Abstract
Background
Ubiquinol-cytochrome C reductase core protein II (QCR2) is essential for mitochondrial functions, yet, its role in cancer development has remained elusive.
Methods
The expression of QCR2 in cancer patients was assessed by immunohistochemistry. The proliferation of cancer cells was assessed by CCK-8 assay, EdU staining and Flow cytometry analysis. The biological function of QCR2 and PHB were determined using western blotting, RT-qPCR, microarray analysis and xenografts. The interactions between proteins and the ubiquitination of p53 were assessed by immunoprecipitation, mass spectrometry analysis and GST pull down. The subcellular location of PHB and QCR2 was assessed by immunoblotting and immunofluorescence.
Finding
The expression of QCR2 is upregulated in multiple human tumors. Suppression of QCR2 inhibits cancer cell growth by activating p53 signaling and inducing p21-dependent cell cycle arrest and senescence. QCR2 directly interacts with PHB in the mitochondria. Overexpression of QCR2 inhibits PHB binding to p53 in the nucleus, and facilitates p53 ubiquitination and degradation, consequently leading to tumorigenesis. Also, increased QCR2 and decreased PHB protein levels are well correlated with decreased expression of p21 in cervical cancer tissues.
Interpretation
These results identify a novel role for QCR2, together with PHB, in negative regulation of p53 stability and activity, thus promote cervical carcinogenesis.
Fund
“973” Program of China, the National Science-technology Supporting Plan Projects, the National Natural Science Foundation of China, National Science and Technology Major Sub-Project and Technical Innovation Special Project of Hubei Province.
Keywords: QCR2, Tumorigenesis, p53, Degradation, PHB, Senescence
Research in context.
Evidence before this study
There has been long-standing interest in finding factors that promote the degradation of p53 in cancer. We systematically searched PubMed on December 29, 2018 with the following terms: “UQCRC2 OR QCR2” AND “p53”. There were no relevant human studies although we did find reports of QCR2 expression changes in high fertility bulls and is involved in the effect of nutlin-3a on Male Fertility.
Added value of this study
We identified QCR2 as a novel p53 suppressor independent of HPV E6 protein from a set of cancer cell lines, an in vivo xenograft model, and clinical specimens. This conclusion was further illustrated by the molecular mechanism that QCR2 facilitates p53 polyubiquitination and degradation by interfering with the interaction between p53 and its chaperone PHB in cancer cells that express wild-type p53. Our findings will accelerate the understanding of the mechanisms of tumorigenesis in cervical cancer and provide a basis for the development of new therapeutic strategies.
Implications of all the available evidence
Our findings, together with the existing evidence, highlight the role of QCR2 in tumorigenesis and p53 regulation. Our study as presented here provides a new insight into the regulation of p53 expression by QCR2 and suggests this mitochondrial protein as a potential anti-cancer target for future exploration.
Alt-text: Unlabelled Box
1. Introduction
Mitochondria play central and multifunctional roles in malignant progression through their distinct bioenergetics and biosynthetic functions [1]. Recently, various components of mitochondrial oxidative phosphorylation have been shown to exert diverse functions that are involved in tumor progression. For example, succinate dehydrogenases (SDHs) act as tumor suppressors, while ubiquinol-cytochrome C reductase binding protein (UQCRB) promotes tumor angiogenesis. Also, ectopic ATP synthase can be relocated to the cell surface, involving tumor-like acidic and hypoxic microenvironment [[1], [2], [3], [4]]. QCR2 is a core protein of mitochondrial complex III (CIII) and indispensable for the functions of this complex [5,6]. Although our previous report showed that QCR2 plays a role in trichostatin A (TSA)-induced apoptosis using the suppression of mortality by antisense rescue technique (SMART) [7], QCR2 is also upregulated in lung adenocarcinoma and breast cancer and binds to NLR family member X1 (NLRX1) and human papilloma virus (HPV)-18 E2. These studies suggest that QCR2 might be involved in tumorigenesis [[8], [9], [10], [11]]. Addressing this possibility would help us better understand the role of QCR2 in tumorigenesis, and also it is important to dissect the underlying mechanism(s). Our initial attempts to address these issues revealed p53 as a possible downstream player of QCR2.
p53 is a well-studied and critical tumor suppressor, as inactivation of this suppressor (encoded by the TP53 gene) is likely attributed to almost all human malignancies, including cervical cancer [12]. Activation of p53 increases p21 (encoded by CDKN1A) mRNA expression and leads to p21-dependent cell cycle arrest and senescence [13]. Also, p53 maintains mitochondrial integrity and accelerates mitochondrial oxidative phosphorylation via upregulation of p53-controlled ribonucleotide reductase (p53R2) and Synthesis of Cytochrome c Oxidase 2 (SCO2) and inhibition malic enzymes ME1 and ME2 [14]. Furthermore, mitochondrial oxidative stress can lead to p53 activation, likely by blocking the MDM2/MDMX-p53 feedback loop [15].
p53 is a short-lived protein that is degraded in most of human cells by its two feedback regulators MDM2 and MDMX via a ubiquitin-dependent 26S proteasomal pathway [16,17]. It can also be degraded via a ubiquitin-independent 20S proteasome [16]. CIII appears to play a role in p53 degradation via the latter, as CIII inhibitors can activate p53 by blocking the dihydroorotate dehydrogenase step of pyrimidine biosynthesis and interfering with its degradation by the 20S proteasome [18]. Mitochondrial reactive oxygen species (ROS), produced during the process of mitochondrial oxidative phosphorylation, increase the production of NAD(P)H quinone dehydrogenase 1, leading to stabilization of p53 through a ubiquitin-independent proteasome pathway [19]. Mitochondrial dysfunction also activates p53 by reducing NAD+/NADH ratios and inducing AMP-activated protein kinase (AMPK) [20] [21]. However, it remains incompletely understood what other mitochondrial proteins might be involved in this p53 degradation pathway.
In this study, we further investigated the role of QCR2 in tumorigenesis. We discovered that this protein is elevated in human tumors and plays a role in the regulation of p53 levels through a newly discovered PHB-dependent pathway. Our study as presented here provides a new insight into the regulation of p53 expression by QCR2 and suggests this mitochondrial protein as a potential anti-cancer target for future exploration.
2. Materials and methods
2.1. Cell culture
HeLa, Caski, SiHa, C33A, A549, HepG2, MCF-7, HCT116, and HEK293 cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured under standard conditions specified by the manufacturer. The hepatic cell line of human origin QSG7701 was purchased from the Type Culture Collection of the Chinese Academy of Sciences (GNHu7, Shanghai, China). The immortalized human cervical keratinocyte cell line S12 was a gift from Kenneth Raj (Centre for Radiation, Chemical and Environmental Hazards, Health Protection Agency, Chilton, Didcot, United Kingdom); the acquisition of this cell line was permitted by the original owner, Margaret Stanley (Department of Pathology, University of Cambridge, Cambridge, United Kingdom) [22]. S12 cells were cultured in a 1:3 mixture of Dulbecco's modified Eagle's medium and Ham F-12 medium supplemented with 5% fetal bovine serum (FBS; Gibco), 5 μg/mL insulin (Sigma-Aldrich, St. Louis, MO, USA), 8.4 ng/mL cholera toxin (Sigma-Aldrich), 24.3 μg/mL adenine (Sigma-Aldrich), 0.5 μg/mL hydrocortisone (Sigma-Aldrich), and 10 ng/mL epidermal growth factor (Peprotech).
2.2. Western blotting analysis
Cells were harvested into RIPA buffer supplemented with protease inhibitors (cat. no. 04693116001, Roche). Sixty micrograms of whole cell lysate or the immunocomplexes were loaded onto an acrylamide gel and then blotted to PVDF membranes. The membranes were incubated with primary antibodies at 4 °C overnight. Secondary antibodies were as follows: anti-mouse IgG (IRDye 800CW goat anti-mouse IgG, cat. no. 926–32,210, LI-COR or Goat Anti-Rabbit IgG H&L (HRP), cat. no. ab205718, abcam), anti-rabbit IgG (IRDye 800CW goat anti-rabbit IgG, cat. no. 926–32,211, LI-COR or Goat Anti-Mouse IgG H&L (HRP), cat: ab205719, abcam). Signals were detected by Odyssey scanner (LI-COR, Biosciences, USA) or ECL. Analysis of optical density was assessed using ImageJ software.
2.3. Transfection
The siRNAs and vectors were transfected into tumor cells using Lipofectamine 3000 according to standard protocols. QCR2 and PHB-specific siRNAs were obtained from Thermo Scientific, and p53-specific siRNAs were obtained from Ribobio (China). The siRNAs used in Supplementary Fig. 9b except QCR2 siRNA-2 were obtained from Ribobio (China) and a cocktail of three pairs of siRNA was used to knock down gene expression as indicated in each figure. Sequences of siRNAs used are as follows (5′-3′): QCR2 siRNA-1 (GGCAGUAGAUAUGAGGACU), QCR2 siRNA-2 (GGUGGCAAAUUAAGUGUGACCGCAA), PHB siRNA-1 (GAGAGGACUAUGAUGAGCGUGUGCU), PHB siRNA-2 (CCAAGGCAGCUGAGCUGAUUGCCAA), PHB siRNA-3 (GCCAAAGUGUUUGAGUCCAUUGGCA), p53 siRNA (GAAGAAACCACUGGAUGGA), UQCC2 siRNA (GCCUGUCGUUGGAAGAGUA, GGUGUGGAGUCUUGUACAU, CCUUGGCUCUUACUGAUAU), UQCRFS1 siRNA (CCAUGUUGUCGGUAGCAUC, CUCCUAUUUGGUAACUGGA, CACCAGUGACGAUAUGGUG), CYC1 siRNA (CUGCCAACAACGGAGCAUU, ACGGAGGAUGAAGCUAAGG, AGUGGUCAGUCCUGAAGAG), UQCRB siRNA (AUACGAGGAUGAAGAUGUA, CCUUGAACCGUAUCUGAAA, ACAGGAUGUUUCGCAUUAA), UQCRQ siRNA (ACAGCUUGUCACCGUUCGA, GCACGUCUUCACUAAAGGA, GACUGAAGAGUUCGAGAGA), MT-CYB siRNA (GCGCCUCAAUAUUCUUUAU, CCGUGAGGCCAAAUAUCAU, GCAGACCUCCUCAUUCUAA), UQCR10 siRNA (GACGCUAUCUACGACCACA, CGCGACGUUGACUUCGAAA, ACACAUCAAGCACAAGUAU), UQCRH siRNA (GCAGUUGGAGAAAUGUGUA, CCCACAAACUCUUUAACAA, AGAGGAGGAAUUAGUGGAU), LYRM7 siRNA (CAGACCACAAUACACUGAA, ACUCAGAACAUCUGUUAUA, GCACCAACUCAGAAGCAAU), UQCC1 siRNA (GGAUGUGUCUAGUCCGAAU, GGAAGUACAUGUGUCGUAU, GGUAGAGUAUGUGAGGAAA), TTC19 siRNA (GCGAGCCAAGUUGAGCAUU, GGAGGACAAUGCAAUAAUU, GCUUUGAUGAGGCCUAUAU), BCS1L siRNA (GCCAUUACAUGAUCACACU, GUGGCCGCAUUUCCACUAA, AGACGUGGCUACCUGCUUU), PHB2 siRNA-1 (CCAAAGACCUACAGAUGGUGAAUAU), PHB2 siRNA-1 (GCAAGAUUCGAGCAGCCCAGAAUAU), PHB2 siRNA-1 (ACGAUCGCCACAUCACAGAAUCGUA).
