Abstract
MicroRNAs (miRNAs or miRs) contribute to the development of various malignant neoplasms, including glioblastoma multiforme (GBM). The present study aimed to explore the pathogenesis of GBM and to identify latent therapeutic agents for patients with GBM, based on an in silico analysis. Gene chips that provide miRNA expression profiling in GBM were obtained from the Gene Expression Omnibus (GEO) database. Differentially expressed miRNAs (DEMs) were also determined via the RobustRankAggreg algorithm. The target genes of DEMs were predicted and then intersected with GBM-associated genes that were collected from the Gene Expression Profiling Interactive Analysis. Gene Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of the overlapping genes were then performed. Simultaneously, a connectivity map (CMap) analysis was performed to screen for potential therapeutic agents for GBM. A total of 10 DEMs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b, hsa-miR-542-3p, hsa-miR-129-3p, hsa-miR-1224-5p, hsa-miR-876-3p and hsa-miR-770-5p) were obtained from three GEO gene chips (GSE25631, GSE42657 and GSE61710). Then, 1,720 target genes of the 10 miRNAs and 4,185 differently expressed genes in GBM were collected. By intersecting the aforementioned gene clusters, the present study identified 390 overlapping genes. GO and KEGG analyses of the 390 genes demonstrated that these genes were involved in certain cancer-associated biological functions and pathways. Eight genes [(GTPase NRas (NRAS), calcium/calmodulin-dependent protein kinase type II subunit Gamma (CAMK2G), platelet-derived growth factor receptor alpha (PDGFRA), calmodulin 3 (CALM3), cyclin-dependent kinase 6 (CDK6), calcium/calmodulin-dependent protein kinase type II subunit beta (CAMK2B), retinoblastoma-associated protein (RB1) and protein kinase C beta type (PRKCB)] that were centralized in the glioma pathway were selected for CMap analysis. Three chemicals (W-13, gefitinib and exemestane) were identified as putative therapeutic agents for GBM. In summary, the present study identified three miRNA-based chemicals for use as a therapy for GBM. However, more experimental data are needed to verify the therapeutic properties of these latent drugs in GBM.
Keywords: glioblastoma multiforme, miRNA, pathway, connectivity map, therapeutic agent
Introduction
Glioblastoma multiforme (GBM), which is classified by the World Health Organization as a grade IV glioma, exhibits a high morbidity and mortality, comprising 47.1% of all malignant tumors of the central nervous system (1,2). In total, ~13,000 people in America are diagnosed with GBM each year (3). The main treatment of GBM is surgical resection in combination with radiotherapy or chemotherapy. However, the majority of patients relapse within the 7 months following their original diagnoses (4). Furthermore, a resistance to current chemotherapy leads to a heavy tumor burden for patients with GBM. Although novel treatments, including immunotherapy and molecular targeted therapy, have been in development for several years (5,6), the 5-year survival rate is relatively low, with a median survival time of 15 months (4), indicating the urgency of determining novel therapies.
MicroRNAs (miRNAs) are a class of small non-coding RNAs comprising ~19–23 nucleotides (7). By binding to target mRNAs, miRNAs regulate gene expression at the transcriptional or posttranscriptional level, which enables them to serve pivotal roles in various biological processes, including cell growth, apoptosis, invasion and metastasis (8,9). In terms of GBM, it is widely reported that miRNAs participate in various molecular pathways associated with cancer development (10–12). miRNA-targeted therapy has been utilized in cancer treatment by developing miRNA mimetic and anti-miRNA agents (13). The present study aimed to determine latent miRNA-based therapeutic agents for GBM by employing a Connectivity Map (CMap) method. Using >7,000 expression profiles (representing 1,309 compounds), the CMap reveals connections among genes, chemicals and diseases (14). It is extensively used in the exploitation of novel drugs or existing drug applications (15,16). Drugs that are available in CMap are all licensed for human use by the Food and Drug Administration (17). It is an ideal database for probing chemicals that may be applied in the therapy of GBM.
The experimental processes of the present study were as follows (Fig. 1): i) A statistical analysis of differentially expressed miRNAs (DEMs) in GBM was performed using the expression data of gene chips collected from the Gene Expression Omnibus (GEO); ii) the target genes of DEMs were predicted and intersected with the GBM-associated genes and the resulting genes were defined as miRNA-associated differentially expressed genes (DEGs); iii) Gene Ontology (GO) functional annotations and a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed, and the genes participating in the ‘glioma pathway’ were selected for further analysis; iv) a CMap analysis was applied to probe for potential therapeutic chemicals for GBM; and v) a molecular docking approach was adopted to assess the affinity of the selected chemicals to their target genes.
Figure 1.
Experimental flowchart of the present study. GEO, Gene Expression Omnibus; DEMs, differently expressed miRNAs; miRNA, microRNA; GEPIA, Gene Expression Profiling Interactive Analysis; DEGs, differently expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of genes and genomes; PPI, protein-protein interaction.
Materials and methods
Identification of DEMs in GBM
A search of miRNA-associated microarray datasets in GBM was performed using the GEO database (18) with the following screening criteria: i) the organism must be restricted to ‘homo sapiens’; ii) a complete expression profile determination of the miRNA had to have been performed; and iii) cancerous and noncancerous samples needed to contain at least five samples. A Bioconductor package ‘Limma’ (19) was applied to screen for DEMs in the available gene chip. The DEMs in the individual gene chips were then integrated and ranked with RobustRankAggreg (20), which is an R package for the comprehensive integration of the gene list.
Collection of miRNA-associated DEGs
With filter conditions of [log2 (fold change)]≥1.5 and an adjusted P<0.05, DEGs in GBM were collected from Gene Expression Profiling Interactive Analysis (GEPIA) (21), an online interactive server for gene sequence analysis that provides a gene expression profile comparison between 163 GBM specimens and 207 normal brain samples from the Cancer Genome Atlas and Genotype-Tissue Expression projects.
The target genes of DEMs were predicted using miRWalk (22), an integrated resource that provides predictions of miRNA- target interactions. Only the target genes that were predicted by at least eight target-predicting algorithms were considered to intersect with the DEGs gathered from GEPIA. Overlapping genes were regarded as miRNA-associated DEGs in GBM.
GO and KEGG enrichment analyses and protein-protein interaction (PPI) network construction
GO and KEGG enrichment analyses were performed using the ClusterProfiler (23), an R package that is used for the systematic analysis of gene clusters. A PPI network was constructed using STRING (24), a database that provides functional interactions among proteins.
Immunohistochemistry
The Human Protein Atlas (https://www.proteinatlas.org/), an interactive web tool that assesses proteins in all major tissues and organs in the human body (25) was used to determine the expression level of proteins. The main clinical characteristic of the patients are presented in Table SII.
