Abstract
Myeloma is characterized by the neoplastic proliferation of monoclonal plasma cells. A diagnosis of myeloma is based on the criteria proposed by the International Myeloma Working Group and the pathological findings.
Myeloma cells are classified into four types: mature, immature, pleomorphic, and plasmablastic. There are three patterns in which myeloma infiltrates bone marrow - nodular, interstitial, and diffuse. Dutcher bodies are highly specific to neoplastic myeloma cells. On immunohistochemical staining, the specificity of CD138 is high for plasma cells. As a clear image is often not obtained from the immunohistochemical staining of the immunoglobulin light chain, in situ hybridization is recommended. Abnormal expression of CD56 is seen in 70-80% of cases by flow cytometry analysis. CD56 expression definitively indicates myeloma, suggesting its high diagnostic value. Evaluation of the infiltration pattern, monoclonality, and abnormal antigen expression of plasma cells is more important than the plasmocytic ratio to determine whether a case is reactive or neoplastic.
Multiple gene abnormalities function in the onset and progression of myeloma. In our department, we analyze CCND1, FGFR3, MAF, and del (17p13) by FISH for all myeloma cases. None of the cases with genetic abnormalities were recognized by G-banding. Therefore, FISH is more effective than G-banding for the evaluation of genetic abnormalities in myeloma.
Keywords: Myeloma, Bone marrow, Pathological finding, FISH method, Genetic abnormality
INTRODUCTION
Myeloma is characterized by the neoplastic proliferation of a single plasma cell clone that produces monoclonal immunoglobulin. In most cases, neoplastic plasma cells proliferate in the bone marrow and result in extensive skeletal destruction with osteolytic lesions. Myeloma often manifests with many clinical symptoms and organ damage, including hypercalcemia, renal insufficiency, anemia, lytic bone lesions, hyper-viscosity, amyloidosis, and infection.
Based on the classification and diagnostic criteria proposed by the International Myeloma Working Group (IMWG), the diagnosis of myeloma is based on the level of M-protein in the serum and/or urine, rate of clonal plasma cells in the bone marrow, and the presence of organ damage.1 However, it is important to determine precisely whether proliferating plasma cells in the bone marrow are neoplastic by pathological diagnosis. Recently, gene analyses are being carried out for prognostic prediction and therapy selection.
In this report, I discuss the pathological and immunohistochemical characteristics of myeloma, and the findings that are key in distinguishing between plasma cells and neoplastic cells. In addition, I introduce the gene analysis process using the FISH method we developed.
PATHOLOGICAL FINDINGS
Morphology of myeloma cells
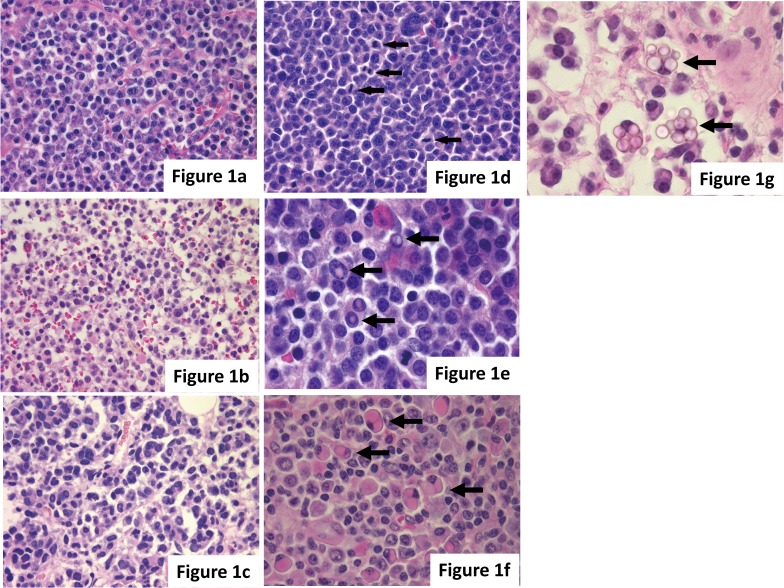
Myeloma cells show various morphologies such as normal plasma cell morphology and large nuclear pleomorphism. Myeloma cells are classified into four types: mature, immature, pleomorphic, and plasmablastic,2,3 and these types are sometimes mixed. Mature myeloma cells are usually indistinguishable from normal cells, with a round eccentric cartwheel nucleus without nucleoli, abundant basophilic cytoplasm, and a perinuclear hof. Immature myeloma cells have an irregular nucleus with more dispersed chromatin, a higher N/C ratio, and usually prominent nucleoli. Pleomorphic myeloma cells have increased nuclear polymorphism, a multinucleated polylobated nucleus, and prominent nucleoli. Plasmablastic myeloma cells are large, with increased nuclear polymorphism and mitotic figures, and resemble diffuse large B-cell lymphoma. These cells exhibit increased cellular atypia, and their presence indicates a poor prognosis.2 For the pleomorphic and plasmablastic types, confirmation by immunohistochemical staining is necessary to determine the plasma cell origin (Fig. 1a-d).