2.4. Use of animals
All animals used in this work were obtained from BEIJING HFK BIOSCIENCE Co., Ltd. (Beijing, China), and experiments were approved by the Committee on the Ethics of Animal Experiments of Tongji Medical College. Mice were maintained in the accredited animal facility of Tongji Medical College.
2.5. Ethical approval
The tissues used in this work were obtained from Tongji Hospital (Wuhan, China) and Xi'an Alena Biotechnology Ltd., Co. The study was approved by the Ethical Committee. The specimens were completely anonymous and had no direct identifiers and no codes or indirect identifiers that link back to subjects.
2.6. Immunohistochemistry (IHC)
The tissue samples were obtained from patients during surgery and details regarding these tissue samples are shown in Supplementary Tables 1, 2 and 3. Manual immunohistochemical staining was performed according to the Proteintech protocol (http://www.ptglab.com/Support/index.aspx). Staining intensity and area were assessed via the generally accepted German semiquantitative scoring system: multiplying intensity (0–3+) and extent (0–100%) of staining via light microscopy (range: 0–12) [23].
2.7. RNA isolation, cDNA microarray analysis, and reverse transcription quantitative PCR (RT-qPCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, USA) according to the manufacturer's instructions and further reverse transcribed into cDNA with M-MLV reverse transcriptase (Fermentas, Thermo Fisher Scientific, USA). The relative quantity of mRNA was determined by qPCR as described previously [24]. Primers used were QCR2 (5′-TTCAGCAATTTAGGAACCACCC-3′, 5′-GGTCACACTTAATTTGCCACCAA-3′), UQCC2 (5′-TCAGATGTACGAGAGCTTAGCG-3′, 5′-TGTACTCTTCCAACGACAGGC-3′), BCS1L (5′-ACCCGTACTCAGCACCTCA-3′, 5′-GTTCTACCCGAATCCATTTCCC-3′), UQCRFS1 (5′- CGTCACCCAGTTCGTTTCCA-3′, 5′-AGGGGTTTGCCTCTCCATTTG-3′), CYC1 (5′-AGCTATCCGTGGTCTCACC-3′, 5′-CCGCATGAACATCTCCCCATC-3′), UQCRB (5′-GGTAAGCAGGCCGTTTCAG-3′, 5′-AGGTCCAGTGCCCTCTTAATG-3′), UQCRQ (5′-CGCGAGTTTGGGAATCTGAC-3′, 5′-TAGTGAAGACGTGCGGATAGG-3′), UQCR10 (5′-ATCGTGGGCGTCATGTTCTTC-3′, 5′-ATGTGGTCGTAGATAGCGTCC-3′), UQCRH (5′-GAGGACGAGCAAAAGATGCTT-3′, 5′-CGAGAGGAATCACGCTCATCA-3′), LYRM7 (5′-GGACGGGCAGTCAAGGTTTTA-3′, 5′-GGTGCATCACAATATGGCACATT-3′), UQCC1 (5′-GGAGAAAACTGACTTCGAGGAAT-3′, 5′-TCCAGACGTGGAGTAGGGTTA-3′), TP53 (5′-GAGGTTGGCTCTGACTGTACC-3′, 5′-TCCGTCCCAGTAGATTACCAC-3′), TTC19 (5′-GCGAGCCAAGTTGAGCATTAT-3′,5′-GCGAGACGAAGAGCGTCAT-3′), CDKN1A (5′-CGATGGAACTTCGACTTTGTCA-3′,5′-GCACAAGGGTACAAGACAGTG-3′), MDM2 (5′-GAATCATCGGACTCAGGTACATC-3′, 5′-TCTGTCTCACTAATTGCTCTCCT-3′), CCNG1 (5′-GAGTCTGCACACGATAATGGC-3′, 5′-GTGCTTGGGCTGTACCTTCA-3′), GAPDH (5′-GGATTTGGTCGTATTGGGCG-3′, 5′-TCCTGGAAGATGGTGATGGGA-3′), CCNB1 (5′-TTGGGGACATTGGTAACAAAGTC-3′, 5′-ATAGGCTCAGGCGAAAGTTTTT-3′), MT-CYB(5′-GCCTGCCTGATCCTCCAAAT-3′, 5′-AAGGTAGCGGATGATTCAGCC-3′), APAF1 (5′-GTCACCATACATGGAATGGCA-3′, 5′-CTGATCCAACCGTGTGCAAA-3′), PERP (5′-CTTCACCCTTCATGCCAACC-3′, 5′-GCCAATCAGGATAATCGTGGC-3′), SESN1 (5′-CTACATTGGAATAATGGCTGCGG-3′, 5′-AGGTCTATGGGCTAACACTTTGTT-3′), RRM2B (5′-AGAGGCTCGCTGTTTCTATGG-3′, 5′-GCAAGGCCCAATCTGCTTTTT-3′), TP53I3 (5′-GCCAGGCTCAGTACGTCAC-3′, 5′-GTCCTGCATGGATTAGCACATAG-3′), GADD45A (5′-GAGAGCAGAAGACCGAAAGGA-3′, 5′-CAGTGATCGTGCGCTGACT-3′), RRM2 (5′-ATTGGGCCTTGCGATGGATAG-3′,5′-GAGTCCTGGCATAAGACCTCT-3′), FAS (5′-AGATTGTGTGATGAAGGACATGG-3′,5′-TGTTGCTGGTGAGTGTGCATT-3′), THBS1 (5′-GCCATCCGCACTAACTACATT-3′, 5′-TCCGTTGTGATAGCATAGGGG-3′), SERPINE (5′-GCACCACAGACGCGATCTT-3′, 5′-ACCTCTGAAAAGTCCACTTGC-3′), SESN2 (5′-AAGGACTACCTGCGGTTCG-3′, 5′-CGCCCAGAGGACATCAGTG-3′), SESN3 (5′-ACCTGCTCTGTACCAACTGC-3′, 5′-GACGACCGGATGTAGAGTATTCT-3′), ZMAT3 (5′-AGAAGCCTTTTGGGCAGGAG-3′, 5′-TGCTGCATAGTAATTTCGGAGTT-3′), PPM1D (5′-AGAAGCAGAAGGGTTTCACCT-3′, 5′-CATTCCGCCAGTTTCTTCCAC-3′) and PMAIP1 (5′-ACCAAGCCGGATTTGCGATT-3′, 5′-ACTTGCACTTGTTCCTCGTGG-3′). Intact RNA extracted from A549 cells transfected with QCR2 siRNA or NC siRNA was analyzed with an Affymetrix Gene Chip array (HTA-2_0).
2.8. Subcellular fractionation
Mitochondria isolation used for immunoprecipitation was performed using a Mitochondria Isolation Kit for Cultured Cells (cat. no. 89874; Thermo Scientific) according to the manufacturer's instructions. Fractions enriched in cytoplasm, nuclear and mitochondrial proteins were obtained from 2.0 × 107 cells as described previously [25].
2.9. Immunoprecipitation and mass spectrometry analysis
Immunoprecipitation was performed using a Pierce Crosslink Immunoprecipitation Kit (cat. no. 26147; Thermo Scientific) according to the manufacturer's instructions. The eluted fractions were subjected to liquid chromatography and tandem mass spectrometry (LC-MS/MS; Shanghai Applied Protein Technology, China).
2.10. GST pull-down assays
GST pull-down assays were performed using a GST protein interaction pull-down kit (cat. no. 21516; Thermo Scientific) according to the manufacturer's instructions. GST-p53, GST-QCR2 or GST-PHB plasmids were introduced into Escherichia coli BL21 strain, and the recombinant proteins were induced by the addition of 1 mM isopropyl-b-d-thiogalactoside at 30 °C for 6 h. HEK293 cells treated with PS-341 for 12 h were harvested, and 2 mg cell lysates were incubated with recombinant proteins bound to sepharose beads.
2.11. EdU proliferation and Cell Counting Kit-8 (CCK-8) assays
EdU labeling was carried out using an EdU Cell Proliferation Assay Kit (C00031; Ribobio, China) according to the manufacturer's instructions. Images were obtained using an Olympus BX53 fluorescence microscope. For CCK-8 assays, cells were seeded in a 96-well plate with cell density of 4 × 104/mL with 100 μL medium in each well. After incubation for the indicated times, CCK-8 reagent (cat. no. CK04; Dojindo Laboratories) was added to each well, and cells were incubated for 1 h at 37 °C. The absorbance was measured using an enzyme-labeled meter at 490 nm to calculate cell growth rate.
2.12. Real-time PCR for mitochondrial DNA
42 tissue samples were obtained from patients during surgery in Tongji Hospital (Wuhan, China) and made into paraffin sections. DNA of cervix cancer tissues was extracted using QIAamp® DNA FFPE Tissue Kit according to manufacturer's instructions (QIAGEN). RT-qPCR was used for the amplification of mtDNA. The mtDNA amplification was determined by the following primers, 5′-ATGGCCAACCTCCTACTCCTCATT-3′ [26]. Quantitative mtDNA amplification data was normalized to GAPDH as an internal reference gene. The RT-qPCR was initiated with 3 min at 95 °C, followed by 45 cycles of 10 s at 95 °C and 30 s at 60 °C.
2.13. Reagents and antibodies
PS-341 (cat. no. 1846-1) was purchased from BioVison. Cycloheximide (CHX, C8030–100) was purchased from Solarbio. Doxorubicin hydrochloride (D1515-10MG) was purchased from Sigma-Aldrich. Dorsomorphin (Compound C) and GSK621 were obtained from Selleck. Antibodies used in this study were listed with the source in parentheses - anti-QCR2 (14742-1-AP, Proteintech), anti-GAPDH (10494-1-AP, Proteintech), anti-p53 (10442–1-AP, Proteintech), anti-p21 (10355-1-AP, Proteintech), anti-Flag (AF0036, Beyotime), anti-PHB (10787-1-AP, Proteintech), anti-Ubiquitin (BML-PW8390-0100, Enzo), anti-PHDA1 (ab110330, Abcam), anti- AMPK (5831T, CST), anti-p-AMPK (2535T, CST), anti-PCNA (10205-2-AP, Proteintech), anti-α-Tubulin (11224-1-AP, Proteintech), anti-PHB2 (12295-1-AP, Proteintech). Flag Agarose (PM020-8) used for immunoprecipitation was obtained from Medical & Biological Laboratories.
