Abstract
Systemic treatment options for soft tissue sarcomas (STSs) have remained unchanged despite the need for novel drug candidates to improve STS outcomes. Drug repurposing involves the application of clinical drugs to different diseases, reducing development time, and cost. It has also become a fast and effective way to identify drug candidates. The present study used a computational method to screen three drug-gene interaction databases for novel drug candidates for the treatment of several common STS histologic subtypes through drug repurposing. STS survival-associated genes were generated by conducting a univariate cox regression analysis using The Cancer Genome Atlas survival data. These genes were then applied to three databases (the Connectivity Map, the Drug Gene Interaction Database and the L1000 Fireworks Display) to identify drug candidates for STS treatment. Additionally, pathway analysis and molecular docking were conducted to evaluate the molecular mechanisms of the candidate drug. Bepridil was identified as a potential candidate for several STS histologic subtype treatments by overlapping the screening results from three drug-gene interaction databases. The pathway analysis with the Kyoto Encyclopedia of Genes and Genomes predicted that Bepridil may target CRK, fibroblast growth factor receptor 4 (FGFR4), laminin subunit β1 (LAMB1), phosphoinositide-3-kinase regulatory subunit 2 (PIK3R2), WNT5A, cluster of differentiation 47 (CD47), elastase, neutrophil expressed (ELANE), 15-hydroxyprostaglandin dehydrogenase (HPGD) and protein kinase cβ (PRKCB) to suppress STS development. Further molecular docking simulation suggested a relatively stable binding selectivity between Bepridil and eight proteins (CRK, FGFR4, LAMB1, PIK3R2, CD47, ELANE, HPGD, and PRKCB). In conclusion, a computational method was used to identify Bepridil as a potential candidate for the treatment of several common STS histologic subtypes. Experimental validation of these in silico results is necessary before clinical translation can occur.
Keywords: soft tissue sarcoma, drug repositioning, bepridil, molecular docking, The Cancer Genome Atlas
Introduction
Soft tissue sarcoma (STS) is a group of rare malignant tumors that occur in the connective tissue and account for ~10% of cancers in children and <1% of all adult solid malignant cancers (1). These tumors can originate throughout the human body, particularly in the extremities and the trunk. Although STS accounts for no >1% of all malignancies, it appears to be part of a heterogeneous disease, as >50 different histological subtypes have been identified (2,3). Recurrence occurs in ~50% of high-grade, advanced, and metastatic STSs. For patients in relapse or with a delayed diagnosis, few long-term effective agents are available. Advanced STS presents poor overall survival (OS) with the median OS being no >2 years (4,5). Meanwhile, molecular biomarkers to optimize therapy strategies and overcome disease resistance remain unknown (6). Multidisciplinary approaches and specific molecular targeting therapies are recommended for STS patients, but the clinical impact of these therapies in certain common STS subtypes are unclear. Additional molecular pathway targets and treatment strategies are required to improve the outcomes of this confounding disease.
Drug repurposing involves the application of clinical drugs to different diseases in order to reduce the development time and cost. It has become a fast and effective way to identify drug candidates (7). Drug repurposing can be facilitated by an established and searchable database that collects drug-gene interactions from various sources (8–10).
The Connectivity Map (CMap; http://portals.broadinstitute.org/cmap/) is a web-based computational drug-repurposing tool that allows users to screen bioactive small molecules through genome-wide transcription expression data (8). The current version (build 02) of CMap includes >7,000 gene expression profiles from five human cell lines and reflects treatment with 1,309 bioactive small molecules at different doses (11). Screening is achieved by comparing the reference profiles stored in the database with the significantly differentially expressed genes of the user's targeted disease using a pattern-matching algorithm. The screening results in a list of small molecule connectivity scores that range from −1 to +1. A small molecule-expression profile that presents a strong negative correlation to a disease-relevant gene expression signature indicates that the molecule may have a potentially therapeutic effect on the disease (12). CMap has successfully identified bioactive small molecules and combination therapies that have shown promise in the treatment of a variety of diseases (13–19).
The Drug Gene Interaction Database (DGIdb; http://www.dgidb.org), another drug repurposing web-based application, contains >40,000 genes and >10,000 drugs that are involved in >15,000 drug-gene interactions (9). These drug-gene interactions were collected using expert curation and text-mining from the drug-related databases DrugBank, Therapeutic Target Database, PharmGKB, Guide to Pharmacology, and ClinicalTrials.gov. In addition, bioinformatic analyses, such as gene ontology and pathway analysis, were used to categorize potentially druggable genes.
The L1000 Fireworks Display (L1000 FWD; http://amp.pharm.mssm.edu/L1000FWD) is another web-based application that provides interactive visualization of >16,000 drug and gene expression signatures (10). In this database, potential drugs or bioactive small molecules can be easily identified when users enter the differentially expressed gene sets of a specific disease into the search box.
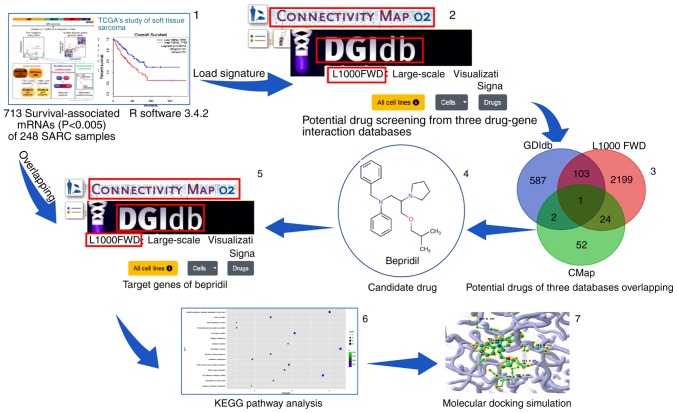
The present study sought to identify novel drug candidates for the treatment of several common histologic subtypes of STS (58 dedifferentiated liposarcoma, 99 leiomyosarcoma, 8 malignant peripheral nerve sheath tumors, 25 myxofibrosarcoma, 10 synovial sarcoma and 49 undifferentiated pleomorphic sarcoma). According to previous studies (6,20–23), these six common STS subtypes may have collective biological targets that can be recognized by one or more drugs. STS survival-associated genes were identified using The Cancer Genome Atlas (TCGA) data and then applied to the CMap, DGIdb and L1000 FWD databases. The repetitions from the predicted drugs obtained from these three drug-gene interaction databases were removed and over-lapped in order to identify the final drug candidates for the STS histologic subtypes, thus providing more reliable results. A pathway analysis and molecular docking were conducted to evaluate the molecular mechanism of the candidate drug. Fig. 1 presents a schematic of the study design concept.
Figure 1.
Schematic depicting a flow diagram of the screening and analysis conducted in the study. Panels 1, 2 and 5 present screenshots from The Cancer Genome Atlas database and three drug-gene interaction database homepages. Panels 3, 4, 6 and 7 present certain results of the present study. SARC, soft tissue sarcoma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Materials and methods
STS survival-associated mRNA screening
mRNA-sequence data (Level 3) was downloaded from the STS samples in the TCGA database (http://cancergenome.nih.gov/). A total of 261 STS samples were collected (including 59 dedifferentiated liposarcoma, 105 leiomyosarcoma, 9 malignant peripheral nerve sheath tumors, 25 myxofibrosarcoma, 10 synovial sarcoma and 51 undifferentiated pleomorphic sarcomas, as well as 2 desmoid tumors). Subsequently, edgeR, a BioconductoR software package, was used to normalize the expression profile (24). To obtain more reliable data, 2 desmoid tumors and 9 tumor samples with survival times <90 days were excluded. Then, a univariate Cox regression was conducted using R software (3.4.2 version; R Foundation for Statistical Computing, Vienna, Austria) to obtain the STS survival-associated genes. The eligible genes were divided into two groups (risk and protective) according to their hazard ratio values. The cut-off value was 1.
