Abstract
Due to their beneficial effects on human health, antioxidant peptides have attracted much attention from researchers. However, the structure-activity relationships of antioxidant peptides have not been fully understood. In this paper, quantitative structure-activity relationships (QSAR) models were built on two datasets, i.e., the ferric thiocyanate (FTC) dataset and ferric-reducing antioxidant power (FRAP) dataset, containing 214 and 172 unique antioxidant tripeptides, respectively. Sixteen amino acid descriptors were used and model population analysis (MPA) was then applied to improve the QSAR models for better prediction performance. The results showed that, by applying MPA, the cross-validated coefficient of determination (Q2) was increased from 0.6170 to 0.7471 for the FTC dataset and from 0.4878 to 0.6088 for the FRAP dataset, respectively. These findings indicate that the integration of different amino acid descriptors provide additional information for model building and MPA can efficiently extract the information for better prediction performance.
Keywords: quantitative structure-activity relationship, QSAR, antioxidant tripeptides, model population analysis, amino acid descriptors
1. Introduction
Bioactive peptides, usually containing 2–20 amino acid residues, are typically derived from the enzymatic hydrolysis of proteins [1]. They are inactive within the sequence of proteins, but they can exert various physiological functions after release. Antioxidant peptides are one of the most important groups of bioactive peptides, which can prevent oxidative stress and they have notable contributions to human health [2]. Antioxidant peptides have been isolated and purified from sources, such as cereals, milk, meat, and fish [3]. The methods to assess the antioxidant capacities of peptides include the Trolox equivalent antioxidant capacity (TEAC), the ferric ion reducing antioxidant power (FRAP), the 2,2-diphenyl-1-picrylhydrazyl radical-scavenging capacity (DPPH), the oxygen radical absorbance capacity (ORAC), the total radical trapping antioxidant parameter (TRAP), etc. [4]. However, it is impossible to test all of the peptides to find valid antioxidants, when considering the large number of theoretical possible peptides, i.e., 400 dipeptides, 8000 tripeptides, 160,000 tetrapeptides, etc.
The activities of peptides are determined by the amino acid compositions, sequences, and structures. Quantitative structure-activity relationship (QSAR), which is a well-recognized tool for estimating chemical activities, has been widely applied for bioactive peptides prediction [5]. The QSAR models have been successfully built on ACE-inhibitory peptides [6], antimicrobial peptides [7], antioxidant peptides [8,9,10], antitumor peptides [11], bitter peptides [12], and etc. The QSAR study of antioxidant peptides mainly focused on di and tripeptides, because they can be absorbed intact from the intestinal lumen into the bloodstream and then produce biological effects at the tissue level [13]. When compared to dipeptides, tripeptides were reported to exhibit higher levels of antioxidant activity [14]. Besides, tripeptides had much larger structural diversity than dipeptides, which is a good property for developing multifunctional food additives [15].
The prediction performances need to be further improved, although plenty of QSAR models have been built on antioxidant peptides. The relationship between peptide structure and antioxidant activity is still unclear. This may be due to the restriction of model building methods. Model population analysis (MPA) provides a new strategy of model building, which is to use multi-models instead of a single model to improve prediction ability and interpretability [16,17]. Previous studies showed that, through the application of MPA strategy, the performance of regression models could be improved [6,18].
In this study, we built QSAR models based on two antioxidant tripeptides datasets. The first dataset contains 214 artificially designed tripeptides and the second dataset contains 172 β-Lactoglobulin derived tripeptides, which represent designed or food originated tripeptides, respectively. 16 amino acid descriptors were used to construct sophisticated data for the comprehensive information of peptides. The MPA strategy was applied to extract useful information from the data and to optimize the models. The aim of this study is not to build a new set of descriptors, but to integrate different descriptors under the framework of MPA for better QSAR model performance on antioxidant tripeptides data. The improved method for QSAR modelling will help in discovering new antioxidant tripeptides for future drugs or food additives.
2. Results
2.1. FTC Dataset
The results of QSAR models on the FTC dataset are displayed in Table 1. Before outlier elimination, the largest Q2 value of 0.4901 is obtained on the VSW descriptor. After outlier elimination, the HSEHPCSV descriptor showed the largest Q2 value of 0.6170 among the 16 amino acid descriptors. The integration of 16 descriptors gave rise to an improvement of the model performance (Q2 = 0.6818). Finally, the model prediction performance was further improved (Q2 = 0.7471) after variable selection while using the BOSS method.
Table 1.
Comparisons among different quantitative structure-activity relationships (QSAR) models on ferric thiocyanate (FTC) dataset a.
| Descriptors | Before Outlier Elimination | After Outlier Elimination | |||||
|---|---|---|---|---|---|---|---|
| Q2 | R2 | optPC | Q2 | R2 | optPC | Outlier | |
| HSEHPCSV | 0.3861 | 0.5781 | 4 | 0.6170 | 0.7338 | 20 | 183, 182, 181, 134 |
| ST-scale | 0.4268 | 0.5733 | 12 | 0.5993 | 0.6844 | 13 | 183, 182, 181, 134 |
| HESH | 0.4091 | 0.5366 | 2 | 0.5968 | 0.7047 | 10 | 183, 181, 182, 134, 129 |
| VSW | 0.4901 | 0.5771 | 3 | 0.5925 | 0.6768 | 5 | 181, 183, 182, 134, 151 |
| G-scale | 0.4516 | 0.5527 | 6 | 0.5843 | 0.6574 | 9 | 181, 183, 182, 134, 118 |
| FASGAI | 0.4814 | 0.5457 | 5 | 0.5544 | 0.6130 | 6 | 129, 181, 128 |
| DPPS | 0.4740 | 0.5637 | 7 | 0.5379 | 0.6278 | 8 | 181, 182, 183, 134 |
| E-scale | 0.4956 | 0.5451 | 4 | 0.5144 | 0.5582 | 4 | 181, 182, 183, 112 |
| 5Z-scale | 0.3903 | 0.4626 | 12 | 0.3974 | 0.4653 | 9 | 181, 182, 183, 172 |
| VHSE | 0.4265 | 0.5432 | 12 | 0.3974 | 0.514 | 8 | 181, 182, 183, 172 |
| T-scale | 0.3280 | 0.4215 | 9 | 0.3728 | 0.4362 | 9 | 181, 182, 183 |
| V-scale | 0.3371 | 0.3785 | 5 | 0.3070 | 0.3458 | 6 | 181, 183, 182 |
| Z-scale | 0.2814 | 0.3398 | 4 | 0.2678 | 0.3415 | 4 | 181 |
| ISA-ECI | 0.1493 | 0.1916 | 6 | 0.1572 | 0.1836 | 6 | 183, 182, 181 |
| MS-WHTM1 | 0.0736 | 0.1488 | 3 | 0.1036 | 0.1678 | 3 | 181, 183, 182 |
| MS-WHTM2 | 0.0775 | 0.1445 | 3 | 0.0882 | 0.1617 | 3 | 181, 182, 183 |
| Integrated descriptors | 0.4811 | 0.5843 | 3 | 0.6818 | 0.7964 | 8 | 181, 183, 182, 134, 151, 153, 188 |
| BOSS | 0.7471 ± 0.0032 | 0.7931 ± 0.0062 | 9.72 ± 3.2199 | ||||
a R2 is the coefficient of determination; Q2 is the cross-validated R2; optPC is optimal principal components for PLS regression model; the results of BOSS are shown in the form of mean value ± standard deviation in 100 runs; the top ranked Q2 scores were marked in bold.