Fig. 1.
(a-d) Cytological features of myeloma cells exhibiting characteristics of the (a) mature type, (b) immature type, (c) pleomorphic type to (d) plasmablastic type. Mitotic figures (arrows) are shown in myeloma cells. (e-g) Arrows indicate (e) Dutcher bodies, (f) Russel bodies, and (g) Mott cells in myeloma cells. (a-g) HE staining.
Characteristic structures, such as Dutcher bodies and Russel bodies, are often found in myeloma cells. A Dutcher body is an inclusion body structure in the nucleus that is highly specific to neoplastic myeloma cells, and there is little doubt regarding the neoplastic characteristics if observed. A Russel body is an inclusion body structure in the cytoplasm formed by immunoglobulin deposition, and a cell with abundant grape-like cytoplasmic inclusions of immunoglobulin deposits is termed a Mott cell. Unlike Dutcher bodies, Russel bodies are sometimes found in reactive plasma cells; therefore, their presence is not definitive for neoplasticity (Fig. 1e-g).
Several studies have reported the correlation between the morphological and morphometrical characteristics of myeloma cells and clinical outcome, including prognosis. Seili-Bekafigo et al. demonstrated the following significant morphological characteristics that indicated short survival; 1) atypical plasma cells, defined by an enlarged nucleus, dispersed chromatin, visible nucleoli, and scant cytoplasm, and if plasma cells constitute more than 15% in the bone marrow aspirate, 2) the mean maxND/maxCD (largest nuclear diameter/largest cytoplasmic diameter) of plasma cells is 0.65 or above, 3) anisocytosis, which is expressed as the standard deviation of the max CD, is more than 4.2 um. They also found that plasma cells with irregular nuclei lacking the perinuclear hof indicate a more advanced stage of disease and worse prognosis.4 Furthermore, the presence of plasmablastic myeloma cells demonstrates a poor prognosis.2 These findings suggest that myeloma cell morphology is a potential prognostic factor.
Although there are several diagnostic methods for myeloma, the diagnosis process begins with a morphological evaluation of the bone marrow aspirate or biopsy. The association between the morphological findings of myeloma cells and clinical process, including prognosis has not been investigated; however, it is important that clinicians and pathologists diagnosing myeloma recognize the relationship between morphological findings and clinical processes.
Immunohistochemical findings
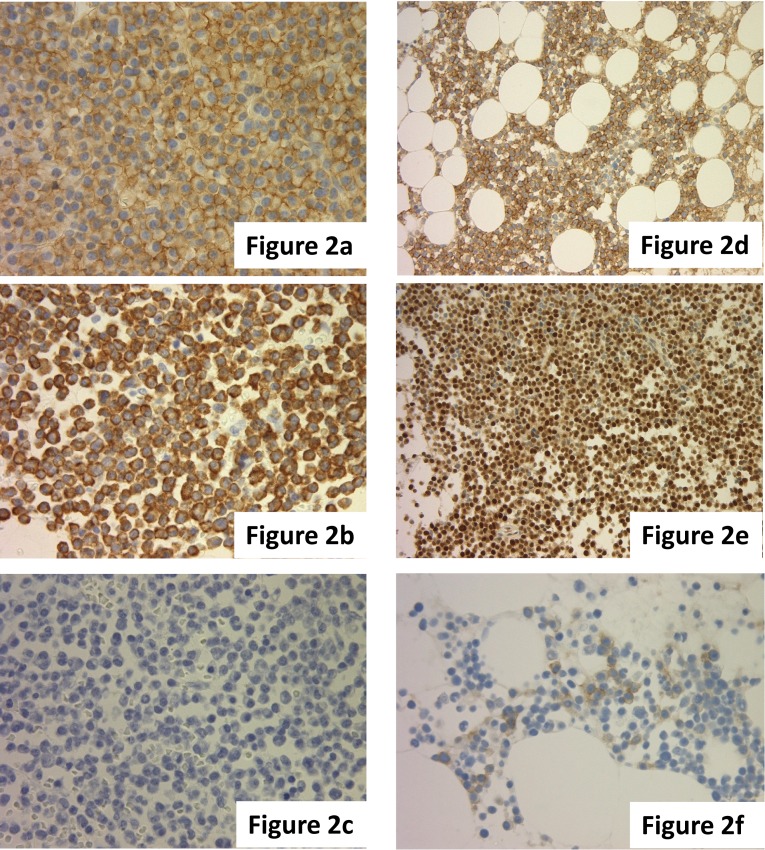