2.14. Plasmids and lentiviral constructs
For overexpression of QCR2, a recombinant adenovirus vector expressing QCR2 (GenBank accession number NM_003366) or empty pcDNA control was provided by Vigene Biosciences (China). For overexpression of PHB, the full-length cDNA-encoding PHB whose c-terminal was fused with a cDNA fragment encoding flag was inserted into pcDNA3.1 vector (Invitrogen). A series of plasmids that encode different fragments of p53, QCR2 or PHB were constructed by inserting fragments generated by PCR and cloned into pGEX-4 T-1. For stable transfection of QCR2, pre-designed shRNA lentiviral particles were obtained from Genechem, the shRNA sequence (the targeting sequence: 5′-CAGACTCATGTCATTGAAA-3’) was inserted into GV344 (hU6-MCS-Ubiquitin-Luc_firefly-IRES-puromycin). For stable transfection of PHB, pre-designed shRNA lentiviral particles were obtained from Genechem, and the shRNA sequence (the targeting sequence: 5′-CAGAAATCACTGTGAAATT-3′) was inserted into GV344 (hU6-MCS-Ubiquitin-Luc_firefly-IRES-puromycin). For the control lentiviral, the sequence of 5′-TTCTCCGAACGTGTCACGT-3′ was inserted into GV344 (hU6-MCS-Ubiquitin-Luc_firefly-IRES-puromycin).
2.15. Senescence-associated-β-Galactosidase (SA-β-Gal) staining
SA-β-Gal staining was performed using a Senescence β-Galactosidase Staining Kit (C0602, Beyotime Biotechnology, China) according to the manufactory's protocols.
2.16. Cell synchronization and cell cycle analysis
Cells were serum-starved for 12 h and then re-stimulated with 10% FBS and paclitaxel containing-medium for the indicated time points. Cell cycle distribution was determined as previously described [27].
2.17. Immunofluorescence
HeLa cells were transfected with NC siRNA or QCR2 siRNA-2 for 96 h, and incubated with MitoTracker™ Red FM (cat. no. M22425, Thermo Fisher Scientific) for 45 min under standard conditions. Then cells were fixed in 4% PFA, permeabilized in 0.2% Triton X-100 for 15 min at room temperature, and stained with a rabbit anti-PHB antibody at 4 °C overnight, followed by secondary antibody labeling with an anti-rabbit AlexaFluor-488 for 60 min at room temperature. Then cells were stained with 4′, 6-diamidino-2-phenylindole at room temperature. Images were acquired using an Olympus FV1000 confocal laser scanning microscope.
2.18. Cell apoptosis
Floating cells and attached cells were collected together and then cell apoptosis was determined by an Annexin V FITC Apoptosis Detection Kit (cat. no. 556547, BD Pharmingen) using flow cytometry according to manufacturer's instructions. Cells were analyzed using a BD FACSCalibur flow cytometer (San Jose, CA, USA).
2.19. Statistics
Data were presented as the mean ± standard deviations (SDs). Statistical analyses between experimental groups were conducted using Student's t-tests. The correlation between QCR2/PHB expression and p21 expression was determined by Pearson's correlation tests.
2.20. Data sets
Microarray data have been deposited in the Gene Expression Omnibus under accession no. GSE87850.
3. Results
3.1. QCR2 expression is elevated in human cancer tissues
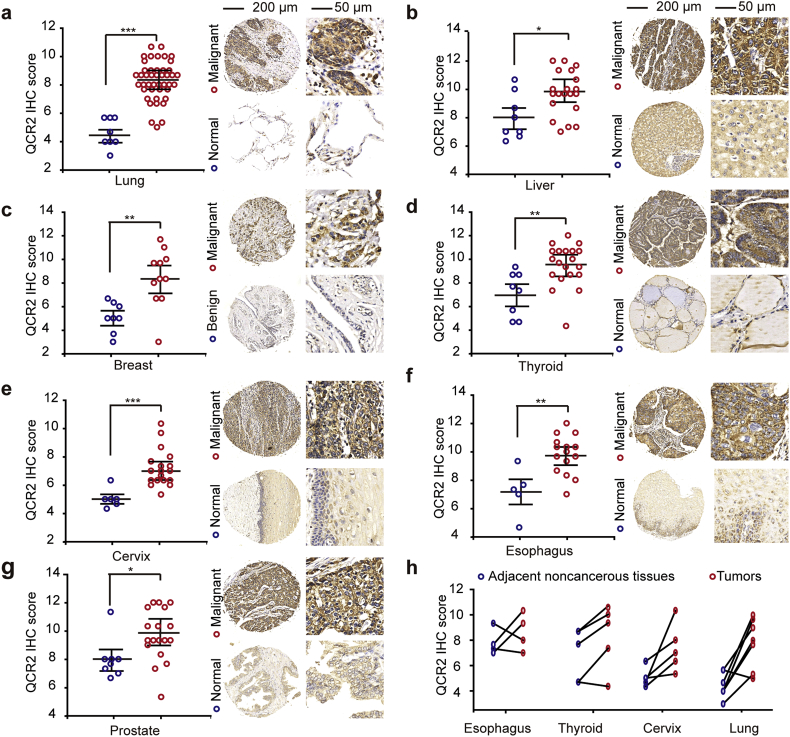
Our previous study showed that QCR2 is required for TSA-induced apoptosis in tumor cells using SMART [7]. In line with the previous conclusion, QCR2 was located to mitochondria (Fig. S1a and b). However, it remained unclear if this mitochondrial protein might play a role in tumorigenesis. To address this issue, we first used immunohistochemistry (IHC) in tissue microarrays to determine whether QCR2 levels are altered in human tumors. We found that QCR2 was located in cytoplasm in the tumor specimens (Fig. S1c). And QCR2 protein levels were significantly higher in human lung, liver, breast, thyroid, cervical, esophageal and prostate tumor specimens when compared to normal specimens (Fig. 1a–g). Further analysis of 22 pairs of human tumor tissues and their adjacent normal tissues from the esophagus, thyroid, cervix, and lungs showed that the protein level of QCR2 was elevated in >80% of cancer specimens (two of four cases of esophageal cancer, four of five cases of thyroid cancer, five of five cases of cervical cancer, and seven of eight cases of lung cancer; Fig. 1h). These results demonstrate that overexpression of QCR2 is a general event in several types of human cancers and suggest that it might play an oncogenic role.
Fig. 1.
QCR2 protein is upregulated in seven different types of human cancers. (a–g) The protein expression of QCR2 in normal/benign and malignant tissues was assessed by IHC using a tissue microarray consisting of various normal and malignant tissues (*p < .05, **p < .01, ***p < .001, student's t-test). Left: scot graph showing the statistical results; right: representative IHC images (100×, scale bar = 200 μm, 400×, scale bar = 50 μm). (h) QCR2 protein expression in pairs of tumors and adjacent noncancerous tissues derived from the esophagus, thyroid, cervix, and lungs.
Mitochondria number is loss in several types of human cancers and contributes to cancer progression [28]. And the amount of mitochondrial DNA per individual mitochondrion is essentially constant [29]. To find the relation between QCR2 expression and mitochondria number, mtDNA copy numbers of resected specimens from 42 cervical cancer patients were measured by quantitative real-time PCR analyses. As shown in Fig. S2a, there was no relation between QCR2 expression and mitochondria number in cervical cancer tissues in which QCR2 expression was evaluated by IHC (Fig. S2b). The results indicate that QCR2 overexpression is not caused by mitochondrial number loss.
3.2. Silencing of QCR2 inhibits cancer cell growth by inducing G0/G1 arrest and cellular senescence
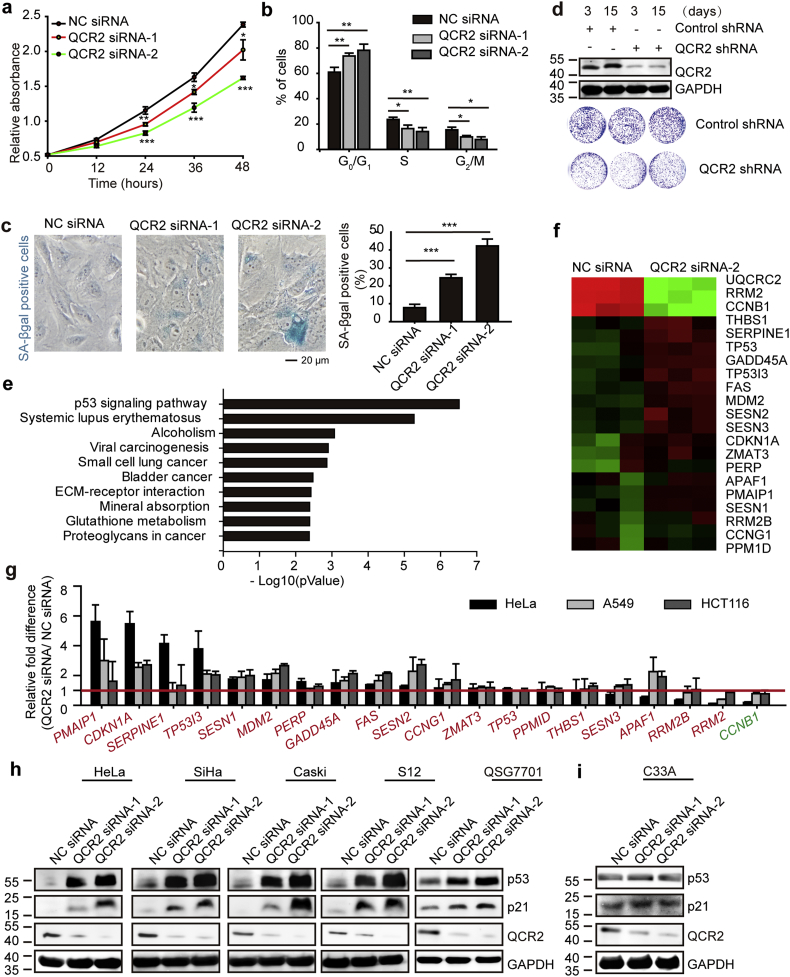
To test the possible oncogenic role of QCR2, we first determined if QCR2 is required for cancer cell survival and proliferation by silencing its expression via siRNA. Indeed, silencing of QCR2 in human cervical carcinoma HeLa cells significantly inhibited cell viability, as assessed using CCK-8 (Fig. 2a) and EdU proliferation assays (Fig. S3a), respectively. Consistently, knockdown of QCR2 also induced profound G0/G1-phase cell cycle arrest, regardless of the asynchronous or synchronous nature of cancer cell cultures (Fig. 2b and Fig. S3b) and marked senescence (Fig. 2c) as detected by morphology and quantified by fluorescence-assisted cell sorting (FACS) analysis (Fig. S4) as well. In line with these results, knockdown of QCR2 drastically reduced HeLa cell colony formation (Fig. 2d). Similar results were obtained in S12 cells (an immortalized human cervical keratinocyte cell line containing HPV DNA as integrated copies), HCT116 cells (colorectal cancer), A549 cells (lung cancer), and MCF-7 cells (breast cancer) as well as SiHa and Caski cells (cervical cancer) (Fig. S5a–f). Also, upregulation of QCR2 promoted cell growth in HCT116 and A549 cells (Fig. S6a and b). These results demonstrate that QCR2 is required for the proliferation and viability of a variety of human cancer cells.