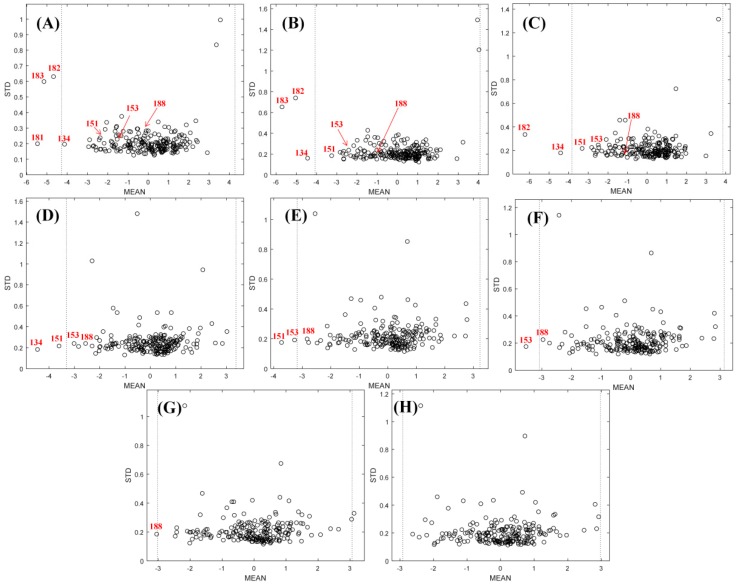
In this study, an MPA-based outlier elimination procedure [19] was carried out to remove outliers one by one (Figure 1). For the integrated data, samples of no. 181, 183, 182, 134, 151, 153, and 188 were removed in sequence. Finally, all of the samples were within the range according to the three-sigma rule after outlier removal (Figure 1H, dashed line).
Figure 1.
The process of model population analysis (MPA)-based outlier elimination on the FTC dataset of integrated descriptors. The dashed line is defined as the boundary for outliers, which is mean ± 3× standard deviation of prediction errors. (A) No outlier was eliminated, (B) sample No. 181 was eliminated, (C) sample No. 183 was eliminated, (D) sample No. 182 was eliminated, (E) sample No. 134 was eliminated, (F) sample No. 151 was eliminated, (G) sample No. 153 was eliminated, and (H) sample No. 188 was eliminated and all of the outliers were removed.
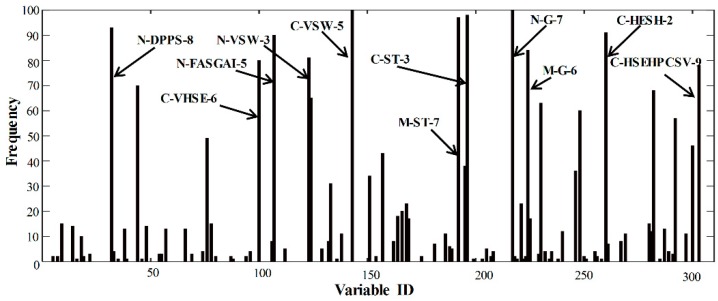
Figure 2 showed the selected variables by the BOSS method in 100 runs. The variables being selected more frequently reflect high variable importance. The top 11 variables (frequency>75), in descending order, were as follows: C-VSW-5 = N-G-7 > C-ST-3 > M-ST-7 > N-DPPS-8 > C-HESH-2 > N-FASGAI-5 > M-G-6 > N-VSW-3 > C-VHSE-6 >C-HSEHPCSV-9, which are marked on Figure 2. All the top 11 variables originated from the best preformed amino acid descriptors, i.e., HSEHPCSV, ST-scale, HESH, G-scale, FASGAI, and DPPS (Table 1). It showed that the ultimate model has the merit of the best performed models that were constructed by single amino acid descriptors.
Figure 2.
Frequency of variables selected by the bootstrapping soft shrinkage (BOSS) method on the FTC dataset in 100 runs. The higher frequency denotes higher variable importance. The top 11 variables with frequency larger than 75 were marked in the figure.
2.2. FRAP Dataset
The results of QSAR models on FRAP dataset are displayed in Table 2. Before logarithmic transformation of response vector Y, the largest Q2 value of 0.1408 is obtained on 5Z-scale descriptor. The low Q2 value indicated that the tripeptide structures and their antioxidant activities that were evaluated by FRAP assay did not share a linear relationship. After logarithmic transformation, the VHSE descriptor showed the largest Q2 value of 0.4878. Through integrating the 16 descriptors, the Q2 value was increased slightly to 0.4953. The prediction performance of the model was promoted after variable selection using the BOSS method (Q2 = 0.6088). It indicated that a linear relationship between the structures and the activities was built after the logarithmic transformation of Y and the MPA strategy was efficient in improving the model.
Table 2.
Comparisons among different QSAR models on FRAP dataset a.
| Descriptors | Before Logarithmic Transformation | After Logarithmic Transformation | ||||
|---|---|---|---|---|---|---|
| Q2 | R2 | optPC | Q2 | R2 | optPC | |
| VHSE | 0.0042 | 0.2655 | 3 | 0.4878 | 0.6122 | 6 |
| 5Z-scale | 0.1408 | 0.3177 | 2 | 0.4809 | 0.5568 | 3 |
| DPPS | 0.0059 | 0.2290 | 3 | 0.4147 | 0.5463 | 4 |
| ST-scale | 0.0263 | 0.3220 | 8 | 0.3968 | 0.5410 | 9 |
| FASGAI | 0.0470 | 0.2753 | 2 | 0.3735 | 0.5006 | 4 |
| E-scale | 0.0560 | 0.2521 | 1 | 0.3714 | 0.4734 | 5 |
| HESH | 0.0444 | 0.2818 | 10 | 0.3668 | 0.5290 | 3 |
| HSEHPCSV | 0.0259 | 0.2475 | 7 | 0.3624 | 0.4952 | 3 |
| G-scale | 0.1066 | 0.2334 | 5 | 0.2836 | 0.3850 | 1 |
| VSW | 0.0130 | 0.3071 | 1 | 0.2382 | 0.4361 | 2 |
| MS-WHTM2 | 0.0342 | 0.0370 | 3 | 0.1728 | 0.2594 | 3 |
| MS-WHTM1 | 0.0452 | 0.0329 | 9 | 0.1207 | 0.1941 | 4 |
| T-scale | 0.0682 | 0.0706 | 2 | 0.0750 | 0.2129 | 10 |
| V-scale | 0.0293 | 0.0748 | 4 | 0.0699 | 0.1495 | 1 |
| Z-scale | 0.0052 | 0.1445 | 1 | 0.0301 | 0.1456 | 6 |
| ISA-ECI | 0.0242 | 0.0141 | 1 | 0.0071 | 0.0411 | 1 |
| Integrated descriptors | 0.1069 | 0.4212 | 3 | 0.4953 | 0.6423 | 3 |
| BOSS | 0.6088 ± 0.0041 | 0.6655 ± 0.0094 | 3.5100 ± 2.5086 | |||
a R2 is the coefficient of determination; Q2 is the cross-validated R2; optPC is optimal principal components for PLS regression model; the results of BOSS are shown in the form of mean value ± standard deviation in 100 runs, the top ranked Q2 scores were marked in bold.