Potential drug predictions
The STS survival-associated genes were separated into two files (risk factors and protective factors) in order to query the CMap, DGIdb, and L1000 FWD databases. As the total number of upload tags cannot be >1,000 for CMap, only STS survival-associated genes with P-values <0.005 were selected. Prior to querying CMap, all of the gene symbols were converted to Affymetrix probe IDs using the Affymetrix site (https://www.affymetrix.com/site/mainPage.affx). A total of 535 genes with a negative correlation with STS survival, and 178 genes with a positive correlation with STS survival were separately uploaded to CMap. In the CMap database, drugs with significantly negative scores were expected to be putative novel therapeutic indications for STS. The connectivity scores were calculated using the gene-set-enrichment analysis algorithm (9). A mean score of ≤0.65 was used to identify potential drug candidates.
For DGIdb, all of the STS survival-associated gene symbols were pasted directly into the search box using the ‘Search Drug-Gene Interactions’ function button, and then the predictive drug list, TSV, was downloaded. For the L1000 FWD database, all of the STS survival-associated gene symbols were divided into differential expression gene sets (up and down) before querying the potential drugs. As with the CMap database, drugs with negative scores were expected to have putative novel therapeutic indications for STS.
To increase the reliability of the results, the candidate drugs were selected by overlapping the results from the three databases.
Identified target genes of the candidate drug
Genes regulated by the candidate drug were subsequently identified in the three databases using an in silico analysis. For the CMap database, the identified gene symbols were converted into Affymetrix probe identifiers and then tagged with the ‘up’ and ‘down’ files in. grp format prior to being uploaded to the CMap ‘quick query’ separately (25). Significantly aberrantly expressed probes with amplitude values ≤0.67 or >0.67 were selected (an amplitude of ±0.67 l represents a two-fold change between the treatment and the control). All of the predicted targets were included for the DGIdb and L1000 FWD databases as no threshold was provided.
Pathways analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed for the significantly aberrantly expressed probes of the candidate drugs using the WebGestalt database (http://www.webgestalt.org/option.php) (26). The P-value of each pathway was adjusted using the Hochberg (BH) procedure (27), and pathways with P<0.05 were considered significant. Furthermore, target genes in pathways that previously been reported to be involved in tumor genesis or progression were uploaded to cBioPortal database (http://www.cbioportal.org/) to analyze their genetic alterations.
Molecular docking
Molecular docking is an efficient computational method which can rapidly calculate the binding potential of a small molecule (drug candidate) to a target protein. It has been widely used in computer-aided drug discovery due to its speed and low cost (28,29). SystemsDock (http://systemsdock.unit.oist.jp/) is a web server for network pharmacology-based prediction and analysis, which employs two machine learning systems (Machine Learning Systems A and B) and integrates curated signaling networks, bioinformatics databases and molecular virtual docking simulation to comprehensively and rapidly evaluate potential binding affinities of drug candidates against target proteins (30). Compared with other docking programs (31,32), it provides a major advance in quality and reliability of assessing protein-ligand interaction. However, systemsDock taking protein structure availability and binding site certainty into consideration, and the protein residues involved in the binding interaction are automatically identified by exploring the position where the biggest native ligand is bound. Ducking score, the indication of binding strength, is a negative logarithm of the experimental dissociation/inhibition constant (pKd/pKi) that ranges from 0–10 (i.e., from weak to strong). A good accuracy level (80–83%) was observed when the cut-off scores were in the range of 4.82–6.11 (pKd), which is conventionally used to classify ligand binding activity.
In the present study, molecular docking was singly performed on the proteins of several KEGG pathways with the candidate drug using systemsDock to check whether the candidate drug may have an anti-STS function. The docking simulation was carried out in three steps: i) Specifying the proteins and binding sites by uploading the names or Protein Data Bank (PDB; http://www.rcsb.org/) IDs of the proteins; ii) preparing the small molecules (drugs) for the test by uploading their structure files in 2D/3D SDF, Mol2 or SMILES formats; and iii) clicking the ‘run’ button.
Results
Identification of STS survival-associated genes in the TCGA database
A total of 2,842 survival-associated genes were extracted from 248 sample (58 dedifferentiated liposarcoma, 99 leiomyosarcoma, 7 malignant peripheral nerve sheath tumors, 25 myxofibrosarcoma, 10 synovial sarcoma and 49 undifferentiated pleomorphic sarcomas) mRNA profiles in the TCGA database using a univariate Cox regression. As the CMap database could not process >1,000 uploaded tags, SPS survival-associated genes with P<0.005 were selected to screen the small molecules. Among those genes, 535 were demonstrated to be risk factors for STS, while 178 were protective factors.
STS-targeted screening for candidate drugs
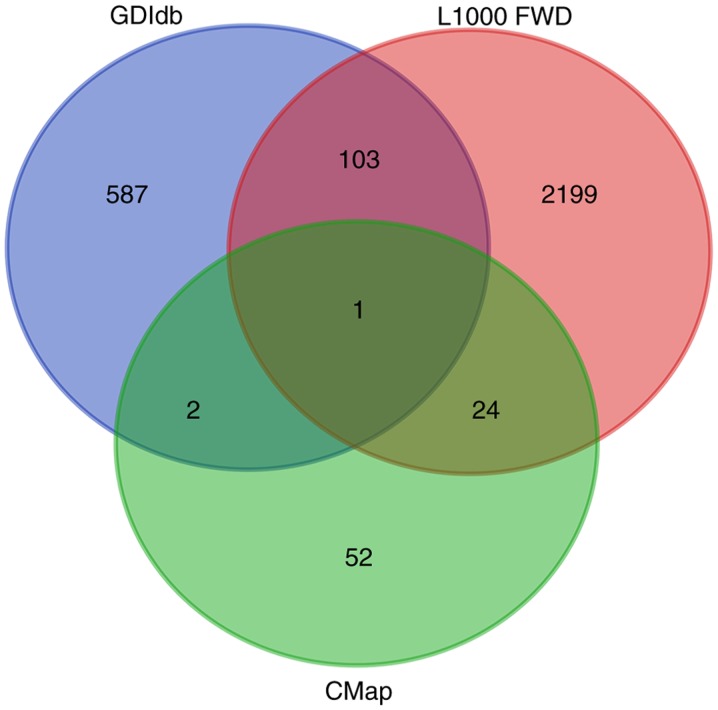
According to the screening conditions mentioned in the methods above, 79, 693 and 2,327 potential drugs were screened from the CMap, DGIdb and L1000 FWD databases, respectively. When the results of this screening were overlapped, only Bepridil was identified in all three databases simultaneously (Fig. 2). For the CMap database, gene-expression changes in three kinds of human cell lines that were treated with Bepridil partially matched those in the STS survival-associated genes. For the L1000 FWD database, similar findings were identified for two human cell lines (Table I). In the DGIdb database, Bepridil was identified by mapping the STS survival-associated genes with the Guide to Pharmacology database.
Figure 2.
Venn diagram depicting potential drugs in the CMap, DGIdb and L1000 FWD databases. CMap, Connectivity Map; DGIdb, Drug-gene interaction database; L1000FWD, L1000 Fireworks Display.
Table I.
Cell lines treated with Bepridil in the CMap and L1000 FWD databases.
| Database | Mean score | Cell line | Dose (µm) | Score | n | P-value |
|---|---|---|---|---|---|---|
| CMap | −0.659 | 4 | 0.071 | |||
| MCF7 | 10 | −0.677 | 2 | 0.101 | ||
| PC3 | 10 | −0.756 | 1 | – | ||
| HL60 | 10 | −0.525 | 1 | – | ||
| L1000FWD | −0.0167 | 2 | – | |||
| HCC515 | 10 | N/A | 1 | 0.001 | ||
| VCaP | 10 | N/A | 1 | 0.002 |
CMap, Connectivity Map; L1000FWD, L1000 Fireworks Display; MCF7, human breast cancer cell line; PC3, human prostate cancer cell line; HL60, human leukemia cell line; HCC515, human lung adenocarcinoma cell line; VCaP, human prostate cancer cell line; N, number of cell lines.
