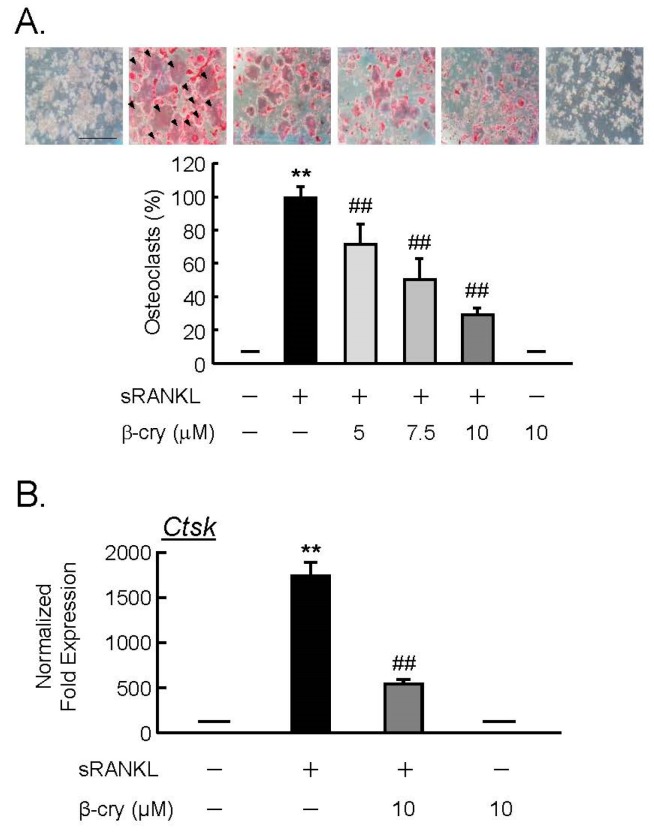
In order to test the direct action of β-cry on osteoclast precursor cells, RAW264.7 cells were cultured for four days with sRANKL (100 ng/mL) and β-cry (5–10 µM). β-cry suppressed the RANKL-induced osteoclast differentiation in a dose-dependent manner, as seen in Figure 4A. The mRNA expression of cathepsin K (Ctsk) was elevated by RANKL in RAW264.7 cells, but this increase was repressed by adding β-cry to the real-time PCR analysis, as seen in Figure 4B.
Figure 4.
β-cry inhibits the soluble RANK ligand (sRANKL)-induced osteoclast differentiation and expression of cathepsin K (Ctsk) mRNA in RAW264.7 cells. RAW264.7 cells, which are mouse macrophages, were cultured with sRANKL (100 ng/mL) and β-cryptoxanthin (5–10 µM) for four days. (A) Osteoclasts were stained for tartrate-resistant acid phosphatase (TRAP), and the percentage of TRAP-positive multinucleated cells was measured. Data are expressed as the mean ± SD for eight independent wells. Arrowheads indicate TRAP-positive multinucleated osteoclasts. Bar = 500 µm. (B) Total RNA was extracted and reverse transcribed into cDNA. cDNA was amplified, and the mRNA expression of cathepsin K (Ctsk) was measured by RT-qPCR. Data are expressed as the mean ± SD of three replicated wells in triplicate. Significant differences are indicated, ** p < 0.01 vs. control; ## p < 0.01 vs. sRANKL.
4. Discussion
β-cry is known to be abundantly found in Japanese mandarin oranges and has been reported to exert various beneficial effects, such as antioxidant and cancer-preventive effects [10,11,12]. Japanese mandarin oranges (Citrus unshiu) contain high levels of β-cry and are produced in Japan. Sugiura et al. [27] reported that a daily intake of one or more Citrus unshiu increased the concentration between 0.5 to 3.5 µM of serum β-cry in healthy Japanese individuals. They also reported that β-cry is easily accumulated in various tissues compared with β-carotene [28]. Several reports have shown that β-cry exhibited preventive effects on estrogen deficiency-induced bone loss, rheumatoid arthritis, and osteoarthritis in in vivo studies [14,29,30]. Our previous study showed that β-cry inhibited LPS-induced osteoclast differentiation in cocultures of mouse bone marrow cells and POBs and restored the bone loss of mandibular alveolar bone induced by local injection of LPS in a mouse model of periodontitis [22]. This study also showed that 5 µM of β-cry did not exhibit cell cytotoxicity in osteoblasts [22]. In the present study, we showed that β-cry inhibited the COX-2- and mPGES-1-mediated PGE2 production in osteoblasts and further suggested that IKKβ was a novel molecular target of β-cry in LPS-induced osteoclast differentiation. Therefore, it is possible that β-cry may regulate inflammatory response in various cells, but further studies are needed to clarify this point.