It is necessary to confirm the monoclonality of plasma cells by immunohistochemical staining for a definitive diagnosis of myeloma. The specificity of CD138 is high for plasma cells. As the background is sometimes stained due to the presence of serum light chain and because a clear image is often not obtained from the immunohistochemical staining of the immunoglobulin light chain (IgL), evaluation by in situ hybridization (ISH) is recommended. Monoclonality can be confirmed even for the non-secretory type of myeloma using ISH analysis (Fig. 2a-c).
Fig. 2.
(a-c) Slide showing (a) CD138(+), (b) kappa-ISH (+), and (c) lambda-ISH (-) myeloma cells. (d-f) Slide showing the abnormal expression of (d) CD56, (e) cyclin D1, and (f) CD117 in myeloma cells. (a, d-f) Immunohistochemical staining. (b, c) ISH.
The abnormal expression of antigens, including CD56, CD20, CD117, and CD10, is found in approximately 90% of myeloma cases by flow cytometry analysis, and these antigens are generally not expressed by normal plasma cells.5 The expression of CD56 in particular is observed in 70-80% of cases by flow cytometry analysis, and compared with other antibodies, its expression rate is commonly high. If CD56 is positive, it is definitive for myeloma, demonstrating the high diagnostic value of CD56.6,7 Conversely, the absence of CD56 expression is associated with plasma cell leukemia.8 CD117 expression is correlated with a better prognosis of myeloma.9 The expression of CD20 and cyclin D1 correlates with the presence of (IGH/CCND1) t(11;14)(q13;q32) translocation (Fig. 2d-f).10,11
Infiltration pattern of myeloma cells
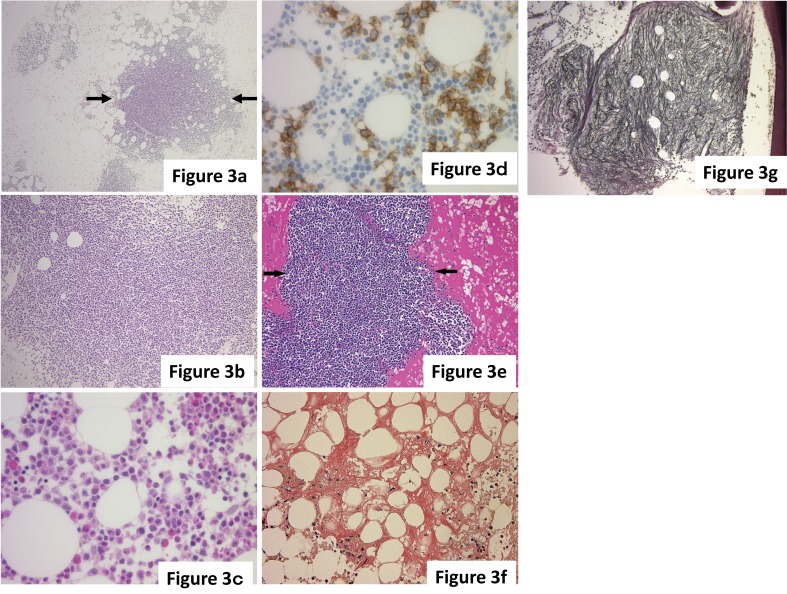
The infiltration pattern of myeloma is classified as one of three patterns – nodular, interstitial, or diffuse, and these patterns are often mixed.3 In the nodular pattern, myeloma cells displace normal hematopoietic cells and fat cells, and form a clear nodular lesion in the bone marrow. The distribution of nodular lesions is unequal in the bone marrow. This indicates an increase in the neoplastic pattern. In the interstitial pattern, myeloma cells form a small cluster and infiltrate individual spaces between normal hematopoietic cells. As the identification of a myeloma cell by HE staining is often difficult, evaluation by immunohistochemical staining is necessary to identify neoplastic plasma cells. In the diffuse pattern, a large number of myeloma cells diffusely infiltrate the bone marrow, and the expansive areas are replaced by myeloma cells. The diffuse pattern progresses from a localized nodule to an interstitial pattern. In the nodular and interstitial patterns, hematopoietic cells are relatively maintained, but in the diffuse pattern, hematopoietic cells are highly reduced and hematopoiesis is markedly suppressed (Fig. 3a-d).
