Abstract
Several studies have indicated that microRNAs (miRs) mediate multiple pathways associated with tumorigenesis and progression. Our preliminary study experimentally verified that miR-146a-5p has a role in the biological behavior of non-small cell lung cancer (NSCLC) cells. To perform further investigation of miR-146a-5p, the present study evaluated miR-146a-5p by targeting its downstream gene tumor collagenase stimulatory factor (TCSF) to influence cell viability, proliferation and apoptosis in NSCLC. Online sequence prediction, a thorough search of the open source database The Cancer Genome Atlas (TCGA), immunohistochemistry (IHC) of TCSF in clinical lung cancer tissues, and a dual-luciferase assay, as well as assays to test viability, proliferation and apoptosis in vitro, were conducted to explain the targeted regulation association between miR-146a-5p and TCSF in NSCLC. The miRanda and TargetScanHuman database revealed that TCSF and miR-146a-5p had target binding sites. A luciferase reporter assay demonstrated that miR-146a-5p and TCSF did have complementary sequences (P<0.05). From the TCGA database, TCSF was highly expressed in lung adenocarcinoma and lung squamous cell carcinoma tissues when compared with normal lung tissues (P<0.05). Furthermore, the protein level of TCSF in cancerous lung tissues was determined by IHC, and it was concluded that TCSF protein was also upregulated in NSCLC tissues (P<0.001). A significant difference was identified following in vitro experiments for the NSCLC cell line A549, which revealed that miR-146a-5p and TCSF regulated cell viability, proliferation and apoptosis. In conclusion, the present study verified the target action association between TCSF and miR-146a-5p with high throughput data analysis and experimental results in NSCLC.
Keywords: tumor collagenase stimulatory factor, target, cell, microRNA-146a-5p, non-small cell lung cancer
Introduction
Lung cancer is the main type of tumor associated with mortality worldwide and is becoming increasingly more prevalent worldwide (1). Lung cancer is a severe disease that poses a threat to the health of the Chinese population. It includes small cell lung cancer and non-small cell lung cancer (NSCLC); NSCLC represents ~85% of lung cancer (2). Furthermore, a large percentage of the patient population has a very late stage of the disease when they are first diagnosed with lung cancer, which increases the difficulty of treating this disease (3–7). Many chemotherapy drugs and targeted medicines have been applied in the clinic, including Bevacizumab, Pemetrexed, Gemcitabine and Platinum (8). EGFR mutations are easily identified in female patients who do not smoke, and the anaplastic lymphoma kinase receptor tyrosine kinase (ALK) fusion gene is also common in patients with lung cancer (9–12). Fortunately, there are targeted medicines for these patients, including Gefitinib, Erlotinib and Icotinib for patients with the EGFR mutation, and Crizotinib for patients with the ALK fusion gene. According to a report including 741 patients with Chinese lung adenocarcinoma, patients with EGFR mutation account for ~50.2% of the population (13). Another report revealed that the patients with ALK gene rearrangements account for ~8.1% of Chinese NSCLC patients, the majority of whom are younger patients (14). Therefore, gene testing is required for lung cancer patients and may also support future treatment. The search for novel targets of cancerous diseases is also imperative for developing new disease therapeutics.
MicroRNA (miRNA/miR) is a class of non-coding single- stranded RNA with a length of 22 nucleotides. miRNA has multiple target genes that regulate the downstream pathways. Once miRNA combines with target mRNA, the expression of mRNA is downregulated as miRNA inhibits protein translation or mRNA is degraded directly. Previous studies have shown that miRNAs have abnormal levels in many cancerous diseases (15–19). Our previous study concluded that low expression of miR-146a in NSCLC cells inhibited cell proliferation and metastasis, and induced apoptosis through the epidermal growth factor receptor (EGFR) signaling pathway (20). Thus, miRNA serves an important role in tumor biological function through targeting downstream mRNAs and regulating their expression.
Currently, many high-throughput experimental studies and array analyses are based on the mechanism of miRNA binding to hundreds of mRNAs. Various mechanisms involving the recruitment of the miRNA-induced silencing complex (miRISC) have been associated with specific mRNA targets and a reduction in the levels of target proteins (21,22). At the 5′ end of miRNA, a 6–8 nucleotide region interacts with target mRNA sites. These sites are closely associated with the miRNA-dependent degradation of mRNA. These sites can be the primary focus of miRNA target gene prediction databases in silico, such as TargetScan (23) or miRbase (24). When the expression level of miRNA is altered, the majority of the predicted targeted genes either undergo some changes at the RNA or protein levels, or exert phenotypic consequences through biogenesis pathways.
Tumor collagenase stimulatory factor (TCSF) is a type of transmembrane glycoprotein that is a part of the immunoglobulin superfamily. It is also known as Basigin, cluster of differentiation 147 (CD147), HAB18G or extracellular matrix metalloproteinase inducer. TCSF is closely associated with monocarboxylate transporters (MCTs) and supports their cell surface translocation and activities (25). As a chaperone for some MCT isoforms, TCSF also serves important roles in cancer pathogenesis and its upregulated expression level functions in tumor metabolic transformation. With the acceleration of lactate efflux and aerobic glycolysis, TCSF provides the cancerous cells with an acidic microenvironment that makes the cells metabolic and invasive (26). TCSF takes part in various biological processes, including growth, invasion, glycolysis and metastasis of malignant cells. Additionally, angiogenesis, anoikis resistance and multidrug resistance are also associated with the function of TCSF (27). Cell viability, proliferation and apoptosis can be induced by TCSF-mediation of vascular endothelial growth factor (VEGF), whereas stimulation of tumor cell invasion is associated with TCSF-induced matrix metalloproteinases (MMPs). Therefore, TCSF has the potential to be a novel target of cancer therapy. As reported recently, TCSF silencing could suppress the progression of malignant melanoma (28).
In the present study, the sequences of miR-146a-5p and TCSF were determined using bioinformatics prediction and their complementary relationships were verified by a dual-luciferase reporter system. The expression levels of miR-146a-5p and TCSF also demonstrated their reverse regulation mechanism. Furthermore, an in vitro experiment revealed that miR-146a-5p had a role in cell viability, proliferation and apoptosis by regulating the target gene TCSF.
Materials and methods
Targeted gene prediction
The microRNA database miRanda, (www.microrna.org/), the miRDB database (www.mirdb.org/), the TargetScanHuman release 7.1 database (www.targetscan.org/vert_71/) and miRwalk 2.0 (zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/micrornapredictedtarget.html) (29–33) were used to predict the miRNAs and were searched using the targeted gene symbol. These databases can list the targeted mRNAs of miRNAs and show the binding sites with different algorithms.
In order to investigate the function of potential target genes, Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation were summarized using the Database for Annotation, Visualization, and Integrated Discovery online tool (DAVID; version 6.8; david.ncifcrf.gov/home.jsp). The 10 most significant GO terms following enrichment analysis and 10 KEGG pathways (all P<0.05) were chosen for subsequent analysis. The software Cytoscape 3.5.1 (cytoscape.org) was used analyze the results of the enrichment analysis of biological process (BP), cellular component (CC) and molecular function (MF).
Expression of miR-146a-5p and TCSF from The Cancer Genome Atlas (TCGA)
TCGA (cancergenome.nih.gov/) program was started in 2006 and is a collaboration of the National Cancer Institute and the National Human Genome Research Institute. It contains information on key genomic changes in 33 types of cancers. In the present study, the ‘Transcriptome Profiling’ and ‘miRNA’ files for lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) were downloaded from TCGA (32,34–37). Then, the miRNA and mRNA expression levels of miR-146a-5p and TCSF were extracted and standardized. The expression of mature miR-146a-5p in TCGA from University of California Santa Cruz Xena (xena.ucsc.edu/) was also downloaded. Two datasets, TCGA LUAD miRNA mature strand expression by RNAseq (IlluminaHiseq, n=495) and TCGA LUSC miRNA mature strand expression by RNAseq (IlluminaHiseq; n=380), were obtained from UCSC Xena. In addition, the expression profiling by arrays were searched within Gene Expression Omnibus (GEO) DataSets (www.ncbi.nlm.nih.gov/gds/).
Detection of TCSF protein expression in clinical tissues by immunohistochemistry
The present study obtained 371 lung cancer patient tissues (age, 53.58±10.9 years) and 30 non-cancerous tissues (age, 54.03±12.2 years) from the Pathology Department, First Affiliated Hospital of Guangxi Medical University (Guangxi, China) (n=395; male/female ratio, 3.1:1). The tissues were collected between January 2010 and February 2014; the inclusion criteria included any tissues that contained adenocarcinoma, squamous carcinoma, adenosquamous carcinoma, undifferentiated carcinoma, large cell carcinoma or small cell carcinoma. The experiments were approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University and written informed consent was signed by each participant. Hematoxylin and eosin staining was applied to observe the pathological histology of lung cancer tissues. Briefly, lung tissues were immersed in 4% paraformaldehyde for 4 h at room temperature and transferred to 70% ethanol. Individual lung tissue biopsy material was placed in processing cassettes, dehydrated through a serial alcohol gradient and embedded in paraffin wax blocks. Prior to staining, lung tissues were sliced into 5 µm thick, then dewaxed in xylene, rehydrated through decreasing concentrations of ethanol (from absolute ethyl alcohol to 75% ethanol) and washed in distilled water. The sections were stained with hematoxylin for 10 min and eosin for 2 min at room temperature, and then dehydrated through increasing concentrations of ethanol and xylene.