Osteoblasts perform dual roles in bone formation and bone resorption. The expression of RANKL is induced by inflammatory molecules, such as LPS, and is essential for inflammatory bone resorption. NF-κB is the key transcription factor associated with the regulation of COX-2 and mPGES1 after LPS stimulation [31,32]. We previously reported that the lack of PGE2 production in mPGES1-deficient mice (mPges1−/−) attenuated the LPS-induced inflammatory bone destruction of alveolar bone [5]. In the present study, we found that β-cry suppressed the mRNA expression of PGE2 synthases, COX-2, and mPGES-1, as well as PGE2 production via the attenuation of NF-κB activity in osteoblasts, leading to the downregulation of RANKL expression. Along with our previous finding that β-cry inhibited LPS-induced osteoclast differentiation in cocultures of mouse bone marrow cells and POBs [22], the present results suggest that β-cry inhibits osteoclast differentiation and bone resorption by downregulating the PGE2-mediated RANKL expression in osteoblasts and by direct action on osteoclast precursors to suppress osteoclast differentiation through inhibiting NF-κB activity. However, it was also reported that β-cry stimulated cell growth, differentiation, and mineralization via the upregulation of bone formation genes, including Runx2, in the mouse osteoblastic cell line MC3T3-E1 [15,16,17]. Therefore, β-cry may act on both bone resorption and bone formation in bone tissues.
Uchiyama et al. [20,21] reported that β-cry suppressed the formation of osteoclast-like cells induced by bone-resorbing factors, such as parathyroid hormone (PTH), PGE2, 1,25-dihydroxyvitamin D3, LPS, and TNFα, and induced apoptotic cell death through the upregulation of apoptosis protease-associating factor (Apaf)-2 and caspase-3 in mouse marrow cultures and osteoclastic cell cultures. Ozaki et al. [14] reported that β-cry inhibited sRANKL-induced osteoclastogenesis, downregulated the mRNA expression of TRAP, and attenuated the activity of NF-κB but not AP-1 and NFATc1 in Raw264.7 cells. In the present study, we found that β-cry suppressed sRANKL-induced osteoclast differentiation and downregulated the mRNA expression of cathepsin K in cultures of RAW264.7 cells. Therefore, β-cry may act directly on osteoclast precursor cells to inhibit their differentiation into mature osteoclasts.
Since β-cry is a provitamin A, the effects of β-cry might be attributed to that of vitamin A (retinol). However, a previous study showed that retinol was not detected in RAW264.7 cells treated with β-cry [11]. Uchiyama et al. [15] suggested that MAPK or protein kinase C (PKC) are potential targets of β-cry in osteoblasts and speculated that β-cry could bind to orphan receptors to exert its biological functions. We previously suggested that lutein, a carotenoid, could directly inhibit the kinase activity of IKK [33]. IKK complex, an enzyme complex consisting of the catalytic kinases IKKα and IKKβ as well as a regulatory subunit NEMO, is a signal component of NF-κB signaling. IKK complex stimulates NF-κB translocation by the phosphorylation and degradation of inhibitor of NF-κB (IκB). The IKK complex is activated by various inflammatory responses, including proinflammatory cytokines, bacterial LPS, RANKL–RANK signaling, viral infection, and stress-induced responses [34,35,36]. We examined the effects of β-cry on IKKβ activity with an in vitro assay and found that β-cry directly suppressed the IKKβ activity. Kim et al. [26] suggested that luteolin, a flavonoid, could bind to the ATP-binding pocket of IKKβ to inhibit its activity and attenuate NF-κB activation. Our present study indicated that excess ATP could competitively inhibit the effects of β-cry on IKKβ activity. These findings suggest that β-cry inhibits IKKβ activity via competition with ATP-binding pocket, leading to the attenuation of NFκB activation and osteoclast differentiation. Sahin et al. [12] have shown that β-cry suppressed nuclear localization of NFκB in various cells in liver and adipose tissues. Further studies are needed to define the effects of β-cry on the nuclear localization of NFκB in both osteoblasts and osteoclasts.