Bepridil is a long-acting, non-selective calcium-channel blocker with significant antianginal activity. As Bepridil has been demonstrated to cause ventricular arrhythmias, it is no longer used in clinical practice (33). To determine whether Bepridil could be repurposed for the treatment of STS, specific targets were identified for further analysis. As a result, 57, 19 and 434 genes were reported to be regulated by Bepridil in the CMap, DGIdb and L1000 FWD databases, respectively. In total, 510 potential Bepridil targets were gathered for subsequent pathway analysis.
KEGG functional pathway analysis of Bepridil targets
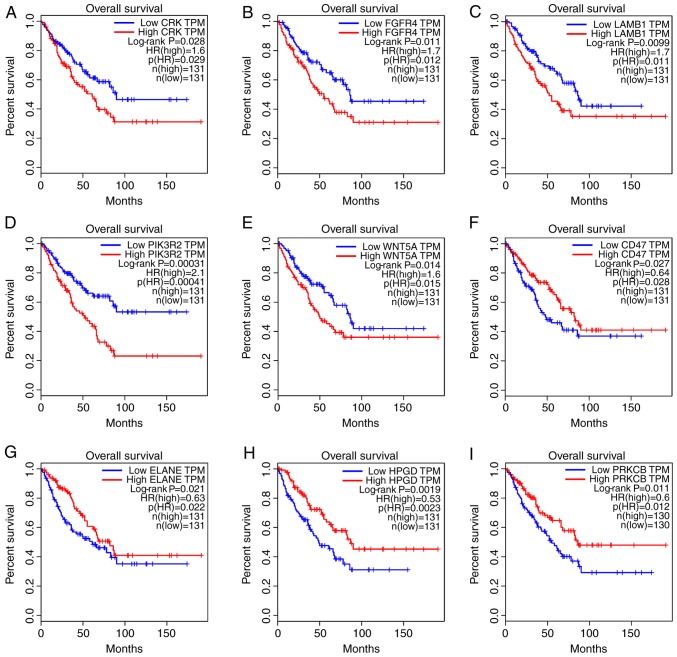
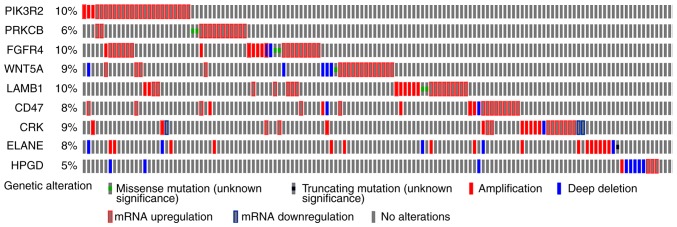
Next, the KEGG pathways in which the Bepridil target genes were likely involved were explored. WebGestalt, a functional enrichment analysis web tool, was used to conduct the pathway analysis. Thirty pathways were identified as associated with the Bepridil targets (Table II). Several of these pathways were previously reported to be involved in tumor genesis or progression (34–36), including epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor resistance (hsa01521), signaling pathways regulating stem cell pluripotency (hsa04550), extracellular matrix (ECM)-receptor interaction (hsa04512), focal adhesion (hsa04510), transcriptional misregulation in cancer (hsa05202) and the phosphatidylinositol 3 kinase-protein kinase B (PI3K-Akt) signaling pathway (hsa04151). Overall survival analysis of the genes clustered in the above-noted pathways revealed that CRK, fibroblast growth factor receptor 4 (FGFR4), laminin subunit β1 (LAMB1), phosphoinositide-3-kinase regulatory subunit 2 (PIK3R2) and WNT5A were likely STS risk factors, while overexpression of cluster of differentiation 47 (CD47), elastase, neutrophil expressed (ELANE), 15-hydroxyprostaglandin dehydrogenase (HPGD) and protein kinase cβ (PRKCB) was correlated with a better prognosis in STS tissues (Fig. 3). The genetic alteration of these 9 genes was also evaluated using one TCGA dataset (with 265 STS samples) in order to explore how they may function in STS. The results demonstrated that the alterations in the other genes mainly appeared as mRNA upregulation (Fig. 4), except for the alterations in ELANE and HPGD, which were either amplifications or deep deletions. This suggests that changes in gene expression may allow these 9 genes to function in STS. In addition, mutually exclusive analysis revealed 18 gene pairs with mutually exclusive alterations, and 18 gene pairs with co-occurrent alterations; however, they were all non-significant (adjusted P>0.05; data not shown).
Table II.
KEGG functional annotation of the Bepridil targets.
| Gene set | Description | Count | P-value | Gene |
|---|---|---|---|---|
| hsa00480 | Glutathione metabolism | 7 | 0.0022 | GPX1; GPX4; ANPEP; GSTA1; GSTA4; GSTP1; ODC1 |
| hsa04964 | Proximal tubule bicarbonate reclamation | 5 | 0.0029 | ATP1A1; PCK1; PCK2; CA2; SLC4A4 |
| hsa04514 | Cell adhesion molecules | 13 | 0.0040 | VCAN; CADM1; HLADQB1; ITGA4; ITGAV; CD99; NCAM1; CLDN18; PTPRM; SDC2; SDC4; CD58; CDH1 |
| hsa05206 | microRNAs in cancer | 14 | 0.0058 | SPRY2; CDKN1B; CRK; E2F2; DICER1; TNC; MARCKS; ABCB1; SERPINB5; PIK3R2; PRKCB; SOX4; THBS1; CCND2 |
| hsa04924 | Renin secretion | 8 | 0.0069 | ADCYAP1R1; CLCA1; AGT; PDE1A; PDE1C; PDE1B; PTGER4; CLCA2 |
| hsa04972 | Pancreatic secretion | 10 | 0.0077 | CLCA1; SLC26A3; KCNQ1; ATP1A1; PLA2G2A; PRKCB; SLC12A2; CA2; SLC4A4; CLCA2 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 9 | 0.0136 | FGFR2; GAS6; NRG1; IGF1R; PIK3R2; PRKCB; AXL; PDGFC; BCL2L1 |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 12 | 0.0161 | FZD10; FGFR2; FGFR4; ID3; ID4; IGF1R; INHBB; SMAD3; PIK3R2; WNT5A; NANOG; KLF4 |
| hsa04614 | Renin-angiotensin system | 4 | 0.0176 | CTSG; AGT; ANPEP; CTSA |
| hsa00010 | Glycolysis/gluconeogenesis | 7 | 0.0225 | ALDH2; ALDOB; HK2; PCK1; PCK2; PFKP; PGK1 |
| hsa04512 | ECM-receptor interaction | 8 | 0.0259 | COL6A3; TNC; ITGA4; ITGAV; LAMB1; SDC4; THBS1; CD47 |
| hsa01230 | Biosynthesis of amino acids | 7 | 0.0289 | ALDOB; IDH3G; PFKP; PGK1; PSPH; BCAT1; PYCRL |
| hsa00330 | Arginine and proline metabolism | 5 | 0.0332 | ALDH2; OAT; ODC1; SMOX; PYCRL |
| hsa05032 | Morphine addiction | 8 | 0.0343 | GABRP; PDE1A; PDE1C; PDE2A; PDE1B; PRKCB; GNG12; CACNA1A |
| hsa00910 | Nitrogen metabolism | 3 | 0.0409 | CA1; CA2; CA12 |
| hsa05215 | Prostate cancer | 8 | 0.0531 | CDKN1B; E2F2; FGFR2; GSTP1; HSP90AA1; IGF1R; PIK3R2; PDGFC |
| hsa04974 | Protein digestion and absorption | 7 | 0.0585 | COL5A2; COL6A3; COL10A1; COL13A1; KCNQ1; ATP1A1; SLC1A1 |
| hsa04310 | Wnt signaling pathway | 10 | 0.0609 | FZD10; DKK1; SMAD3; PRKCB; SFRP1; SFRP4; TBL1X; WNT5A; TBL1XR1; CCND2 |