Drug discovery in CMap
Query genes were uploaded to the CMap web tool (26), comparing >7,000 gene expression profiles following treatment with 1,309 active chemicals in human cell lines. The link between the query genes and the 1,309 chemicals was measured via a connectivity score provided by the CMap tool, which ranged valued from −1 to 1. A positive score implied a stimulative effect, while a negative score indicated a suppressive effect of the chemical on the given signatures.
Molecular docking analysis
Molecular docking between the proteins encoded by miRNA-associated DEGs and filtered chemicals was performed using Sybyl-X (27). Protein crystal structures were downloaded from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) (28) and chemical structures were obtained from PubChem (29). First, the protein crystal structure was imported into the Sybyl-X 2.1.1 software on the Surflex-Dock interface. Following the removal of irrelevant water molecules and ions, the repair of side chains, the charging of terminal groups and the addition of polar hydrogen atoms, the proteins were prepared for docking. Protomols, which are active pockets that ligands are aligned to, were generated with the use of an automatic mode. Compounds in the mol2 format were then imported into the software on the Docking interface and protein-ligand docking was run under the surflex-dock geom mode, after which a total score was exported, with these scores being directly proportional to the binding affinity. Other parameters were set by default. The binding mode was visualized by the use of PyMOL (30).
Results
Screening of 10 DEMs in GBM by the RobustRankAggreg method
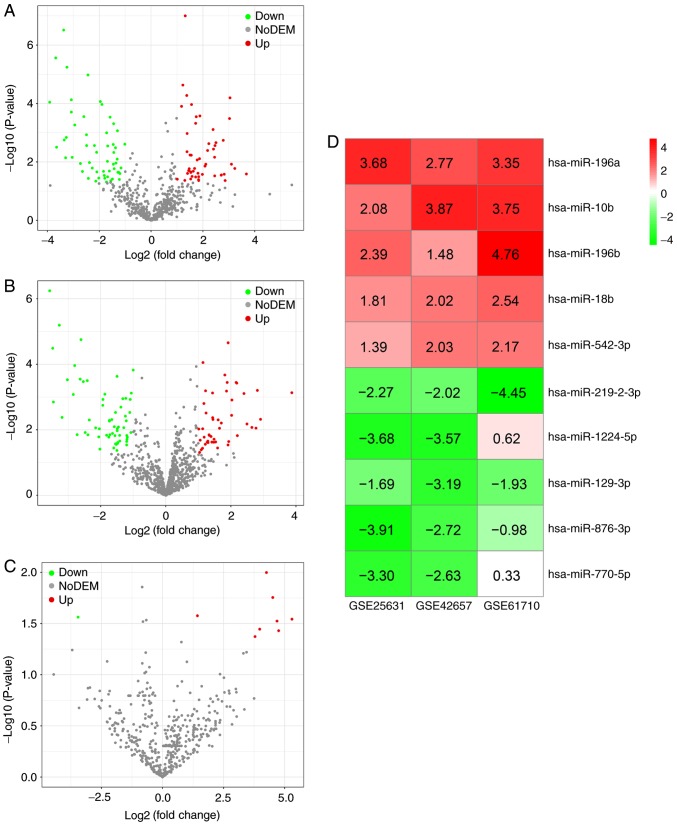
Three gene chips [GSE25631 (31), GSE42657 (32) and GSE61710 (33)], were collected from GEO. The basic information of the three microarray datasets are presented in Table I and the main clinicopathological characteristics of the samples in the three gene chips are presented in supplementary Table SI with the screening criteria of [log2 (fold change)] >1 and P<0.05, 51 upregulated miRNAs and 56 downregulated miRNAs were obtained in gene chip GSE25631 (Fig. 2A); 47 upregulated miRNAs and 61 downregulated miRNAs were obtained in gene chip GSE42657 (Fig. 2B); and eight upregulated miRNAs and one downregulated miRNA were identified in gene chip GSE61710 (Fig. 2C). Following robust rank aggregation, an ordered list of all of the DEMs in the three datasets was acquired, among which the top five upregulated DEMs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b and hsa-miR-542-3p) and the top five downregulated DEMs (hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p) were determined as being miRNAs associated with GBM (Fig. 2D). The expression of the 10 DEMs in each dataset is presented in Fig. 3. Published studies focusing on the associations between the 10 DEMs and GBM are included in Table II.
Table I.
Basic information of the three gene chips obtained from Gene Expression Omnibus.
| First author (publication year) | Country | Data source | Platform | Sample size (T/N) | (Refs.) |
|---|---|---|---|---|---|
| Zhang et al (2012) | China | GSE25631 | GPL8179 | 82/5 | (31) |
| Jones et al (2015) | United Kingdom | GSE42657 | GPL8179 | 5/7 | (32) |
| Piwecka et al (2015) | Poland | GSE61710 | GPL10656 | 10/5 | (33) |
T, tumor; N, normal control; Ref., reference.
Figure 2.
DEMs in GBM based on data from the Gene Expression Omnibus. (A) Volcano plots for DEMs in GBM based on the datasets for (A) GSE25631, (B) GSE42657 and (C) GSE61710. (D) Heatmap for the five top upregulated and five top downregulated miRNAs in GBM using the RobustRankAggreg method with a value of P<0.05. DEMs, differentially expressed miRNAs; GBM, glioblastoma multiforme; Down, downregulated; Up, upregulated; miR, microRNA.
Figure 3.
Violin plots for the expression of the 10 DEMs in the 3 gene chips. (A) Expression of upregulated miRNAs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b and hsa-miR-542-3p) in GSE25631. (B) Expression of five downregulated miRNAs (hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p) in GSE25631. (C) Expression of upregulated miRNAs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b and hsa-miR-542-3p) in GSE42657. (D) Expression of downregulated miRNAs (hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p) in GSE42657. (E) Expression of upregulated miRNAs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b and hsa-miR-542-3p) in GSE61710. (F) Expression of downregulated miRNAs (hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p) in GSE61710. DEMs, differentially expressed miRNAs; miR, microRNA; GBM, glioblastoma multiforme.
Table II.
