Abstract
Acute otitis media (AOM) is one of the most common bacterial infections in children. Empiric antibiotherapy leads to increasing antimicrobial resistance rates among otopathogens and may impair the correct development of the microbiota in early life. In this context, probiotics seem to be an attractive approach for preventing recurrent AOM (rAOM) through the restoration of the middle ear and nasopharyngeal microbiota. The aim of this study was the selection of a probiotic strain (Lactobacillus salivarius PS7), specifically tailored for its antagonism against otopathogens. Since L. salivarius PS7 was safe and displayed a strong antimicrobial activity against otopathogens, its efficacy in preventing rAOM was assessed in a trial involving 61 children suffering from rAOM. Children consumed daily ~1 × 109 CFU of L. salivarius PS7, and the number of AOM episodes were registered and compared with that observed in the previous 6 and 12 months. The microbiota of samples collected from the external auditory canal samples was quantitatively and qualitatively assessed. The number of AOM episodes during the intervention period decreased significantly (84%) when compared to that reported during the 6 months period before the probiotic intervention. In conclusion, L. salivarius PS7 is a promising strain for the prevention of rAOM in infants and children.
Keywords: Lactobacillus salivarius, otitis, probiotic, bacteriocin, prevention
1. Introduction
Acute otitis media (AOM) is one of the most common diseases in infancy and childhood, and the leading cause for seeking medical care and for the prescription of antibiotics, both in high-income and low-income countries [1,2]. Approximately 70% of infants experience at least one otitis episode by the age of 2 years, and 20–30% suffer from recurrent AOM (rAOM) [3]. rAOM is a very relevant issue in clinical practice since it causes pain and discomfort in children, causing a strong impact on their families and a relevant economic burden on society [3].
Middle ear and nasopharyngeal colonization with multiple bacterial otopathogens (mainly Streptococcus pneumoniae, Streptococcus pyogenes, Haemophilus influenzae and Moraxella catarrhalis) is considered to be the main risk for both AOM and rAOM [4,5]. Recent microbiome studies have revealed that Alloiococcus otitidis may also play a relevant role in the pathogenesis of otitis media [6,7], while other species frequently isolated from the middle ear fluid of children experiencing this condition include Staphylococcus aureus, Staphylococcus epidermidis, Pseudomonas aeruginosa and Escherichia coli [8].
In practice, eradicating or decreasing the concentration of such species through empiric systemic and topical antibiotherapy is usually considered as the main (and, often, unique) approach for the treatment of rAOM and, also, for providing prophylaxis for this condition [9]. However, a routine antibiotherapy may have negative consequences. First of all, it drives the selection of resistant otopathogens [9,10]. Although rAOM episodes occurring within one month from the completion of an antibiotic therapy may be the result of either a relapse or a new infection, antibiotic pressure seems to be essential for selecting the causal agent(s) in both circumstances [11,12]; in fact, the incidence of antibiotic-resistant S. pneumoniae and H. influenzae strains in nasopharyngeal samples is higher among children with rAOM than among healthy controls [13,14]. In addition, otopathogens, such as H. influenzae, S. pneumoniae, M. catarrhalis and A. otitidis, form polymicrobial biofilms within the middle ear [15,16,17], and bacteria within these structures have an increased antibiotic resistance [18]. Recently, it was shown that A. otitidis promoted H. influenzae survival in mixed biofilms by decreasing its antibiotic susceptibility and enhancing its growth under adverse conditions [17]. A Cochrane review [2] aimed to assess the effects of antibiotics for children with AOM revealed that, although they may be useful in children under two years of age with bilateral AOM and otorrhea, their global effect on health outcomes associated to this condition is limited. The same review encouraged the weighing of the benefits of antibiotics against possible harms, including adverse events (such as vomiting, diarrhea or rashes), and suggested that clinical management should provide a limited role for antibiotics, and that a search for new preventive and treatment strategies should be stimulated.
The collateral damage that antibiotics exert on the host’s health by eliminating prominent (but sensitive) members of the microbiota must also be taken into account. Our relationship with our microbiota is especially important during the first years of life, when the microbiome is still forming and any strong disturbance can have short, medium and long-term consequences for homeostasis and health [19,20,21,22,23,24]. The microbiome of the middle ear, ear canal and nasopharynx of healthy children with no history of AOM seems to be characterized by the presence of potentially protective commensal bacteria and the absence or low abundance of classic otopathogens [25].