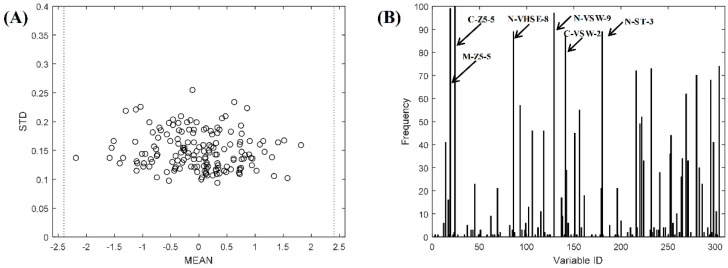
Similarly, an MPA-based outlier elimination procedure was carried out on the FRAP dataset. No outlying sample was detected, since all of the samples gather within the range according to the three-sigma rule (Figure 3A, dashed line). The important variables that were selected by BOSS are displayed in Figure 3B. The six most important variables (frequency > 75) are C-Z5-5, M-Z5-5, N-VSW-9, N-VHSE-8, N-ST-3, and C-VSW-2, respectively. Most of the important variables originated from three well performed descriptors, i.e., VHSE, 5Z-scale, and ST-scale. However, there still some variables selected from the poorly performed descriptor, such as VSW. It suggested that descriptors with poor performance also contained useful information for model building.
Figure 3.
The result of QSAR model building on the FRAP dataset. (A) The result of MPA-based outlier detection on the FRAP dataset of integrated descriptors. No outlier was detected. (B) Frequency of variables selected by the BOSS method on the FRAP dataset in 100 runs. The higher frequency denotes higher variable importance. The six top variables with frequency larger than 75 are marked in the figure.
3. Discussion
3.1. Comparison with the Reported Models
For the FTC dataset, our method showed higher prediction accuracy (Q2 = 0.7471), when compared to the previous report (Q2 = 0.6310) [20]. Note that 41 sample were eliminated as outliers in the previous study, while only seven outliers were eliminated in this study. A much larger number of samples was used in our model, which is more representative. It showed that our method exhibited a model with higher prediction performance and the relatively larger applicability domain.
Similarly, for FRAP dataset, our method showed a higher prediction accuracy (Q2 = 0.6008) when compared to the previous report (Q2 = 0.5410) [21]. It should be noted that, in the previous study, five samples with the highest activities and 14 inactive samples were removed, while in our study, only inactive samples were removed. Thus, our model showed improved prediction accuracy and enlarged applicability domain.
3.2. Relationship between Antioxidant Activities and Peptide Structures
Previous studies showed that the N-terminus and C-terminus amino acids are important in relating to antioxidant activities [20]. Our results are in agreement with the previous findings that most of the important variables that were selected by BOSS originated from the N-terminus or C-terminus (Figure 2 and Figure 3B). In addition, studies showed that tripeptides containing Cys (C), Trp (W), and Tyr (Y) residues exhibited strong antioxidant activities [8,10]. Tripeptides YHY and LTC, for the two datasets, respectively, having the highest antioxidant activities is confirmed by our study.
On the FTC dataset, a linear relationship between antioxidant activities and peptide structures was constructed. However, on the FRAP dataset, the relationship was only built on the log-transformed activities and structure properties. It indicates that the antioxidant activity and peptide structures on the FRAP dataset exhibits a non-linear relationship. Data transformation is crucial before model building on this kind of data. The different performance of the two datasets may be attributed to the structure diversities of peptides. In the FTC dataset, tripeptides contain either the His or Tyr residue, which have similar structures, while the structure diversity in the FRAP dataset is much larger.
3.3. The Integration of Amino Acid Descriptors
A number of amino acid descriptors have been developed and applied in the QSAR studies of bioactive peptides. Each descriptor has its merits and demerits. Our study shows that an optimal descriptor does not exist. Instead, all of the descriptors are data dependent, which means that each descriptor performs well on different datasets. It makes the researches difficult to select descriptors. By integrating different descriptors, each one can contribute particular information to the model and create a new possibility for further improvement of the model. Subsequently, the next question has become how to efficiently extract information from different descriptors and to get rid of the redundancy of the data? Model population analysis (MPA) may provide a solution for that. It uses multi-models instead of a single model for prediction. Each sub-model contains a random combination of different descriptors. Through statistical analysis of the sub-model outcomes, the informative variables from the descriptors are extracted and an optimized descriptor combination is obtained [22]. Finally, the optimized model performs better than any of the single descriptor model, as it is shown in Table 1 and Table 2. To summarize, the aim of this study is not to build a new set of descriptors, but to provide a general framework to integrate different descriptors. The framework can take in any newly developed descriptor and fit on different datasets. The more diverse the integrated descriptors are, the better performance the model can be.
4. Materials and Methods
4.1. Data Collection
4.1.1. Ferric Thiocyanate (FTC) Dataset
A dataset of 214 antioxidant tripeptides that contain either His or Tyr residue was obtained from the published literatures [20,23]. All of the tripeptides were chemically synthesized using solid phase Fmoc Chemistry and their antioxidant activities were measured by the FTC method [23]. Test samples (500 μg) in 0.5 mL of deionized water were mixed with linoleic acid emulsion (1.0 mL, 50 mM) and phosphate buffer (1.0 mL, 0.1 M) in glass test tubes (5 mL). The tubes were sealed with silicon rubber caps and then kept at 60 °C in the dark. 50 μL reaction mixtures were taken out at different intervals during incubation. The degree of oxidation was measured by sequentially adding ethanol (2.35 mL, 75%), ammonium thiocyanate (50 μL, 30%), and ferrous chloride (50 μL, 20 mM in 3.5% HCl). After the mixture had stood for 3 min, the absorbance of the solution was measured at 500 nm with a Jasco model Ubest 30 spectrophotometer (Tokyo, Japan). A control was performed containing the same contents with test sample but without the peptides. The number of days that was taken to attain the absorbance of 0.3 was defined as the induction period. The relative activities were calculated by dividing the induction period of test samples by that of the control (Table 3). All of the experiments were carried out in triplicate and averaged.