Fig. 2.
Knockdown of QCR2 inhibits cancer cell proliferation. (a–c) HeLa cells were transfected with NC siRNA, QCR2 siRNA-1, or QCR2 siRNA-2. Equal numbers of cells were plated in 96-well plates in triplicate, and viable cell proliferation was assessed using CCK-8 assays (*p < .05, **p < .01, ***p < .001, student's t-test) (a). Cell cycle distribution of asynchronous cells obtained from FACS analysis: cells were transfected with NC siRNA, QCR2 siRNA-1, or QCR2 siRNA-2, and then cultured under standard conditions. After 72 h, the cells were collected and analyzed by FACS. The bar graph represents quantification analyses of cell cycle distribution analysis (*p < .05, ***p < .001, student's t-test) (b). SA-β-gal staining of cells (scale bar = 20 μm) and the statistical result (***p < .001, student's t-test) (c). (d) Western blotting for QCR2 in HeLa cells expressing a lentiviral vector harboring QCR2 shRNA or control shRNA for 3 and 15 days (upper panels). Cells (1 × 103) with or without QCR2 knockdown were plated in 6-cm dishes (lower panels), cultured for 10 days, and stained with crystal violet. (e, f) The transcriptional profiles of A549 cells transfected with NC siRNA or QCR2 siRNA-2 were analyzed by a cDNA microarray. The bar graph represents the specific categories of genes within KEGG, for which P values were <0.005 (e). The heat map shows the representative genes belonging to the p53 signaling pathway within KEGG that were differently expressed in QCR2-knockdown cells (f). (g)The different expression of 20 genes belonging to the p53 signaling pathway within KEGG was tested by RT-qPCR. The red words were the genes activated by p53 within KEGG. The green words were the gene inhibited by p53 within KEGG, (h, i) Western blotting for p53 and p21 after transfection of cells with NC siRNA, QCR2 siRNA-1, or QCR2 siRNA-2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To explore the underlying molecular events and mechanisms for QCR2-dependent cell proliferation and survival, we analyzed gene expression profiles of A549 cells transfected with nontargeted control (NC) siRNA or QCR2 siRNA-2 using a cDNA microarray. Interestingly, bioinformatics analysis of the gene expression profiles revealed that a number of genes in the p53-responsive pathway were differentially expressed in siQCR2 cells, compared to control siRNA cells (Kyoto Encyclopedia of Genes and Genomes [KEGG]; Fig. 2e). The heat map showed that 20 genes that belong to the p53 signaling pathway within KEGG were differentially expressed when QCR2 was knockdown (Fig. 2f). This result was validated by RT-qPCR analysis in A549, HeLa, and HCT116 cells, all of which contain wild type p53 (Fig. 2g). In particular, p21, a gene involved in G0/G1-phase cell cycle arrest and cellular senescence, was markedly overexpressed (Fig. 2g) [30]. Accordingly, the protein levels of p53 and p21 were also upregulated when QCR2 was downregulated in HeLa, SiHa, Caski, S12, and QSG7701 cells (a hepatic cell line of human origin), as well as four other cancer cell lines harboring wild-type p53, including A549, HepG2, HCT116, and MCF-7 cells (Fig. 2h and Fig. S7a). Consistent with these findings, overexpression of QCR2 via adenovirus (QCR2-Flag Ad) resulted in downregulation of p53 and p21 proteins (Fig. S7b). These data strongly suggest that QCR2 could inhibit p53 activity. In contrast, knockdown of QCR2 did not alter levels of p53 and p21 in a mutant p53 (p53R273C)-carrying C33A cervical cancer cell line (Fig. 2i). Together, these results demonstrate that knockdown of QCR2 leads to the activation of the p53 pathway, while overexpression of QCR2 inhibits this pathway, and also suggest that QCR2 may regulate cell cycle and proliferation by controlling the p53 pathway.
3.3. QCR2 negatively acts on p53 by promoting its ubiquitination and degradation
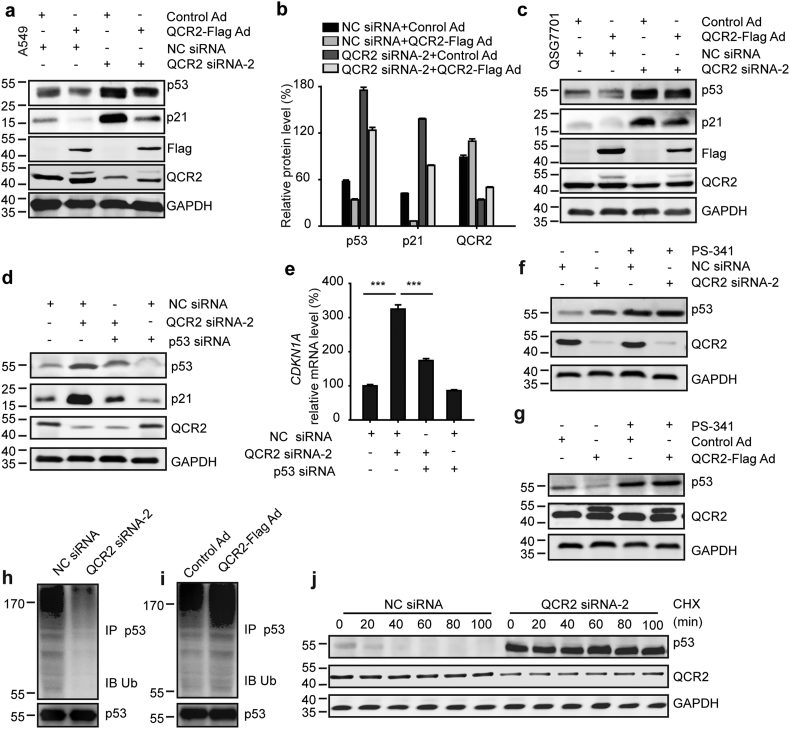
To understand how QCR2 might act on p53 expression, we conducted the following set of experiments. First, we tested if overexpression of QCR2 can reverse the induction of p53 by knocking down this mitochondrial protein. Indeed, this was the case as overexpression of Flag-QCR2 markedly reduced not only the basal levels, but also the increased levels of endogenous p53 and p21 in response to knockdown of endogenous QCR2 (Fig. 3a and b). Similar results were also obtained with QSG7701 (Fig. 3c). The p21 induction by QCR2 knockdown was p53-dependent, as knockdown of p53 in cancer cells impaired the induction of p21's protein and mRNA levels (Fig. 3d and e). By contrast, QCR2 had no effects on p21 levels in C33A cells expressing p53R273C or p53-null H1299 cells (Fig. 2i and Fig. S8a–c). Together, these results demonstrate that knockdown of QCR2 indeed leads to the activation of p53 and its pathway, and suggest that QCR2 might be involved in regulation of p53 level and activity in normal and cancer cells.
Fig. 3.
QCR2 is associated with p53 destabilization. (a) Western blotting for p53 and p21 in A549 cells transfected or infected with NC siRNA and Control Ad, NC siRNA and QCR2-Flag Ad, QCR2 siRNA-2 and Control Ad, or QCR2 siRNA-2 and QCR2-Flag Ad. Control Ad: adenovirus with an empty pcDNA vector control; QCR2-Flag Ad: adenovirus with a vector harboring QCR2-Flag. (b) Quantitative analysis of p53, p21, and QCR2 protein levels from the western blotting results shown in (a) using ImageJ. (c) Western blotting for indicated proteins in the hepatic cell line QSG7701 cells treated as described in (a). (d) Western blotting for p53, p21, and QCR2 in A549 cells transfected with NC siRNA, NC siRNA and QCR2 siRNA-2, QCR2 siRNA-2 and p53 siRNA, or NC siRNA and p53 siRNA. (e) RT-qPCR analysis for CDKN1A mRNA in cells treated as described in (c) (***p < .001, student's t-test). (f, g) A549 cells were transfected with NC siRNA or QCR2 siRNA-2 (f) and Control Ad or QCR2-Flag Ad (g) in the presence or absence of a proteasome inhibitor, PS-341. The transfected cells were analyzed by western blotting for p53 and QCR2. (h, i) A549 cells were transfected with NC siRNA or QCR2 siRNA-2 (h) and Control Ad or QCR2-Flag Ad (i). The cell lysates were immunoprecipitated with an anti-p53 antibody, and immunocomplexes were analyzed by western blotting for p53 and ubiquitin (Ub). (j) A549 cells were treated with 1 μg/μL of a protein synthesis inhibitor, cycloheximide (CHX), at the indicated time points after transfection with QCR2 siRNA-2 or NC siRNA for 72 h. Proteins were extracted and subjected to western blotting for p53, QCR2, and GAPDH.
It has been shown that p53 is mainly regulated at its posttranslational level via ubiquitination-dependent proteasomal mechanisms [31]. Our results also showed that QCR2 knockdown or overexpression leads to the increase of p53 protein, but not mRNA (Figs. S9a and S9b), suggesting that QCR2 might have an effect on p53 stability. Also, treatment of A549 cells with PS-341, a 26S proteasome inhibitor, nullified the effects of QCR2 manipulation on the p53 protein levels, suggesting that QCR2 reduces p53 levels via the ubiquitin-proteasome pathway (Fig. 3f and g) [32,33]. Furthermore, the abundance of polyubiquitinated p53 was decreased following knockdown of QCR2 in A549 and HCT116 cells (Fig. 3h and Fig. S9c). Additionally, p53 protein appeared to be heavily ubiquitinated when QCR2 was overexpressed in A549 and HCT116 cells (Fig. 3i and Fig. S9d). Also, the half-life of endogenous p53 was remarkably prolonged when QCR2 was knocked down in A549 and HeLa cells (Fig. 3j and Fig. S9e). Besides, in HeLa cells harboring low level of p53, QCR2-Flag Ad reduced the upregulated p53 expression and increased the downregulated ubiquitination induced by DDP (Figs. S10a and S10b). Taken together, these results demonstrate that QCR2 can promote p53 polyubiquitination and proteasomal turnover in cells.
3.4. QCR2 interacts with PHB, a chaperone of p53
QCR2 is a component of the ubiquinol-cytochrome c reductase complex (complex III, CIII) and required for the CIII assembly. Next, we examined effects of downregulation of nine subunits of CIII, as well as five other subunits (UQCC1, UQCC2, TTC19, LYRM7, and BSC1L) involved in the integrated activity of mitochondrial complex III, on p53 levels in HeLa and A549 cells [34,35]. Surprisingly, knockdown of any of these proteins, other than QCR2, did not alter p53 protein levels (Fig. S11a), suggesting that the regulation of p53 stability is specific to QCR2.