In this context, probiotics seem to be an attractive approach for preventing rAOM through the restoration of the middle ear and nasopharyngeal microbiota. The lack of specificity of the probiotics used for this target may be one of the main reasons for the limited and contradictory results obtained so far [25,26]. Therefore, the aim of this study was the characterization of a probiotic strain specifically selected for its antagonism against otopathogens. In addition, other potentially properties related to its probiotic potential and safety were investigated, including the assessment of its acute and repeated-doses oral toxicity in a rat model. Finally, the efficacy of the selected strain (Lactobacillus salivarius PS7) in preventing rAOM in infants and children was assessed in a pilot clinical trial.
2. Materials and Methods
2.1. Isolation and Identification of the Strain
Strain PS7 was isolated in a de Man, Rogosa, and Sharpe (MRS, Oxoid, Basingstoke, UK) agar plate within the framework of a previous study to evaluate the bacterial diversity of milk from healthy women. Initially, the identification of the strain was performed by PCR amplification and the sequencing of the 16S rRNA gene using the primers pbl16 (5′-AGAGTTTGATCCTGGCTCAG-3′) and mbl16 (5′-GGCTGCTGGCACGTAGTTAG-3′) [27]. The identification was confirmed by Matrix Assisted Laser Desorption Ionization-Time of Flight (MALDI-TOF) mass spectrometry using a Vitek-MS™ instrument (BioMérieux, Marcy l’Etoile, France) [28]. The strain could be differentiated from other L. salivarius strains of our own collection by genotyping using randomly amplified polymorphic DNA (RAPD) analyses, as previously described [29].
2.2. Survival after In Vitro Exposure to Saliva and Gastrointestinal-Like Conditions
The survival of the strain to conditions resembling those found in the human digestive tract (saliva, human stomach and small intestine) was assessed in the in vitro system described by Marteau et al. [30], with the modifications reported by Martín et al. [31]. For this purpose, the strain was vehiculated in UHT-treated milk (25 mL) at a concentration of 109 CFU/mL. The values of the pH curve in the stomach-like compartment were those recommended by Conway et al. [32]. Different fractions were taken at 20, 40, 60, and 80 min from this compartment, and exposed for 120 min to a solution with a composition similar to that of human duodenal juice [30]. The survival rate of the strain was determined by culturing the samples on MRS agar plates, which were incubated at 37 °C for 48 h.
2.3. Adhesion to Caco-2/HT-29 Cells
The ability of the strain to adhere to HT-29 and Caco-2 cells was evaluated as described by Coconnier et al. [33] with the modifications reported by Martín et al. [31]. HT-29 and Caco-2 were cultured to confluence in 2 mL of DMEM medium (PAA, Linz, Austria) containing 25 mM of glucose, 1 mM of sodium pyruvate and supplemented with 10% heat-inactivated foetal calf serum, 2 mM of L-glutamine and 1% of a non-essential amino acid preparation. At day 10 after the confluence, 1 mL of the medium was replaced with 1 mL of DMEM containing 108 CFU/mL of the PS7 strain. The adherence was measured as the number of lactobacilli adhering to the cells in 20 random microscopic fields. The assay was performed by triplicate.
2.4. Production of Riboflavin, Folate and Cyanocobalamin
The riboflavin, folate and cyanocobalamin production by strain PS7 was determined using the microbiological assays described by Juarez del Valle et al. [34], Laiño et al. [35], and Horwitz [36], respectively. Lactobacillus rhamnosus ATCC 7469, L. rhamnosus NCIMB 10463 and 7469, and Lactobacillus delbrueckii B12 were used as the indicator organism for the biosynthesis of the respective vitamins. The riboflavin production in the riboflavin-free medium was confirmed by an HPLC analysis following a procedure described previously [34].