Five top upregulated miRNAs and five top downregulated miRNAs from Gene Expression Omnibus.
| Literature retrieval | |||||
|---|---|---|---|---|---|
| Author | miRNA | RobustRankAggreg | Expression | Function | (Refs.) |
| Dou et al | hsa-miR-196a | Up | Polymorphism | (37) | |
| Yang et al | Up | Tumor growth | (38) | ||
| Guan et al | Up | None | (39) | ||
| Yang et al | Up | None | (40) | ||
| Sasayama et al | hsa-miR-10b | Up | Up | Cell invasion | (41) |
| Guessous et al | Up | Cell invasion, cell migration, tumor growth | (42) | ||
| Gabriely et al | Up | Cell proliferation, cell death | (43) | ||
| Ji et al | Up | None | (44) | ||
| Guan et al | hsa-miR-196b | Up | Up | None | (45) |
| Lakomy et al | Up | None | (46) | ||
| Ma et al | Up | None | (47) | ||
| You et al | Up | None | (48) | ||
| Karsy M et al | Up | None | (49) | ||
| hsa-miR-18b | Up | None | No ref. | ||
| Cai et al | hsa-miR-543-3p | Up | Down | Cell invasion | (50)a |
| hsa-miR-219-2-3p | Down | None | No ref. | ||
| Qian et al | hsa-miR-1224-5p | Down | Down | Cell proliferation, cell invasion, cell apoptosis | (51) |
| Ouyang et al | hsa-miR-129-3p | Down | Down | Cell proliferation, tumor growth | (52) |
| Fang et al | Down | Cell viability, cell growth | (53) | ||
| hsa-miR-876-3p | Down | None | No ref. | ||
| hsa-miR-770-5p | Down | None | No ref. | ||
The expression of hsa-miR-542-3p was inconsistent with our result. Ref., reference.
Collection of 390 miRNA-associated DEGs in GBM
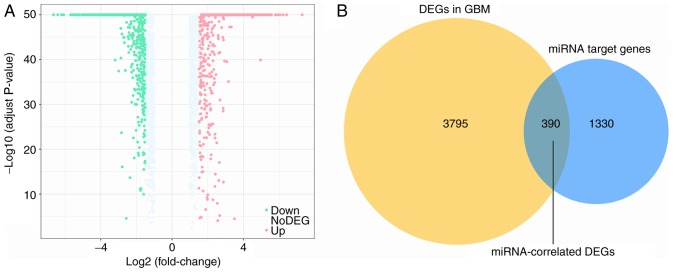
A total of 1,720 genes were identified as targets of the aforementioned 10 DEMs. Additionally, 4,185 DEGs containing 2,719 upregulated genes and 1,466 downregulated genes in GBM were collected from GEPIA (Fig. 4A). An intersection between the 1,720 target genes and the 4,185 DEGs was performed, revealing 390 overlapping miRNA-associated DEGs (Fig. 4B).
Figure 4.
Identification of 390 miRNA-associated differently expressed genes. (A) Volcano plot for DEGs in GBM from the Gene Expression Profiling Interactive Analysis. The volcano plot was generated using the R package ‘ggplot2’. (B) Venn diagram of overlapping genes from the DEGs in GBM and the target genes of the 10 differently expressed miRNAs. miRNA, microRNA; DEGs, differentially expressed genes; GBM, glioblastoma multiforme.
GO and KEGG enrichment analyses of 390 miRNA-associated DEGs, and PPI analysis of eight GBM-associated genes
GO functional annotations were performed to determine the potential molecular mechanisms employed by the 390 miRNA-associated DEGs. The top 10 biological processes (BP), cellular components (CC) and molecular functions (MF) are listed in Fig. 5A-C. The highly enriched BP terms were ‘cell growth’, ‘signal release’ and ‘regulation of cell growth’. The markedly enriched CC terms were ‘presynapse’, ‘axon’ and ‘synaptic membrane’. The predominantly enriched MF terms were ‘transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding’, ‘transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding’ and ‘transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding’.
Figure 5.
Significantly enriched Gene Ontology enrichment annotations and KEGG pathways of the 390 miRNA-associated differently expressed genes in glioblastoma multiforme. (A) Top 10 biological process terms (B) Top 10 cellular component terms. (C) Top 10 molecular function terms. (D) Significantly enriched KEGG pathways (P<0.05). The GO and KEGG pathway analyses were performed using the ClusterProfiler package in R. The GO terms were visualized using the R package ‘GOplot’ and the KEGG terms were visualized using the enrichMap function in the ClusterProfiler package. The enriched terms color changes gradually from deep to light in ascending order according to the P-values. KEGG, Kyoto Encyclopedia of genes and genomes.
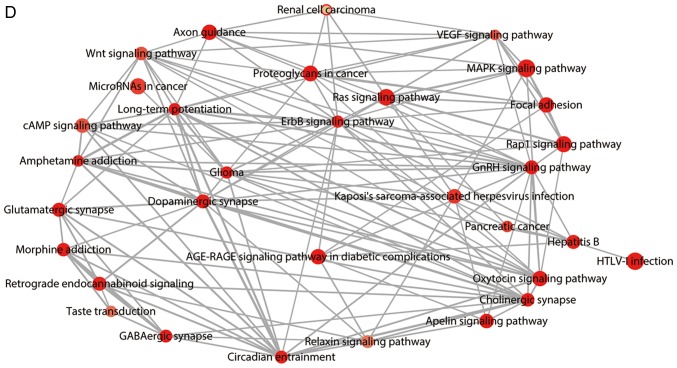
The KEGG pathway was utilized to further probe the underlying pathological pathways that the 390 genes are involved in during the inception and progression of GBM. With an adjusted value of P<0.05, 32 pathways were enriched by the 390 genes (Fig. 5D), many of which were tumor-associated pathways, including the ‘mitogen activated protein kinase (MAPK) signaling pathway’, the ‘gonadotropin-releasing hormone (GnRH) signaling pathway’ and the ‘oxytocin signaling pathway’. In addition, eight genes [cyclin-dependent kinase-6 (CDK6), retinoblastoma-associated protein (RB1), calcium/calmodulin-dependent protein kinase type II subunit gamma (CAMK2G), calcium/calmodulin-dependent protein kinase type II subunit beta (CAMK2B), GTPase NRas (NRAS), protein kinase C beta type (PRKCB), platelet derived growth factor receptor alpha (PDGFRA) and calmodulin 3 (CALM3)] were determined to be centralized in the glioma pathway (Fig. 6). In this pathway, the eight genes were revealed to participate in cell growth and proliferation, G1/S progression, cell migration and mitosis by interacting with upstream or downstream genes. The expression of the eight genes in GBM and normal controls were compared based on the data from GEPIA using a Student's t-test presented as scatter-box plots (Fig. 7). Among them, CDK6, RB1, NRAS, and PDGFRA were upregulated in GBM, and CAMK2G, CAMK2B, PRKCB, and CALM3 were downregulated in GBM tissues. Additionally, the expression patterns of their encoded proteins in GBM and normal brain tissues were validated using The Human Protein Atlas (https://www.proteinatlas.org/) as presented in Fig. 8. The PPI network indicated close associations among the eight genes (Fig. 9).