In the present study, we have suggested that IKKβ might be the novel target of β-cry in osteoclast differentiation. Since the IKK complex is involved in both LPS–TLR4–IKKs–NFκB signaling and RANKL–RANK–IKKs–NFκB signaling, β-cry may inhibit the LPS-induced RANKL expression in osteoblasts and RANKL-induced osteoclast differentiation in osteoclast precursor cells. These data are helpful for understanding the molecular mechanism of action of β-cry, which may be a potential natural compound for the prevention of inflammatory bone loss.
Author Contributions
N.H., R.I., C.M., and M.I. conceived and designed the experiments; N.H., R.I., T.T., K.T., K.W., and S.M. performed the experiments; R.I., T.T., C.M., M.H., K.S., and F.M.W.G. analyzed the data; C.M., M.I., K.S., and M.H. contributed reagents/materials/analysis tools; R.I., N.H., and T.T. wrote the paper; C.M., M.I., and F.M.W.G. reviewed and improved the manuscript.
Funding
This work is partly supported by Institute of Global Innovation Research in TUAT (M. I., F. G.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., et al. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. USA. 1998;95:3597–3602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyaura C., Inada M., Suzawa T., Sugimoto Y., Ushikubi F., Ichikawa A., Narumiya S., Suda T. Impaired bone resorption to prostaglandin E2 in prostaglandin E receptor EP4-knockout mice. J. Biol. Chem. 2000;275:19819–19823. doi: 10.1074/jbc.M002079200. [DOI] [PubMed] [Google Scholar]
- 3.Suzawa T., Miyaura C., Inada M., Maruyama T., Sugimoto Y., Ushikubi F., Ichikawa A., Narumiya S., Suda T. The role of prostaglandin E receptor subtypes (EP1, EP2, EP3, and EP4) in bone resorption: An analysis using specific agonists for the respective EPs. Endocrinology. 2000;141:1554–1559. doi: 10.1210/endo.141.4.7405. [DOI] [PubMed] [Google Scholar]
- 4.Miyaura C., Inada M., Matsumoto C., Ohshiba T., Uozumi N., Shimizu T., Ito A. An Essential Role of Cytosolic Phospholipase A2α in Prostaglandin E2–mediated Bone Resorption Associated with Inflammation. J. Exp. Med. 2003;197:1303–1310. doi: 10.1084/jem.20030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inada M., Matsumoto C., Uematsu S., Akira S., Miyaura C. Membrane-bound prostaglandin E synthase-1-mediated prostaglandin E2 production by osteoblast plays a critical role in lipopolysaccharide-induced bone loss associated with inflammation. J. Immunol. 2006;177:1879–1885. doi: 10.4049/jimmunol.177.3.1879. [DOI] [PubMed] [Google Scholar]
- 6.Hirata M., Kobayashi M., Takita M., Matsumoto C., Miyaura C., Inada M. Hyaluronan inhibits bone resorption by suppressing prostaglandin E synthesis in osteoblasts treated with interleukin-1. Biochem. Biophys. Res. Commun. 2009;381:139–143. doi: 10.1016/j.bbrc.2009.01.146. [DOI] [PubMed] [Google Scholar]
- 7.Laulederkind S., Kirtikara K., Raghow R., Ballou L. The regulation of PGE(2) biosynthesis in MG-63 osteosarcoma cells by IL-1 and FGF is cell density-dependent. Exp. Cell. Res. 2000;258:409–416. doi: 10.1006/excr.2000.4961. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the Lps gene product. J. Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 9.Tominari T., Matsumoto C., Watanabe K., Hirata M., Grundler F., Miyaura C., Inada M. Epigallocatechin gallate (EGCG) suppresses lipopolysaccharide-induced inflammatory bone resorption, and protects against alveolar bone loss in mice. FEBS Open Bio. 2015;5:522–527. doi: 10.1016/j.fob.