2.5. Antimicrobial Activity of Strain PS7 against AOM-Related Pathogens
Initially, an overlay method was used as previously described [37] to determine the ability of strain PS7 to inhibit the growth of a spectrum of bacterial strains previously isolated from clinical cases of AOM (own collection of the Complutense University of Madrid) including: A. otitidis MP02, S. pneumoniae MP07, S. pyogenes MP03, Enterococcus faecalis MP64, S. aureus MP29, S. epidermidis MP33, H. influenzae MP04, M. catarrhalis MP08, P. aeruginosa MP24 and E. coli MP69. Brain Heart Infusion (BHI, Oxoid, Basingstoke, UK), Columbia Nalidixic Acid (CNA, Biomerieux, Marcy-l’Étoile, France) or Trypticase Soy (TSA, Oxoid) agar plates (depending on the indicator strain) were overlaid with bacterial indicators, incubated at 37 °C for 48 h, and they were examined for zones of inhibition around the PS7 streaks.
2.6. Production of Specific Antimicrobials (Bacteriocins, Lactic Acid, Acetic Acid, Hydrogen Peroxide) by Strain PS7
Bacteriocin production was assayed using an agar diffusion method as described by Dodd et al. [38] and modified by Martín et al. [31], using the Gram-positive strains employed for the overlay method as indicator bacteria. Since, at that stage, it was already known that strain PS7 belongs to the Lactobacillus salivarius species, the strain was tested by PCR for the presence of the structural genes encoding salivaricins. More specifically, the primer couples used in this study were: (i) SalB-for (5′-TGATAAGAAAGAATTGGCACATATAATTG-3′) and SalB-rev (5′-TCTGTTTAACTACAAATATTTTGATTTGAATG-3′) for salivaricin B [39], and (ii) Abp118A-for (5′-AAACGTGGTCCTAACTGTGTAGG-3′) and Abp118B-rev (5′-AACGGCAACTTGTAAAACCACCAG-3′) for bacteriocin Abp-118 [40]. The PCR reactions were carried out as indicated in the respective articles.
L-and D-lactic acid and acetic acid production by the strain PS7 was quantified in MRS cultures using enzymatic kits (Roche Diagnostics, Mannheim, Germany), as previously reported [31]. The assays were performed in triplicate and the values were expressed as the mean ± SD. The pH values of the supernatants were also measured. Finally, hydrogen peroxide production by the strain PS7 was assayed using the procedures described by Song et al. [41] and by Yap and Gilliland [42]. L. johnsonii La1 was used as a positive control in both assays.
2.7. Coagregation Assays
The ability of the strain to aggregate with cells of the otitis-related strains cited above was investigated following the procedure of Reid et al. [43], as adapted by Younes et al. [44].
2.8. Co-Culture Studies
Broth co-cultures of strain PS7 and some of the otitis-related strains cited above were performed in a BHI broth, since it was observed that this medium allowed most of their growth. The tubes were initially inoculated at a concentration of ~1 × 108 CFU/mL for each of the bacterial strains (PS7 and the corresponding otitis-related strain) and incubated overnight at 37 °C in aerobic conditions. BHI monocultures of each of the strains used in these assays were performed as control cultures. After incubation, samples of all the co-cultures and monocultures were seeded onto MRS, CNA, TSA and BHI agar plates for a selective enumeration based on the ability of the strains to growth and to display differential colony morphologies when inoculated on such media.
2.9. Sensitivity to Antibiotics
The sensitivity of the strain PS7 to antibiotics was tested using the lactic acid bacteria susceptibility test medium (LSM) [45] and the microtiter VetMIC plates for lactic acid bacteria (National Veterinary Institute of Sweden, Uppsala, Sweden), as described previously [46]. Parallel, minimum inhibitory concentrations (MICs) were also determined by the E-test (AB BIODISK, Solna, Sweden) following the instructions of the manufacturer. The results were compared to the cut-off levels proposed by the European Food Safety Authority [47].
2.10. Formation of Biogenic Amines and Degradation of Mucin
The capacity of the strain PS7 to synthesize biogenic amines (tyramine, histamine, putrescine and cadaverine) from their respective precursor amino acids (tyrosine, histidine, ornithine and lysine; Sigma-Aldrich) was evaluated using the method described by Bover-Cid and Holzapfel [48]. The potential of the strain to degrade gastric mucine (HGM; Sigma, St. Louis, MO, USA) was evaluated in vitro as indicated by Zhou et al. [49].