In the present study, TCSF protein expression was detected by immunohistochemistry (IHC) with the anti-CD147 (TCSF) antibody (cat. no. EPR4052; 1:250 dilution; Abcam, Cambridge, MA, USA). SPlink Detection kits (Biotin-Streptavidin HRP Detection Systems; cat. no. SP-9000; ZSGB-BIO; OriGene Technologies, Inc., Beijing, China) was used in the IHC experiments. Sections were blocked with 10% goat serum (included in the SPlink Detection kit) for 20 min at room temperature. Then the primary antibody was added and incubated for 1 h at room temperature. Then, 100 µl of the secondary antibody (Goat Anti-Rabbit Immunoglobulin G; included in the aforementioned SPlink Detection kit) was added for incubation for 15 min at room temperature, according to the kit instructions. Five random images were captured with a light microscope. The percentage of positive TCSF staining was scored by 0–4, which indicated 0 to ≤10, >10 to ≤25, 25 to ≤50, 50 to ≤75 and 75 to ≤100%, respectively. The weak, moderate and strong intensities of TCSF staining were scored using 1, 2 and 3, respectively. The scoring criteria were examined by two independent pathologists. When the score was >2, positive TCSF staining was confirmed and otherwise the sample was considered negative (38,39).
Cell culture
The human lung adenocarcinoma cell line A549 was purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The A549 cell line was cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life Sciences, Logan, UT, USA) and 100 U/ml penicillin-streptomycin sulfate in the 5% carbon dioxide (CO2) at a temperature of 37°C. When the confluence of cells reached 80–90%, the cells were passaged and frozen at −80°C. Cell media was changed every 3 days.
Vector construction and luciferase reporter assay
The A549 lung cancer cell line was cultured with RPMI-1640 and 10% FBS during these experiments. A luciferase reporter assay kit was purchased from Promega Corporation (Madison, WI, USA). The pGL3 reporter vector was applied to construct the TCSF wild type (TCSF-wt) plasmid and TCSF mutant type (TCSF-mut) plasmid. The miR-146a mimic and miRNA mimic negative control (NC) were synthesized by Ambion (Thermo Fisher Scientific, Inc.). When the cells reached high confluency (>80%), culture medium without FBS or penicillin-streptomycin was placed into 48-well plates. The recombined plasmids and miR-146a mimics/NC were transfected with Lipofectamine 2000™ (Thermo Fisher Scientific, Inc.) into the prepared cells (~1×105/well). Following 6 h of transfection, the cells were cultured with new culture medium supplemented with 10% FBS for 24 h. A GloMax™ 96 Microplate Luminometer was then used to detect the fluorescence intensity values. Firefly luciferase activity and Renilla luciferase activity were detected and the results were normalized to those of Renilla luciferase.
Small interfering (si)-RNA transfection
The primer sequences of the siRNA (synthesized by Ruibiotech, Inc., Beijing, China) used were as follows: TCSF sense, 5′-GGUCAGAGCUACACAUUGA-3′ and antisense, 5′-UCAAUGUGUAG-CUCUGA CC-3′; NC siRNA (the scrambled siRNA control group), sense, 5′-UUCUCCGAACGUGUCACGU-3′ and antisense, 5′-ACGUGACACGUUCGGAGAA-3′. The TCSF and NC siRNAs were transfected with Lipofectamine 2000™ (Thermo Fisher Scientific, Inc.) using 2.5 µg siRNA (20 µM is ~0.26 µg/µl; diluted to 9.62 µl/well of a 6-well plate). Following 6 h of the transfection, the cells were cultured with new culture medium supplemented with 10% FBS at 37°C for 48 h.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA synthesis and RT-qPCR reactions were performed in the LightCycler 1.5 instrument, initially with SYBR-Green detection. The LightCycler FastStart DNA MasterPLUS SYBR-Green kit (Roche Diagnostics, Basel, Switzerland) was used. The thermocycling conditions were as follows: 10 min at 95°C, followed by denaturation at 95°C for 10 sec, annealing at 60°C for 5 sec and extension at 72°C for 5 sec. The primer sequences were as follows: TCSF, sense 5′-CCATGCTGGTCTGCAAGTCAG-3′ and antisense 5′-CCGTTCATGAGGGCCTTGTC-3′; and GAPDH, sense 5′-CTATAAATTGAGCCCGCAGC-3′ and antisense 5′-GACCAAATCCGTTGACTCCG-3′. The original Cq value of GAPDH and TCSF was exported directly from the instrument and the experiment was repeated 3 times. RT-qPCR analyses for TCSF and the normalization control gene GAPDH were performed using the 2−∆∆Cq method (40). When the expression of TCSF in the TCSF siRNA group was significantly decreased compared to the scrambled siRNA control group (P<0.05), the cell viability, proliferation and apoptosis assays proceeded.
Cell viability assay
The CellTiter-Blue Cell Viability Assay kit (cat. no. G8080; Promega Corporation) was used to perform the cell viability assay. A549 cells were cultured and prepared in a 96-well plate (3×104/well). First, the frozen cells were added to 20 ml CellTiter-Blue Reagent and incubated at room temperature for 90 min. Then, the solution was added to a 96-well plate with 20 µl/well and the plate was shaken for 10 sec. The cells in the 96-well plate were cultured in 5% CO2 at 37°C for 2 h. Then, the plate was shaken for 10 sec. The absorbance fluorescence values were measured at 560/590 nm (41–44).
Cell proliferation assay
A Promega MTS kit (cat. no. G3580; Promega Corporation) was applied for the cell proliferation assay. A549 cells were cultured and prepared in a 96-well plate for use (3×104/well). First, thawed CellTiter 96 AQueous One Solution Reagent and cells were incubated at room temperature for 90 min. The mixture was then added to a 96-well plate with 20 µl/well and the plate was shaken for 10 sec. The cells in the 96-well plate were cultured in 5% CO2 at 37°C for 2 h. Then, the plate was shaken for 10 sec. The absorbance fluorescence values were measured at 490 nm (42,43,45–48).
Cell apoptosis analysis
The cell apoptosis analysis experiment was conducted via an Apo-ONE Homogeneous Caspase-3/7 Assay (cat. no. G7790; Promega Corporation). A549 cells were cultured and prepared in a 96-well plate for use. The Caspase Substrate and Apo-ONE Caspase-3/7 Buffer were thawed and mixed to produce the Apo-ONE Caspase-3/7 Reagent, then mixed reagent was added to a 96-well plate containing blanks (only cells), controls (cells transfected with scrambled siRNA) and samples (cells transfected with TCSF siRNA or TCSF siRNA + miR-146a-5p inhibitor/mimic). All of the mixed contents were incubated for 1 h at room temperature. Under a 499-nm excitation wavelength for detection and 521-nm emission maximum, the absorbance fluorescence values were measured (41,42,44,49).
Hoechst 33342 and propidium iodide (PI; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were also used to stain cells to determine the level of cell apoptosis. Cells were stained with a mixture of 5 µl Hoechst 33342 and 5 µl PI at 4°C for 20 min. The stained cells were observed under a fluorescent microscope with a magnification of ×200. The cell membrane can be penetrated by Hoechst 33342 and then combined with double-stranded DNA to emit blue fluorescence. However, PI does not penetrate the membrane of viable cells, but instead stains the nucleus of apoptotic cells red. Normal cells show weak intensity of red and blue fluorescence. Apoptotic cells exhibit weak red fluorescence but strong blue fluorescence. Necrotic cells present a strong intensity of red and blue fluorescence (41,44).
Statistical analysis
Statistical analysis was performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) was applied to plot the results. A pairwise comparison using a method of a Student's t-test and a non-parametric test or one-way analysis variance (ANOVA) with a Bonferroni post hoc test was used for comparisons among groups. Qualitative data were analyzed by Pearson's chi-squared test. When there were ≥3 comparison objects, the χ2 method (Student-Newman-Keuls and Newman-Keuls tests) was used. For correlation analysis in GraphPad Prism 7.0, the expression of mir-146a and TCSF from the TCGA database was listed in two columns. ‘Analyze’ was selected to start the correlation analysis and ‘assume data are sampled from Gaussian populations (Pearson)’ was chosen. Then Pearson r and P-values were produced. The results of cell viability, proliferation and apoptosis analysis were analyzed using Variance Analysis of Randomized Block Design ANOVA and Bonferroni multiple comparisons post hoc test. P<0.05 was considered to indicate a statistically significant difference.
The statistical analysis of miR-146a-5p expression from different databases was performed using Stata Statistical software version 12.0 (StataCorp LP, College Station, TX, USA). Standard mean difference (SMD) with 95% confidence interval (CI) was evaluated for the continuous outcomes. If heterogeneity existed (P<0.05 or I2>50%), the SMDs were pooled by random-effect models (DerSimonian-Laird method). Otherwise, the fixed-effects model (Mantel-Haenszel method) was chosen. In this analysis the random-effect mode was used to calculate the pooled SMD with forest plots. Finally 6 studies were pooled for this analysis, including 4 studies from the GEO database and 2 studies from TCGA database. The SMD with 95% CI of miR-146a-5p expression in each group was calculated and pooled.
Results
Targeted associations of miR-146a-5p and TCSF
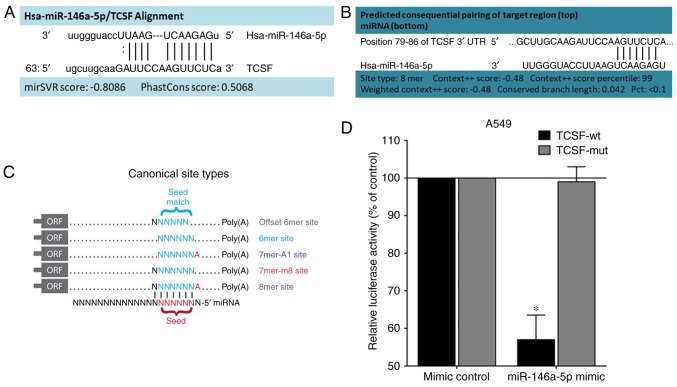
miRanda (Fig. 1A) and TargetScanHuman (Fig. 1B) databases revealed that TCSF and miR-146a-5p had target binding sites, while the miRDB database did not have any associated information. As shown in TargetScanHuman (Fig. 1C), an exact match of TCSF and miR-146a-5p to positions 2–8 of the mature miRNA (the seed sequence) followed by an ‘A’ was defined as an 8mer site.