Table 3.
Sequences and antioxidant activities of tripeptides on ferric thiocyanate (FTC) dataset a.
| No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LHA | 3.918 | 37 | PHA | 5.793 | 73 | RHA | 5.205 | 109 | DHH | 0.9045 | 145 | HHH | 0.0635 | 181 | YHY | 9.886 |
| 2 | LHD | 3.593 | 38 | PHD | 4.622 | 74 | RHD | 3.304 | 110 | EHH | 0.9045 | 146 | HHK | 0.0635 | 182 | YKY | 9.886 |
| 3 | LHE | 6.136 | 39 | PHE | 6.152 | 75 | RHE | 5.096 | 111 | HHH | 0.0000 | 147 | HHR | 0.0635 | 183 | YRY | 9.886 |
| 4 | LHF | 3.628 | 40 | PHF | 3.916 | 76 | RHF | 3.300 | 112 | KHH | 0.0000 | 148 | HHA | 0.0680 | 184 | YAY | 3.607 |
| 5 | LHG | 6.697 | 41 | PHG | 5.197 | 77 | RHG | 5.725 | 113 | AHH | 2.020 | 149 | HHI | 0.0680 | 185 | YIY | 3.607 |
| 6 | LHH | 4.836 | 42 | PHH | 6.051 | 78 | RHH | 3.296 | 114 | IHH | 2.020 | 150 | HHL | 0.0680 | 186 | YLY | 3.607 |
| 7 | LHI | 6.531 | 43 | PHI | 4.916 | 79 | RHI | 4.806 | 115 | FHH | 1.803 | 151 | HHF | 3.612 | 187 | YFY | 2.233 |
| 8 | LHK | 4.225 | 44 | PHK | 3.426 | 80 | RHK | 2.694 | 116 | WHH | 1.803 | 152 | HHW | 3.612 | 188 | YWY | 2.233 |
| 9 | LHL | 5.920 | 45 | PHL | 5.311 | 81 | RHL | 3.501 | 117 | YHH | 1.803 | 153 | HHY | 3.612 | 189 | YYY | 2.233 |
| 10 | LHM | 4.504 | 46 | PHM | 3.714 | 82 | RHM | 3.218 | 118 | GHH | 1.089 | 154 | HHG | 0.3170 | 190 | YGY | 3.366 |
| 11 | LHN | 5.148 | 47 | PHN | 6.061 | 83 | RHN | 5.713 | 119 | NHH | 1.089 | 155 | HHN | 0.3170 | 191 | YNY | 3.366 |
| 12 | LHQ | 4.136 | 48 | PHQ | 3.718 | 84 | RHQ | 3.108 | 120 | QHH | 1.089 | 156 | HHQ | 0.3170 | 192 | YQY | 3.366 |
| 13 | LHR | 5.184 | 49 | PHR | 4.751 | 85 | RHR | 4.302 | 121 | MHH | 2.015 | 157 | HHM | 0.0817 | 193 | YMY | 1.780 |
| 14 | LHS | 4.293 | 50 | PHS | 4.042 | 86 | RHS | 3.386 | 122 | SHH | 1.320 | 158 | HHS | 0.0862 | 194 | YSY | 3.447 |
| 15 | LHT | 5.584 | 51 | PHT | 6.247 | 87 | RHT | 5.987 | 123 | THH | 1.320 | 159 | HHT | 0.0862 | 195 | YTY | 3.447 |
| 16 | LHV | 3.481 | 52 | PHV | 3.335 | 88 | RHV | 3.206 | 124 | CHH | 0.9369 | 160 | HHC | 0.1277 | 196 | YCY | 3.087 |
| 17 | LHW | 6.791 | 53 | PHW | 6.535 | 89 | RHW | 5.878 | 125 | HDH | 1.477 | 161 | DYY | 3.417 | 197 | YYD | 4.116 |
| 18 | LHY | 4.203 | 54 | PHY | 4.227 | 90 | RHY | 3.378 | 126 | HEH | 1.477 | 162 | EYY | 3.417 | 198 | YYE | 4.116 |
| 19 | LWA | 1.192 | 55 | PWA | 1.396 | 91 | RWA | 1.212 | 127 | HHH | 0.0441 | 163 | HYY | 2.257 | 199 | YYH | 5.303 |
| 20 | LWD | 1.717 | 56 | PWD | 1.096 | 92 | RWD | 0.9091 | 128 | HKH | 0.0441 | 164 | KYY | 2.257 | 200 | YYK | 5.303 |
| 21 | LWE | 1.717 | 57 | PWE | 1.096 | 93 | RWE | 1.091 | 129 | HRH | 0.0441 | 165 | RYY | 2.257 | 201 | YYR | 5.303 |
| 22 | LWF | 1.414 | 58 | PWF | 0.9192 | 94 | RWF | 0.9091 | 130 | HAH | 0.9518 | 166 | AYY | 3.071 | 202 | YYA | 3.344 |
| 23 | LWG | 1.313 | 59 | PWG | 2.687 | 95 | RWG | 1.717 | 131 | HIH | 0.9518 | 167 | IYY | 3.071 | 203 | YYI | 3.344 |
| 24 | LWH | 3.212 | 60 | PWH | 1.184 | 96 | RWH | 1.091 | 132 | HLH | 0.9518 | 168 | LYY | 3.071 | 204 | YYL | 3.344 |
| 25 | LWI | 1.111 | 61 | PWI | 1.396 | 97 | RWI | 1.232 | 133 | HFH | 2.026 | 169 | FYY | 1.911 | 205 | YYF | 4.050 |
| 26 | LWK | 1.899 | 62 | PWK | 0.4066 | 98 | RWK | 0.6061 | 134 | HWH | 2.026 | 170 | WYY | 1.911 | 206 | YYW | 4.050 |
| 27 | LWL | 0.6060 | 63 | PWL | 1.096 | 99 | RWL | 3.212 | 135 | HYH | 2.026 | 171 | YYY | 1.911 | 207 | YYY | 4.050 |
| 28 | LWM | 1.394 | 64 | PWM | 0.7955 | 100 | RWM | 0.7273 | 136 | HGH | 0.8318 | 172 | GYY | 5.071 | 208 | YYG | 2.996 |
| 29 | LWN | 1.313 | 65 | PWN | 2.104 | 101 | RWN | 2.404 | 137 | HNH | 0.8318 | 173 | NYY | 5.071 | 209 | YYN | 2.996 |
| 30 | LWQ | 2.505 | 66 | PWQ | 1.202 | 102 | RWQ | 0.6061 | 138 | HQH | 0.8318 | 174 | QYY | 5.071 | 210 | YYQ | 2.996 |
| 31 | LWR | 2.909 | 67 | PWR | 2.705 | 103 | RWR | 2.384 | 139 | HMH | 0.8734 | 175 | MYY | 1.991 | 211 | YYM | 2.103 |
| 32 | LWS | 2.020 | 68 | PWS | 1.096 | 104 | RWS | 0.8081 | 140 | HSH | 0.7304 | 176 | SYY | 3.070 | 212 | YYS | 3.983 |
| 33 | LWT | 2.020 | 69 | PWT | 2.598 | 105 | RWT | 3.818 | 141 | HTH | 0.7304 | 177 | TYY | 3.070 | 213 | YYT | 3.983 |
| 34 | LWV | 1.616 | 70 | PWV | 1.008 | 106 | RWV | 0.6061 | 142 | HCH | 0.9747 | 178 | CYY | 0.4699 | 214 | YYC | 0.6369 |
| 35 | LWW | 3.515 | 71 | PWW | 2.899 | 107 | RWW | 2.707 | 143 | HHD | 0.1877 | 179 | YDY | 3.047 | |||
| 36 | LWY | 2.222 | 72 | PWY | 1.114 | 108 | RWY | 0.8081 | 144 | HHE | 0.1877 | 180 | YEY | 3.047 | □ | □ | □ |
4.1.2. Ferric-reducing Antioxidant Power (FRAP) Dataset