Fig. 3.
(a-d) The infiltration pattern of myeloma. (a) Nodular pattern. Myeloma cells form a nodular lesion (arrows), and the border with the surrounding hematopoietic cells is clear. (b) Diffuse pattern. Myeloma cells exhibit diffused infiltration into the bone marrow, and hematopoietic cells are markedly reduced. (c, d) Interstitial pattern. (c) Myeloma cells are scattered between normal hematopoietic cells with occasional small clusters, but identification of neoplastic cells based on morphology is difficult. (d) CD138(+) myeloma cells are easily identified. (e-g) Secondary changes with myeloma. (e) Interstitial acidophilic change. The stroma exhibits acidophilic changes reflecting hyperproteinemia. Arrows indicate a nodular lesion of myeloma. (f) Amyloid deposition. Amyloid deposition, indicated by the orange stain, is broadly observed in the stroma. (g) Myelofibrosis (grade 2). Reticular fibers are diffusely increased in the nodular lesion of myeloma cells. In this case, the genetic abnormality FGFR3-IGH was identified by FISH. (a-c, e) HE staining. (d) Immunohistochemical staining. (f) Congo red staining. (g) Reticular staining.
Secondary changes with myeloma
With the infiltration of myeloma cells, secondary changes in the bone marrow are sometimes observed in cases of myeloma. Interstitial acidophilic changes that reflect hyperproteinemia, the deposition of amyloid, and myelofibrosis are among the secondary changes (Fig. 3e-g). Myelofibrosis is observed in approximately 10% of myeloma cases. BJP (Bence Jones protein)-type myeloma and marked cellular atypia often cause myelofibrosis. In the myelofibrosis case shown in Fig. 3g, the genetic abnormality FGFR3-IGH was also identified by FISH.
Evaluation of minor lesions
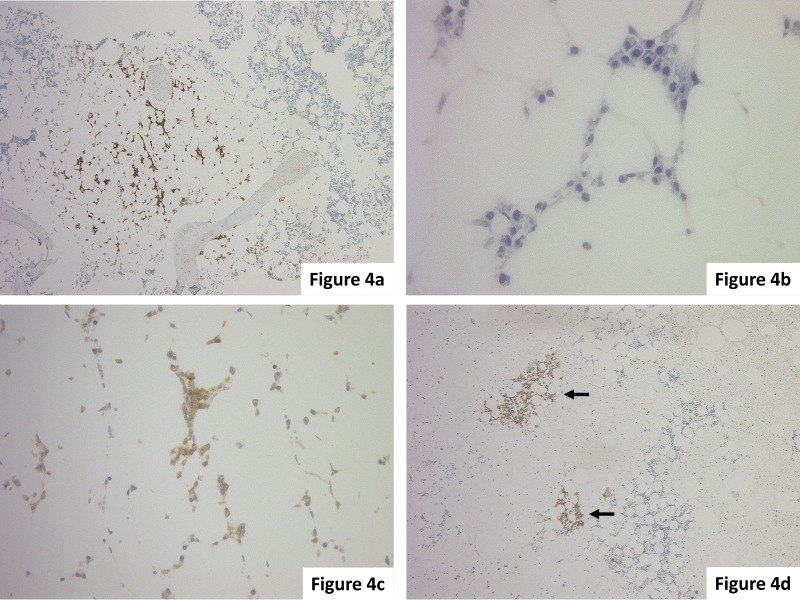
In general, when 30% of the bone marrow space is occupied by plasma cells and the lesions of plasma cells displace the normal bone marrow hematopoietic cells, a diagnosis of myeloma is likely. However, the distribution of lesions in myeloma is heterogeneous, and only a few neoplastic cells may be present depending on the bone marrow sampling site. When there is a small tumor, determining whether it is neoplastic becomes difficult. In this case, for a correct diagnosis, it is important to evaluate cellular atypia, such as nuclear immaturity or polymorphism, the presence of plasmacytic nests replacing hematopoietic cells (Fig. 4d), the presence of infiltration into the inter-fat marrow space (Fig. 4a-c), the monoclonality of IgLs, and abnormal antigen expression such as CD56. A previous report demonstrated that 1) homogeneous nodules of plasma cells occupying at least half of one high-power field, 2) monoclonal aggregates of plasma cells filling at least one inter-fat marrow space, and 3) marked bone marrow plasmacytosis with monoclonal plasma cells were diagnostic indicators of myeloma.12
Fig. 4.