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iskandar A.R., Miao B., Li X., Hu K.-Q., Liu C., Wang X.-D. β-Cryptoxanthin Reduced Lung Tumor Multiplicity and Inhibited Lung Cancer Cell Motility by Downregulating Nicotinic Acetylcholine Receptor α7 Signaling. Cancer Prev. Res. 2016;9:875–886. doi: 10.1158/1940-6207.CAPR-16-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsuura S., Imamura T., Bando N., Yamanishi R. β-Carotene and β-cryptoxanthin but not lutein evoke redox and immune changes in RAW264 murine macrophages. Mol. Nutr. Food Res. 2009;53:1396–1405. doi: 10.1002/mnfr.200800566. [DOI] [PubMed] [Google Scholar]
- 12.Sahin K., Orhan C., Akdemir F., Tuzcu M., Sahin N., Yılmaz I., Juturu V. β-Cryptoxanthin ameliorates metabolic risk factors by regulating NF-κB and Nrf2 pathways in insulin resistance induced by high-fat diet in rodents. Food Chem. Toxicol. 2017;107:270–279. doi: 10.1016/j.fct.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Park G., Horie T., Fukasawa K., Ozaki K., Onishi Y., Kanayama T., Iezaki T., Kaneda K., Sugiura M., Hinoi E. Amelioration of the Development of Osteoarthritis by Daily Intake of β-Cryptoxanthin. Biol. Pharm. Bull. 2017;40:1116–1120. doi: 10.1248/bpb.b17-00161. [DOI] [PubMed] [Google Scholar]
- 14.Ozaki K., Okamoto M., Fukasawa K., Iezaki T., Onishi Y., Yoneda Y., Sugiura M., Hinoi E. Daily intake of β-cryptoxanthin prevents bone loss by preferential disturbance of osteoclastic activation in ovariectomized mice. J. Pharmacol. Sci. 2015;129:72–77. doi: 10.1016/j.jphs.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Uchiyama S., Yamaguchi M. beta-Cryptoxanthin stimulates cell proliferation and transcriptional activity in osteoblastic MC3T3-E1 cells. Int. J. Mol. Med. 2005;15:675–681. doi: 10.3892/ijmm.15.4.675. [DOI] [PubMed] [Google Scholar]
- 16.Uchiyama S., Yamaguchi M. β-cryptoxanthin stimulates cell differentiation and mineralization in osteoblastic MC3T3-E1 cells. J. Cell. Biochem. 2005;95:1224–1234. doi: 10.1002/jcb.20496. [DOI] [PubMed] [Google Scholar]
- 17.Yamaguchi M., Weitzmann M. The bone anabolic carotenoid beta-cryptoxanthin enhances transforming growth factor-beta1-induced SMAD activation in MC3T3 preosteoblasts. Int. J. Mol. Med. 2009;24:671–675. doi: 10.3892/ijmm_00000278. [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi M., Weitzmann M. The bone anabolic carotenoids p-hydroxycinnamic acid and β-cryptoxanthin antagonize NF-κB activation in MC3T3 preosteoblasts. Mol. Med. Rep. 2009;2:641–644. doi: 10.3892/mmr_00000150. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi M. Role of carotenoid β-cryptoxanthin in bone homeostasis. J. Biomed. Sci. 2012;19:36. doi: 10.1186/1423-0127-19-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchiyama S., Yamaguchi M. Inhibitory effect of beta-cryptoxanthin on osteoclast-like cell formation in mouse marrow cultures. Biochem. Pharmacol. 2004;67:1297–1305. doi: 10.1016/j.bcp.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Uchiyama S., Yamaguchi M. Beta-cryptoxanthin stimulates apoptotic cell death and suppresses cell function in osteoclastic cells: Change in their related gene expression. J. Cell. Biochem. 2006;98:1185–1195. doi: 10.1002/jcb.20824. [DOI] [PubMed] [Google Scholar]
- 22.Matsumoto C., Ashida N., Yokoyama S., Tominari T., Hirata M., Ogawa K., Sugiura M., Yano M., Inada M., Miyaura C. The Protective Effects of β-Cryptoxanthin on Inflammatory Bone Resorption in a Mouse Experimental Model of Periodontitis. Biosci. Biotechnol. Biochem. 2013;77:860–862. doi: 10.1271/bbb.120791. [DOI] [PubMed] [Google Scholar]