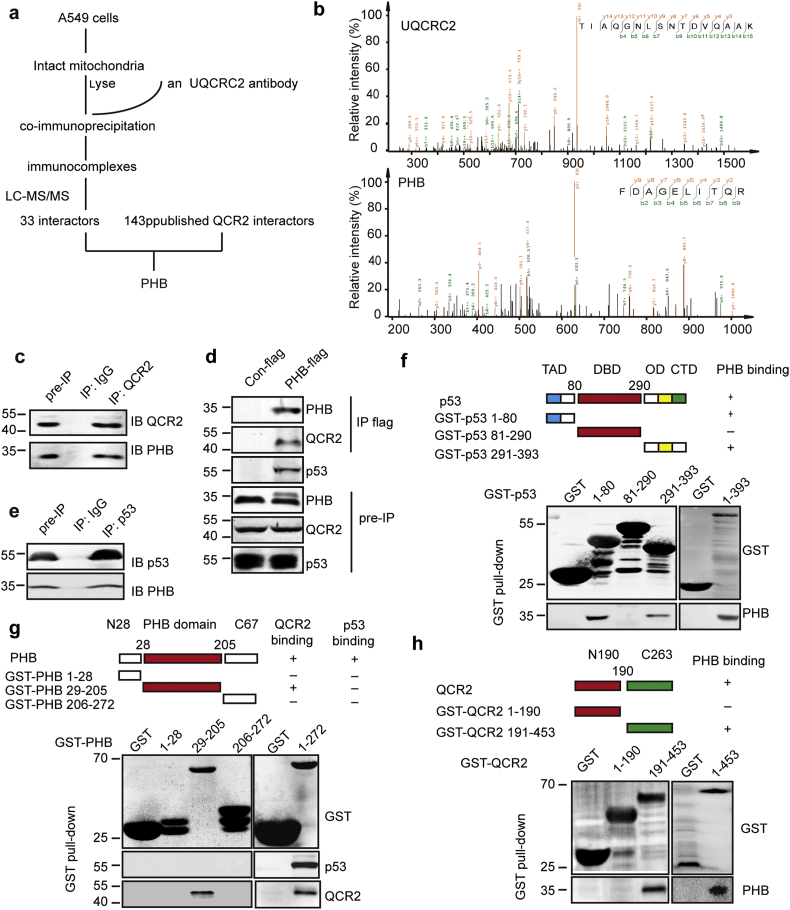
To understand the mechanism for how QCR2 might mediate p53 degradation, we aimed to identify proteins interacting with QCR2. To this end, intact mitochondria were isolated from A549 cells for immunoprecipitation using anti-QCR2 antibodies, and immunocomplexes were analyzed by LC-MS/MS. Thirty-three candidates were identified (Fig. 4a). Interestingly, among them, PHB was the only QCR2-binding protein via bioinformatics analysis of 143 known QCR2-binding proteins (Fig. 4a–b and Fig. S12a), which was previously shown to bind to p53 and enhance its transcriptional activity [[36], [37], [38]].
Fig. 4.
QCR2 interacts with PHB, a p53 chaperone. (a) Flowchart to identify proteins interacting with QCR2: intact mitochondria from A549 cells were isolated, and the mitochondrial lysates were used for co-immunoprecipitation studies using an anti-QCR2 antibody or control IgG. Immunocomplexes were analyzed by LC-MS/MS. Combined with the analysis with 143 published UQCRC2/QCR2 interactors, PHB was identified as a potential QCR2 target protein. (b) The spectra of QCR2 and PHB obtained by LC-MS/MS. (c) Intact mitochondria from A549 cells were isolated, and the mitochondrial lysates were used for co-immunoprecipitation studies using an anti-QCR2 antibody or control IgG. Immunocomplexes were analyzed by western blotting for QCR2 and PHB. (d) Plasmids encoding Con-flag or PHB-flag were transfected into HCT116 cells. Whole cell lysates were then used for co-immunoprecipitation studies using an anti-flag antibody. Cell lysates and immunocomplexes were analyzed using anti-QCR2, anti-p53 and anti-PHB antibodies. (e) HCT116 cells treated with PS-341 for 4 h were isolated, and the lysates were immunoprecipitated using an anti-p53 antibody or control IgG. Immunocomplexes were analyzed by western blotting for p53 and PHB. (f) Mapping the PHB binding domain on p53 by GST pull-down. Purified GST-p53 proteins were incubated with cell extracts prepared from 293 cells and then detected by western blotting for PHB. (g) Mapping the QCR2 binding domain on PHB by GST pull-down. Purified GST-PHB proteins were incubated with cell extracts prepared from 293 cells treated with PS-341 and detected by western blotting for QCR2 and p53. (h) Mapping the PHB binding domain on QCR2 by GST pull-down. Purified GST-QCR2 proteins were incubated with cell extracts prepared from 293 cells and then detected by western blotting for PHB.
The interaction between endogenous QCR2 and PHB in A549 cells was confirmed by co-immunoprecipitation (IP) with an anti-QCR2 antibody (Fig. 4c), which was also reproduced using exogenous Flag-tagged PHB and endogenous QCR2 (Fig. 4d). In addition, the interaction between p53 and PHB was detected by co-IP-WB analysis (Fig. 4e). We then mapped their binding domains by performing a set of GST pull-down assays. PHB was specifically pulled down with the transactivation domain (TAD; amino acids 1–80) and the C-terminal region (amino acids 291–393) of p53 (Fig. 4f). However, none of the PHB fragments, including N28 (amino acids 1–28), PHB domain (amino acids 29–205) and C67 (amino acids 206–272), was found to interact with p53, suggesting that PHB requires its full-length protein to interact with p53 (Fig. 4g). Using the same approach, we also found that QCR2 interacts with the PHB domain (amino acids 29–205) of PHB (Fig. 4g), while PHB interacts with a 191–453 amino acid fragment at C-terminal QCR2 (Fig. 4h). These results demonstrate that the C-terminal domain of QCR2 binds to PHB's central domain directly and validate the interaction between PHB and p53, which requires the entire PHB and the N-terminal domain and C-terminal domain of the latter.
3.5. PHB is necessary for QCR2-induced degradation of p53
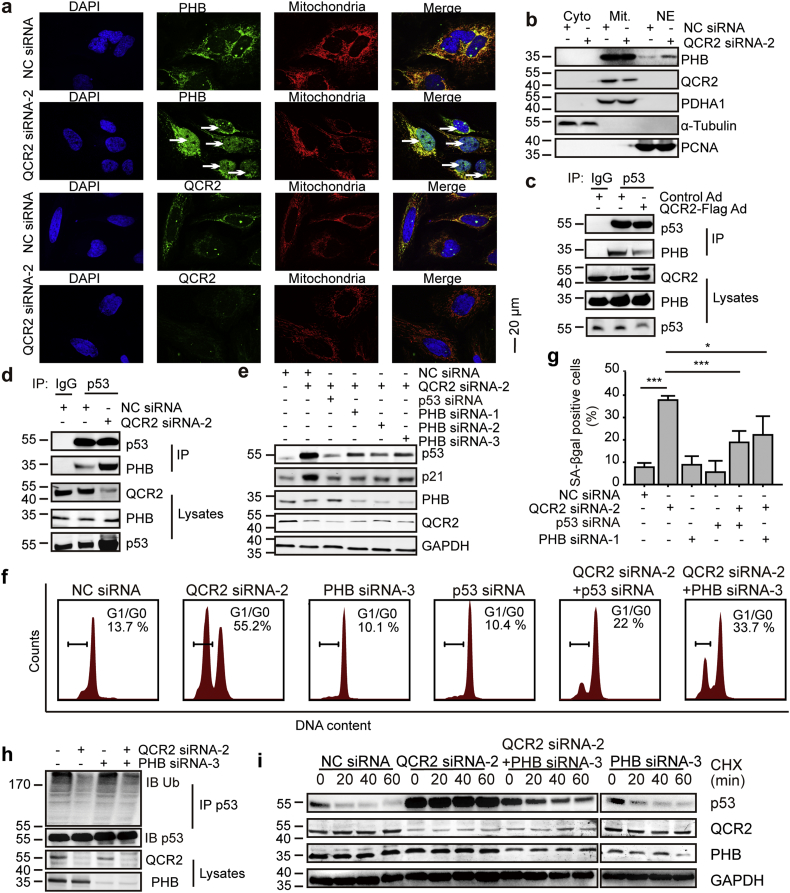
The results as shown above indicated that PHB can form a complex with QCR2 and p53. We therefore tested if PHB might participate in QCR2 regulation of p53. As a protein bind to QCR2, PHB was mainly detected in mitochondrial-enriched extracts (Fig. S13a). Using immunofluorescence studies, we observed that QCR2 was located to mitochondria and knockdown of QCR2 facilitated the transfer of PHB from the mitochondria to the nucleus in HeLa cells and A549 cells (Figs. 5a and S14a). Consistently, more PHB was detected in nuclear-enriched extracts when cells were transfected with QCR2 siRNA-2 (Figs. 5b and Fig. S14b). The interaction between QCR2 and PHB was enhanced when QCR2 was overexpressed (Fig. S15a). Also, overexpression of Flag-QCR2 markedly attenuated the interaction between PHB and p53, while knockdown of QCR2 enhanced the interaction (Fig. 5c and d). Furthermore, silencing of PHB by three individual PHB-specific siRNAs, similar to p53 silencing, abrogated the induction of p53 by QCR2 knockdown (Fig. 5e). Correspondingly, the induced expression of p53 target genes, such as CDKN1A, TP53I3, and MDM2, upon QCR2 silencing was also suppressed by knocking down PHB (Fig. S16a). Consistently, PHB-specific siRNA rescued G0/G1-phase cell cycle arrest and decreased S-phase DNA synthesis induced by QCR2 knockdown (Fig. 5f and Fig. S16b). Either PHB knockdown or p53 knockdown abrogated the senescence caused by QCR2 knockdown (Fig. 5g). These results indicate that PHB is required for p53 activation that is induced by QCR2 signaling. As PHB2 was another subunit of prohibitin complex and deletion of PHB2 results in the absence of PHB [39], knockdown of PHB2, just like knockdown of PHB, also abrogated the effect of QCR2 on p53 (Fig. S17a and b). Given that PHB2 interacted with AMPK which was an important regulator of p53, we detected the activated AMPK in cells transfected with QCR2 siRNA-2, QCR2 siRNA-2 and PHB siRNAs or PHB2 siRNAs. We found QCR2 knockdown not only upregulated the expression of p53, but also activated AMPK. However, the activation of AMPK by QCR2 knockdown was not suppressed by PHB or PHB2 knockdown (Figs. S17b and S17c). At last, we transfected A549 cells with QCR2 siRNA and 24 h later cells were treated with an AMPK activator GSK621 and an AMPK inhibitor Dorsomorphin. The result showed that, neither GSK621 nor Dorsomorphin eliminated the upregulation of p53 by siQCR2 (Fig. S17d), which indicating that the function of PHB on p53 was not dependent on AMPK activity.