(a-c) This case was (a) CD138(+), and a few (c) kappa-ISH (+) plasma cells infiltrated into inter-fat marrow spaces; the diagnosis was myeloma. (d) In this case, small clusters of CD138(+) myeloma cells were identified (arrows). The clusters replaced hematopoietic cells and the distribution is unequal in the bone marrow. (a, d) Immunohistochemical staining. (b) HE staining. (c) ISH.
In the case shown in Fig. 4a-c, although myeloma cells accounted for less than 10% of hematopoietic cells, kappa-ISH positive monoclonal plasma cells had infiltrated into the inter-fat marrow space, and this case was diagnosed as myeloma.
The evaluation of the infiltration pattern, monoclonality of the IgL, and abnormal antigen expression is more important than the plasmocytic ratio to evaluate a case as reactive or neoplastic.
DIFFERENTIATED DIAGNOSIS
Monoclonal gammopathy of undetermined significance (MGUS)
MGUS is an asymptomatic premalignant clonal plasma cell or lymphoplasmacytic proliferation disorder. It is a clinically established disease concept, and is often lacking in specific histological findings.
Reactive plasmacytosis
In reactive plasmacytosis, the plasma cells mainly infiltrate around blood vessels. If the ratio of plasma cells increases, they infiltrate between hematopoietic cells. Multinuclear cells with two or three nuclei are sometimes observed, but the presence of multinuclear cells is not definitive for myeloma. In some cases, myeloma cells account for less than 10% of cells in the bone marrow, whereas in other cases, reactive plasma cells can comprise nearly 30% of marrow hematopoietic cells.11 However, unlike in myeloma, the distribution of reactive plasma cells is equal in normal bone marrow, and monoclonality of the IgL is not observed.
Lymphoplasmacytic lymphoma (LPL)
According to the WHO classification (2017), LPL is defined as a neoplasm of small B-lymphocytes, plasmacytoid lymphocytes, and plasma cells. In LPL, an increase in small B-lymphocytes is the main histological feature, accompanied by a few plasma cells. Thus, it is different from myeloma in which the increased cells are mainly plasma cells.
GENETIC ABNORMALITY
Recently, new gene abnormalities associated with myeloma have been detected using next-generation and whole genome sequencing, revealing that multiple gene abnormalities participate in the onset and progression of myeloma. From the viewpoint of prognostic prediction and treatment options, the specific gene abnormalities in myeloma are important.13
Chromosomal translocations, including the immunoglobulin heavy chain (IGH) domain on 14q32, and hyperdiploidy appear to be involved in the first step of the genesis of myeloma. Seven oncogenes, including CCND1 on 11q13, CCND2 on 12p13, CCND3 on 6p21, MAF on 16q23, MAFA on 8q24, MAFB on 20q11, and FGFR3 on 4p16.3, are involved in the 14q32 translocation. Furthermore, the development of myeloma is associated with the deletion of chromosome 13 (13q14), RAS point mutation, c-myc rearrangement, deletion or mutation of TP53 (17p13), gain of chromosome 1q, loss of 1p, and NF-kappa B activating mutations, among other factors. These gene abnormalities have a complex relationship with the onset and progression of myeloma.14,15