Figure 6.
Glioma pathway from Kyoto Encyclopedia of genes and genomes pathway analysis. The glioma pathway was visualized using the R package ‘pathview’. The color of the eight glioblastoma multiforme associated genes changed gradually from green to red in ascending order of the log2 (fold change) of each gene.
Figure 7.
Box plots for the expression of the eight genes in glioblastoma multiforme determined from the Gene Expression Profiling Interactive Analysis: (A) CDK6, (B) RB1, (C) NRAS, (D) PDGFRA, (E) CAMK2G, (F) CAMK2B, (G) PRKCB and (H) CALM3. Num, number; T, tumor; N, normal; CDK6, cyclin-dependent kinase 6; RB1, retinoblastoma-associated protein; NRAS, GTPase NRas; PDGFRA, platelet-derived growth factor receptor alpha; CAMK2G, calcium/calmodulin-dependent protein kinase type II subunit gamma; CAMK2B, calcium/calmodulin-dependent protein kinase type II subunit beta; PRKCB, protein kinase C beta type; CALM3, calmodulin 3.
Figure 8.
Immunohistochemistry staining of the proteins encoded by the 8 miRNA-associated differently expressed genes in the glioma pathway obtained from The Human Protein Atlas. (A) Low CDK6 expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000105810-CDK6/tissue/cerebral+cortex#img). (B) High CDK6 expression in glioblastoma multiforme (GBM) tissues (https://www.proteinatlas.org/ENSG00000105810-CDK6/pathology/tissue/glioma#img). (C) Low RB1 expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000139687-RB1/tissue/cerebral+cortex#img). (D) Moderate RB1 expression in GBM tissue (https://www.proteinatlas.org/ENSG00000139687-RB1/pathology/tissue/glioma#img). (E) NRAS protein was not detected in normal brain tissues (https://www.proteinatlas.org/ENSG00000213281-NRAS/tissue/cerebral+cortex#img). (F) Moderate NRAS expression in GBM tissue (https://www.proteinatlas.org/ENSG00000213281-NRAS/pathology/tissue/glioma#img). (G) PDGFRA protein was not detected in normal brain tissues (https://www.proteinatlas.org/ENSG00000134853-PDGFRA/tissue/cerebral+cortex#img). (H) Moderate PDGFRA expression in GBM tissues (https://www.proteinatlas.org/ENSG00000134853-PDGFRA/pathology/tissue/glioma#img). (I) High CAMK2G expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000148660-CAMK2G/tissue/cerebral+cortex#img). (J) Moderate CAMK2G expression in GBM tissues (https://www.proteinatlas.org/ENSG00000148660-CAMK2G/pathology/tissue/glioma#img). (K) Moderate CAMK2B expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000058404-CAMK2B/tissue/cerebral+cortex#img). (L) Weak CAMK2B expression in GBM tissues (https://www.proteinatlas.org/ENSG00000058404-CAMK2B/pathology/tissue/glioma#img). (M) Moderate PRKCB expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000166501-PRKCB/tissue/cerebral+cortex#img). (N) Weak PRKCB expression in GBM tissues (https://www.proteinatlas.org/ENSG00000166501-PRKCB/pathology/tissue/glioma#img). (O) Moderate CALM3 expression in normal brain tissues (https://www.proteinatlas.org/ENSG00000160014-CALM3/tissue/cerebral+cortex#img). (P) Low CALM3 expression in GBM tissues (https://www.proteinatlas.org/ENSG00000160014-CALM3/pathology/tissue/glioma#img). Magnification of each, ×100. CDK6, cyclin-dependent kinase 6; RB1, retinoblastoma-associated protein; NRAS, GTPase NRas; PDGFRA, platelet-derived growth factor receptor alpha; CAMK2G, calcium/calmodulin-dependent protein kinase type II subunit gamma; CAMK2B, calcium/calmodulin-dependent protein kinase type II subunit beta; PRKCB, protein kinase C beta type; CALM3, calmodulin 3.
Figure 9.

Protein-protein interaction network of the eight miRNA-associated differently expressed genes enriched in the glioma pathway. CDK6, cyclin-dependent kinase 6; RB1, retinoblastoma-associated protein; NRAS, GTPase NRas; PDGFRA, platelet-derived growth factor receptor alpha; CAMK2G, calcium/calmodulin-dependent protein kinase type II subunit gamma; CAMK2B, calcium/calmodulin-dependent protein kinase type II subunit beta; PRKCB, protein kinase C beta type; CALM3, calmodulin 3.
Identification of three potential chemicals for GBM treatment using CMap analysis
The eight genes, including four that were upregulated (CDK6, RB1, NRAS and PDGFRA) and four that were downregulated (CAMK2G, CAMK2B, PRKCB and CALM3) were submitted to the CMap web tool as up and down tags to acquire latent drugs in the therapy for GBM. By ranking the connectivity score in descending order, the top three chemicals (W-13, gefitinib and exemestane) were identified as being potential treatment options for GBM (Table III). The chemical structures of the three chemicals are presented in Fig. 10.
Table III.
Three chemicals identified as therapeutic agents for glioblastoma multiforme from CMap analysis.
| CMap name | Enrichment | Dose | Cell lines | Up score | Down score |
|---|---|---|---|---|---|
| W-13 | −0.989 | 10 µM | MCF7 | −0.356 | 0.532 |
| Gefitinib | −0.989 | 10 µM | HL60 | −0.289 | 0.479 |
| Exemestane | −0.981 | 10 nM | MCF7 | −0.382 | 0.351 |
CMap, connectivity map.
Figure 10.