Fig. 5.
PHB is indispensable for QCR2-dependent p53 signaling and cell cycle arrest. (a) After transfection of HeLa cells with NC siRNA or QCR2 siRNA-2, the cells were used for immunofluorescence studies for nuclei (DAPI, blue), PHB or QCR2 (green), and mitochondria (MitoTracker, red). Arrows indicate nuclear translocation of PHB (scale bar = 20 μm). (b) Western blots of HeLa extracts fractionated into cytoplasm, nuclear and mitochondrial fractions after transfection with siRNAs for the indicated protein. PCNA as a nuclear marker, PDHA1 as a mitochondria maker and α-tubulin as a cytoplasm marker. (c, d) Intact nucleus from HCT116 cells transfected with Control Ad or QCR2-Flag Ad (c) and NC siRNA or QCR2 siRNA-2 (d) were isolated, and the lysates were immunoprecipitated using an anti-p53 antibody or control IgG. Immunocomplexes were analyzed using anti-p53 and anti-PHB antibodies. (e) HeLa cells were transfected with the indicated siRNAs for 96 h, and cell lysates were analyzed by western blotting for indicated proteins. (f) HeLa cells were transfected with the indicated siRNAs for 96 h, and cells serum-starved for 12 h (starved) and then restimulated with 10% FBS and taxol containing-medium for 24 h were assessed for cell cycle distribution via FACS analysis. (g) The bar graph represents quantification analyses of SA-β-gal positive HeLa cells transfected with the indicated siRNAs (*p < .05, ***p < .001, student's t-test, n = 3). (h) A549 cells were transfected with the indicated siRNAs, and lysates were immunoprecipitated with an anti-p53 antibody. The resulting immunocomplexes were analyzed by western blotting using anti-p53 and anti-Ub antibodies. (i) A549 cells were transfected with siRNAs, followed by treatment with 1 μg/μL CHX for the indicated time points 72 h later. Endogenous p53 expression was examined by western blotting. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Since p53's mRNA levels were not altered in cells transfected with PHB siRNA, regardless of QCR2 silencing (Fig. S18a), and PS-341 blocked the suppression of p53 activation in response to QCR2 knockdown by silencing PHB, the regulation of p53 by PHB upon QCR2 knockdown was at posttranslational, but not transcriptional level, highly likely through regulation of the ubiquitination-dependent proteasome system (Fig. S18b). Indeed, the reduced polyubiquitination of p53 upon QCR2 knockdown was reversed by silencing PHB (Fig. 5h). Also, knockdown of PHB led to the shorter half-life of p53, which was extended upon QCR2 knockdown (Fig. 5i).
Taken together, these results strongly suggest that PHB is involved in the negative regulation of QCR2-promoted p53 proteasomal degradation, though the detailed mechanism(s) remains unclear.
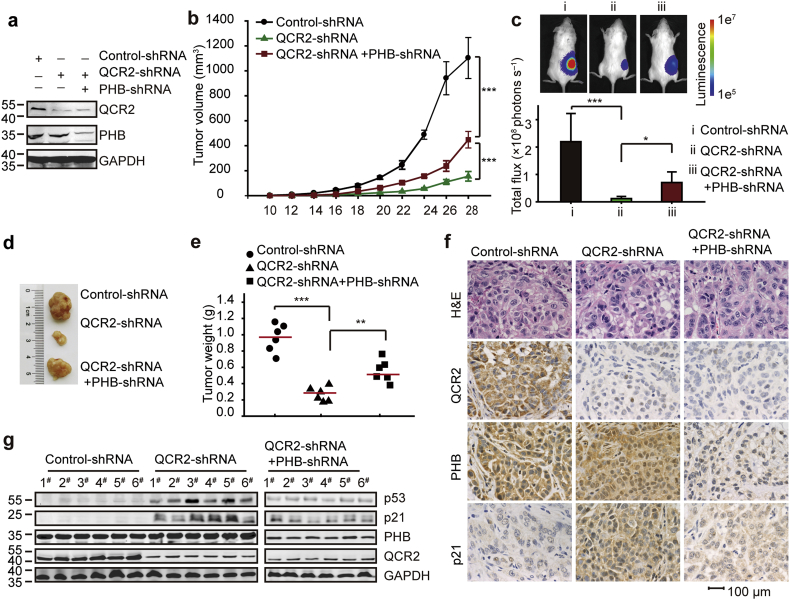
3.6. Silencing of QCR2 leads to PHB-dependent inhibition of tumor growth in vivo
To further determine if QCR2 and PHB might affect tumorigenesis by regulating p53 in vivo, we injected luciferase labeled and wt p53-harboring A549 cells, which were stably transfected with QCR2 shRNA or both QCR2 shRNA and PHB shRNA (Fig. 6a), subcutaneously into severe combined immunodeficiency (SCID) mice, and then monitored tumor growth by measuring subcutaneous tumors periodically. Interestingly, knockdown of QCR2 led to the marked decrease of xenograft tumor growth, which was partially, but significantly, reversed by further knocking down PHB (Fig. 6b–e). The reduction of tumor growth must be due to the induction of p53 and its activity by QCR2 siRNA, as knocking down QCR2 induced the protein levels of p53 and p21, whereas further knockdown of PHB reduced this induction as examined by IHC staining (Fig. 6f) and Western blot (WB) analysis (Fig. 6g). These results demonstrate that QCR2 and PHB play opposite roles in regulation of p53 and tumor growth, which is in line with their functions in regulation of p53 stability and activity as shown above.
Fig. 6.
Effects of QCR2 and PHB expression on cancer cell proliferation in vivo. A total of 1 × 106 luciferase labeled A549 cells expressing QCR2-shRNA, QCR2-shRNA and PHB-shRNA, or empty vector were subcutaneously injected into SCID mice (n = 6 per group). (a) Western blotting analysis of QCR2 and PHB expression levels following stable knockdown for QCR2 and/or PHB. (b) Tumor volumes (mm3) were estimated using calipers for 4 weeks after tumor cell injections (***p < .001, student's t-test, n = 6). (c) All mice were monitored by the in vivo luciferase imaging system (IVIS Lumina) at 28 days after injections. Representative images are shown (top panel), and luciferase intensities (total flux) are shown as means ± SDs (*p < .05, ***p < .001, student's t-test, n = 6). (d) Tumors were excised at 28 days after injections and photographed. (e) Tumor weights (g) were measured. Red bars indicate the means, and data were analyzed using Student's t-tests (**p < .01, ***p < .001, student's t-test, n = 6). (f) Representative images of H&E staining and IHC for QCR2, PHB, and p21 using tumors developed in mice (scale bar = 20 μm). (g) Six tumor samples from each group were analyzed for the protein expression of p53, p21, PHB, QCR2, and GAPDH by western blotting.
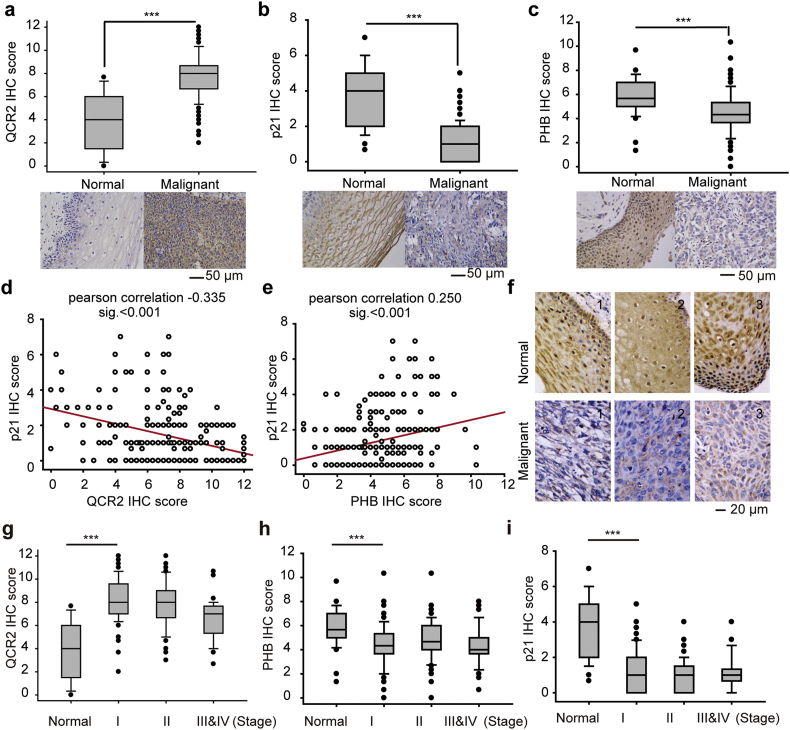
3.7. QCR2 overexpression and PHB downregulation are associated with reduced p21 in cervical cancer
To translate the information gained from the above studies into clinically significance, we enrolled 270 cervical cancer specimens and 34 normal cervical tissues. Because it is difficult to detect p53 owing to its short half-life partially due to HPV infection [40], we examined p21 levels as well as QCR2 and PHB levels by IHC. As a result, we found that QCR2 is highly expressed in cervical cancer tissues compared to normal cervical tissues (p < .001; Fig. 7a). By contrast, p21 and PHB levels were significantly lower in cervical cancer tissues than that in normal cervical tissues (p < .001; Fig. 7b and c). The negative correlation between the elevated QCR2 level and the low or undetectable p21 level was further confirmed by Pearson analysis (r = −0.335, p < .001; Fig. 7d). Conversely, a positive correlation between the expression levels of PHB and p21 was also obtained using the same analysis (r = 0.250, p < .001; Fig. 7e).
Fig. 7.
Differential expression of QCR2, PHB, and p21 in normal cervical tissues and cervical cancer tissues. (a–c) Top panels: quantitative analysis of IHC staining for QCR2 (a), p21 (b), and PHB (c) using normal cervical tissues and cervical cancer tissues. Lower panels: representative IHC staining of normal and cervical cancer tissues for QCR2 (a), p21 (b), and PHB (c) (*p < .05, **p < .01, ***p < .001, student's t-test, scale bar = 50 μm). (d) IHC scores for QCR2 and p21 were used to test the inverse correlations between levels of p53 and QCR2 proteins using Pearson correlation tests. Pearson correlation: −0.335; significance: p < .001. (e) IHC scores for PHB and p21 were used to test the positive correlations between levels of p53 and PHB proteins using Pearson correlation tests. Pearson correlation: 0.250; significance: p < .001. (f) Representative IHC images showing that PHB was located to both the nucleus and the cytoplasm in normal cervical tissues but mainly to the cytoplasm in cervical cancer tissues (scale bar = 20 μm). (g–i) Summary of IHC results showing differential expression of QCR2 (g), PHB (h), and p21 (i) in normal tissues and cervical cancers with different FIGO clinical stages (***p < .001, student's t-test).