Figure 1.
Targeted association between miR-146a-5p and TCSF. (A) The binding sites of miR-146a-5p and TCSF in miRanda. (B) The binding sites of miR-146a-5p and TCSF in TargetScanHuman. (C) miR-146a-5p and TCSF binding involves 8mer sites according to TargetScanHuman. (D) A dual luciferase reporter assay system revealed that miR-146a-5p and TCSF did have complementary sequences. *P<0.05 vs. TCSF-mut. miR/miRNA, microRNA; TCSF, tumor collagenase stimulatory factor; wt, wild-type; mut, mutant; ORF, open reading frame.
In total, 47,131 potential target genes were predicted from the database and only 325 target genes, which were repeatedly predicted in 8 or more prediction programs, were chosen for further study. Further GO term enrichment, which included BP, CC and MF, was used to investigate gene function and the biological pathways of target genes. GO and KEGG analyses were performed using the DAVID online tool. The top 10 of KEGG pathways are shown in Table I. Additionally, the top 10 BP, CC and MF terms are listed in Table II.
Table I.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of miR-146-5p target genes.
| Term | Count | % | P-value | Genes |
|---|---|---|---|---|
| hsa04724:Glutamatergic synapse | 9 | 2.893891 | 6.200×10−4 | PRKCA, SLC1A2, ADCY2, ADCY9, PPP3R2, GRIA3, PRKACB, GRIN3A, SLC1A1 |
| hsa04918:Thyroid hormone synthesis | 7 | 2.250804 | 1.075×10−3 | PRKCA, ADCY2, GPX6, ADCY9, CREB3L1, PRKACB, LRP2 |
| hsa04530:Tight junction | 9 | 2.893891 | 2.047×10−3 | PPP2R1B, PRKCA, NRAS, RAB3B, MAGI3, PRKCI, PARD6G, PRKCE, YES1 |
| hsa04310:Wnt signaling pathway | 9 | 2.893891 | 2.143×10−3 | PRKCA, PRICKLE2, RAC1, SMAD4, PPP3R2, FZD3, PRKACB, NFATC2, DAAM2 |
| hsa04727:GABAergic synapse | 7 | 2.250804 | 2.926×10−3 | PRKCA, GABRG1, GAD2, ADCY2, TRAK2, ADCY9, PRKACB |
| hsa04370:VEGF signaling pathway | 6 | 1.92926 | 3.441×10−3 | PRKCA, NRAS, PTGS2, RAC1, PPP3R2, NFATC2 |
| hsa04390:Hippo signaling pathway | 9 | 2.893891 | 3.745×10−3 | PPP2R1B, BMPR2, SMAD4, PRKCI, YWHAB, TEAD1, FZD3, PARD6G, YAP1 |
| hsa04152:AMPK signaling pathway | 8 | 2.572347 | 4.285×10−3 | PPP2R1B, IRS2, CAB39L, SLC2A4, SCD, ELAVL1, CREB3L1, PRKAA2 |
| hsa05031:Amphetamine addiction | 6 | 1.92926 | 4.838×10−3 | PRKCA, PPP3R2, CREB3L1, GRIA3, PRKACB, GRIN3A |
| hsa04921:Oxytocin signaling pathway | 9 | 2.893891 | 4.926×10−3 | PRKCA, NRAS, ADCY2, ADCY9, PTGS2, PPP3R2, PRKACB, PRKAA2, NFATC2 |
Table II.
Gene Ontology term enrichment analysis for the most significantly associated targets of miR-146-5p.
| Category | Term | Count | % | P-value | Genes |
|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | GO:0006511~ubiquitin-dependent protein catabolic process | 10 | 3.215434 | 3.368×10−3 | USP30, PSMF1, USP6, USP3, UBE2G1, SIAH2, ITCH, USP33, ZNRF3, USP31 |
| GOTERM_BP_DIRECT | GO:0042759~long-chain fatty acid biosynthetic process | 3 | 0.96463 | 3.920×10−3 | MYO5A, SCD, QKI |
| GOTERM_BP_DIRECT | GO:0060071~Wnt signaling pathway, planar cell polarity pathway | 7 | 2.250804 | 4.238×10−3 | PSMF1, PRICKLE2, RAC1, ROR1, FZD3, ZNRF3, PSMB11 |
| GOTERM_BP_DIRECT | GO:0007411~axon guidance | 9 | 2.893891 | 5.041×10−3 | NRP2, NRAS, KLF7, TNR, PTPRA, RAC1, SMAD4, SIAH2, USP33 |
| GOTERM_BP_DIRECT | GO:0061549~sympathetic ganglion development | 3 | 0.96463 | 9.105×10−3 | NRP2, PHOX2B, FZD3 |
| GOTERM_BP_DIRECT | GO:0035329~hippo signaling | 4 | 1.286174 | 9.788×10−3 | YWHAB, TEAD1, YAP1, AMOTL2 |
| GOTERM_BP_DIRECT | GO:0014047~glutamate secretion | 4 | 1.286174 | 1.083×10−2 | SYT1, SLC1A2, PPFIA1, SLC1A1 |
| GOTERM_BP_DIRECT | GO:0046326~positive regulation of glucose import | 4 | 1.286174 | 1.310×10−2 | SLC1A2, IRS2, PRKCI, CREBL2 |
| GOTERM_BP_DIRECT | GO:0003091~renal water homeostasis | 4 | 1.286174 | 1.562×10−2 | ADCY2, ADCY9, PRKACB, AQP1 |
| GOTERM_BP_DIRECT | GO:0010634~positive regulation of epithelial cell migration | 4 | 1.286174 | 1.698×10−2 | DOCK1, BMPR2, CLASP2, PRKCE |
| GOTERM_MF_DIRECT | GO:0005515~protein binding | 177 | 56.91318 | 5.430×10−4 | DLC1, SYT1, PTGS2, XPO5, INTS2, JAG1, SPRTN, GRIN3A, AQP1, AMOTL2, ZNRF3, ERLEC1, IQGAP1, TRAK2, SLC2A4, FBXO28, MAP3K8, MS4A1, CREB3L1, ITCH, etc. |
| GOTERM_MF_DIRECT | GO:0001077~transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding | 13 | 4.180064 | 7.000×10−4 | PHOX2B, KLF13, GABPA, ONECUT2, SMAD4, TEAD1, SIX4, MYBL1, ZIC1, TCF21, CREB3L1, POU3F2, NFATC2 |
| GOTERM_MF_DIRECT | GO:0000978~RNA polymerase II core promoter proximal region sequence-specific DNA binding | 14 | 4.501608 | 7.493×10−3 | PHOX2B, KLF13, MTA2, GABPA, ONECUT2, SMAD4, TEAD1, SIX4, MYBL1, ZIC1, GCFC2, ZNF148, POU3F2, NFATC2 |
| GOTERM_MF_DIRECT | GO:0003700~transcription factor activity, sequence-specific DNA binding | 27 | 8.681672 | 1.215×10−2 | ZNF641, KLF7, ELF2, ZNF532, TFCP2L1, MTA2, ZFX, GABPA, SMAD4, TEAD1, ZNF367, NFIX, PARK2, ZIC1, MYT1, GCFC2, TCF20, HSF5, CREB3L1, ZSCAN25, ZNF540, POU3F2, NFATC2, ZNF605, ZNF449, CREBL2, KLF3 |
| GOTERM_MF_DIRECT | GO:0004697~protein kinase C activity | 3 | 0.96463 | 2.580×10−2 | PRKCA, PRKCI, PRKCE |
| GOTERM_MF_DIRECT | GO:0004970~ionotropic glutamate receptor activity | 3 | 0.96463 | 2.580×10−2 | GRIA3, GRIN3A, GRID1 |
| GOTERM_MF_DIRECT | GO:0015501~glutamate:sodium symporter activity | 2 | 0.643087 | 3.348×10−2 | SLC1A2, SLC1A1 |
| GOTERM_MF_DIRECT | GO:0005234~extracellular-glutamate-gated ion channel activity | 3 | 0.96463 | 3.637×10−2 | GRIA3, GRIN3A, GRID1 |
| GOTERM_MF_DIRECT | GO:0061630~ubiquitin protein ligase activity | 8 | 2.572347 | 3.980×10−2 | TRIM37, XIAP, UBE2G1, UBR1, SIAH2, PARK2, ITCH, ZNRF3 |
| GOTERM_MF_DIRECT | GO:0003723~RNA binding | 16 | 5.144695 | 4.576×10−2 | XPO5, SRSF12, SYNJ1, KIAA0430, ELAVL1, DIS3L, EARS2, LARP6, EIF4G2, TRMT1L, SRSF6, DCP2, QKI, CELF2, SYNJ2, STRBP |
| GOTERM_CC_DIRECT | GO:0005737~cytoplasm | 114 | 36.65595 | 2.000×10−4 | DLC1, ADCY2, PTGS2, XPO5, AQP1, ZIC1, IQGAP1, TRAK2, MAP3K8, ITCH, ZNF641, SIX4, UBR1, DIS3L, MARK1, EARS2, ST13, TRIM37, CCDC6, PDGFRA, etc. |
| GOTERM_CC_DIRECT | GO:0030424~axon | 13 | 4.180064 | 2.790×10−4 | NRP2, CCK, KCNC3, HTT, FZD3, IQGAP1, FAM168B, EIF4G2, GAD2, ADCY9, SYNJ2, OLFM1, DTNA |
| GOTERM_CC_DIRECT | GO:0005829~cytosol | 77 | 24.75884 | 7.070×10−4 | DLC1, PHKB, XPO5, SYNJ1, IQGAP3, TTPA, FERMT1, WWC2, AMOTL2, IQGAP1, PRMT3, TRIM5, SLC2A4, TRIM8, MAP3K8, SYNJ2, YAP1, ITCH, PRKACB, PRKCA, etc. |
| GOTERM_CC_DIRECT | GO:0043005~neuron projection | 11 | 3.536977 | 5.420×10−3 | MYO5A, SYT1, SYNPR, PTGS2, ARHGEF7, SYNJ1, LIN7C, SIAH2, GRIN3A, PARK2, IQGAP1 |
| GOTERM_CC_DIRECT | GO:0005776~autophagosome | 5 | 1.607717 | 2.001×10−2 | C9ORF72, HTT, USP33, ZFYVE1, PIP4K2B |
| GOTERM_CC_DIRECT | GO:0016324~apical plasma membrane | 11 | 3.536977 | 2.058×10−2 | MTDH, BMPR2, PRKCI, STC1, FZD3, JAG1, LRP2, AQP1, AMOTL2, SLC1A1, SLC46A1 |
| GOTERM_CC_DIRECT | GO:0005884~actin filament | 5 | 1.607717 | 2.105×10−2 | MYO5A, ACTC1, RAC1, YES1, IQGAP1 |
| GOTERM_CC_DIRECT | GO:0080008~Cul4-RING E3 ubiquitin ligase complex | 3 | 0.96463 | 3.026×10−2 | DCAF5, DCAF12, DCAF17 |
| GOTERM_CC_DIRECT | GO:0005654~nucleoplasm | 58 | 18.64952 | 3.395×10−2 | C9ORF72, ELF2, ZBTB34, CDC14A, XPO5, FAM20B, YLPM1, INTS2, SPRTN, ZIC1, TRIM8, ZNF148, ZNF540, YAP1, PRKACB, PRKCA, ZNF641, MTA2, GABPA, TIMELESS, etc. |
| GOTERM_CC_DIRECT | GO:0043234~protein complex | 13 | 4.180064 | 3.658×10−2 | NOS1, PTGS2, HTT, ARHGEF7, MTA2, YWHAB, PRKCI, PARK2, IQGAP1, ST13, PDGFRA, PARD6G, DTNA |
GO, Gene Ontology.