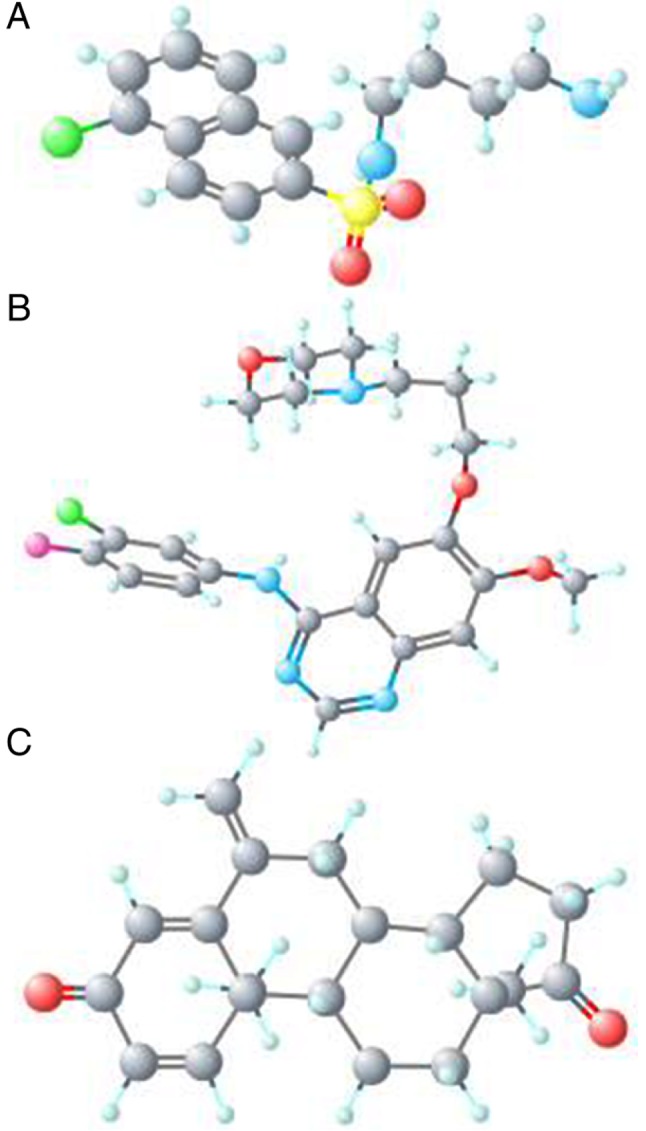
3D-structure of the three compounds identified by the Connective map analysis. (A) W-13, (B) gefitinib and (C) exemestane.
Exploration of the interactions between the three chemicals and the eight genes
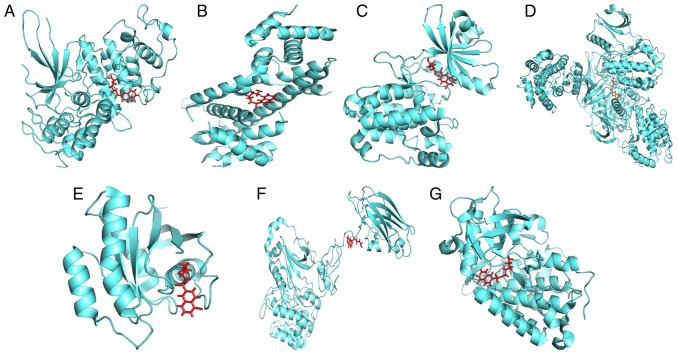
To ascertain whether the three chemicals directly bind to the proteins encoded by the eight genes, a protein-ligand docking analysis was performed. As the crystal structure of protein CALM3 was not identified in the PDB database, it was removed from the molecular docking analysis. The docking results are presented in Table IV and Figs. 11–13. The total docking score ranged from 3.5943 to 6.9781, indicating interactions between the three chemicals and the seven proteins. However, further experiments are required to verify these associations.
Table IV.
Results of protein-ligand docking.
| Gene symbol | Protein name | PDB ID | Chemical | Total score | Crash | Polar |
|---|---|---|---|---|---|---|
| CDK6 | Cyclin-dependent kinase 6 | 1BI7 | W-13 | 6.9158 | −0.6018 | 1.0852 |
| Gefitinib | 4.5461 | −1.5497 | 1.0374 | |||
| Exemestane | 4.2617 | −0.6835 | 1.2109 | |||
| RB1 | Retinoblastoma-associated protein | 2QDJ | W-13 | 6.1141 | −0.7408 | 2.1589 |
| Gefitinib | 5.1361 | −3.0039 | 1.0429 | |||
| Exemestane | 4.5673 | −1.2303 | 2.2315 | |||
| CAMK2G | Calcium/calmodulin-dependent protein kinase type II subunit gamma | 2V7O | W-13 | 6.5899 | −0.9631 | 2.1155 |
| Gefitinib | 5.5314 | −0.8239 | 4.6009 | |||
| Exemestane | 5.5085 | −2.4882 | 2.8500 | |||
| CAMK2B | Calcium/calmodulin-dependent protein kinase type II subunit beta | 3BHH | W-13 | 6.9781 | −0.8004 | 2.9012 |
| Gefitinib | 5.4665 | −1.7396 | 3.6928 | |||
| Exemestane | 4.7782 | −1.8678 | 4.6573 | |||
| NRAS | GTPase NRas | 3CON | W-13 | 6.9580 | −0.7966 | 3.5558 |
| Gefitinib | 5.1191 | −1.6695 | 2.1113 | |||
| Exemestane | 3.7527 | −1.3243 | 1.6902 | |||
| PRKCB | Protein kinase C beta type | 3PFQ | W-13 | 5.9087 | −0.7642 | 4.3292 |
| Gefitinib | 5.9886 | −0.9356 | 2.5291 | |||
| Exemestane | 3.5943 | −0.6226 | 2.3138 | |||
| PDGFRA | Platelet-derived growth factor receptor alpha | 5K5X | W-13 | 6.7711 | −1.3263 | 4.9966 |
| Gefitinib | 4.0157 | −1.0638 | 1.1911 | |||
| Exemestane | 4.8780 | −0.9005 | 2.6185 |
PDB ID: protein data bank identification.
Figure 11.
Binding mode of W-13 and proteins, (A) cyclin-dependent kinase 6, (B) retinoblastoma-associated protein, (C) calcium/calmodulin-dependent protein kinase type II subunit gamma, (D) calcium/calmodulin-dependent protein kinase type II subunit beta, (E) GTPase NRas, (F) protein kinase C beta type and (G) platelet-derived growth factor receptor alpha.
Figure 13.
Binding mode of exemestane and proteins, (A) cyclin-dependent kinase 6, (B) retinoblastoma-associated protein, (C) calcium/calmodulin-dependent protein kinase type II subunit gamma, (D) calcium/calmodulin-dependent protein kinase type II subunit beta, (E) GTPase NRas, (F) protein kinase C beta type and (G) platelet-derived growth factor receptor alpha.
Discussion
Since the identification of the first miRNA, an increasing number of studies have focused on the action and clinical application of miRNAs, particularly in terms of neoplasm treatment (34). It has been demonstrated that miRNAs participate in multiple biological processes involved in tumorigenesis (35). With the advantage of targeting genes that are involved in multiple pathological pathways, drug developments based on miRNAs have received increasing attention (36).