The most important genetic abnormalities associated with a high risk of myeloma are the deletion of TP53 (17p13) including the p53 domain, IGH-MAF t(14;16), and IGH-MAFB t(14;20). On the other hand, myeloma with the IGH-CCND1 t(11;14) genetic abnormality has a relatively good prognosis, and is classified in the standard risk group. The deletion of 17p13 is the gene abnormality associated with the poorest prognosis, and the merge rate of extramedullary lesions, CNS involvement, and plasma cell leukemia are high in myeloma with this genetic abnormality. We did not observe an association of p53 expression by immunohistochemical staining with the deletion of 17p13 by FISH analysis. IGH-FGFR3 t(4;14) was previously reported to indicate a poor prognosis, but it is currently classified in the intermediate risk group. Recently, it was confirmed that long-term bortezomib was effective for cases with IGH-FGFR3. Myeloma with multiple genetic factors indicative of poor prognosis has a worse prognosis.16,17
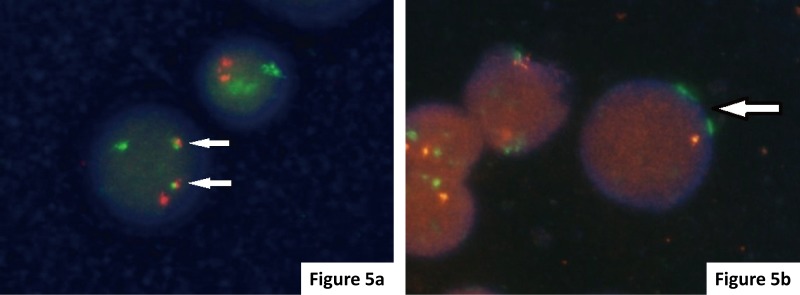
The IMWG recommends t(4;14)(p16;q32), t(14;16)(q32;q23), and del (17p13) as a minimal panel to evaluate gene abnormalities in myeloma.17 In our department, we have analyzed CCND1, FGFR3, MAF, and del (17p13) by FISH for all myeloma cases since 2011 (Table 1, Fig. 5). Table 2 presents the confirmed myeloma cases with gene abnormalities from 2011 to 2016. None of these cases with genetic abnormalities were recognized by G-banding. As the growth of myeloma cells is slow, the frequency of genetic abnormalities that can be detected by G-banding is approximately 15%. Analysis by FISH is more effective than G-banding for the evaluation of genetic abnormalities in myeloma.
Table 1. Myeloma-related FISH items used in the department.
| · IGH/CCND1 t(11;14) |
| LSI IgH/CCND1 Dual Color Dual Fusion Translocation Probe |
| · IGH/FGFR3 t(4;14) |
| LSI IgH/FGFR3 Dual Color Dual Fusion Translocation Probe |
| · IGH/MAF t(14;16) |
| LSI IgH/MAF Dual Color Dual Fusion Translocation Probe |
| · TP53 deletion del (17p13) |
| LSI TP53(17p13.1)/CEP17(17p11.1-q11.1) Probe |
Fig. 5.
FISH analysis of myeloma cells. (a) FGFR3-IGH: The presence of t(4;14) was noted in this case showing 1 red FGFR3 signal, 1 green IGH signal, and 2 yellow fusion signals (arrows). (b) TP53 deletion: The presence of del (17p) was identified in this case showing 1 red TP53 signal and 2 green CEP 17 signals (arrows).
Table 2. Myeloma gene abnormality in our hospital (2011 - 2016 / 7).
| Gene abnormality | FGFR3 | MAF | CCND1 | del(17) |
|---|---|---|---|---|
| Case number (%) |
8/113 (7.1%) |
3/113 (2.7%) |
15/113 (13.3%) |
7/113 (6.2%) |
CONCLUSION
In this report, I described the pathological findings of myeloma and gene analyses performed at our department. Regarding the pathological findings, a diagnosis of myeloma is possible even if less than 10% of cells are myeloma cells by evaluating the infiltration pattern, abnormal antigen expression, and monoclonality of the neoplastic cells. There are also secondary changes related to myeloma, such as myelofibrosis and amyloid deposition, and the evaluation of pathological findings can provide information that cannot be obtained from a smear. The frequency of detecting gene abnormalities associated with myeloma using G-banding is low; therefore, gene evaluation by FISH is useful in myeloma diagnosis.
Research on gene abnormalities in myeloma will advance and new gene abnormalities will be discovered. For prognostic prediction and therapy selection, pathological diagnosis, including genetic evaluation, will become more important in the future.