| hsa04510 | Focal adhesion | 14 | 0.0629 | COL6A3; CRK; TNC; IGF1R; ITGA4; ITGAV; LAMB1; MYLK; PIK3R2; PRKCB; PDGFC; ACTB; THBS1; CCND2 |
| hsa04068 | FoxO signaling pathway | 10 | 0.0730 | CDKN1B; PRMT1; IGF1R; SMAD3; FOXO4; PCK1; PCK2; PIK3R2; S1PR4; CCND2 |
| hsa04350 | TGF-β signaling pathway | 7 | 0.0742 | ID3; ID4; INHBB; SMAD3; BMP6; TFDP1; THBS1 |
| hsa05205 | Proteoglycans in cancer | 14 | 0.0751 | FZD10; ANK1; IGF1R; ITGAV; PIK3R2; PRKCB; PTCH1; ACTB; SDC2; SDC4; THBS1; TWIST1; WNT5A; NANOG |
| hsa05020 | Prion diseases | 4 | 0.0764 | HSPA1A; NCAM1; PRNP; C7 |
| hsa05130 | Pathogenic Escherichia coli infection | 5 | 0.0849 | ARPC1A; KRT18; ACTB; TUBB6; CDH1 |
| hsa05222 | Small cell lung cancer | 7 | 0.0874 | CDKN1B; CKS2; E2F2; ITGAV; LAMB1; PIK3R2; BCL2L1 |
| hsa04971 | Gastric acid secretion | 6 | 0.0877 | KCNQ1; MYLK; ATP1A1; PRKCB; ACTB; CA2 |
| hsa05202 | Transcriptional misregulation in cancer | 12 | 0.0906 | CDKN1B; ELANE; FUT8; NUPR1; HPGD; IGF1R; LMO2; MMP3; MPO; BCL2A1; BCL2L1; CCND2 |
| hsa04151 | PI3K-Akt signaling pathway | 20 | 0.0920 | CDKN1B; COL6A3; FGFR2; FGFR4; HSP90AA1; TNC; IGF1R; IL2RG; ITGA4; ITGAV; LAMB1; PCK1; PCK2; PIK3R2; GNG12; PDGFC; BCL2L1; THBS1; YWHAB; CCND2 |
| hsa03320 | PPAR signaling pathway | 6 | 0.0930 | PLIN2; FABP1; FABP5; ACSL1; PCK1; PCK2 |
| hsa04110 | Cell cycle | 9 | 0.0986 | CDKN1B; CDKN1C; E2F2; SMAD3; MCM4; TFDP1; YWHAB; CCND2; CDC20 |
KEGG, Kyoto Encyclopedia of Genes, and Genomes; EGFR, epidermal growth factor receptor; ECM, extracellular matrix; Fox, forkhead box; TGF, transforming growth factor; PI3K-Akt, phosphatidylinositol 3 kinase-protein kinase B; PPAR, peroxisome proliferator-activated receptor.
Figure 3.
Significant prognostic values of genes that are reportedly involved in tumor genesis or progression pathways. (A) CRK (B) FGFR4. (C) LAMB1. (D) PIK3R2. (E) WNT5A. (F) CD47. (G) ELANE. (H) HPGD. (I) PRKCB. FGFR4, fibroblast growth factor receptor 4; LAMB1, laminin subunit β1; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; CD47, cluster of differentiation 47; ELANE, elastase, neutrophil expressed; HPGD, 15-hydroxyprostaglandin dehydrogenase; PRKCB, protein kinase cβ; TPM, trans per million; HR, hazard ratio.
Figure 4.
Genetic alteration analysis of soft-tissue sarcoma survival-associated genes from The Cancer Genome Atlas dataset. FGFR4, fibroblast growth factor receptor 4; LAMB1, laminin subunit β1; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; CD47, cluster of differentiation 47; ELANE, elastase, neutrophil expressed; HPGD, 15-hydroxyprostaglandin dehydrogenase; PRKCB, protein kinase cβ.
Molecular docking
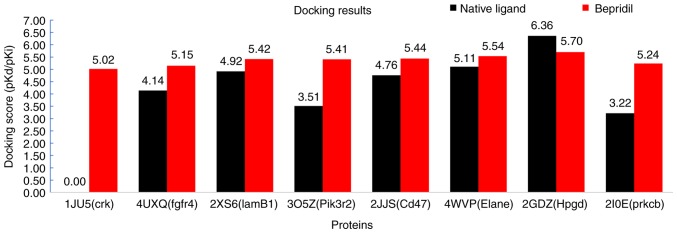
Finally, to explore how Bepridil suppresses or prevents carcinogenic progression, the proteins of 8 STS survival-associated genes (CRK, FGFR4, LAMB1, PIK3R2, WNT5A, CD47, ELANE, HPGD and PRKCB) were analyzed further using molecular docking. Notably, compared with the cut-off scores (4.82–6.11), except the protein of WNT5A, another 8 proteins all exhibited stable docking (medium-to-good binding) results with Bepridil. The lowest docking score was 5.017 for CRK, while the other protein-ligand interaction scores were >5.1. The highest docking simulation was found between HPGD and Bepridil (docking score, 5.702). The PDB IDs, docking compound, and docking scores are presented in Table III and Fig. 5. The detailed protein-ligand interactions of the docking pose (3D and 2D structures) are presented in Figs. 6 and 7. Finally, a network was constructed to display the association between the pathways, targets and candidate drug Bepridil (Fig. 8).
Table III.
Docking simulation results for Bepridil and proteins that are reportedly involved in tumor genesis or progression.
| Gene/protein name | PDB ID | Docking scores (pKd/pKi) |
|---|---|---|
| CRK/CRK | 1JU5 | 5.017 |
| FGFR4/FGFR4 | 4UXQ | 5.146 |
| LAMB1/LAMB1 | 5XAU | 5.417 |
| PIK3R2/PIK3R2 | 2XS6 | 5.414 |
| CD47/CD47 | 2JJS | 5.436 |
| ELANE/ELANE | 4WVP | 5.539 |
| HPGD/HPGD | 2GDZ | 5.702 |
| PRKCB/PRKCB | 2I0E | 5.195 |
PDB ID, Protein data bank ID; FGFR4, fibroblast growth factor receptor 4; LAMB1, laminin subunit β1; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; CD47, cluster of differentiation 47; ELANE, elastase, neutrophil expressed; HPGD, 15-hydroxyprostaglandin dehydrogenase; PRKCB, protein kinase cβ.
Figure 5.
Docking prediction results displayed in an interactive histogram. Docking scores for Bepridil and the native ligand are provided for each protein. X-axis labels are presented as Protein Data Bank ID (protein name). FGFR4, fibroblast growth factor receptor 4; LAMB1, laminin subunit β1; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; CD47, cluster of differentiation 47; ELANE, elastase, neutrophil expressed; HPGD, 15-hydroxyprostaglandin dehydrogenase; PRKCB, protein kinase cβ.
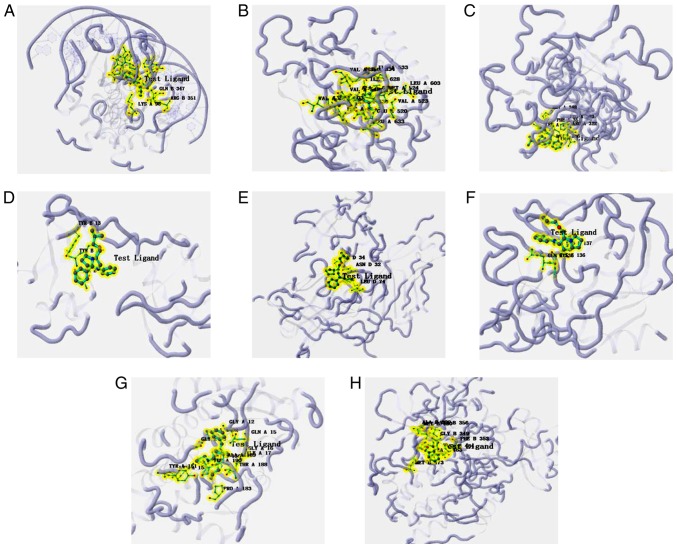
Figure 6.