A dataset of 172 antioxidant tripeptides were derived from β-Lactoglobulin, where all possible tripeptides were collected based on its amino sequence [21]. All of the tripeptides were chemically synthesized while using solid phase Fmoc Chemistry and their antioxidant activities were evaluated using the FRAP assay [24]. Ten microliters of 100 mmol/mL tripeptide solution were incubated at 37 °C with 100 μL of FRAP reagent, containing 10 mmol/L of 2,4,6-tripyridyl-s-triazine and 20 mmol/L of FeCl3. The absorption values were read at a wavelength of 570 nm using a microplate reader (Model 680, Bio-Rad, Hercules, CA, USA) after 10 min reaction. Aqueous Fe2+ solutions at concentrations that ranged from 10 to 1000 μmol/L were used to produce a calibration curve. The results were expressed as micromoles Fe2+ equivalents per mole of the sample based on the standard curve. All of the experiments were carried out in triplicate and then averaged. The activities were logarithmic transformed prior to modeling, where 14 inactive peptides (activity = 0) were removed (Table 4). The measured activities before logarithmic transformation were displayed in Table S1.
Table 4.
Sequences and activities of tripeptides on ferric ion reducing antioxidant power (FRAP) dataset a.
| No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity | No. | Sequence | Activity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LTC | 2.83 | 30 | LPM | 1.04 | 59 | YKK | 0.25 | 88 | NGE | −0.30 | 117 | ELK | −0.66 | 146 | KIP | −1.15 |
| 2 | CQC | 2.53 | 31 | TDY | 1.01 | 60 | AQA | 0.22 | 89 | QSA | −0.33 | 118 | PEQ | −0.66 | 147 | LLD | −1.22 |
| 3 | GTW | 2.52 | 32 | QCH | 1.00 | 61 | LRV | 0.20 | 90 | DAQ | −0.34 | 119 | IDA | −0.68 | 148 | DLE | −1.22 |
| 4 | LFC | 2.07 | 33 | TWY | 0.96 | 62 | PTP | 0.18 | 91 | ENS | −0.34 | 120 | LLA | −0.70 | 149 | PEV | −1.22 |
| 5 | CLV | 2.06 | 34 | RVY | 0.95 | 63 | ALN | 0.18 | 92 | ENG | −0.37 | 121 | ALA | −0.72 | 150 | LKP | −1.40 |
| 6 | QKW | 2.03 | 35 | KWE | 0.90 | 64 | LEI | 0.16 | 93 | NSA | −0.37 | 122 | GLD | −0.72 | 151 | ALE | −1.52 |
| 7 | CME | 1.99 | 36 | CLL | 0.89 | 65 | LVR | 0.13 | 94 | EKT | −0.38 | 123 | DIS | −0.72 | 152 | TQL | −1.52 |
| 8 | YLL | 1.91 | 37 | LAM | 0.85 | 66 | HIR | 0.12 | 95 | EQS | −0.38 | 124 | PEG | −0.72 | 153 | LEE | −1.52 |
| 9 | QCL | 1.69 | 38 | YSL | 0.81 | 67 | KKI | 0.11 | 96 | AMA | −0.41 | 125 | LDI | −0.74 | 154 | LEK | −1.70 |
| 10 | LAC | 1.69 | 39 | MKG | 0.80 | 68 | SFN | 0.07 | 97 | KID | −0.41 | 126 | AEP | −0.74 | 155 | DAL | −2.00 |
| 11 | GEC | 1.64 | 40 | QTM | 0.80 | 69 | SLL | 0.06 | 98 | GAQ | −0.43 | 127 | ALI | −0.77 | 156 | EVD | −2.00 |
| 12 | EQC | 1.52 | 41 | LAL | 0.76 | 70 | PAV | 0.04 | 99 | PLR | −0.44 | 128 | LDA | −0.77 | 157 | VDD | −2.00 |
| 13 | FCM | 1.51 | 42 | QAL | 0.73 | 71 | RLS | 0.04 | 100 | ILL | −0.46 | 129 | VFK | −0.77 | 158 | DEA | −2.00 |
| 14 | CHI | 1.45 | 43 | MEN | 0.73 | 72 | AGT | 0.04 | 101 | VRT | −0.46 | 130 | ALK | −0.77 | 159 | ALT | - |
| 15 | ACQ | 1.38 | 44 | MKC | 0.72 | 73 | LLF | 0.02 | 102 | IAE | −0.49 | 131 | AQK | −0.82 | 160 | KGL | - |
| 16 | EEL | 1.33 | 45 | LSF | 0.69 | 74 | PMH | 0.00 | 103 | QSL | −0.49 | 132 | IIA | −0.82 | 161 | IQK | - |
| 17 | WEN | 1.31 | 46 | TCG | 0.67 | 75 | EEQ | −0.01 | 104 | KTK | −0.51 | 133 | LIV | −0.85 | 162 | QKV | - |
| 18 | VYV | 1.19 | 47 | SLA | 0.65 | 76 | LVL | −0.02 | 105 | ASD | −0.52 | 134 | EGD | −0.85 | 163 | GDL | - |
| 19 | MHI | 1.16 | 48 | TMK | 0.64 | 77 | QLE | −0.05 | 106 | APL | −0.52 | 135 | QKK | −0.85 | 164 | EIL | - |
| 20 | CAQ | 1.12 | 49 | LDT | 0.62 | 78 | FDK | −0.07 | 107 | AQS | −0.57 | 136 | IPA | −0.85 | 165 | KII | - |
| 21 | WYS | 1.12 | 50 | EKF | 0.54 | 79 | LLL | −0.08 | 108 | ENK | −0.57 | 137 | SDI | −0.89 | 166 | NKV | - |
| 22 | KYL | 1.08 | 51 | VLV | 0.53 | 80 | SAP | −0.08 | 109 | TPE | −0.59 | 138 | VEE | −0.89 | 167 | DTD | - |
| 23 | CGA | 1.08 | 52 | MAA | 0.44 | 81 | LLQ | −0.12 | 110 | RTP | −0.59 | 139 | DDE | −0.89 | 168 | EPE | - |
| 24 | KKY | 1.08 | 53 | PTQ | 0.44 | 82 | NPT | −0.17 | 111 | VLD | −0.62 | 140 | KVL | −0.92 | 169 | EAL | - |
| 25 | NEN | 1.08 | 54 | VAG | 0.41 | 83 | FNP | −0.20 | 112 | IRL | −0.62 | 141 | KFD | −0.92 | 170 | DKA | - |
| 26 | ECA | 1.07 | 55 | ALP | 0.37 | 84 | LNE | −0.24 | 113 | AAS | −0.64 | 142 | IVT | −0.96 | 171 | KAL | - |
| 27 | DYK | 1.06 | 56 | AVF | 0.36 | 85 | SAE | −0.26 | 114 | LQK | −0.64 | 143 | VTQ | −0.96 | 172 | LKA | - |
| 28 | KCL | 1.05 | 57 | KVA | 0.31 | 86 | KPT | −0.28 | 115 | FKI | −0.64 | 144 | AEK | −0.96 | |||
| 29 | YVE | 1.05 | 58 | TQT | 0.26 | 87 | DIQ | −0.30 | 116 | ISL | −0.66 | 145 | TKI | −1.10 | □ | □ | □ |
a The data containing 172 antioxidant tripeptides was collected from the literature of Tian et al. [21]. Antioxidant activities of tripeptides were measured by the FRAP assay and were logarithmic transformed. Fourteen inactive peptides were removed before model building.