Footnotes
CONFLICT OF INTEREST: The author declares no conflict of interest in this manuscript.
REFERENCES
- 1.Rajkumar SV, Dimopoulos MA, Palumbo A, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014; 15: e538-e548. 10.1016/S1470-2045(14)70442-5 [DOI] [PubMed] [Google Scholar]
- 2.Greipp PR, Leong T, Bennett JM, et al. Plasmablastic morphology--an independent prognostic factor with clinical and laboratory correlates: Eastern Cooperative Oncology Group (ECOG) myeloma trial E9486 report by the ECOG Myeloma Laboratory Group. Blood. 1998; 91: 2501-2507. [PubMed] [Google Scholar]
- 3.Bartl R, Frisch B, Fateh-Moghadam A, et al. Histologic classification and staging of multiple myeloma. A retrospective and prospective study of 674 cases. Am J Clin Pathol. 1987; 87: 342-355. 10.1093/ajcp/87.3.342 [DOI] [PubMed] [Google Scholar]
- 4.Seili-Bekafigo I, Valković T, Babarović E, et al. Myeloma cell morphology and morphometry in correlation with clinical stages and survival. Diagn Cytopathol. 2013; 41: 947-954. 10.1002/dc.22986 [DOI] [PubMed] [Google Scholar]
- 5.Lin P, Owens R, Tricot G, et al. Flow cytometric immunophenotypic analysis of 306 cases of multiple myeloma. Am J Clin Pathol. 2004; 121: 482-488. 10.1309/74R4TB90BUWH27JX [DOI] [PubMed] [Google Scholar]
- 6.Pozdnyakova O, Morgan EA, Li B, et al. Patterns of expression of CD56 and CD117 on neoplastic plasma cells and association with genetically distinct subtypes of plasma cell myeloma. Leuk Lymphoma. 2012; 53: 1905-1910. 10.3109/10428194.2012.676174 [DOI] [PubMed] [Google Scholar]
- 7.Bataille R, Jégo G, Robillard N, et al. The phenotype of normal, reactive and malignant plasma cells. Identification of “many and multiple myelomas” and of new targets for myeloma therapy. Haematologica. 2006; 91: 1234-1240. [PubMed] [Google Scholar]
- 8.Fernández de Larrea C, Kyle RA, Durie BGM, et al. International Myeloma Working Group Plasma cell leukemia: consensus statement on diagnostic requirements, response criteria and treatment recommendations by the International Myeloma Working Group. Leukemia. 2013; 27: 780-791. 10.1038/leu.2012.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bataille R, Pellat-Deceunynck C, Robillard N, et al. CD117 (c-kit) is aberrantly expressed in a subset of MGUS and multiple myeloma with unexpectedly good prognosis. Leuk Res. 2008; 32: 379-382. 10.1016/j.leukres.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 10.Specht K, Haralambieva E, Bink K, et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood. 2004; 104: 1120-1126. 10.1182/blood-2003-11-3837 [DOI] [PubMed] [Google Scholar]
- 11.Kumar S, Kimlinger T, Morice W. Immunophenotyping in multiple myeloma and related plasma cell disorders. Best Pract Res Clin Haematol. 2010; 23: 433-451. 10.1016/j.beha.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sukpanichnant S, Cousar JB, Leelasiri A, et al. Diagnostic criteria and histologic grading in multiple myeloma: histologic and immunohistologic analysis of 176 cases with clinical correlation. Hum Pathol. 1994; 25: 308-318. 10.1016/0046-8177(94)90204-6 [DOI] [PubMed] [Google Scholar]
- 13.Lohr JG, Stojanov P, Carter SL, et al. Multiple Myeloma Research Consortium Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014; 25: 91-101. 10.1016/j.ccr.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zingone A, Kuehl WM. Pathogenesis of monoclonal gammopathy of undetermined significance and progression to multiple myeloma. Semin Hematol. 2011; 48: 4-12. 10.1053/j.seminhematol.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehl WM, Bergsagel PL. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest. 2012; 122: 3456-3463. 10.1172/JCI61188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chesi M, Bergsagel PL. Molecular pathogenesis of multiple myeloma: basic and clinical updates. Int J Hematol. 2013; 97: 313-323. 10.1007/s12185-013-1291-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009; 23: 2210-2221. 10.1038/leu.2009.174 [DOI] [PMC free article] [PubMed] [Google Scholar]