Molecular binding interactions of Bepridil and the proteins that are reportedly involved in tumor genesis or progression displayed in 3D structures. (A) CRK. (B) Fibroblast growth factor receptor 4. (C) Laminin subunit β1. (D) Phosphoinositide-3-kinase regulatory subunit 2. (E) Cluster of differentiation 47. (F) Elastase, neutrophil expressed. (G) 15-hydroxyprostaglandin dehydrogenase. (H) Protein kinase cβ. All the protein residues presented in figures were the one that identified to involve in the binding interaction.
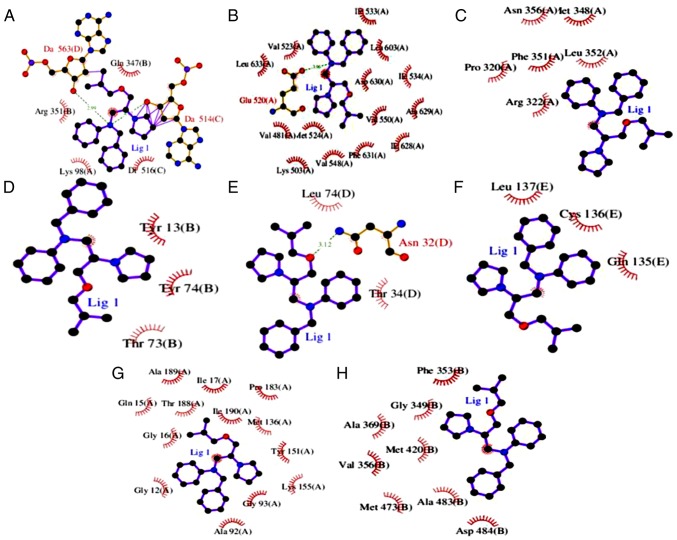
Figure 7.
Molecular binding interactions of Bepridil, and the proteins that are reportedly involved in tumor genesis or progression displayed in 2D structures. (A) CRK. (B) Fibroblast growth factor receptor 4. (C) Laminin subunit β1. (D) Phosphoinositide-3-kinase regulatory subunit 2. (E) Cluster of differentiation 47. (F) Elastase, neutrophil expressed. (G) 15-hydroxyprostaglandin dehydrogenase. (H) Protein kinase cβ. All protein residues presented were involved in the binding interaction.
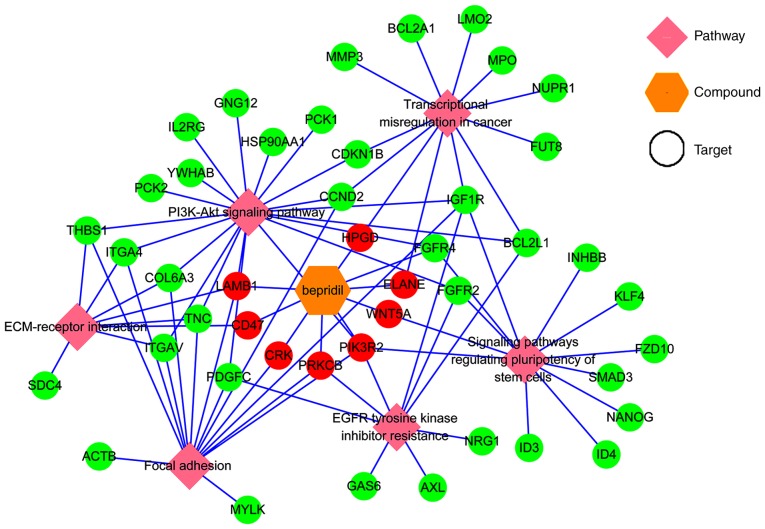
Figure 8.
A network visualization of the association between the identified pathways, targets and candidate drug. Green circles represent the genes clustered in different pathways, red circles represent both the genes clustered in different pathways and the Bepridil targets. FGFR4, fibroblast growth factor receptor 4; LAMB1, laminin subunit β1; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; CD47, cluster of differentiation 47; ELANE, elastase, neutrophil expressed; HPGD, 15-hydroxyprostaglandin dehydrogenase; PRKCB, protein kinase cβ.
Discussion
Recent studies have tended to rely on a panel of cancer-related genes to predict potential drugs for diseases (12,14,17); however, this has not yet been successfully performed for STS. Therefore, the present study used a panel of cancer-related genes to predict potential drugs for several STS subtypes. Previous studies using this methodology have yielded several novel candidates and several clinical drugs have been applied successfully to different diseases using the drug-repurposing strategy. To identify potential drug candidates, studies typically focus on the genomic signatures that are available in electronic patient records, as well as targets of drugs. For example, Paik et al (37) used computational methods to predict that terbutaline sulfate, an anti-asthmatic, could be used to treat amyotrophic lateral sclerosis. Similarly, Dudley et al (38) used an in silico computational approach to discover a novel drug therapy for inflammatory bowel disease that uses the public gene-expression profiles of the disease and the potential targets of drugs. These results suggested that using cancer-related genes to predict potential drugs may be a promising method for drug repurposing. Changes in gene expression are a significant factor in disease genesis and progression Analyzing these changes can identify potential targets for improving therapeutic intervention. In the present study, survival-correlated gene expression signatures were used to make in silico predictions of new indications for approved drugs. Then, three drug-gene interaction databases (CMap, DGIdb, and L1000 FWD) were used to identify potential drugs for STS histologic subtypes. Linking disease-drug profiles based on gene expression signatures is a well-established modality for drug repurposing (8,9,10). CMap used several human tumor cell lines to generate gene-expression profiles of >1,300 compounds, and the predicted drugs were ranked by considering multiple parameters, such as the mean score, enrichment score, match specificity and P-value, to enhance the stability and reliability of the predicted results. The developers of CMap and other groups (26,39) have demonstrated that CMap is a potent tool for identifying compounds with medicinal benefits for a wide range of diseases. L1000 FWD is a similar drug-gene database, but its prediction results only provide the drug name and similarity score. While drug-gene interactions in the DGIdb database were collected using expert curation and text-mining of data from the drug-related database DrugBank, the Therapeutic Target Database, PharmGKB, and ClinicalTrials.gov provide an alternative method to predict drugs for STS histologic subtypes. Therefore, the predicted drugs in these three databases were combined in order to obtain more stable and reliable results. After mapping the targets of these molecules in CMap and querying the DGIdb and L1000 FWD databases with STS survival-associated genes, Bepridil was discovered to be a drug candidate for several common histologic subtypes of STS, which appeared simultaneously in all three databases.
Bepridil is a long-acting, non-selective, calcium-channel blocker that was once used as an antianginal treatment. It induces significant coronary vasodilation and has modest peripheral effects (40). However, Bepridil is no longer used in clinical practice as it has been implicated in ventricular arrhythmias. As novel drug discovery is time consuming and risky, identifying novel indications for known drugs (drug repurposing) has become an effective and innovative method for disease therapy. Therefore, it was explored in the present study whether Bepridil could be repurposed for tumor therapy. A literature search revealed that in the 1990s, Bepridil was revealed to be effective for treating different types of tumors. For example, an early study conducted by van Kalken et al (41) demonstrated that Bepridil combined with anthracyclines could reverse anthracycline resistance in cancer patients. Later, Lee et al (42) evaluated the effects of Bepridil in vitro and similarly suggested that it could be combined with Benzamil to effectively inhibit the growth of human brain tumor cells. More recently, a study conducted by Baldoni et al (43) demonstrated that Bepridil could be used for anti-NOTCH1 targeted therapy for patients with chronic lymphocytic leukemia. However, Bepridil has not been reported as an anticancer agent for STS. The present study combined the screening results of three drug-gene interaction databases that used different algorithms. All three databases indicated that Bepridil may act as an anticancer agent for several common histologic subtypes of STS. If it was truly confirmed that Bepridil could inhibit the proliferation of tumor cells in in vitro STS cell lines and in vivo orthotopic PDX animal models, or if it were used for similar clinical in the future, further studies should be conducted to rationalize the potential negative side-effects of Bepridil, such as by combining antiarrhythmic agents and Bepridil. Further studies would then be required to identify an appropriate method to overcome the potential side-effects of Bepridil. As Bepridil has previously been used in clinical practice, the time and cost of reintroducing this compound into the market may be substantially reduced.