Interestingly, PHB was detected in both the nucleus and the cytoplasm of normal cervical tissues, but only in the cytoplasm of cervical cancer specimens. These results suggest that PHB might be imported to the nucleus, where it functions to protect p53 in normal cells, but translocated to the cytoplasm in response to QCR2 upregulation during tumorigenesis (Fig. 7f). Intriguingly, the upregulation of QCR2 and downregulation of PHB and p21 appeared to occur during the early stages of cervical cancer progression (Figs. 7g–i). These results, consistent with the cellular and xenograft results (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6), suggest that QCR2 and PHB might play opposite roles in regulation of the growth of human cervical carcinoma, perhaps by controlling p53 activity as presented by the expression of p21.
4. Discussion
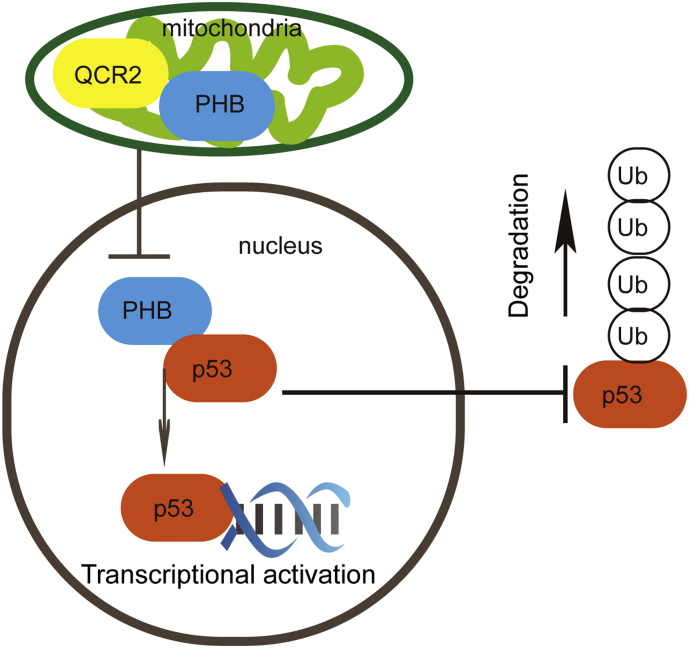
Although p53 has been reported to modulate mitochondrial activity, it has been seldom investigated whether mitochondrial proteins might regulate p53 level and activity [16,41]. In this study, we uncovered QCR2 as the first mitochondrial protein that can regulate p53 stability and activity, which can be reversed by another mitochondrial protein PHB, though the latter was previously shown as a p53 chaperone (Fig. 8). QCR2 might directly binds to PHB in the mitochondria, preventing PHB from binding to p53. Conversely, QCR2 knockdown leads to PHB translocation from the mitochondria to the nucleus where PHB might activate p53 by binding to and stabilizing it. Therefore, our results reveal the inverse regulations of p53 stability and activity by QCR2 and PHB.
Fig. 8.
Working model of QCR2 regulation of p53.
It has been shown that p53 maintains the balance between cell proliferation and death, acting as a barrier to cancer progression [42]. Our study also showed that the p53 signature pathway is negatively regulated by QCR2 through cDNA microarray analysis. This conclusion is further supported by our results obtained from a set of cancer cell lines, an in vivo xenograft model, and clinical specimens. HPV infection is an important factor leading to p53 degradation via the interaction between E6 oncoprotein that is encoded by the HPV viral gene and p53 in cervical cancer [43]. The negative regulation of p53 stability by QCR2 must be independent of E6, as this was shown in a panel of p53-containing cancer cell lines regardless of the HPV status. Among all of the cell lines tested in this study, there were HPV-positive cervical carcinoma cells, such as HeLa, SiHa, and Caski cells, and HPV-negative cancer and normal cells, including A549, HepG2, HCT116, and MCF-7 cells as well as the hepatic cell line of human origin QSG770 and the immortalized human cervical keratinocyte cell line S12, which contained integrated copies of HPV DNA. Also, in line with the results from these cellular experiments, the xenograft mouse model and clinical correlation analysis showed a negative correlation between QCR2 and p21, revealing the physiological role of QCR2 in the p53 pathway [13]. In contrast, QCR2 failed to regulate mutant p53 or p21 levels in either mutant p53-haboring C33A cells or p53-deficient H1299 cells. Hence, our studies as presented here reveal QCR2 as a novel suppressor of p53, playing an oncogenic role in tumorigenesis.
Although it still remains unclear how exactly QCR2 negatively acts on p53 stability, we identified PHB as a novel QCR2-binding protein that might play a part in this regulation through co-IP-coupled proteomic analysis. This interaction was confirmed by co-IP, co-immunofluorescence staining and GST-pull down assays (Fig. 4, Fig. 5). We also mapped the binding domains for the PHB-QCR2 interaction, though it remains to learn if their direct binding is required for regulation of p53 stability and activity. Interestingly, PHB has been reported to interact with p53 in the nucleus of cancer cells and acts as an activator and chaperone of p53, increasing its transcriptional activity [36,44,45]. We showed here that silencing of PHB impairs the induction of p53 upon QCR2 knockdown, suggesting that PHB is required for p53 activation in response to QCR2 knockdown, or alternatively QCR2 might block PHB activation of p53. It was difficult to map the p53-binding domain of PHB, as neither of its short fragments was able to bind to p53 (Fig. 4g), suggesting that the intact PHB protein is probably necessary for its interaction with p53. But, interestingly, PHB bound to the N-terminal and the C-terminal domains of p53 (Fig. 4f). This suggests that PHB might protect p53 from QCR2-facilitated ubiquitination and proteasomal turnover by binding to these domains, though this conjecture needs to be further examined.
Identification of PHB as a suppressor of QCR2 in regulation of p53 stability and activity is biologically significant. PHB was initially identified as a protein with antiproliferative activities and shown to be localized in the mitochondria, where it functions to stabilize mitochondrial proteins [46,47]. We speculate QCR2 may strengthen the interaction with PHB under some conditions, such as inflammations or lack of nutrition, which need future studies. The Human Protein Atlas analysis showed that PHB is downregulated in several cancers, including cervical cancer, glioma, lung cancer, melanoma, prostate cancer, thyroid cancer, and urothelial cancer [48], although its level was also elevated in colorectal carcinoma, thyroid carcinoma, and transitional cell bladder cancer and decreased in the context of tumor dedifferentiation and gastric cancer initiation [[49], [50], [51], [52]]. Our study as presented here showed the marked reduction of PHB levels in human cervical cancer and a positive correlation between PHB and p21 levels, suggesting that PHB might play a tumor suppression role in this type of cancer (Fig. 7). In line with previous studies showing that PHB exhibits differential subcellular localization [53,54], we found that knockdown of QCR2 leads to the nuclear import of PHB from the cytoplasm (Fig. 5a and b), suggesting that PHB might bind to and protect p53 in the nucleus. Consistent with this speculation, comparison of PHB levels between human cervical cancer and normal tissues revealed the reverse correlation between QCR2 level and PHB or p21 levels (Fig. 7). These results suggest that PHB might positively regulate p53 activity as measured by p21 expression by negating QCR2 function toward p53.
In summary, our study as presented here identify the mitochondrial protein QCR2 as a novel suppressor of p53 to negatively regulate its stability and activity, and this suppression can be potentially reversed by another mitochondrial protein PHB. Their regulations of p53 must occur in the nucleus as most of p53 molecules reside and functions as a transcriptional factor in the nucleus. This cellular study was also validated in a xenograft lung cancer model and in primary human cervical cancers. This QCR2-PHB-p53 regulation might be generally applicable to most of the cancer types, because knockdown of QCR2 leads to p53 activation in a large spectrum of cancer cells regardless of their tissue origins. Although it remains to investigate how exactly QCR2 regulates p53 stability and how PHB inhibits this regulation, our study provides new information suggesting that QCR2 might act as an oncogenic player and serve as a potential molecule target for future development of anti-cancer therapy.
Funding sources
This work was supported by the “973” Program of China (grant no. 2015CB553903), the National Science-technology Supporting Plan Projects (grant no. 2015BAI13B05), the National Natural Science Foundation of China (grant nos. 81702573, 81772787, 81402159, 81472783, 81372806, 81372801, and 81572570), National Science and Technology Major Sub-Project (2018ZX10301402-002) and Technical Innovation Special Project of Hubei Province (2018ACA138).
Declaration of interests
The authors declare that there are no conflicts of interest with this work.
Author contributions
QLG planned the project, interpreted the data and drafted the article. HL revised the article and provided advice. T.I., M. A. M., B. P–C. C, YZ provided advice. YYH and PW designed the research and conducted data analysis. ZW, YYH, ZYZ, SG, JL and PPG performed experiments. SJS analyzed human microarray data and performed GEO submission. JT, PBC and QQM provided technical support. JBH, SXW and JFZ supervised the study.
Acknowledgments
The authors thank Prof. Kenneth Raj (Health Protection Agency, Didcot, United Kingdom) for providing the S12 cell line, which was permitted by the primary owner Prof. Margaret Stanley (Division of Virology, National Institute for Medical Research, London, United Kingdom). The authors thank Prof. Ding Ma for his guidance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.002.