- 23.Liu S., Misquitta Y.R., Olland A., Johnson M.A., Kelleher K.S., Kriz R., Lin L.L., Stahl M., Mosyak L. Crystal structure of a human IκB kinase b asymmetric dimer. J. Biol. Chem. 2013;288:22758–22767. doi: 10.1074/jbc.M113.482596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2011;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J.S., Jobin C. The flavonoid luteolin prevents lipopolysaccharide-induced NF-kappaB signalling and gene expression by blocking IkappaB kinase activity in intestinal epithelial cells and bone-marrow derived dendritic cells. Immunology. 2005;115:375–387. doi: 10.1111/j.1365-2567.2005.02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiura M., Kato M., Matsumoto H., Nagao A., Yano M. Serum concentration of β-cryptoxanthin in Japan reflects the frequency of Satsuma mandarin (Citrus Unshiu Marc.) consumption. J. Health Sci. 2002;48:350–353. doi: 10.1248/jhs.48.350. [DOI] [Google Scholar]
- 28.Sugiura M., Ogawa K., Yano M. Absorption, storage and distribution of β-cryptoxanthin in rat after chronic administration of Satsuma mandarin (Citrus unshiu MARC.) juice. Biol. Pharm. Bull. 2013;36:147–151. doi: 10.1248/bpb.b12-00836. [DOI] [PubMed] [Google Scholar]
- 29.Imada K., Tsuchida A., Ogawa K., Sofat N., Nagase H., Ito A., Sato T. Anti-arthritic actions of β-cryptoxanthin against the degradation of articular cartilage in vivo and in vitro. Biochem. Biophys. Res. Commun. 2016;476:352–358. doi: 10.1016/j.bbrc.2016.05.126. [DOI] [PubMed] [Google Scholar]
- 30.Pattison D.J., Symmons D.P., Lunt M., Welch A., Bingham S.A., Day N.E., Silman A.J. Dietary beta-cryptoxanthin and inflammatory polyarthritis: Results from a population-based prospective study. Am. J. Clin. Nutr. 2005;82:451–455. doi: 10.1093/ajcn/82.2.451. [DOI] [PubMed] [Google Scholar]
- 31.D’Acquisto F., Iuvone T., Rombolà L., Sautebin L., Di Rosa M., Carnuccio R. Involvement of NF-kappaB in the regulation of cyclooxygenase-2 protein expression in LPS-stimulated J774 macrophages. FEBS Lett. 1997;418:175–178. doi: 10.1016/S0014-5793(97)01377-X. [DOI] [PubMed] [Google Scholar]
- 32.Díaz-Muñoz M.D., Osma-García I.C., Cacheiro-Llaguno C., Fresno M., Iñiguez M.A. Coordinated up-regulation of cyclooxygenase-2 and microsomal prostaglandin E synthase 1 transcription by nuclear factor kappa B and early growth response-1 in macrophages. Cell Signal. 2010;22:1427–1436. doi: 10.1016/j.cellsig.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Takeda H., Tominari T., Ichimaru R., Matsumoto C., Watanabe K., Hirata M., Inada M., Miyaura C. Lutein, a carotenoid, inhibits lipopolysaccharide-induced alveolar bone loss associated with inflammation in a mouse model of periodontitis. Curr. Top. Biochem. Res. 2016;17:71–76. [Google Scholar]
- 34.Jacobs M.D., Harrison S.C. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/S0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 35.Adli M., Merkhofer E., Cogswell P., Baldwin A.S. IKKalpha and IKKbeta each function to regulate NF-kappaB activation in the TNF-induced/canonical pathway. PLoS ONE. 2010;5:e9428. doi: 10.1371/journal.pone.0009428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian F., Zhou P., Kang W., Luo L., Fan X., Yan J., Liang H. The small-molecule inhibitor selectivity between IKKα and IKKβ kinases in NF-κB signaling pathway. J. Recept. Signal Transduct. Res. 2015;35:307–318. doi: 10.3109/10799893.2014.980950. [DOI] [PubMed] [Google Scholar]