A luciferase reporter assay revealed that the fluorescence intensity of the TCSF-wt group was significantly decreased when compared with the TCSF-mut group (P<0.05; Fig. 1D), which provided strong evidence for the bioinformatics prediction and revealed that miR-146a-5p and TCSF did have binding sites.
Expression levels of miR-146a-5p and TCSF in TCGA
From the miRNA file of TCGA dataset, many of the 478 tumor tissues and 45 normal lung tissues were involved in LUSC and many of the 521 tumor tissues and 46 normal lung tissues were involved in LUAD. The detection of miRNAs in TCGA targeted the miRNA precursor (pre-miRNA), thus the present study determined the expression of the pre-miRNA has-mir-146a for the mature strand miR-146a-5p. The expression level of miR-146a-5p in LUSC patient tissues was significantly lower when compared with non-cancerous lung tissue (P<0.0001), while the expression level in LUAD was not statistically significant between the cancerous tissue and non-cancerous lung tissue (Fig. 2). From the transcriptome profiling of the TCGA dataset, many of the 501 tumor tissues and 51 normal lung tissues were involved in LUSC, and many of the 514 tumor tissues and 59 normal lung tissues were involved in LUAD. The expression levels of TCSF in LUSC and LUAD revealed that TCSF was markedly upregulated in cancerous tissues when compared with normal lung tissues (P<0.05; Fig. 3).
Figure 2.
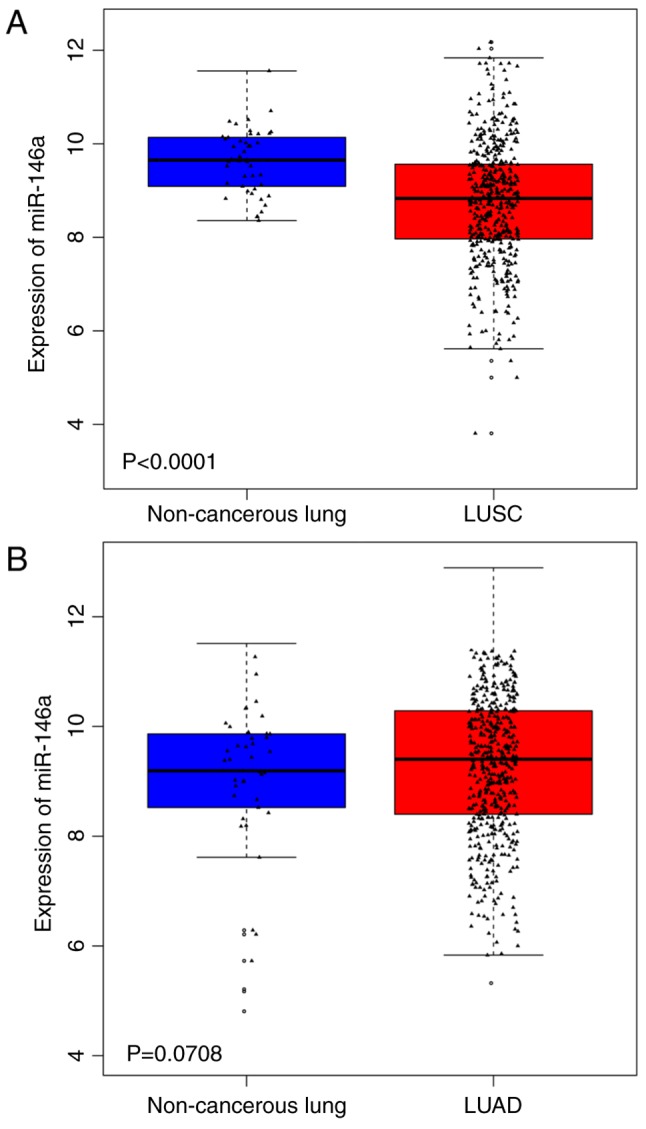
Expression of mir-146a in NSCLC from The Cancer Genome Atlas. (A) LUSC: n=478 cancerous tissues and n=45 normal non-cancerous tissues. (B) LUAD: n=521 cancerous tissues and n=46 normal non-cancerous tissues. miR, microRNA; TCSF, tumor collagenase stimulatory factor; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma.
Figure 3.
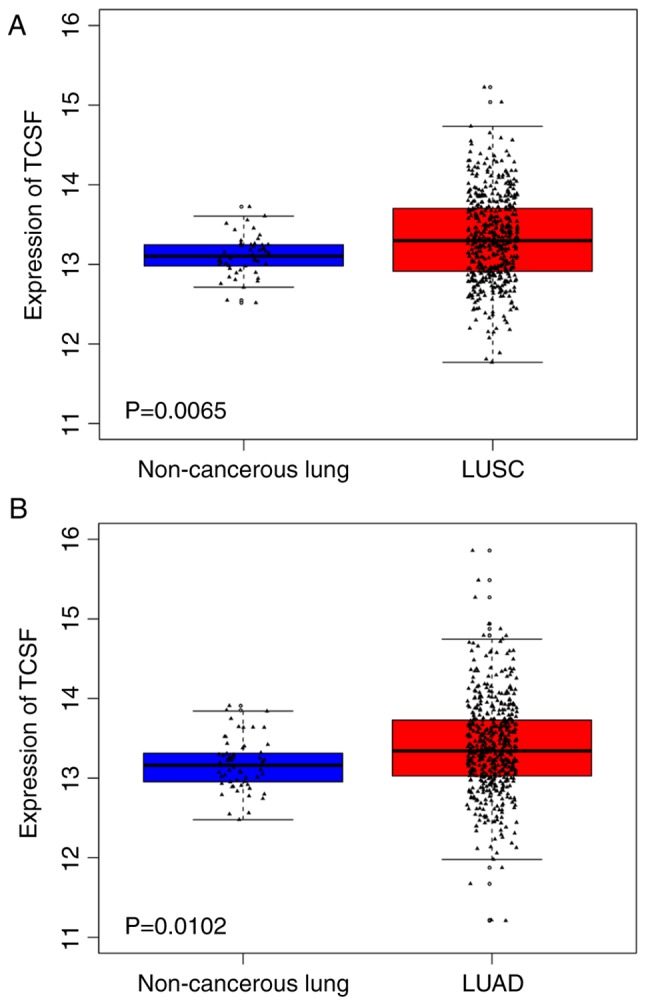
Expression of TCSF in LUSC and LUAD from The Cancer Genome Atlas database. (A) LUSC: n=501 cancerous tissues and n=51 normal non-cancerous tissues. (B) LUAD: n=514 cancerous tissues and n=59 normal non-cancerous tissues. TCSF, tumor collagenase stimulatory factor; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma.
Pearson's correlation analysis was conducted to detect the association between miR-146a-5p and TCSF in LUSC and LUAD. It was concluded that there was a negative correlation between the two in LUSC and LUAD (both Pearson r<0; Figs. 4 and 5). However, there was no significant difference and further investigation with more tissue samples is required.
Figure 4.