Appendix A. Supplementary data
Supplementary material
References
- 1.Wallace D.C. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma Z., Cao M., Liu Y., He Y., Wang Y., Yang C. Mitochondrial F1Fo-ATP synthase translocates to cell surface in hepatocytes and has high activity in tumor-like acidic and hypoxic environment. Acta Biochim Biophys Sin. 2010;42(8):530–537. doi: 10.1093/abbs/gmq063. [DOI] [PubMed] [Google Scholar]
- 3.Lu Z.J., Song Q.F., Jiang S.S., Song Q., Wang W., Zhang G.H. Identification of ATP synthase beta subunit (ATPB) on the cell surface as a non-small cell lung cancer (NSCLC) associated antigen. BMC Cancer. 2009;9:16. doi: 10.1186/1471-2407-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung H.J., Shim J.S., Lee J., Song Y.M., Park K.C., Choi S.H. Terpestacin inhibits tumor angiogenesis by targeting UQCRB of mitochondrial complex III and suppressing hypoxia-induced reactive oxygen species production and cellular oxygen sensing. J Biol Chem. 2010;285(15):11584–11595. doi: 10.1074/jbc.M109.087809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyake N., Yano S., Sakai C., Hatakeyama H., Matsushima Y., Shiina M. Mitochondrial complex III deficiency caused by a homozygous UQCRC2 mutation presenting with neonatal-onset recurrent metabolic decompensation. Hum Mutat. 2013;34(3):446–452. doi: 10.1002/humu.22257. [DOI] [PubMed] [Google Scholar]
- 6.Duncan A.M., Ozawa T., Suzuki H., Rozen R. Assignment of the gene for the core protein II (UQCRC2) subunit of the mitochondrial cytochrome bc1 complex to human chromosome 16p12. Genomics. 1993;18(2):455–456. doi: 10.1006/geno.1993.1500. [DOI] [PubMed] [Google Scholar]
- 7.Wu P., Tian Y., Chen G., Wang B., Gui L., Xi L. Ubiquitin B: an essential mediator of trichostatin A-induced tumor-selective killing in human cancer cells. Cell Death Differ. 2010;17(1):109–118. doi: 10.1038/cdd.2009.142. [DOI] [PubMed] [Google Scholar]
- 8.Xu H., Ma J., Wu J., Chen L., Sun F., Qu C. Gene expression profiling analysis of lung adenocarcinoma. Braz J Med Biol Res. 2016;49(3) doi: 10.1590/1414-431X20154861. = Revista brasileira de pesquisas medicas e biologicas/Sociedade Brasileira de Biofisica [et al] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Owens K.M., Kulawiec M., Desouki M.M., Vanniarajan A., Singh K.K. Impaired OXPHOS complex III in breast cancer. PLoS One. 2011;6(8) doi: 10.1371/journal.pone.0023846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnoult D., Soares F., Tattoli I., Castanier C., Philpott D.J., Girardin S.E. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122:3161–3168. doi: 10.1242/jcs.051193. Pt 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai D., Tan C.L., Gunaratne J., Quek L.S., Nei W., Thierry F. Localization of HPV-18 E2 at mitochondrial membranes induces ROS release and modulates host cell metabolism. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0075625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soussi T., Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1(3):233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 13.el-Deiry W.S., Tokino T., Velculescu V.E., Levy D.B., Parsons R., Trent J.M. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 14.Bieging K.T., Mello S.S., Attardi L.D. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toledo F., Wahl G.M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 16.Tsvetkov P., Reuven N., Shaul Y. Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 2010;17(1):103–108. doi: 10.1038/cdd.2009.67. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khutornenko A.A., Roudko V.V., Chernyak B.V., Vartapetian A.B., Chumakov P.M., Evstafieva A.G. Pyrimidine biosynthesis links mitochondrial respiration to the p53 pathway. Proc Natl Acad Sci U S A. 2010;107(29):12828–12833. doi: 10.1073/pnas.0910885107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venugopal R., Jaiswal A.K. Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc Natl Acad Sci U S A. 1996;93(25):14960–14965. doi: 10.1073/pnas.93.25.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiley C.D., Velarde M.C., Lecot P., Liu S., Sarnoski E.A., Freund A. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 2016;23(2):303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He G., Zhang Y.W., Lee J.H., Zeng S.X., Wang Y.V., Luo Z. AMP-activated protein kinase induces p53 by phosphorylating MDMX and inhibiting its activity. Mol Cell Biol. 2014;34(2):148–157. doi: 10.1128/MCB.00670-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley M.A., Browne H.M., Appleby M., Minson A.C. Properties of a non-tumorigenic human cervical keratinocyte cell line. Int J Cancer. 1989;43(4):672–676. doi: 10.1002/ijc.2910430422. [DOI] [PubMed] [Google Scholar]
- 23.Pan X., Zhou T., Tai Y.H., Wang C., Zhao J., Cao Y. Elevated expression of CUEDC2 protein confers endocrine resistance in breast cancer. Nat Med. 2011;17(6):708–714. doi: 10.1038/nm.2369. [DOI] [PubMed] [Google Scholar]
- 24.Liu D., Zhang X.X., Wan D.Y., Xi B.X., Ma D., Wang H. Sine oculis homeobox homolog 1 promotes alpha5beta1-mediated invasive migration and metastasis of cervical cancer cells. Biochem Biophys Res Commun. 2014;446(2):549–554. doi: 10.1016/j.bbrc.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Cardamone M.D., Tanasa B., Cederquist C.T., Huang J., Mahdaviani K., Li W. Mitochondrial retrograde signaling in mammals is mediated by the transcriptional cofactor GPS2 via direct mitochondria-to-nucleus translocation. Mol Cell. 2018;69(5):757–72 e7. doi: 10.1016/j.molcel.2018.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szolnoki Z., Szekeres M., Szaniszlo I., Balda G., Bodor A., Kondacs A. Decreased number of mitochondria in leukoaraiosis. Arch Med Res. 2015;46(8):604–608. doi: 10.1016/j.arcmed.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Sun S., Han Y., Liu J., Fang Y., Tian Y., Zhou J. Trichostatin a targets the mitochondrial respiratory chain, increasing mitochondrial reactive oxygen species production to trigger apoptosis in human breast cancer cells. PLoS One. 2014;9(3):e91610. doi: 10.1371/journal.pone.0091610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu F., Sanin D.E., Wang X. Mitochondrial DNA in lung cancer. Adv Exp Med Biol. 2017;1038:9–22. doi: 10.1007/978-981-10-6674-0_2. [DOI] [PubMed] [Google Scholar]
- 29.Robin E.D., Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136(3):507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- 30.Dotto G.P. p21(WAF1/Cip1): more than a break to the cell cycle? Biochim Biophys Acta. 2000;1471(1):M43–M56. doi: 10.1016/s0304-419x(00)00019-6. [DOI] [PubMed] [Google Scholar]
- 31.Brooks C.L., Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15(2):164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 32.Steinberg D. Ubiquitin-mediated proteolytic system plays diverse roles in human disease. Scientist. 1998;12(15) (10-+) [Google Scholar]
- 33.Bonvini P., Zorzi E., Basso G., Rosolen A. Bortezomib-mediated 26S proteasome inhibition causes cell-cycle arrest and induces apoptosis in CD-30+ anaplastic large cell lymphoma. Leukemia. 2007;21(4):838–842. doi: 10.1038/sj.leu.2404528. [DOI] [PubMed] [Google Scholar]
- 34.Mordaunt D.A., Jolley A., Balasubramaniam S., Thorburn D.R., Mountford H.S., Compton A.G. Phenotypic variation of TTC19-deficient mitochondrial complex III deficiency: a case report and literature review. Am J Med Genet A. 2015;167(6):1330–1336. doi: 10.1002/ajmg.a.36968. [DOI] [PubMed] [Google Scholar]
- 35.Yadav N., Chandra D. Mitochondrial DNA mutations and breast tumorigenesis. Biochim Biophys Acta. 2013;1836(2):336–344. doi: 10.1016/j.bbcan.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi B., Rastogi S., Morris M., Carastro L.M., DeCook C., Seto E. Differential regulation of human YY1 and caspase 7 promoters by prohibitin through E2F1 and p53 binding sites. Biochem J. 2007;401(1):155–166. doi: 10.1042/BJ20060364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Havugimana P.C., Hart G.T., Nepusz T., Yang H., Turinsky A.L., Li Z. A census of human soluble protein complexes. Cell. 2012;150(5):1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Artal-Sanz M., Tavernarakis N. Opposing function of mitochondrial prohibitin in aging. Aging (Albany NY) 2010;2(12):1004–1011. doi: 10.18632/aging.100246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havre P.A., Yuan J., Hedrick L., Cho K.R., Glazer P.M. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res. 1995;55(19):4420–4424. [PubMed] [Google Scholar]
- 41.Vaseva A.V., Moll U.M. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787(5):414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine A.J., Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storey A., Thomas M., Kalita A., Harwood C., Gardiol D., Mantovani F. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature. 1998;393(6682):229–234. doi: 10.1038/30400. [DOI] [PubMed] [Google Scholar]
- 44.Kathiria A.S., Neumann W.L., Rhees J., Hotchkiss E., Cheng Y., Genta R.M. Prohibitin attenuates colitis-associated tumorigenesis in mice by modulating p53 and STAT3 apoptotic responses. Cancer Res. 2012;72(22):5778–5789. doi: 10.1158/0008-5472.CAN-12-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fusaro G., Dasgupta P., Rastogi S., Joshi B., Chellappan S. Prohibitin induces the transcriptional activity of p53 and is exported from the nucleus upon apoptotic signaling. J Biol Chem. 2003;278(48):47853–47861. doi: 10.1074/jbc.M305171200. [DOI] [PubMed] [Google Scholar]
- 46.Artal-Sanz M., Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metabol. 2009;20(8):394–401. doi: 10.1016/j.tem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 47.McClung J.K., Danner D.B., Stewart D.A., Smith J.R., Schneider E.L., Lumpkin C.K. Isolation of a cDNA that hybrid selects antiproliferative mRNA from rat liver. Biochem Biophys Res Commun. 1989;164(3):1316–1322. doi: 10.1016/0006-291x(89)91813-5. [DOI] [PubMed] [Google Scholar]
- 48.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 49.Wu T.F., Wu H., Wang Y.W., Chang T.Y., Chan S.H., Lin Y.P. Prohibitin in the pathogenesis of transitional cell bladder cancer. Anticancer Res. 2007;27(2):895–900. [PubMed] [Google Scholar]
- 50.Franzoni A., Dima M., D'Agostino M., Puppin C., Fabbro D., Loreto C.D. Prohibitin is overexpressed in papillary thyroid carcinomas bearing the BRAF(V600E) mutation. Thyroid. 2009;19(3):247–255. doi: 10.1089/thy.2008.0235. [DOI] [PubMed] [Google Scholar]
- 51.Chen D., Chen F., Lu X., Yang X., Xu Z., Pan J. Identification of prohibitin as a potential biomarker for colorectal carcinoma based on proteomics technology. Int J Oncol. 2010;37(2):355–365. doi: 10.3892/ijo_00000684. [DOI] [PubMed] [Google Scholar]
- 52.Leal M.F., Cirilo P.D., Mazzotti T.K., Calcagno D.Q., Wisnieski F., Demachki S. Prohibitin expression deregulation in gastric cancer is associated with the 3′ untranslated region 1630 C>T polymorphism and copy number variation. PLoS One. 2014;9(5):e98583. doi: 10.1371/journal.pone.0098583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang P., Xiang Y., Wang Y.J., Li S.M., Wang Y., Hua H.R. Differential expression and subcellular localization of Prohibitin 1 are related to tumorigenesis and progression of non-small cell lung cancer. Int J Clin Exp Pathol. 2013;6(10):2092–2101. [PMC free article] [PubMed] [Google Scholar]
- 54.Shi S.L., Li Q.F., Liu Q.R., Xu D.H., Tang J., Liang Y. Nuclear matrix protein, prohibitin, was down-regulated and translocated from nucleus to cytoplasm during the differentiation of osteosarcoma MG-63 cells induced by ginsenoside Rg1, cinnamic acid, and tanshinone IIA (RCT) J Cell Biochem. 2009;108(4):926–934. doi: 10.1002/jcb.22324. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material