The two datasets are representative for artificially designed or food protein originated tripeptides, respectively. Both of the datasets have been used for building QSAR models before. Thus, it is suitable for model comparison.
4.2. Data Processing
The tripeptide sequences were transformed into X-matrices using 16 amino acid descriptors, respectively, while the dependent variable Y-vectors represents the relative activities of peptides. These descriptors include Z-scale, 5Z-scale, DPPS, MS-WHIM, ISA-ECI, VHSE, FASGAI, VSW, T-scale, ST-scale, E-scale, V-scale, G-scale, HESH, and HSEHPCSV, as is shown in Table 5. They are the most frequently used amino acid descriptors in the QSAR study of bioactive peptides. The peptide structure is characterized by describing amino acids within the sequence. For example, Z-scale descriptor, containing three parameters (Z1, Z2, and Z3), would generate nine variables (3 parameters × 3 amino acids) for tripeptides. To clearly label each variable, we used a unified rule to name them. The amino acid at the N-terminus was designated as N, the C-terminus amino acid was designated as C, and the middle amino acid was designated as M. Thus, the nine variables that were generated by Z-scale descriptor were labeled as N-Z-1, N-Z-2, N-Z-3, M-Z-1, M-Z-2, M-Z-3, C-Z-1, C-Z-2, and C-Z-3, respectively. The 16 descriptors were integrated to build an X-matrix, which contained 306 variables (V1-V306), with the correspondence, as follows: Z-scale (V1-V9), 5Z-scale (V10-V24), DPPS (V25-V54), MS-WHIM1 (V55-V63), MS-WHIM2 (V64-V72), ISA-ECI (V73-V78), VHSE (V79-V102), FASGAI (V103-V120), VSW (V121-V147), E-scale (V148-V162), T-scale (V163-V177), ST-scale (V178-V201), V-scale (V202-V210), G-scale (V211-V234), HESH (V235-V270), and HSEHPCSV (V271-V306), respectively.
Table 5.
Parameters of 16 amino acid descriptors.
| Descriptor | No. of Physicochemical Property | No. of Extracted Variable | Scope of Variable |
|---|---|---|---|
| Z-scale [25] | 29 | 3 | Electronic property, steric property and hydrophobic property |
| 5Z-scale [26] | 26 | 5 | Electronic property, steric property and hydrophobic property |
| DPPS [27] | 119 | 10 | Electronic property, steric property, hydrophobic property and hydrogen bond |
| MS-WHIM [28] | 36 | 3 | Surface charge distribution, size and charge over shape dependence |
| ISA-ECI [29] | / | 2 | Isotropic surface area and electronic charge index |
| VHSE [30] | 50 | 8 | Electronic property, steric property and hydrophobic property |
| FASGAI [31] | 335 | 6 | Hydrophobic property, alpha and turn property, bulky property, electronic property, compositional characteristics, local flexibility |
| VSW [32] | 99 | 9 | Molecular size, shape, symmetry and atom distribution |
| T-scale [33] | 67 | 5 | Topological property |
| ST-scale [34] | 827 | 8 | Molecular constitutional, topological, geometrical, hydrophobic, electronic and steric property |
| E-scale [35] | 237 | 5 | Hydrophobic property, size, preferences for amino acids to occur in α-helices, number of degenerate triplet codons and the frequency of occurrence of amino acid residues in β-strands |
| V-scale [36] | / | 3 | Van Der Wall’s volume, net charge index and hydrophobic parameter of side chains |
| G-scale [37] | 457 | 8 | Electronic property, steric property and hydrophobic property |
| HESH [38] | 171 | 12 | Electronic property, steric property, hydrophobic property and hydrogen bond |
| HSEHPCSV [39] | 95 | 12 | Hydrophobic, steric, electronic properties and hydrogen bond |
4.3. QSAR Model Building
Partial least squares (PLS) regression [40] was used to build the connection between the peptide structure descriptions (variables, X-matrices) and the relative activities (responses, Y-vectors). It was implemented using MATLAB software (Version R2015a, the MathWorks, Inc., Natick, MA, USA). All of the variables were auto-scaled to unit variance and all of the responses were mean-centered prior to model building. The models were validated using cross-validation and the optimal number of PLS components were chosen based on a statistic, called Q2, which is the cross-validated R2, referring to the predictive ability of the model. R2 is the coefficient of determination, providing an estimate of the model fit.
MPA was applied to optimize the model through outlier elimination and variable selection. It is a framework for model building that utilizes multiple models instead of a single model to construct results [16,17]. Generally, it worked, as follows: (1) firstly, a random resampling procedure was applied to obtain sub-datasets; (2) then, sub-models were built based on the sub-datasets; and, (3) finally, a statistical analysis was used to extract useful information from the outcome of sub-models. In the present study, MPA was utilized for outlier detection and variable selection.
The MPA-based outlier detection method [19] was applied to remove the outlying samples from measured data. To begin with, 1000 sub-datasets were generated through random reselecting of 80% samples in sample space. Subsequently, for each sub-dataset, a PLS regression model was built and the prediction error for each sample was recorded. The mean of prediction errors was used as the basis for outlier detection and a three-sigma rule was applied to define the boundary, as it is reported previously [6]. The bootstrapping soft shrinkage (BOSS) method [18] was applied to select informative variables from the pool of descriptors. It is also based on the idea of MPA. Firstly, 1000 sub-datasets were obtained using bootstrap resampling in the variable space. Afterwards, 1000 PLS models were built based on the sub-datasets and the regression coefficients were extracted. In the next step, weighted bootstrap resampling was used to regenerate sub-datasets and to rebuild sub-model. The resampling procedure was repeated until all of the uninformative variables were eliminated.