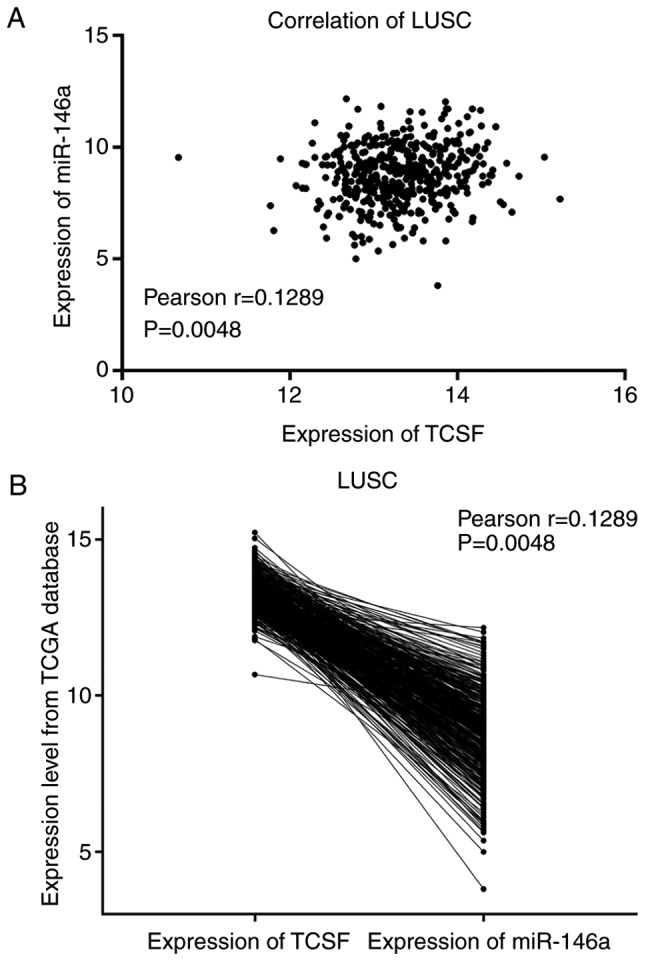
Negative correlation between mir-146a and TCSF expression in LUSC from TCGA database. (A) Correlation curve and (B) expression of mir-146a and TCSF. miR, microRNA; TCSF, tumor collagenase stimulatory factor; LUSC, lung squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
Figure 5.
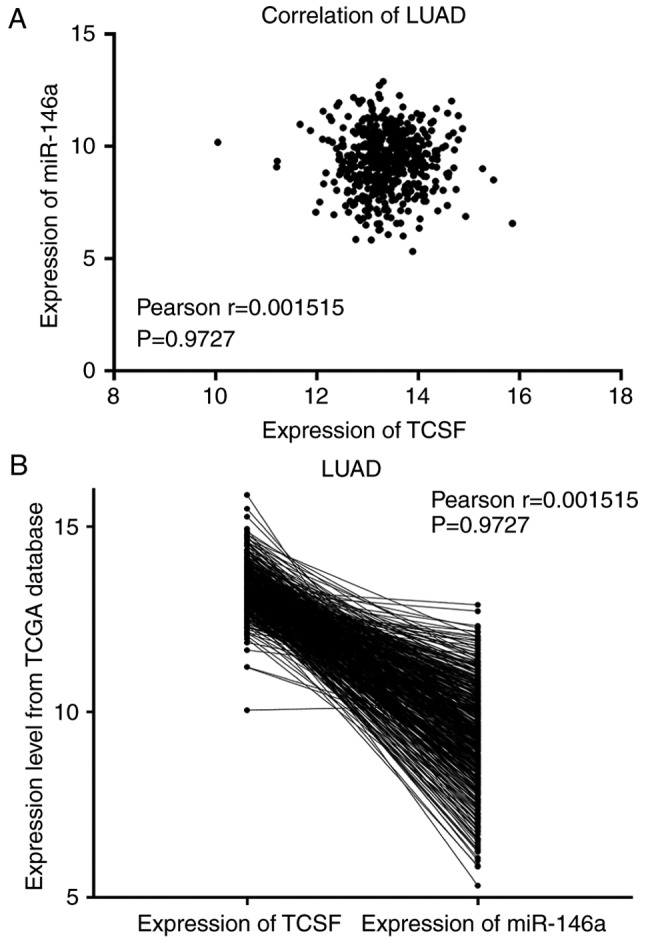
Negative correlation between mir-146a and TCSF expression in LUAD from TCGA database. (A) Correlation curve and (B) expression of mir-146a and TCSF. miR, microRNA; TCSF, tumor collagenase stimulatory factor; LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas.
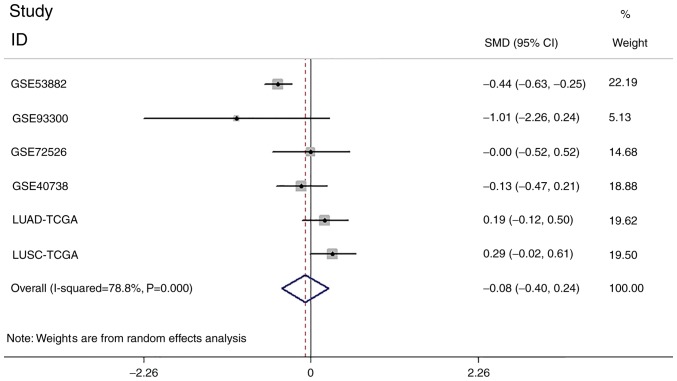
The mature miR-146a-5p expressions in the GEO series, together with data in TCGA, were extracted to perform a meta-analysis of the continuous variable SMDs. Four series of data from GEO and the mature miR-146a-5p expression in LUSC and LUAD from TCGA were inputted in Stata software to generate a forest plot (Fig. 6). It can be concluded that miR-146a-5p has the tendency to be downregulated in lung cancer but with no significance (z=0.48, P=0.626) and heterogeneity (I2=78.8%, P<0.001) More samples of lung cancer tissue should be studied to obtain a credible conclusion.
Figure 6.
Forest plot of microRNA-146a-5p expression levels in patients with lung cancer from the Gene Expression Omnibus and TCGA databases. SMD, Standard mean difference; CI, confidence intervals; TCGA, The Cancer Genome Atlas; LUSC, lung squamous cell carcinoma; LUAD, lung adenocarcinoma.
Experimental evidence of TCSF at the protein level by IHC
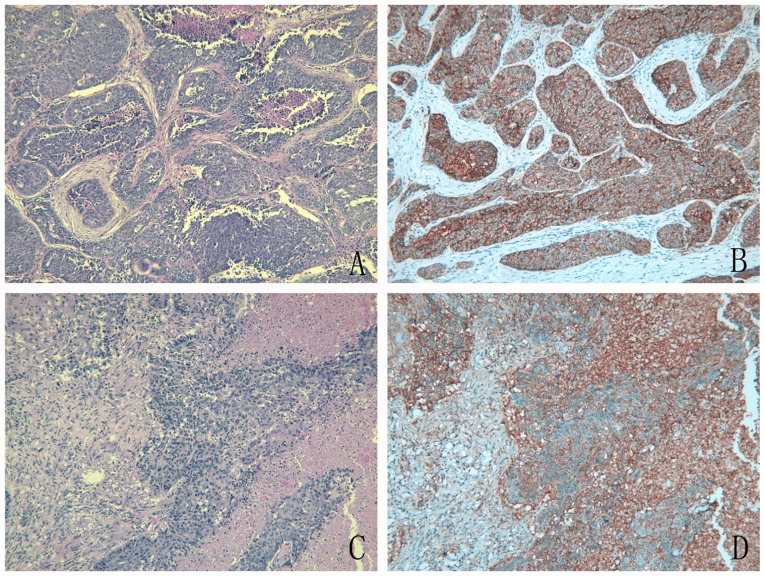
The staining status of the TCSF antibody was evaluated by an IHC assay (Table III) and the images of IHC are presented in Fig. 7. In 371 lung cancer tissues, the number of positive TCSF tissues (52.3%) was higher than the negative tissues (47.7%). In the 30 non-cancerous tissues, the number of negative TCSF tissues (86.7%) was greater than the number of positive tissues (13.3%). A significant difference in TCSF expression was observed between lung cancer and non-cancerous lung tissues (P<0.001) as determined by a non-parametric rank test, which showed that TCSF protein was upregulated in lung cancer tissues. According to an analysis of TCSF expression and the clinicopathological parameters, a greater TCSF expression was significantly associated with a higher pathological grade (P<0.001), later tumor-node-metastasis stage (P<0.001), distant metastasis (P=0.003), lymphatic metastasis (P<0.001) and larger tumor size (P=0.023).
Table III.
Association between tumor collagenase stimulatory factor expression and clinicopathological parameters by immunohistochemistry.