The present study obtained 10 DEMs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b, hsa-miR-542-3p, hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p) using a robust rank aggregation, which is a recognized method for integrating genes from diverse resources that are free of outliers, noise and errors (20). Among these literatures, four studies corroborated overexpressed hsa-miR-196a in GBM and its contribution to the development of GBM (37–40). Additionally, four studies indicated that a high expression of hsa-miR-10b promoted the progression of GBM (41–44). Five studies focused on hsa-miR-196b and demonstrated its high expression in GBM (45–49) and one study, performed by Cai et al (50), focused on hsa-miR-542-3p. The authors of these studies determined that hsa-miR-542-3p was downregulated in glioblastoma cell lines, which was not consistent with the gene chip results from the present study. Given the differences in sample sources, RNA extraction and detection, more studies are required to further assess the role of hsa-miR-542-3p in GBM. Additionally, the decreased expression of hsa-miR-1224-5p (51) and hsa-miR-129-3p (52,53), as well as their inhibitory effects on GBM, have been verified in previous studies. Furthermore, the association of hsa-miR-18b, hsa-miR-219-2-3p, hsa-miR-876-3p and hsa-miR-770-5p with GBM have not yet been reported. Since miRNAs control tumor development by regulating their downstream target genes (54), the present study collected the target genes of the aforementioned 10 DEMs to elucidate how these DEMs mediate the pathophysiological processes of GBM. The DEGs in GBM were also obtained and an intersection between the target genes and the DEGs was performed to determine miRNA-associated DEGs. Gene functional and pathway enrichment analyses of the miRNA-associated DEGs revealed that these genes were involved with multiple tumor-associated biological processes and signaling pathways, including the ‘cell growth’, the ‘MAPK signaling pathway’, the ‘GnRH signaling pathway’ and the ‘oxytocin signaling pathway’. Furthermore, eight genes (CDK6, RB1, CAMK2G, CAMK2B, NRAS, PRKCB, PDGFRA and CALM3) of the miRNA-associated DEGs were enriched in the glioma pathway, indicating their important roles in GBM. In the glioma pathway, the eight genes primarily participated in cell growth and proliferation, G1/S progression, and cell migration and mitosis, indicating that these genes participate in the development of GBM by mediating cell proliferation and metastasis. Each of the eight genes was targeted by more than one miRNA and one miRNA targeted more than one gene. For example, the PDGFRA gene was targeted by hsa-miR-770-5p and hsa-miR-196a. hsa-miR-196a targeted NRAS, PDGFRA and CALM3. miRNAs primarily exert effects via destabilization or translational repression by targeting the 3′ untranslated region of mRNA transcripts in the cytoplasm (7). However, an increasing number of studies have indicated that miRNAs positively regulate gene transcription by targeting promoter elements (55–57). The present study revealed that certain miRNAs were negatively associated with their target genes (hsa-miR-1224-5p and CDK6; hsa-miR-196a and CALM3), while other miRNAs were positively associated with their target genes (hsa-miR-196a and NRAS; hsa-miR-1224-5p and PRKCB). However, further studies are required to assess the regulatory mechanisms of the 10 miRNAs and their target genes.
CMap is a practical tool for the exploration of novel drugs and for the repurposing of existing drugs, and its efficiency has been supported by numerous studies (58,59). Aramadhaka et al (58) identified Gila monster venom and Byetta® as being therapeutic drugs for the treatment of type-2 diabetes using CMap analysis. Wang et al (59) demonstrated that via cell apoptosis, prenylamine could be a candidate agent for the treatment of hepatocellular carcinoma. The present study selected the aforementioned eight genes for CMap analysis. Following this analysis, three chemicals (W-13, gefitinib and exemestane) were determined as latent therapeutic agents for GBM. As a calmodulin antagonist, W-13 has been demonstrated to inhibit cell growth (60) and to induce cell apoptosis (61). However, few studies on W-13 have assessed its anti-GBM effects. The results from the molecular docking analysis performed in the present study revealed that W-13 could bind to proteins CDK6, RB1, NRAS, PDGFRA, CAMK2G, CAMK2B and PRKCB, exhibiting high binding scores and indicating that W-13 could exert its anti-GBM effects by acting on these GBM-associated genes. The present study provides a theoretical basis for the application of W-13 in patients with GBM, but further studies are required to corroborate this conclusion.
The inhibitory effect of gefitinib on GBM, which is an epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor, has been demonstrated in previous studies (62,63). However, its clinical application is limited due to gefitinib resistance (62). A study by Aljohani et al (64) revealed that PDGFRA was significantly upregulated in gefitinib-resistant GBM cells and that overexpressed PDGFRA regulated gefitinib resistance. The present study identified that PDGFRA was a target gene of the 10 DEMs, suggesting the potential of the 10 DEMs in enhancing gefitinib sensitivity in patients with GBM.
Exemestane is a widely used drug in the prevention and treatment of breast cancer due to its aromatase inhibitory role in the production of oestrogen (65–67). However, to the best of our knowledge, its antitumor effect on GBM has not yet been elucidated. A study by Kritikou et al (68) revealed that the combination of exemestane and erlotinib significantly inhibited EGFR mitochondrial translocation. The EGFR mitochondrial translocation event serves important roles in tumor progression (69) and contributes to drug resistance (70). A study by Dasari et al (71) indicated that the inhibition of the EGFR mitochondrial translocation event in GBM may be a therapeutic strategy. Additionally, the translocation of EGFR into mitochondria contributes to EGFR inhibitor drug resistance (70). The combined use of exemestane and the EGFR inhibitor, gefitinib, in patients with GBM may therefore increase gefitinib sensitivity by inhibiting the translocation of EGFR into the mitochondria. However, further in-depth in vitro and in vivo experiments are essential to verify the anti-GBM effects and the synergistic antitumor effects of these compounds.
In the present study, the identification of W-13, gefitinib and exemestane were made on the basis of the eight GBM-associated genes (CDK6, RB1, CAMK2G, CAMK2B, NRAS, PRKCB, PDGFRA and CALM3), which were the target genes of the 10 DEMs (hsa-miR-196a, hsa-miR-10b, hsa-miR-196b, hsa-miR-18b, hsa-miR-542-3p, hsa-miR-219-2-3p, hsa-miR-1224-5p, hsa-miR-129-3p, hsa-miR-876-3p and hsa-miR-770-5p). Thus, the present study hypothesizes that the 10 DEMs may produce synergistic or antagonistic effects on the three chemicals by targeting these genes. However, more experiments are necessary to validate this conjecture.
In conclusion, by employing an integrated strategy of data mining and computational biology, the present study obtained 10 DEMs that may participate in the development of GBM. Furthermore, three candidate agents (gefitinib, W-13 and exemestane) in the treatment of GBM were identified following CMap analysis. Since the results are based on In silico analysis, further in-depth studies are necessary to add to the validity of these results.
Supplementary Material
Figure 12.