5. Conclusions
In this study, we have constructed QSAR models on two datasets of antioxidant tripeptides, i.e., FTC dataset and FRAP dataset. After the integration of 16 amino acid descriptors and utilization of the MPA strategy for model building, the Q2 values were enlarged from 0.6170 to 0.7471 and from 0.4878 to 0.6088, respectively. The results show that the MPA framework is powerful in QSAR model building on antioxidant tripeptides data. The framework can also be applied to investigate the structure and activity relationships of other types of bioactive peptides and to integrate more different molecular descriptors.
Abbreviations
| BOSS | the bootstrapping soft shrinkage method |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl radical-scavenging capacity |
| FRAP | ferric-reducing antioxidant power |
| FTC | ferric thiocyanate |
| MPA | model population analysis |
| ORAC | oxygen radical absorbance capacity |
| PLS | partial least squares |
| QSAR | quantitative structure-activity relationships |
| TEAC | Trolox equivalent antioxidant capacity |
| TRAP | total radical trapping antioxidant parameter |
Supplementary Materials
The following are available online at https://www.mdpi.com/1422-0067/20/4/995/s1.
Author Contributions
B.D. conceived and designed the study; H.L., T.T. and X.N. collected the data; H.L., J.C., G.Y. carried out the model building; F.Z., R.C., D.C., M.Z. and L.Y. analyzed the data; B.D. and H.L. wrote the paper.
Funding
This research was funded by the Nation K&D Program of China (2018YFD0500603), the National Natural Science Foundation of China (Grant Nos. 31790411 and 31872985), and the Natural Science Foundation of Guangdong Province (Grant Nos. 2017A030310410) for supporting of the projects. The studies meet with the approval of the university’s review board.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chakrabarti S., Guha S., Majumder K. Food-Derived Bioactive Peptides in Human Health: Challenges and Opportunities. Nutrients. 2018;10:1738. doi: 10.3390/nu10111738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenzo J.M., Munekata P.E.S., Gomez B., Barba F.J., Mora L., Perez-Santaescolastica C., Toldra F. Bioactive peptides as natural antioxidants in food products—A review. Trends Food Sci. Technol. 2018;79:136–147. doi: 10.1016/j.tifs.2018.07.003. [DOI] [Google Scholar]
- 3.Sila A., Bougatef A. Antioxidant peptides from marine by-products: Isolation, identification and application in food systems. A review. J. Funct. Foods. 2016;21:10–26. doi: 10.1016/j.jff.2015.11.007. [DOI] [Google Scholar]
- 4.MacDonald-Wicks L.K., Wood L.G., Garg M.L. Methodology for the determination of biological antioxidant capacity in vitro: A review. J. Sci. Food Agric. 2006;86:2046–2056. doi: 10.1002/jsfa.2603. [DOI] [Google Scholar]
- 5.Zou T.B., He T.P., Li H.B., Tang H.W., Xia E.Q. The Structure-Activity Relationship of the Antioxidant Peptides from Natural Proteins. Molecules. 2016;21:72. doi: 10.3390/molecules21010072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng B., Ni X., Zhai Z., Tang T., Tan C., Yan Y., Deng J., Yin Y. New Quantitative Structure-Activity Relationship Model for Angiotensin-Converting Enzyme Inhibitory Dipeptides Based on Integrated Descriptors. J. Agric. Food Chem. 2017;65:9774–9781. doi: 10.1021/acs.jafc.7b03367. [DOI] [PubMed] [Google Scholar]
- 7.Vishnepolsky B., Gabrielian A., Rosenthal A., Hurt D.E., Tartakovsky M., Managadze G., Grigolava M., Makhatadze G.I., Pirtskhalava M. Predictive Model of Linear Antimicrobial Peptides Active against Gram-Negative Bacteria. J. Chem. Inf. Model. 2018;58:1141–1151. doi: 10.1021/acs.jcim.8b00118. [DOI] [PubMed] [Google Scholar]
- 8.Chen N., Chen J., Yao B., Li Z.G. QSAR Study on Antioxidant Tripeptides and the Antioxidant Activity of the Designed Tripeptides in Free Radical Systems. Molecules. 2018;23:1407. doi: 10.3390/molecules23061407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao W.Z., Gu L.J., Zheng Y.M., Zhu Z.S., Zhao M.M., Liang M., Ren J.Y. Analysis of the quantitative structure-activity relationship of glutathione-derived peptides based on different free radical scavenging systems. MedChemComm. 2016;7:2083–2093. doi: 10.1039/C6MD00006A. [DOI] [Google Scholar]
- 10.Zheng L., Zhao Y., Dong H., Su G., Zhao M. Structure-activity relationship of antioxidant dipeptides: Dominant role of Tyr, Trp, Cys and Met residues. J. Funct. Foods. 2016;21:485–496. doi: 10.1016/j.jff.2015.12.003. [DOI] [Google Scholar]
- 11.Radman A., Gredicak M., Kopriva I., Jeric I. Predicting Antitumor Activity of Peptides by Consensus of Regression Models Trained on a Small Data Sample. Int. J. Mol. Sci. 2011;12:8415–8430. doi: 10.3390/ijms12128415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S., Qi W., Su R., Li T., Lu D., He Z. CoMFA and CoMSIA analysis of ACE-inhibitory, antimicrobial and bitter-tasting peptides. Eur. J. Med. Chem. 2014;84:100–106. doi: 10.1016/j.ejmech.2014.07.015. [DOI] [PubMed] [Google Scholar]
- 13.Miner-Williams W.M., Stevens B.R., Moughan P.J. Are intact peptides absorbed from the healthy gut in the adult human? Nutr. Res. Rev. 2014;27:308–329. doi: 10.1017/S0954422414000225. [DOI] [PubMed] [Google Scholar]
- 14.Chen H.-M., Muramoto K., Yamauchi F., Nokihara K. Antioxidant activity of designed peptides based on the antioxidative peptide isolated from digests of a soybean protein. J. Agric. Food Chem. 1996;44:2619–2623. doi: 10.1021/jf950833m. [DOI] [Google Scholar]
- 15.Wang J.H., Liu Y.L., Ning J.H., Yu J., Li X.H., Wang F.X. Is the structural diversity of tripeptides sufficient for developing functional food additives with satisfactory multiple bioactivities? J. Mol. Struct. 2013;1040:164–170. doi: 10.1016/j.molstruc.2013.03.004. [DOI] [Google Scholar]