| Clinicopathological parameter | Total number | TCSF (−), n (%) | TCSF (+), n (%) | Z score | P-value |
|---|---|---|---|---|---|
| Tissue | −4.1 | P<0.001d | |||
| Cancerous | 371 | 177 (47.7) | 194 (52.3) | ||
| Non-cancerous | 30 | 26 (86.7) | 4 (13.3) | ||
| Pathological type | 2.393 | 0.122 | |||
| Lung adenocarcinoma | 127 | 64 (50.4) | 63 (49.6) | ||
| Lung squamous cell carcinoma | 175 | 88 (50.3) | 87 (49.7) | ||
| Lung adenosquamous carcinoma | 28 | 11 (39.3) | 17 (60.7) | ||
| Undifferentiated carcinoma | 8 | 3 (37.5) | 5 (62.5) | ||
| Small cell lung cancer | 26 | 10 (38.5) | 16 (61.5) | ||
| Large cell lung cancer | 1 | 1 (100) | 0 (0) | ||
| Other lung cancer | 6 | 0 (0) | 6 (100) | ||
| Sex | −1.017 | 0.309 | |||
| Male | 299 | 158 (52.8) | 141 (47.2) | ||
| Female | 96 | 45 (46.9) | 51 (53.1) | ||
| Age, years | −0.416 | 0.678 | |||
| ≤60 | 218 | 114 (52.3) | 104 (47.7) | ||
| >60 | 177 | 89 (50.3) | 88 (49.7) | ||
| Pathological gradea | 19.065 | P<0.001d | |||
| I | 39 | 27 (69.2) | 12 (30.8) | ||
| II | 92 | 53 (57.6) | 39 (42.4) | ||
| III | 131 | 46 (35.1) | 85 (64.9) | ||
| TNM stage | −3.775 | P<0.001d | |||
| I–II | 299 | 159 (53.2) | 140 (46.8) | ||
| III–IV | 63 | 17 (27.0) | 46 (73.0) | ||
| Distant metastasis | −2.953 | 0.003c | |||
| No | 346 | 174 (50.3) | 172 (49.7) | ||
| Yes | 16 | 2 (12.5) | 14 (87.5) | ||
| Lymphatic metastasis | −7.519 | P<0.001d | |||
| No | 234 | 148 (63.2) | 86 (36.8) | ||
| Yes | 128 | 28 (21.9) | 100 (78.1) | ||
| Tumor size, cm | −2.272 | 0.023b | |||
| ≤7 | 314 | 160 (51.0) | 154 (49.0) | ||
| >7 | 48 | 16 (33.3) | 32 (66.7) |
Pathological grade group pairwise comparison: Pathology grade I vs. Pathology grade III (Pearson Chi-Square=14.276, P<0.001) and Pathology grade II vs. Pathology grade III (Pearson Chi-Square=11.078, P=0.001) were significantly different. Pathology grade I vs. Pathology grade II (Pearson Chi-Square=1.556, P=0.244) was not significantly different.
P<0.05
P<0.01
P<0.001. TNM, tumor-node-metastasis; TCSF, tumor collagenase stimulatory factor.
Figure 7.
H&E and IHC staining of tumor collagenase stimulatory factor expression in non-small cell lung cancer. (A) H&E staining of LUAD (magnification, ×100). (B) IHC of LUAD (magnification, ×100). (C) H&E staining of LUSC (magnification, ×100). (D) IHC of LUSC (magnification, ×100). H&E, hematoxylin and eosin; IHC, immunohistochemistry; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Effects of TCSF and miR-146a-5p interactions on viability, proliferation and apoptosis in A549 cells
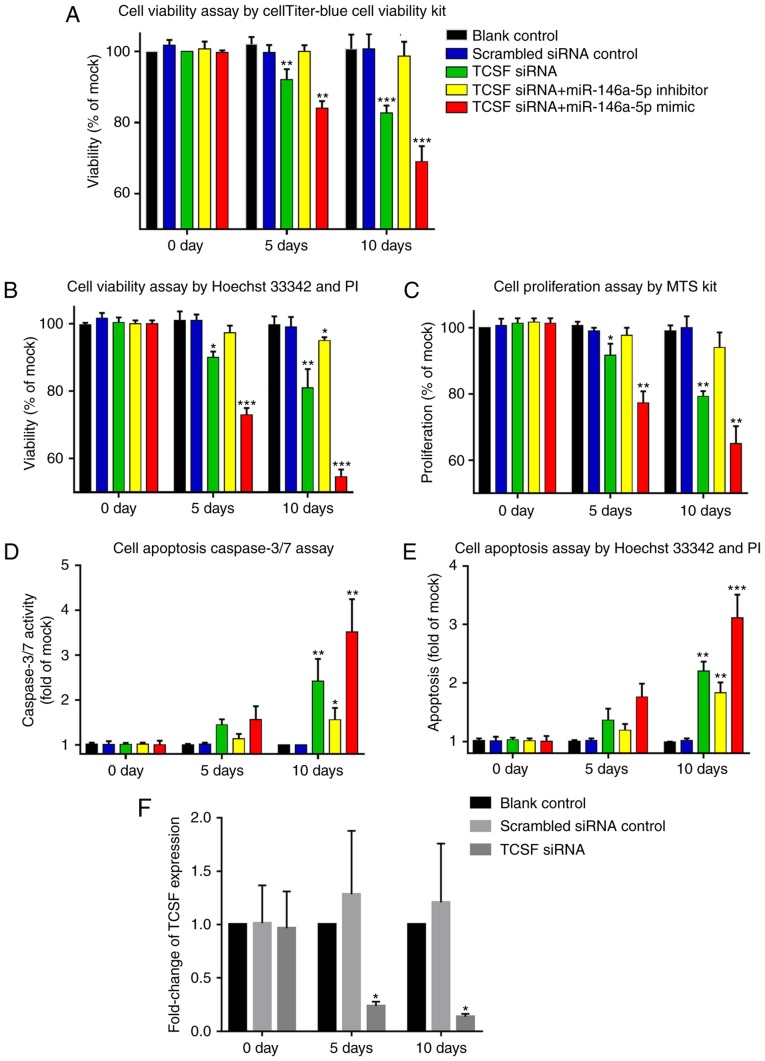
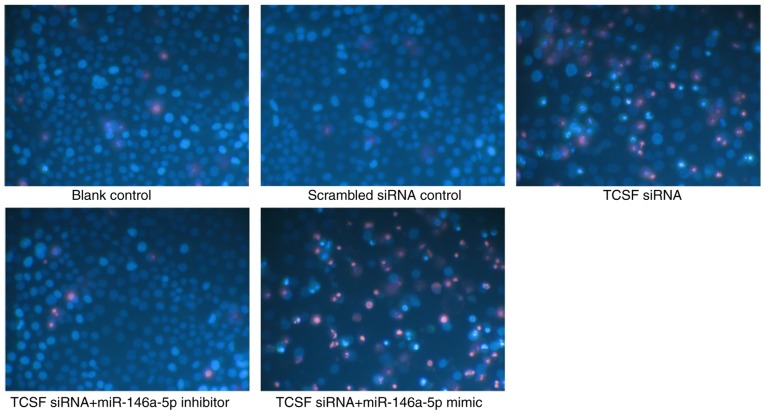
The present study also conducted an in-depth investigation into the biological behavior of A549 cells with TCSF and miR-146a-5p. The influence on cell viability was determined with the CellTiter-Blue Cell Viability Assay kit, as well as a Hoechst 33342 and PI assay. The assay methods all demonstrated that when compared with the scrambled siRNA control group, the cell viability of the group that was transfected with TCSF siRNA was significantly weakened on day 5 and day 10 (P<0.05). When TCSF siRNA and the miR-146a-5p inhibitor were simultaneously transfected into A549 cells, the cell viability was not significantly different when compared with the siRNA control group. Furthermore, significant differences in cell viability were obtained between the TCSF siRNA and miR-146a-5p mimic group and the scrambled siRNA control group at the two of the monitored time points (P<0.05; Fig. 8A and B). The proliferation assay using the MTS kit demonstrated a similar trend. Compared with the scrambled siRNA control group, the group that was transfected with TCSF siRNA only and the TCSF siRNA and miR-146a-5p mimic combined transfection group exhibited significantly increased cell proliferation on days 5 and 10 (P<0.05; Fig. 8C). For the combined TCSF siRNA and miR-146a-5p inhibitor transfection group, the cell proliferation was not significantly different when compared with the scrambled siRNA control group (Fig. 8C) The cell apoptosis assay with the Caspase-3/7 kits revealed that the cell apoptosis of the TCSF siRNA only, TCSF siRNA and miR-146a-5p mimic, and the TCSF siRNA and the miR-146a-5p inhibitor transfection groups were all increased when compared with the control group on day 10 (P<0.05), but there was no difference on day 5 (Fig. 8D). As for the Hoechst 33342 and PI apoptosis assay, a significant increase was observed in the TCSF siRNA only, TCSF siRNA and miR-146a-5p mimic, and the TCSF siRNA and the miR-146a-5p inhibitor transfection groups on day 5 (P<0.05), but not on day 10, when compared with the scrambled siRNA control group (Fig. 8E). Thus, the results suggested that there was no effect of adding miR-146a-5p inhibitor or mimic on cell apoptosis. Knockout using TCSF siRNA at different time points revealed that the expression levels of TCSF were significantly lower when compared with the scrambled control group on days 5 and 10 (Fig. 8F). The apoptosis of A549 cells detected by Hoechst 33324 and PI double fluorescent staining under a fluorescent microscope also indicated a similar conclusion to the previous results obtained in the present study (magnification, ×200; Fig. 9).
Figure 8.
Detection of the effects of miR-146a-5p and TCSF on lung cancer A549 cell viability, proliferation and apoptosis. Cell viability was determined by (A) CellTiter-Blue and (B) Hoechst 33342. (C) Cell proliferation was analyzed using an MTS kit. Cell apoptosis was assessed via (D) Caspaes3/7 assays and (E) Hoechst 33342. (F) The knockout effect of TCSF siRNA at different time points. *P<0.05, **P<0.01 and ***P<0.001 vs. scrambled siRNA. miR, microRNA; TCSF, tumor collagenase stimulatory factor; siRNA, small interfering RNA.
Figure 9.
Apoptosis of lung cancer A549 cells was detected by Hoechst 33342 and propidium iodide double fluorescent staining under a fluorescent microscope (magnification, ×200). miR, microRNA; TCSF, tumor collagenase stimulatory factor; siRNA, small interfering RNA.
Discussion
In conclusion, the present study verified the target action association of TCSF and miR-146a-5p via high throughput data analysis and experimental findings for NSCLC. From the binding sites determined through online prediction it was concluded that TCSF may be one of the targeted genes of miR-146a-5p. The results of a dual-luciferase assay with statistically significant results provided evidence to support this conclusion. In addition, miR-146a-5p and TCSF had opposite effects on cell viability, proliferation and apoptosis, which revealed that they may have roles in NSCLC development.