Potential Bepridil targets and pathways were predicted in order to identify potential correlations between Bepridil and other common therapies for STS histologic subtypes. This analysis resulted in the identification of 510 targets and 30 pathways, several of which were reported to be involved in cancer occurrence or progression. Using the TCGA data, the overall survival analysis of the genes clustered in these pathways revealed that CRK, FGFR4, LAMB1, PIK3R2 and WNT5A (characterized by focal adhesion, signaling pathways regulating stem cell pluripotency, EGFR tyrosine kinase inhibitor resistance, ECM-receptor interaction and the PI3K-Akt signaling pathway) were likely risk factors for STS, while CD47, ELANE, HPGD and PRKCB (characterized by ECM-receptor interaction, transcriptional misregulation in cancer, focal adhesion and EGFR tyrosine kinase inhibitor resistance pathways) were likely protective factors in STS tissues.
These 9 genes were reported to serve several roles in malignances. For example, CRK is an adaptor protein that can affect cancer cell migration and invasion (44,45). The dysregulation of CRK expression has been implicated in various aggressive human malignances, including synovial sarcoma, bladder cancer and breast cancer (46–48). These findings suggest that CRK may have targeted-therapy potential in a wide range of tumors. The FGFR protein family is primarily involved in angiogenesis, the activation of which regulates various oncogenic processes (49). FGFR4, one of the FGF receptors, was also demonstrated to be involved in several important signaling pathways, including the WNT signaling pathway, the MAPK signaling pathway and the PI3K-Akt signaling pathway (50). FGFR-targeted therapy is also considered a promising strategy in refractory cancer treatment (51). Lin et al (52) reported that LAMB1 performed better as a diagnostic antigen rather than a carcino-embryonic antigen for colorectal cancer and may serve as a potential serological biomarker for digestive cancer diagnosis. However, the role of LAMB1 in STS has not been reported. In the present study, it was demonstrated that overexpression of LAMB1 was correlated with poor prognosis in patients with STS. PIK3R2 is involved in cell proliferation, migration and survival (53). PIK3R2 was recently reported to be targeted by microRNA-126-3p and to suppress Kaposi's sarcoma cell proliferation (54). WNT5A belongs to the WNT gene family, which encodes secreted signaling proteins and has been implicated in oncogenesis and several developmental processes (55,56). Ye et al (57) reported that CD47 may be a prognostic marker for oral squamous cell carcinoma. ELANE has been correlated with neutropenia outcomes (58). HPGD has been demonstrated to participate in a variety of physiological and pathological metabolic processes. It has also been identified as a novel therapeutic target in prostate cancer (59,60) and is involved in colorectal cancer progression (61). When these nine genes were queried in the cBioPortal database, it was reported that nearly all were genetically altered in patients with STS.
SystemsDock is a web-based tool for network pharmacology-based prediction and analysis. Hsin et al (30) applied the receiver operating characteristic (ROC) to demonstrate that systemsDock possesses a well-designed scoring function [area under the ROC curve (AUC)=0.84] for molecular docking to evaluate protein-ligand binding activity. According to the conventional rating, 0.9≤AUC≤1 is considered excellent, 0.80≤AUC<0.9 is good, 0.70≤AUC<0.8 is fair, 0.50≤AUC<0.7 is poor and AUC<0.5 represents failure. This tool also considers protein structure availability and binding site certainty, which allows the docking simulation of the described proteins and their ligand selectivity to occur. According to the software description, a cut-off score in the range of 4.82–6.11 indicates reliable accuracy (80–83%) when evaluating protein-ligand binding activity. When systemsDock was applied to test the protein-ligand binding activity of Bepridil and screen out the 9 candidate proteins from the KEGG pathways, it was demonstrated that the lowest docking score was 5.017, which suggested that Bepridil had relatively stable binding selectivity with all of the tested proteins. These results indicate that Bepridil has the potential to become an STS candidate drug. This theory should be confirmed with further in vitro and in vivo experiments. If Bepridil could be repurposed for STS treatment, this may provide a significant benefit to STS patients, as well as reduce development time and cost.
In conclusion, the present study used a computational method that combined three drug-gene interaction databases and a gene-expression signature to explore drug repositioning. As a result, Bepridil was identified as a potential candidate for the treatment of STS. A KEGG pathway analysis predicted that Bepridil may target CRK, FGFR4, LAMB1, PIK3R2, CD47, ELANE, HPGD and PRKCB to suppress STS development. The pathways associated with these targets were demonstrated to serve crucial roles in cancer or cancer treatments. Molecular docking simulations suggested that these proteins could combine with Bepridil in a stable manner. These findings provide rationale that Bepridil may be developed for the treatment of STS. However, the present study has some limitations. For instance, it derives STS survival-associated genes from the TCGA database alone; therefore, it would be desirable to increase the power of the analysis by including additional datasets. Furthermore, the present study was simply based on computational approaches without using STS cell lines and PDX models for further validation. Additionally, when evaluating molecular docking, a control experiment by assessing the binding strength of a known inhibitor and Bepridil was not set, as inhibition constant was built-in in the systemsDock database. The present authors intend to remedy this limitation in the future using in vitro and in vivo methodologies. Although drug repurposing using the computational approach does not definitively measure the effects of a treatment, it allows the safety profiling stage to be bypassed, which results in lower costs and better time efficiency. Therefore, an initial genomics-based recommendation may be made rapidly, and refinements or changes can be made when further in vitro and in vivo results become available.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Guangxi Medical University Training Program for Distinguished Young Scholars (GC), the Medical Excellence Award funded by the Creative Research Development Grant from the First Affiliated Hospital of Guangxi Medical University (GC), the Guangxi Zhuang Autonomous Region Health, the Family Planning Commission Self-Financed Scientific Research Project (grant. no. Z20180979), and the Innovation Project of Guangxi Graduate Education (grant no. YCBZ2018038).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XY, JM and GC were involved in the study design. XY contributed to the preparation of the manuscript and data analysis. WTH, HYW, RQH and AGL contributed to the data analysis and prepared the figures and tables. JM and GC supervised and corrected the manuscript. All authors read and approved the final manuscript and agree to be accountable for all aspects of the work.