- 16.Deng B.-C., Yun Y.-H., Liang Y.-Z. Model population analysis in chemometrics. Chemom. Intell. Lab. Syst. 2015;149:166–176. doi: 10.1016/j.chemolab.2015.08.018. [DOI] [Google Scholar]
- 17.Deng B.C., Lu H.M., Tan C.Q., Deng J.P., Yin Y.L. Model population analysis in model evaluation. Chemom. Intell. Lab. Syst. 2018;172:223–228. doi: 10.1016/j.chemolab.2017.11.016. [DOI] [Google Scholar]
- 18.Deng B.C., Yun Y.H., Cao D.S., Yin Y.L., Wang W.T., Lu H.M., Luo Q.Y., Liang Y.Z. A bootstrapping soft shrinkage approach for variable selection in chemical modeling. Anal. Chim. Acta. 2016;908:63–74. doi: 10.1016/j.aca.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Cao D.S., Liang Y.Z., Xu Q.S., Li H.D., Chen X. A new strategy of outlier detection for QSAR/QSPR. J. Comput. Chem. 2010;31:592–602. doi: 10.1002/jcc.21351. [DOI] [PubMed] [Google Scholar]
- 20.Li Y.W., Li B., He J.G., Qian P. Quantitative structure-activity relationship study of antioxidative peptide by using different sets of amino acids descriptors. J. Mol. Struct. 2011;998:53–61. doi: 10.1016/j.molstruc.2011.05.011. [DOI] [Google Scholar]
- 21.Tian M., Fang B., Jiang L., Guo H.Y., Cui J.Y., Ren F.Z. Structure-activity relationship of a series of antioxidant tripeptides derived from beta-Lactoglobulin using QSAR modeling. Dairy Sci. Technol. 2015;95:451–463. doi: 10.1007/s13594-015-0226-5. [DOI] [Google Scholar]
- 22.Deng B.C., Yun Y.H., Liang Y.Z., Yi L.Z. A novel variable selection approach that iteratively optimizes variable space using weighted binary matrix sampling. Analyst. 2014;139:4836–4845. doi: 10.1039/C4AN00730A. [DOI] [PubMed] [Google Scholar]
- 23.Saito K., Jin D.H., Ogawa T., Muramoto K., Hatakeyama E., Yasuhara T., Nokihara K. Antioxidative properties of tripeptide libraries prepared by the combinatorial chemistry. J. Agric. Food Chem. 2003;51:3668–3674. doi: 10.1021/jf021191n. [DOI] [PubMed] [Google Scholar]
- 24.Benzie I.F.F., Strain J.J. The ferric reducing ability of plasma (FRAP) as a measure of “Antioxidant power”: The FRAP assay. Anal. Biochem. 1996;239:70–76. doi: 10.1006/abio.1996.0292. [DOI] [PubMed] [Google Scholar]
- 25.Hellberg S., Sjöström M., Skagerberg B., Wold S. Peptide quantitative structure-activity relationships, a multivariate approach. J. Med. Chem. 1987;30:1126–1135. doi: 10.1021/jm00390a003. [DOI] [PubMed] [Google Scholar]
- 26.Sandberg M., Eriksson L., Jonsson J., Sjöström M., Wold S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. J. Med. Chem. 1998;41:2481–2491. doi: 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- 27.Tian F., Yang L., Lv F. In silico quantitative prediction of peptides binding affinity to human MHC molecule:an intuitive quantitative structure-activity relationship approach. Amino Acids. 2009;36:535–554. doi: 10.1007/s00726-008-0116-8. [DOI] [PubMed] [Google Scholar]
- 28.Zaliani A., Gancia E. ChemInform Abstract: MS-WHIM Scores for Amino Acids: A New 3D-Description for Peptide QSAR and QSPR Studies. J. Chem. Inf. Model. 1999;39:525–533. doi: 10.1002/chin.199932266. [DOI] [Google Scholar]
- 29.Collantes E.R., Rd D.W. Amino acid side chain descriptors for quantitative structure-activity relationship studies of peptide analogues. J. Med. Chem. 1995;38:2705–2713. doi: 10.1021/jm00014a022. [DOI] [PubMed] [Google Scholar]
- 30.Mei H., Liao Z.H., Zhou Y., Li S.Z. A new set of amino acid descriptors and its application in peptide QSARs. Biopolymers. 2005;80:775–786. doi: 10.1002/bip.20296. [DOI] [PubMed] [Google Scholar]
- 31.Liang G., Li Z. Factor analysis scale of generalized amino acid information as the source of a new set of descriptors for elucidating the structure and activity relationships of cationic antimicrobial peptides. QSAR Comb. Sci. 2007;26:754–763. doi: 10.1002/qsar.200630145. [DOI] [Google Scholar]
- 32.Tong J.B., Liu S.L., Zhou P., Wu B.L., Li Z.L. A novel descriptor of amino acids and its application in peptide QSAR. J. Theor. Biol. 2008;253:90–97. doi: 10.1016/j.jtbi.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 33.Tian F.F., Zhou P., Li Z.L. T-scale as a novel vector of topological descriptors for amino acids and its application in QSARs of peptides. J. Mol. Struct. 2007;830:106–115. doi: 10.1016/j.molstruc.2006.07.004. [DOI] [Google Scholar]
- 34.Yang L., Shu M., Ma K.W., Mei H., Jiang Y.J., Li Z.L. ST-scale as a novel amino acid descriptor and its application in QSAM of peptides and analogues. Amino Acids. 2010;38:805–816. doi: 10.1007/s00726-009-0287-y. [DOI] [PubMed] [Google Scholar]
- 35.Venkatarajan M.S., Braun W. New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical–chemical properties. Mol. Model. Annu. 2001;7:445–453. [Google Scholar]
- 36.Lin Z.H., Long H.X., Bo Z., Wang Y.Q., Wu Y.Z. New descriptors of amino acids and their application to peptide QSAR study. Peptides. 2008;29:1798–1805. doi: 10.1016/j.peptides.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 37.Wang X., Wang J., Lin Y., Ding Y., Wang Y.Q., Cheng X., Lin Z. QSAR study on angiotensin-converting enzyme inhibitor oligopeptides based on a novel set of sequence information descriptors. J. Mol. Model. 2011;17:1599–1606. doi: 10.1007/s00894-010-0862-x. [DOI] [PubMed] [Google Scholar]
- 38.Shu M., Mei H., Yang S.B., Liao L.M., Li Z.L. Structural Parameter Characterization and Bioactivity Simulation Based on Peptide Sequence. Qsar Comb. Sci. 2009;28:27–35. doi: 10.1002/qsar.200710169. [DOI] [Google Scholar]
- 39.Shu M., Huo D.Q., Mei H., Liang G.Z., Zhang M., Li Z.L. New Descriptors of Amino Acids and Its Applications to Peptide Quantitative Structure-activity Relationship. Chin. J. Struct. Chem. 2008;27:1375–1383. [Google Scholar]
- 40.Wold S., Sjöström M., Eriksson L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2008;58:109–130. doi: 10.1016/S0169-7439(01)00155-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.