To the best of our knowledge, miRNA negatively regulates targeted mRNA by following two main mechanisms. One is through complementary base sequences of miRNA and mRNA. Through this approach, the binding domain is always located in the coding region or Open Reading Frame of mRNA and leads to full degradation of mRNA, which requires a small portion of the miRNA regulatory mechanism (50–52). The other approach is incomplete complementation of miRNA and mRNA sequences. The binding area is often located in the 3′-untranslated region (non-coding region) of mRNA. These circumstances can lead to the suppression of the protein translational level or influence targeted mRNA stability. This regulatory approach occurs in the majority of mammals. A single miRNA has multiple targeted mRNAs, and one mRNA could also be regulated by multiple miRNAs in reverse (21,22,50–52).
In our previous study, 3 targeted genes of miR-146a were researched and reported: EGFR (20), interleukin-1 receptor-associated kinase 1 (IRAK1) (53) and tumor necrosis factor receptor-associated factor 6 (TRAF6) (54). Inducing miR-146a could enhance the proliferation capacity of NSCLC cells via drugs that targeted the EGFR (20). IRAK1 and TRAF6 were confirmed to have an association with the progression of NSCLC (53,54). In addition, their effects on miR-146a-5p in NSCLC have been validated by our group. Furthermore, more targeted genes of miR-146a-5p were studied and the miRTarBase (mirtarbase.mbc.nctu.edu.tw/php/index.php) database was used to collect the experimentally validated miRNA-target interactions in various cancers (55). In the miRTarBase database, until now, only 70 targeted genes of hsa-miR-146a-5p have been listed with results from a reporter assay, western blotting, qPCR, microarray, next-generation sequencing technology, amino acids in cell culture or through other methods. Among these genes, the present study consulted the literature to confirm the results of miRTarBase. For example, miR-146a has a critical role in the apoptosis of androgen-independent prostate cancer cells in which it targets Rho associated coiled-coil containing protein kinase 1 (ROCK1) through regulation of the ROCK/Caspase 3 signaling pathway (56). VEGFA, EGFR2, phosphatase and tensin homolog, and C-X-C chemokine receptor type 4 may serve a significant role in the pathogenesis of endometriosis by being a target for miR-146a-5p and other miRNAs (57). The Fanconi Anemia/BRCA pathway in chronic myeloid leukemia involves the target genes ATR serine/threonine kinase (ATR) and the ATR interacting protein of miR-146a-5p (58). The present study verified a novel target gene of miR-146a-5p, which was TCSF.
Bioinformatic prediction serves an increasingly significant role in the research into miRNA, and prediction databases have become more and more capable of performing accurate predictions. The predictions mainly include machine algorithms and experimental validation. The miRNA miRanda database, the first miRNA targeted gene prediction database established in 2003, can list the targeted mRNAs of miRNAs and show the binding sites by selecting alignment details with an algorithm similar to Smith-Waterman (29). The miRDB database can also predict the targeted genes of miRNAs and the possible miRNAs that bind to mRNA genes (30). From the TargetScanHuman database one can download the predicted mRNAs of miRNAs similar to the other two databases (32,33). Due to the different predicted algorithms applied to different databases, the targeted genes and quantity outcomes are different from each other. Taking the intersection of the genes from different databases could enhance the credibility of the prediction.
The present study predicted the targeted genes in silico and combined the reports from previous studies, then selected TCSF as one of the targets of miR-146a-5p. Following further investigation, a dual luciferase reporter assay system provided evidence supporting the association between TCSF and miR-146a-5p. As is reported by Zhang et al (59), in hepatocellular carcinoma (HCC) miR-146a inhibited the invasion and metastasis of HCC cells by targeting HAb18G to downregulate NF-κB p65 and repress VEGF expression via the upregulation of adenomatosis polyposis coli tumor suppressor through multiple pathways. Furthermore, the present study investigated the function of TCSF and miR-146a-5p in NSCLC cells at the mRNA and protein expression levels in vitro. When the miR-146a-5p inhibitor and TCSF siRNA were transfected into the cells concurrently, the influence of cell viability and proliferation was weaker than that observed when cells were transfected with TCSF siRNA only. Additionally, when the miR-146a-5p mimic and TCSF siRNA were transfected in combination, the effect on cell viability and proliferation was stronger than the single treatment with TCSF siRNA. However, in the apoptosis experiments, TCSF siRNA could markedly enhance cell apoptosis on day 10, while no effect was observed following transfection with miR-146a-5p inhibitor or mimic. All of these phenotypes demonstrated that miR-146a-5p could weaken the viability and proliferation effects of TCSF in the NSCLC cells. When miR-146a-5p was removed, TCSF had a stronger impact on NSCLC cells. Since the present study transfected TCSF siRNA only as a comparison, NSCLC cell viability and proliferation were downregulated. It could be concluded that TCSF promoted NSCLC cell growth and repressed its demise. Conversely, the downregulation of miR-146a-5p negatively could regulate the upregulation of TCSF in NSCLC. However, the associated regulatory mechanism underlying this signaling pathway requires further study. As a result, they worked in NSCLC together to regulate tumor infiltration, invasion, metastasis and apoptosis.
In conclusion, the present study verified the association between TCSF and miR-146a-5p, and explored their roles in NSCLC cells. However, further research is required to investigate the signaling pathway through which miR-146a-5p targets TCSF in NSCLC. Therefore, TCSF may be a novel clinical therapeutic target for NSCLC in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science Foundation of China (grant nos. NSFC 81560469 and NSFC 81360327), Natural Science Foundation of Guangxi, China (grant nos. 2017GXNSFAA198016 and 2015GXNSFCA139009), Innovation Project of Guangxi Graduate Education (grant no. YCBZ2018038), Guangxi Medical University Training Program for Distinguished Young Scholars, the Promoting Project of Basic Capacity for Young and Middle-aged University Teachers in Guangxi (grant no. KY2016YB090) and the International Communication of Guangxi Medical University Graduate Education.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
XHH and GC conceived and designed the study, and critically revised the manuscript. WTH and RQH designed and performed the experiments, and analyzed the data. WTH was a major contributor in writing the manuscript. XJL contributed to the acquisition of data and the experiments. JM, ZGP and JCZ contributed to the analysis of experimental data. GC supervised the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
All clinical data were reviewed and approved by the Ethical Committee of the First Affiliated Hospital of Guangxi Medical University (Guangxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Miller K, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR-1252/FOXR2 axis. J Cell Biochem. 2019;120:3765–3772. doi: 10.1002/jcb.27657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiba M, Togashi Y, Tomida S, Mizuuchi H, Nakamura Y, Banno E, Hayashi H, Terashima M, De Velasco MA, Sakai K, et al. MEK inhibitors against MET-amplified non-small cell lung cancer. Int J Oncol. 2016;49:2236–2244. doi: 10.3892/ijo.2016.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He C, Liu Y, Xu Z, Dai P, Chen X, Jin D. Astragaloside IV enhances cisplatin chemosensitivity in non-small cell lung cancer cells through inhibition of B7-H3. Cell Physiol Biochem. 2016;40:1221–1229. doi: 10.1159/000453175. [DOI] [PubMed] [Google Scholar]
- 5.Ma N, Zhang W, Qiao C, Luo H, Zhang X, Liu D, Zang S, Zhang L, Bai J. The tumor suppressive role of MiRNA-509-5p by targeting FOXM1 in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:1435–1446. doi: 10.1159/000443086. [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Chen Z, An L, Wang Y, Zhang Z, Guo Y, Liu C. Analysis of long non-coding RNA expression profiles in non-small cell lung cancer. Cell Physiol Biochem. 2016;38:2389–2400. doi: 10.1159/000445591. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Huang Y, Zhuo W, Zhu Y, Zhu B, Chen Z. Fisetin, a dietary phytochemical, overcomes Erlotinib-resistance of lung adenocarcinoma cells through inhibition of MAPK and AKT pathways. Am J Transl Res. 2016;8:4857–4868. [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider BJ, Kalemkerian GP, Gadgeel SM, Valdivieso M, Hackstock DM, Chen W, Heilbrun LK, Ruckdeschel JC, Wozniak AJ. Phase II trial of dose-dense pemetrexed, gemcitabine, and bevacizumab in patients with advanced, non-small-cell lung cancer. Clin Lung Cancer. 2017;18:299–302. doi: 10.1016/j.cllc.2016.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He Y, Zhang X, Wang L, Tian Z, Liu Q, Yao J, Liu Y, Li C, Min L, Shan B. Detection of cancer specific mutations in early-stage non-small cell lung cancer using cell-free DNA by targeted sequencing. Int J Oncol. 2016;49:2351–2358. doi: 10.3892/ijo.2016.3731. [DOI] [PubMed] [Google Scholar]