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hoang NT, Acevedo LA, Mann MJ, Tolani B. A review of soft-tissue sarcomas: Translation of biological advances into treatment measures. Cancer Manag Res. 2018;10:1089–1114. doi: 10.2147/CMAR.S159641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burningham Z, Hashibe M, Spector L, Schiffman JD. The epidemiology of sarcoma. Clin Sarcoma Res. 2012;2:14. doi: 10.1186/2045-3329-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Koliou P, Karavasilis V, Theochari M, Pollack SM, Jones RL, Thway K. Advances in the treatment of soft tissue sarcoma: Focus on eribulin. Cancer Manag Res. 2018;10:207–216. doi: 10.2147/CMAR.S143019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourcier K, Italiano A. Newer therapeutic strategies for soft-tissue sarcomas. Pharmacol Ther. 2018;188:118–123. doi: 10.1016/j.pharmthera.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Recine F, Bongiovanni A, Riva N, Fausti V, De Vita A, Mercatali L, Liverani C, Miserocchi G, Amadori D, Ibrahim T. Update on the role of trabectedin in the treatment of intractable soft tissue sarcomas. Onco Targets Ther. 2017;10:1155–1164. doi: 10.2147/OTT.S127955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira IM, Borges A, Borges F, Simoes M. Repurposing ibuprofen to control Staphylococcus aureus biofilms. Eur J Med Chem. 2019;166:197–205. doi: 10.1016/j.ejmech.2019.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Qu XA, Rajpal DK. Applications of connectivity map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 9.Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. DGIdb 3.0: A redesign, and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46:D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Lachmann A, Keenan AB, Ma'ayan A. L1000FWD: Fireworks visualization of drug-induced transcriptomic signatures. Bioinformatics. 2018;34:2150–2152. doi: 10.1093/bioinformatics/bty060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 12.Hurle MR, Yang L, Xie Q, Rajpal DK, Sanseau P, Agarwal P. Computational drug repositioning: From data to therapeutics. Clin Pharmacol Ther. 2013;93:335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 13.Zhong Y, Chen EY, Liu R, Chuang PY, Mallipattu SK, Tan CM, Clark NR, Deng Y, Klotman PE, Ma'ayan A, et al. Renoprotective effect of combined inhibition of angiotensin-converting enzyme, and histone deacetylase. J Am Soc Nephrol. 2013;24:801–811. doi: 10.1681/ASN.2012060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karube K, Tsuzuki S, Yoshida N, Arita K, Kato H, Katayama M, Ko YH, Ohshima K, Nakamura S, Kinoshita T, et al. Comprehensive gene expression profiles of NK cell neoplasms identify vorinostat as an effective drug candidate. Cancer Lett. 2013;333:47–55. doi: 10.1016/j.canlet.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 15.Dyle MC, Ebert SM, Cook DP, Kunkel SD, Fox DK, Bongers KS, Bullard SA, Dierdorff JM, Adams CM. Systems-based discovery of tomatidine as a natural small molecule inhibitor of skeletal muscle atrophy. J Biol Chem. 2014;289:14913–14924. doi: 10.1074/jbc.M114.556241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao L, Zhao G, Fang JS, Yuan TY, Liu AL, Du GH. Discovery of the neuroprotective effects of alvespimycin by computational prioritization of potential anti-Parkinson agents. FEBS J. 2014;281:1110–1122. doi: 10.1111/febs.12672. [DOI] [PubMed] [Google Scholar]
- 17.Chen YT, Xie JY, Sun Q, Mo WJ. Novel drug candidates for treating esophageal carcinoma: A study on differentially expressed genes, using connectivity mapping, and molecular docking. Int J Oncol. 2019;54:152–166. doi: 10.3892/ijo.2018.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drullion C, Marot G, Martin N, Desle J, Saas L, Salazar-Cardozo C, Bouali F, Pourtier A, Abbadie C, Pluquet O. Pre-malignant transformation by senescence evasion is prevented by the PERK and ATF6alpha branches of the Unfolded protein response. Cancer Lett. 2018;438:187–196. doi: 10.1016/j.canlet.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Tang X, Xu Y, Lu L, Jiao Y, Liu J, Wang L, Zhao H. Identification of key candidate genes, and small molecule drugs in cervical cancer by bioinformatics strategy. Cancer Manag Res. 2018;10:3533–3549. doi: 10.2147/CMAR.S171661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christinat A, Leyvraz S. Role of trabectedin in the treatment of soft tissue sarcoma. Onco Targets Ther. 2009;2:105–113. doi: 10.2147/ott.s4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demetri GD, von Mehren M, Jones RL, Hensley ML, Schuetze SM, Staddon A, Milhem M, Elias A, Ganjoo K, Tawbi H, et al. Efficacy and safety of trabectedin or dacarbazine for metastatic liposarcoma or leiomyosarcoma after failure of conventional chemotherapy: Results of a Phase III randomized multicenter clinical trial. J Clin Oncol. 2016;34:786–793. doi: 10.1200/JCO.2015.62.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie L, Guo W, Wang Y, Yan T, Ji T, Xu J. Apatinib for advanced sarcoma: Results from multiple institutions' off-label use in China. BMC Cancer. 2018;18:396. doi: 10.1186/s12885-018-4303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu B, Li J, Xie Q, Diao L, Gai L, Yang W. Efficacy and safety of apatinib monotherapy in advanced bone and soft tissue sarcoma: An observational study. Cancer Biol Ther. 2018;19:198–204. doi: 10.1080/15384047.2017.1416275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Chen X, Liu X, Yu Y, Pan H, Haak R, Schmidt J, Ziebolz D, Schmalz G. Complex integrated analysis of lncRNAs-miRNAs-mRNAs in oral squamous cell carcinoma. Oral Oncol. 2017;73:1–9. doi: 10.1016/j.oraloncology.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 25.Brum AM, van de Peppel J, Nguyen L, Aliev A, Schreuders- Koedam M, Gajadien T, van der Leije CS, van Kerkwijk A, Eijken M, van Leeuwen JPTM, et al. Using the connectivity map to discover compounds influencing human osteoblast differentiation. J Cell Physiol. 2018;233:4895–4906. doi: 10.1002/jcp.26298. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible, and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoav Benjamini, Yosef Hochberg. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- 28.Vilar S, Sobarzo-Sanchez E, Santana L, Uriarte E. Molecular docking and drug discovery in beta-Adrenergic receptors. Curr Med Chem. 2017;24:4340–4359. doi: 10.2174/0929867324666170724101448. [DOI] [PubMed] [Google Scholar]
- 29.Eswari JS, Dhagat S, Kaser S, Tiwari A. Homology modeling and molecular docking studies of bacillomycin and iturin synthetases with novel ligands for the production of therapeutic lipopeptides. Curr Drug Discov Technol. 2018;15:132–141. doi: 10.2174/1570163814666170816112536. [DOI] [PubMed] [Google Scholar]
- 30.Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K, Watanabe T, Kawaoka Y, Kitano H. systemsDock: A web server for network pharmacology-based prediction and analysis. Nucleic Acids Res. 2016;44:W507–W513. doi: 10.1093/nar/gkw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain AN. Surflex: Fully automatic flexible molecular docking using a molecular similarity-based search engine. J Med Chem. 2003;46:499–511. doi: 10.1021/jm020406h. [DOI] [PubMed] [Google Scholar]
- 32.Zsoldos Z, Reid D, Simon A, Sadjad BS, Johnson AP. eHiTS: An innovative approach to the docking and scoring function problems. Curr Protein Pept Sci. 2006;7:421–435. doi: 10.2174/138920306778559412. [DOI] [PubMed] [Google Scholar]
- 33.Ma F, Takanari H, Masuda K, Morishima M, Ono K. Short- and long-term inhibition of cardiac inward-rectifier potassium channel current by an antiarrhythmic drug bepridil. Heart Vessels. 2016;31:1176–1184. doi: 10.1007/s00380-015-0762-1. [DOI] [PubMed] [Google Scholar]