Binding mode of gefitinib and proteins, (A) cyclin-dependent kinase 6, (B) retinoblastoma-associated protein, (C) calcium/calmodulin-dependent protein kinase type II subunit gamma, (D) calcium/calmodulin-dependent protein kinase type II subunit beta, (E) GTPase NRas, (F) protein kinase C beta type and (G) platelet-derived growth factor receptor alpha.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
DDX designed the present study, collected miRNA-associated datasets, screened differently expressed miRNAs and wrote the manuscript. WQX collected miRNA-associated datasets and wrote the manuscript. RQH and YWD performed GO and KEGG analyses and construed the PPI network. GC performed CMap analysis and checked all data. DZL designed the experiments and wrote manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella- Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(Suppl_5):v1–v88. doi: 10.1093/neuonc/nox158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 4.Alifieris C, Trafalis DT. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol Ther. 2015;152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 6.Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422–442. doi: 10.1038/s41571-018-0003-5. [DOI] [PubMed] [Google Scholar]
- 7.Bartel BP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: Function, detection, and bioanalysis. Chem Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 9.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 10.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godlewski J, Nowicki MO, Bronisz A, Nuovo G, Palatini J, De Lay M, Van Brocklyn J, Ostrowski MC, Chiocca EA, Lawler SE. MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell. 2010;37:620–632. doi: 10.1016/j.molcel.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu J, Sun T, Wang H, Chen Z, Wang S, Yuan L, Liu T, Li HR, Wang P, Feng Y, et al. MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell. 2016;29:49–60. doi: 10.1016/j.ccell.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu XA, Rajpal DK. Applications of connectivity map in drug discovery and development. Drug Discov Today. 2012;17:1289–1298. doi: 10.1016/j.drudis.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Chien W, Sun QY, Lee KL, Ding LW, Wuensche P, Torres-Fernandez LA, Tan SZ, Tokatly I, Zaiden N, Poellinger L, et al. Activation of protein phosphatase 2A tumor suppressor as potential treatment of pancreatic cancer. Mol Oncol. 2015;9:889–905. doi: 10.1016/j.molonc.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lamb J. The connectivity map: A new tool for biomedical research. Nat Rev Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- 18.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. doi: 10.1007/978-1-4939-3578-9_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolde R, Laur S, Adler P, Vilo J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics. 2012;28:573–580. doi: 10.1093/bioinformatics/btr709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dweep H, Gretz N, Sticht C. miRWalk database for miRNA- target interactions. Methods Mol Biol. 2014;1182:289–305. doi: 10.1007/978-1-4939-1062-5_25. [DOI] [PubMed] [Google Scholar]
- 23.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 26.Musa A, Ghoraie LS, Zhang SD, Glazko G, Yli-Harja O, Dehmer M, Haibe-Kains B, Emmert-Streib F. A review of connectivity map and computational approaches in pharmacogenomics. Brief Bioinform. 2017;18:903. doi: 10.1093/bib/bbx023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guedes IA, de Magalhaes CS, Dardenne LE. Receptor-ligand molecular docking. Biophys Rev. 2014;6:75–87. doi: 10.1007/s12551-013-0130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, et al. The RCSB protein data bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43:D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander N, Woetzel N, Meiler J. bcl::Cluster: A method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int Conf Comput Adv Bio Med Sci. 2011;2011:13–18. doi: 10.1109/ICCABS.2011.5729867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang W, Zhang J, Hoadley K, Kushwaha D, Ramakrishnan V, Li S, Kang C, You Y, Jiang C, Song SW, et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol. 2012;14:712–719. doi: 10.1093/neuonc/nos089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones TA, Jeyapalan JN, Forshew T, Tatevossian RG, Lawson AR, Patel SN, Doctor GT, Mumin MA, Picker SR, Phipps KP, et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-kB pathways. Acta Neuropathol Commun. 2015;3:86. doi: 10.1186/s40478-015-0266-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piwecka M, Rolle K, Belter A, Barciszewska AM, Żywicki M, Michalak M, Nowak S, Naskret-Barciszewska MZ, Barciszewski J. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol Oncol. 2015;9:1324–1340. doi: 10.1016/j.molonc.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Onco. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 35.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dou T, Wu Q, Chen X, Ribas J, Ni X, Tang C, Huang F, Zhou L, Lu D. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol. 2010;136:1853–1859. doi: 10.1007/s00432-010-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang G, Han D, Chen X, Zhang D, Wang L, Shi C, Zhang W, Li C, Chen X, Liu H, et al. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IκBα both in vitro and in vivo. Neuro Oncol. 2014;16:652–661. doi: 10.1093/neuonc/not307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guan Y, Chen L, Bao Y, Qiu B, Pang C, Cui R, Wang Y. High miR-196a and low miR-367 cooperatively correlate with unfavorable prognosis of high-grade glioma. Int J Clin Exp Pathol. 2015;8:6576–6588. [PMC free article] [PubMed] [Google Scholar]
- 40.Yang JP, Yang JK, Li C, Cui ZQ, Liu HJ, Sun XF, Geng SM, Lu SK, Song J, Guo CY, Jiao BH. Downregulation of ZMYND11 induced by miR-196a-5p promotes the progression and growth of GBM. Biochem Biophys Res Commun. 2017;494:674–680. doi: 10.1016/j.bbrc.2017.10.098. [DOI] [PubMed] [Google Scholar]
- 41.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–1413. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 42.Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B, Abounader R. Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol. 2013;112:153–163. doi: 10.1007/s11060-013-1047-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy. 2011;7:1384–1386. doi: 10.4161/auto.7.11.17371. [DOI] [PubMed] [Google Scholar]
- 44.Ji Y, Wei Y, Wang J, Gong K, Zhang Y, Zuo H. Correlation of microRNA-10b upregulation and poor prognosis in human gliomas. Tumour Biol. 2015;36:6249–6254. doi: 10.1007/s13277-015-3310-9. [DOI] [PubMed] [Google Scholar]