- 10.Liu M, Zhou C, Zheng J. Cigarette smoking impairs the response of EGFR-TKIs therapy in lung adenocarcinoma patients by promoting EGFR signaling and epithelial-mesenchymal transition. Am J Transl Res. 2015;7:2026–2035. [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, Li S, Ma R, Sharma A, Bai S, Dun B, Cao H, Jing C, She J, Feng J. Tumor profiling of co-regulated receptor tyrosine kinase and chemoresistant genes reveal different targeting options for lung and gastroesophageal cancers. Am J Transl Res. 2016;8:5729–5740. [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, Zeng Y, Du W, Zhu J, Shen D, Liu Z, Huang JA. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int J Oncol. 2016;49:1360–1368. doi: 10.3892/ijo.2016.3632. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y, Li J, Zhang S, Wang M, Yang S, Li N, Wu G, Liu W, Liao G, Cai K, et al. Molecular epidemiology of EGFR mutations in asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology-mainland china subset analysis of the PIONEER study. PLoS One. 2015;10:e0143515. doi: 10.1371/journal.pone.0143515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong S, Fang W, Hu Z, Zhou T, Yan Y, Qin T, Tang Y, Ma Y, Zhao Y, Xue C, et al. A large-scale cross-sectional study of ALK rearrangements and EGFR mutations in non-small-cell lung cancer in Chinese Han population. Sci Rep. 2014;4:7268. doi: 10.1038/srep07268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shyamasundar S, Lim J, Bay B. miR-93 inhibits the invasive potential of triple-negative breast cancer cells in vitro via protein kinase WNK1. Int J Oncol. 2016;49:2629–2636. doi: 10.3892/ijo.2016.3761. [DOI] [PubMed] [Google Scholar]
- 16.Turconi G, Scaldaferri D, Fabbri M, Monti L, Lualdi M, Pedrini E, Gribaldo L, Taramelli R, Acquati F. RNASET2 silencing affects miRNAs and target gene expression pattern in a human ovarian cancer cell model. Int J Oncol. 2016;49:2637–2646. doi: 10.3892/ijo.2016.3763. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y, Li F, Shi J, Li S, Cai J, Jiang Y. MiR-146a regulates inflammatory infiltration by macrophages in polymyositis/dermatomyositis by targeting TRAF6 and affecting IL-17/ICAM-1 pathway. Cell Physiol Biochem. 2016;40:486–498. doi: 10.1159/000452563. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Huang L, Chen G, Feng Z. Potential targets and clinical value of MiR-224-5p in cancers of the digestive tract. Cell Physiol Biochem. 2017;44:682–700. doi: 10.1159/000485281. [DOI] [PubMed] [Google Scholar]
- 19.Zhi Y, Pan J, Shen W, He P, Zheng J, Zhou X, Lu G, Chen Z, Zhou Z. Ginkgolide B inhibits human bladder cancer cell migration and invasion through MicroRNA-223-3p. Cell Physiol Biochem. 2016;39:1787–1794. doi: 10.1159/000447878. [DOI] [PubMed] [Google Scholar]
- 20.Chen G, Umelo I, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Grève J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions-beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. doi: 10.1038/nrg3765. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi K. Anatomy of RISC: How do small RNAs and chape-rones activate Argonaute proteins? Wiley Interdiscip Rev RNA. 2016;7:637–660. doi: 10.1002/wrna.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman R, Farh K, Burge C, Bartel D. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozomara A, Griffiths-Jones S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muramatsu T. Basigin (CD147), a multifunctional transmembrane glycoprotein with various binding partners. J Biochem. 2016;159:481–490. doi: 10.1093/jb/mvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, Yu X, Dai D, Song X, Xu W. The altered glucose metabolism in tumor and a tumor acidic microenvironment associated with extracellular matrix metalloproteinase inducer and monocarboxylate transporters. Oncotarget. 2016;7:23141–23155. doi: 10.18632/oncotarget.8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao C, Lu C, Chen J. Biological characteristics of cluster of differentiation 147 and its relationship with tumour. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2016;38:589–593. doi: 10.3881/j.issn.1000-503X.2016.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Hu X, Su J, Zhou Y, Xie X, Peng C, Yuan Z, Chen X. Repressing CD147 is a novel therapeutic strategy for malignant melanoma. Oncotarget. 2017;8:25806–25813. doi: 10.18632/oncotarget.15709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. 2016;32:1316–1322. doi: 10.1093/bioinformatics/btw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal V, Bell G, Nam J, Bartel D. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromm B, Billipp T, Peck L, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao X, Wu Y, Yu W, Li H. Identification of a seven-miRNA signature as prognostic biomarker for lung squamous cell carcinoma. Oncotarget. 2016;7:81670–81679. doi: 10.18632/oncotarget.13164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia W, Chen Q, Wang J, Mao Q, Dong G, Shi R, Zheng Y, Xu L, Jiang F. DNA methylation mediated silencing of microRNA-145 is a potential prognostic marker in patients with lung adenocarcinoma. Sci Rep. 2015;5:16901. doi: 10.1038/srep16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie K, Ma H, Liang C, Wang C, Qin N, Shen W, Gu Y, Yan C, Zhang K, Dai N, et al. A functional variant in miR-155 regulation region contributes to lung cancer risk and survival. Oncotarget. 2015;6:42781–42792. doi: 10.18632/oncotarget.5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xie K, Wang C, Qin N, Yang J, Zhu M, Dai J, Jin G, Shen H, Ma H, Hu Z. Genetic variants in regulatory regions of microRNAs are associated with lung cancer risk. Oncotarget. 2016;7:47966–47974. doi: 10.18632/oncotarget.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L, Zhong J, Huang W, Dang Y, Kang M, Chen G. Integrative analysis of BSG expression in NPC through immunohistochemistry and public high-throughput gene expression data. Am J Transl Res. 2017;9:4574–4592. [PMC free article] [PubMed] [Google Scholar]
- 39.Huang W, Gao L, He R, Ma J, Wei K, Chen G. Over-expressed CD147 may be a reminder of tumor progression in cervical lesions: An immunohistochemical investigation with 494 cases. Int J Clin Exp Pathol. 2017;10:3189–3198. [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.He R, Yang L, Lin X, Chen X, Lin X, Wei F, Liang X, Luo Y, Wu Y, Gan T, et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8:15632–15641. [PMC free article] [PubMed] [Google Scholar]
- 42.Li JJ, Luo J, Lu JN, Liang XN, Luo YH, Liu YR, Yang J, Ding H, Qin GH, Yang LH, et al. Relationship between TRAF6 and deterioration of HCC: An immunohistochemical and in vitro study. Cancer Cell Int. 2016;16:76. doi: 10.1186/s12935-016-0352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang R, Zhong T, Dang Y, Zhang X, Li P, Chen G. Association between downexpression of MiR-203 and poor prognosis in non-small cell lung cancer patients. Clin Transl Oncol. 2016;18:360–368. doi: 10.1007/s12094-015-1377-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Huang S, Leng Y, Chen X, Liu T, Wang H, Wei F, Luo D, Chen G, Wei Z. Effect of DcR3-specific siRNA on cell growth suppression and apoptosis induction in glioma cells via affecting ERK and AKT. Onco Targets Ther. 2016;9:5195–5202. doi: 10.2147/OTT.S108395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang W, Wang H, Yang H, Ren FH, Luo YH, Huang CQ, Liang YY, Liang HW, Chen G, Dang YW. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: A clinicopathological and in silico study. Onco Targets Ther. 2016;9:5163–5180. doi: 10.2147/OTT.S108828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li CY, Pang YY, Yang H, Li J, Lu HX, Wang HL, Mo WJ, Huang LS, Feng ZB, Chen G. Identification of miR-101-3p targets and functional features based on bioinformatics, meta-analysis and experimental verification in hepatocellular carcinoma. Am J Transl Res. 2017;9:2088–2105. [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Chen WJ, Gan TQ, Zhang XL, Xie ZC, Ye ZH, Deng Y, Wang ZF, Cai KT, Li SK, et al. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: A study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. doi: 10.1038/s41598-017-05856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong DN, Luo YH, Mo WJ, Zhang X, Tan Z, Zhao N, Pang SM, Chen G, Rong MH, Tang W. High expression of long non-coding HOTAIR correlated with hepatocarcinogenesis and metastasis. Mol Med Rep. 2018;17:1148–1156. doi: 10.3892/mmr.2017.7999. [DOI] [PubMed] [Google Scholar]
- 49.He R, Gao L, Ma J, Peng Z, Zhou S, Yang L, Feng Z, Dang Y, Chen G. The essential role of MTDH in the progression of HCC: A study with immunohistochemistry, TCGA, meta-analysis and in vitro investigation. Am J Transl Res. 2017;9:1561–1579. [PMC free article] [PubMed] [Google Scholar]
- 50.Achkar NP, Cambiagno DA, Manavella PA. miRNA bioge-nesis: A dynamic pathway. Trends Plant Sci. 2016;21:1034–1044. doi: 10.1016/j.tplants.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 51.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 52.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Dang Y, Li P, Rong M, Chen G. Expression of IRAK1 in lung cancer tissues and its clinicopathological significance: A microarray study. Int J Clin Exp Pathol. 2014;7:8096–8104. [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang XL, Dang YW, Li P, Rong MH, Hou XX, Luo DZ, Chen G. Expression of tumor necrosis factor receptor-associated factor 6 in lung cancer tissues. Asian Pac J Cancer Prev. 2014;15:10591–10596. doi: 10.7314/APJCP.2014.15.24.10591. [DOI] [PubMed] [Google Scholar]
- 55.Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL, Lee WH, Yang CD, Hong HC, Wei TY, Tu SJ, et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu B, Huang Y, Niu X, Tao T, Jiang L, Tong N, Chen S, Liu N, Zhu W, Chen M. Hsa-miR-146a-5p modulates androgen-independent prostate cancer cells apoptosis by targeting ROCK1. Prostate. 2015;75:1896–1903. doi: 10.1002/pros.23068. [DOI] [PubMed] [Google Scholar]
- 57.Yang RQ, Teng H, Xu XH, Liu SY, Wang YH, Guo FJ, Liu XJ. Microarray analysis of microRNA deregulation and angiogenesis-related proteins in endometriosis. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15027826. [DOI] [PubMed] [Google Scholar]
- 58.Yap E, Norziha Z, Simbun A, Tumian NR, Cheong SK, Leong CF, Wong CL. MicroRNAs that affect the Fanconi Anemia/BRCA pathway are downregulated in imatinib-resistant chronic myeloid leukemia patients without detectable BCR-ABL kinase domain mutations. Leuk Res. 2017;59:32–40. doi: 10.1016/j.leukres.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Z, Zhang Y, Sun X, Ma X, Chen Z. microRNA-146a inhibits cancer metastasis by downregulating VEGF through dual pathways in hepatocellular carcinoma. Mol Cancer. 2015;14:5. doi: 10.1186/1476-4598-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.