- 34.Vitiello PP, Cardone C, Martini G, Ciardiello D, Belli V, Matrone N, Barra G, Napolitano S, Della Corte C, Turano M, et al. Receptor tyrosine kinase-dependent PI3K activation is an escape mechanism to vertical suppression of the EGFR/RAS/MAPK pathway in KRAS-mutated human colorectal cancer cell lines. J Exp Clin Cancer Res. 2019;38:41. doi: 10.1186/s13046-019-1035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge W, Wang SH, Sun B, Zhang YL, Shen W, Khatib H, Wang X. Melatonin promotes Cashmere goat (Capra hircus) secondary hair follicle growth: A view from integrated analysis of long non-coding and coding RNAs. Cell Cycle. 2018;17:1255–1267. doi: 10.1080/15384101.2018.1471318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Doi T, Yang JC, Shitara K, Naito Y, Cheng AL, Sarashina A, Pronk LC, Takeuchi Y, Lin CC. Phase I study of the focal adhesion kinase inhibitor BI 853520 in Japanese and Taiwanese patients with advanced or metastatic solid tumors. Target Oncol. 2019 Feb 6; doi: 10.1007/s11523-019-00620-0. (Epub ahead of print). doi: 10.1007/s11523-019-00620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paik H, Chung AY, Park HC, Park RW, Suk K, Kim J, Kim H, Lee K, Butte AJ. Repurpose terbutaline sulfate for amyotrophic lateral sclerosis using electronic medical records. Sci Rep. 2015;5:8580. doi: 10.1038/srep08580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudley JT, Sirota M, Shenoy M, Pai RK, Roedder S, Chiang AP, Morgan AA, Sarwal MM, Pasricha PJ, Butte AJ. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci Transl Med. 2011;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Musa A, Ghoraie LS, Zhang SD, Glazko G, Yli-Harja O, Dehmer M, Haibe-Kains B, Emmert-Streib F. A review of connectivity map, and computational approaches in pharmacogenomics. Brief Bioinform. 2018;19:506–523. doi: 10.1093/bib/bbw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaspar T, Kis B, Snipes JA, Lenzsér G, Mayanagi K, Bari F, Busija DW. Neuronal preconditioning with the antianginal drug, bepridil. J Neurochem. 2007;102:595–608. doi: 10.1111/j.1471-4159.2007.04501.x. [DOI] [PubMed] [Google Scholar]
- 41.van Kalken CK, van der Hoeven JJ, de Jong J, Giaccone G, Schuurhuis GJ, Maessen PA, Blokhuis WM, van der Vijgh WJ, Pinedo HM. Bepridil in combination with anthracyclines to reverse anthracycline resistance in cancer patients. Eur J Cancer. 1991;27:739–744. doi: 10.1016/0277-5379(91)90178-G. [DOI] [PubMed] [Google Scholar]
- 42.Lee YS, Sayeed MM, Wurster RD. Intracellular Ca2+ mediates the cytotoxicity induced by Bepridil, and benzamil in human brain tumor cells. Cancer Lett. 1995;88:87–91. doi: 10.1016/0304-3835(94)03619-T. [DOI] [PubMed] [Google Scholar]
- 43.Baldoni S, Del Papa B, Dorillo E, Aureli P, De Falco F, Rompietti C, Sorcini D, Varasano E, Cecchini D, Zei T, et al. Bepridil exhibits anti-leukemic activity associated with NOTCH1 pathway inhibition in chronic lymphocytic leukemia. Int J Cancer. 2018;143:958–970. doi: 10.1002/ijc.31355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C, Zeng X, Liu Z, Li F, Wang K, Wu B. BDNF VAL66MET polymorphism elevates the risk of bladder cancer via MiRNA-146b in Micro-Vehicles. Cell Physiol Biochem. 2018;45:366–377. doi: 10.1159/000486908. [DOI] [PubMed] [Google Scholar]
- 45.Gong XH, Chen C, Hou P, Zhu SC, Wu CQ, Song CL, Ni W, Hu JF, Yao DK, Kang JH, et al. Overexpression of miR-126 inhibits the activation, and migration of HSCs through targeting CRK. Cell Physiol Biochem. 2014;33:97–106. doi: 10.1159/000356653. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe T, Tsuda M, Tanaka S, Ohba Y, Kawaguchi H, Majima T, Sawa H, Minami A. Adaptor protein Crk induces Src-dependent activation of p38 MAPK in regulation of synovial sarcoma cell proliferation. Mol Cancer Res. 2009;7:1582–1592. doi: 10.1158/1541-7786.MCR-09-0064. [DOI] [PubMed] [Google Scholar]
- 47.Matsumoto R, Tsuda M, Wang L, Maishi N, Abe T, Kimura T, Tanino M, Nishihara H, Hida K, Ohba Y, et al. Adaptor protein CRK induces epithelial-mesenchymal transition, and metastasis of bladder cancer cells through HGF/c-Met feedback loop. Cancer Sci. 2015;106:709–717. doi: 10.1111/cas.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar S, Lu B, Davra V, Hornbeck P, Machida K, Birge RB. Crk tyrosine phosphorylation regulates PDGF-BB-inducible Src activation, and breast tumorigenicity, and metastasis. Mol Cancer Res. 2018;16:173–183. doi: 10.1158/1541-7786.MCR-17-0242. [DOI] [PubMed] [Google Scholar]
- 49.Xu M, Chen S, Yang W, Cheng X, Ye Y, Mao J, Wu X, Huang L, Ji J. FGFR4 links glucose metabolism, and chemotherapy resistance in breast cancer. Cell Physiol Biochem. 2018;47:151–160. doi: 10.1007/s11010-017-3219-2. [DOI] [PubMed] [Google Scholar]
- 50.Katoh M, Nakagama H. FGF receptors: Cancer biology, and therapeutics. Med Res Rev. 2014;34:280–300. doi: 10.1002/med.21288. [DOI] [PubMed] [Google Scholar]
- 51.Schelch K, Kirschner MB, Williams M, Cheng YY, van Zandwijk N, Grusch M, Reid G. A link between the fibroblast growth factor axis, and the miR-16 family reveals potential new treatment combinations in mesothelioma. Mol Oncol. 2018;12:58–73. doi: 10.1002/1878-0261.12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Q, Lim HS, Lin HL, Tan HT, Lim TK, Cheong WK, Cheah PY, Tang CL, Chow PK, Chung MC. Analysis of colorectal cancer glyco-secretome identifies laminin β-1 (LAMB1) as a potential serological biomarker for colorectal cancer. Proteomics. 2015;15:3905–3920. doi: 10.1002/pmic.201500236. [DOI] [PubMed] [Google Scholar]
- 53.Qi L, Sun K, Zhuang Y, Yang J, Chen J. Study on the association between PI3K/AKT/mTOR signaling pathway gene polymorphism, and susceptibility to gastric cancer. J BUON. 2017;22:1488–1493. [PubMed] [Google Scholar]
- 54.Wu XJ, Zhao ZF, Kang XJ, Wang HJ, Zhao J, Pu XM. MicroRNA-126-3p suppresses cell proliferation by targeting PIK3R2 in Kaposi's sarcoma cells. Oncotarget. 2016;7:36614–36621. doi: 10.18632/oncotarget.9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kobayashi Y, Kadoya T, Amioka A, Hanaki H, Sasada S, Masumoto N, Yamamoto H, Arihiro K, Kikuchi A, Okada M. Wnt5a-induced cell migration is associated with the aggressiveness of estrogen receptor-positive breast cancer. Oncotarget. 2018;9:20979–20992. doi: 10.18632/oncotarget.24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang L, Yao M, Fang M, Zheng WJ, Dong ZZ, Pan LH, Zhang HJ, Yao DF. Expression of hepatic Wnt5a, and its clinicopathological features in patients with hepatocellular carcinoma. Hepatobiliary Pancreat Dis Int. 2018;17:227–232. doi: 10.1016/j.hbpd.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Ye X, Wang X, Lu R, Zhang J, Chen X, Zhou G. CD47 as a potential prognostic marker for oral leukoplakia, and oral squamous cell carcinoma. Oncol Lett. 2018;15:9075–9080. doi: 10.3892/ol.2018.8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makaryan V, Zeidler C, Bolyard AA, Skokowa J, Rodger E, Kelley ML, Boxer LA, Bonilla MA, Newburger PE, Shimamura A, et al. The diversity of mutations, and clinical outcomes for ELANE-associated neutropenia. Curr Opin Hematol. 2015;22:3–11. doi: 10.1097/MOH.0000000000000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vainio P, Gupta S, Ketola K, Mirtti T, Mpindi JP, Kohonen P, Fey V, Perälä M, Smit F, Verhaegh G, et al. Arachidonic acid pathway members PLA2G7, HPGD, EPHX2, and CYP4F8 identified as putative novel therapeutic targets in prostate cancer. Am J Pathol. 2011;178:525–536. doi: 10.1016/j.ajpath.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi X, Wang Y, Hou J, Huang Y. A Single nucleotide polymorphism in HPGD gene is associated with prostate cancer risk. J Cancer. 2017;8:4083–4086. doi: 10.7150/jca.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pereira C, Queiros S, Galaghar A, Sousa H, Pimentel-Nunes P, Brandão C, Moreira-Dias L, Medeiros R, Dinis-Ribeiro M. Genetic variability in key genes in prostaglandin E2 pathway (COX-2, HPGD, ABCC4, and SLCO2A1), and their involvement in colorectal cancer development. PLoS One. 2014;9:e92000. doi: 10.1371/journal.pone.0092000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request.