- 45.Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, et al. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clin Cancer Res. 2010;16:4289–4297. doi: 10.1158/1078-0432.CCR-10-0207. [DOI] [PubMed] [Google Scholar]
- 46.Lakomy R, Sana J, Hankeova S, Fadrus P, Kren L, Lzicarova E, Svoboda M, Dolezelova H, Smrcka M, Vyzula R, et al. MiR-195, miR-196b, miR-181c, miR-21 expression levels and O−6-methylguanine-DNA methyltransferase methylation status are associated with clinical outcome in glioblastoma patients. Cancer Sci. 2011;102:2186–2190. doi: 10.1111/j.1349-7006.2011.02092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma R, Yan W, Zhang G, Lv H, Liu Z, Fang F, Zhang W, Zhang J, Tao T, You Y, et al. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS One. 2012;7:e38096. doi: 10.1371/journal.pone.0038096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You G, Yan W, Zhang W, Wang Y, Bao Z, Li S, Li S, Li G, Song Y, Kang C, et al. Significance of miR-196b in tumor-related epilepsy of patients with gliomas. PLoS One. 2012;7:e46218. doi: 10.1371/journal.pone.0046218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karsy M, Arslan E, Moy F. Current progress on understanding MicroRNAs in glioblastoma multiforme. Genes Cancer. 2012;3:3–15. doi: 10.1177/1947601912448068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai J, Zhao J, Zhang N, Xu X, Li R, Yi Y, Fang L, Zhang L, Li M, Wu J, et al. MicroRNA-542-3p suppresses tumor cell invasion via targeting AKT pathway in human astrocytoma. J Biol Chem. 2015;290:24678–24688. doi: 10.1074/jbc.M115.649004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian J, Li R, Wang YY, Shi Y, Luan WK, Tao T, Zhang JX, Xu YC, You YP. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403:33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- 52.Ouyang Q, Chen G, Zhou J, Li L, Dong Z, Yang R, Xu L, Cui H, Xu M, Yi L. Neurotensin signaling stimulates glioblastoma cell proliferation by upregulating c-Myc and inhibiting miR-29b-1 and miR-129-3p. Neuro Oncol. 2016;18:216–226. doi: 10.1093/neuonc/nov114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang DZ, Wang YP, Liu J, Hui XB, Wang XD, Chen X, Liu D. MicroRNA-129-3p suppresses tumor growth by targeting E2F5 in glioblastoma. Eur Rev Med Pharmacol Sci. 2018;22:1044–1050. doi: 10.26355/eurrev_201802_14387. [DOI] [PubMed] [Google Scholar]
- 54.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 55.Matsui M, Chu Y, Zhang H, Gagnon KT, Shaikh S, Kuchimanchi S, Manoharan M, Corey DR, Janowski BA. Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res. 2013;41:10086–10109. doi: 10.1093/nar/gkt777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Majid S, Dar AA, Saini S, Yamamura S, Hirata H, Tanaka Y, Deng G, Dahiya R. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao M, Li J, Li W, Wang Y, Wu F, Xi Y, Zhang L, Ding C, Luo H, Li Y, et al. miRNA and cancer; computational and experimental approaches. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017;14:1326–1334. doi: 10.1080/15476286.2015.1112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aramadhaka LR, Prorock A, Dragulev B, Bao Y, Fox JW. Connectivity maps for biosimilar drug discovery in venoms: The case of Gila monster venom and the anti-diabetes drug Byetta®. Toxicon. 2013;69:160–167. doi: 10.1016/j.toxicon.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Li M, Wang Y, Liu X. Integrating subpathway analysis to identify candidate agents for hepatocellular carcinoma. Onco Targets Ther. 2016;9:1221–1230. doi: 10.2147/OTT.S97211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strobl JS, Peterson VA. Tamoxifen-resistant human breast cancer cell growth: Inhibition by thioridazine, pimozide and the calmodulin antagonist, W-13. J Pharmacol Exp Ther. 1992;263:186–193. [PubMed] [Google Scholar]
- 61.Takadera T, Ohyashiki T. Calmodulin inhibitor-induced apoptosis was prevented by glycogen synthase kinase-3 inhibitors in PC12 cells. Cell Mol Neurobiol. 2007;27:783–790. doi: 10.1007/s10571-007-9172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mu L, Wang T, Chen Y, Tang X, Yuan Y, Zhao Y. β-Elemene enhances the efficacy of gefitinib on glioblastoma multiforme cells through the inhibition of the EGFR signaling pathway. Int J Oncol. 2016;49:1427–1436. doi: 10.3892/ijo.2016.3626. [DOI] [PubMed] [Google Scholar]
- 63.Parker JJ, Dionne KR, Massarwa R, Klaassen M, Foreman NK, Niswander L, Canoll P, Kleinschmidt-Demasters BK, Waziri A. Gefitinib selectively inhibits tumor cell migration in EGFR-amplified human glioblastoma. Neuro Oncol. 2013;15:1048–1057. doi: 10.1093/neuonc/not053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aljohani H, Koncar RF, Zarzour A, Park BS, Lee SH, Bahassi el M. ROS1 amplification mediates resistance to gefitinib in glioblastoma cells. Oncotarget. 2015;6:20388–20395. doi: 10.18632/oncotarget.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barton MK. Exemestane is effective for the chemoprevention of breast cancer. CA Cancer J Clin. 2011;61:363–364. doi: 10.3322/caac.20131. [DOI] [PubMed] [Google Scholar]
- 66.Pagani O, Regan MM, Francis PA. Exemestane with ovarian suppression in premenopausal breast cancer. N Engl J Med. 2014;371:1358–1359. doi: 10.1056/NEJMoa1404037. [DOI] [PubMed] [Google Scholar]
- 67.Van Asten K, Neven P, Lintermans A, Wildiers H, Paridaens R. Aromatase inhibitors in the breast cancer clinic: Focus on exemestane. Endocr Relat Cancer. 2014;21:R31–R49. doi: 10.1530/ERC-13-0269. [DOI] [PubMed] [Google Scholar]
- 68.Kritikou I, Giannopoulou E, Koutras AK, Labropoulou VT, Kalofonos HP. The combination of antitumor drugs, exemestane and erlotinib, induced resistance mechanism in H358 and A549 non-small cell lung cancer (NSCLC) cell lines. Pharm Biol. 2013 Nov 5; doi: 10.3109/13880209.2013.841718. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 69.Che TF, Lin CW, Wu YY, Chen YJ, Han CL, Chang YL, Wu CT, Hsiao TH, Hong TM, Yang PC. Mitochondrial translocation of EGFR regulates mitochondria dynamics and promotes metastasis in NSCLC. Oncotarget. 2015;6:37349–37366. doi: 10.18632/oncotarget.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cao X, Zhu H, Ali-Osman F, Lo HW. EGFR and EGFRvIII undergo stress- and EGFR kinase inhibitor-induced mitochondrial translocalization: A potential mechanism of EGFR-driven antagonism of apoptosis. Mol Cancer. 2011;10:26. doi: 10.1186/1476-4598-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dasari VR, Velpula KK, Alapati K, Gujrati M, Tsung AJ. Cord blood stem cells inhibit epidermal growth factor receptor translocation to mitochondria in glioblastoma. PLoS One. 2012;7:e31884. doi: 10.1371/journal.pone.0031884. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.