2.11. Acute and Repeated Dose (4-Weeks) Oral Toxicity Studies in a Rat Model
Wistar male and female rats (Charles River Inc., Marget, Kent, UK) were used to study the acute and repeated dose (4-weeks) oral toxicity of the strain PS7 in a rat model. The acclimatization, housing and management (including feeding) of the rats was performed as previously described [50]. The rats were 56-days old at the initiation of treatment. Acute (limit test) and repeated dose (4 weeks) studies were conducted in accordance with the European Union guidelines (EC Council Regulation No. 440) and authorized by the Ethical Committee on Animal Research of the Complutense University of Madrid (protocol 270111).
In the acute (limit test) study, 24 rats (12 males, 12 females) were distributed into two groups of 6 males and 6 females each. After an overnight fasting, each rat received skim milk (500 µL) orally (control group or Group 1), or a single oral dose of 1 × 1010 CFU of L. salivarius PS7 dissolved in 500 µL of skim milk (treated group or Group 2). The doses of the test and control products were administered by gavage. At the end of a 14-day observation period, the rats were weighed, euthanized by CO2 inhalation, exsanguinated, and necropsied.
The repeated dose (4 weeks) (limit test) study was conducted in 48 rats (24 males, 24 females) divided in four groups of 6 males and 6 females each (control group or Group 3; treated group or Group 4; satellite control group or Group 5; and satellite treated group or Group 6). Rats either received a daily oral dose of skim milk (Groups 3 and 5) or of 1 × 109 CFU of L. salivarius PS7 dissolved in 500 µL of skim milk (Groups 4 and 6) over 4 weeks. All rats of Groups 3 and 4 were deprived of food for 18 h, weighed, euthanized by CO2 inhalation, exsanguinated, and necropsied on day 29. All animals of the satellite groups (Groups 5 and 6) were kept for a further 14-day period without treatment to detect delayed occurrence, persistence or recovery from potential toxic effects. All rats from Groups 5 and 6 were deprived of food for 18 h, weighed, euthanized by CO2 inhalation, exsanguinated, and necropsied on day 42.
The behavior and clinical observations, blood biochemistry and haematology analysis, organ weight ratios and histopathological analysis were conducted as described previously [50]. The bacterial translocation to the blood, liver or spleen, and the total liver glutathione (GSH) concentration was evaluated following the methods described by Lara-Villoslada et al. [51].
2.12. Pilot Clinical Trial: Prevention of rAOM in Children
In this prospective pilot clinical assay, 64 children (aged 10 months to 6 years) with a previous history of rAOM were recruited between September 2012 and April 2015. The primary outcome variables were the occurrence and duration of the AOM episodes. The secondary end point was the frequency of an otitis-related pathogen carriage in the external auditory canal. The inclusion criteria were at least four episodes of AOM during the preceding 12 months or at least three episodes during the preceding 6 months [52]. The exclusion criteria included chronic medication, chronic illnesses, lip or palatal cleft, programmed tympanostomy or adenoidectomy during the study, and lactose intolerance or cow’s milk protein allergy (because of the excipient used to administer the strain). Those children who had undergone tympanostomy or adenoidectomy during the preceding 6 months were also excluded. The data on the occurrence of AOM episodes from a similar population (age, gender) of otitis-prone children (n = 60), who were attended by the same pediatrician but did not receive the tested strain, were used for comparison purposes. Written informed consent was obtained from the parents or legal tutors in accordance with the Declaration of Helsinki. The protocol was approved by the Ethical Committee on Clinical Research of Hospital Universitario Clínico San Carlos (Madrid, Spain), under protocol B12/262.
During the 6-month intervention period, the recruited children consumed daily ~1 × 109 CFU of L. salivarius PS7. The parents recorded whether the child had or had not received the daily dose. Compliance was expressed as the percentage of days in which the child received the dose. The use of antibiotics (except for the treatment of an AOM episode) or other probiotics was not allowed during the study. A physical examination was performed by a pediatrician for each suspected AOM episode. AOM was diagnosed according to defined clinical criteria [52,53], including evidence of middle ear effusion, inflammation of the tympanic membrane and any other sign of an acute infection (fever, ear ache, otorrhoea, etc.). The number of AOM episodes and the duration of each episode were also recorded and compared to the same data obtained in the 6 months preceding the probiotic treatment.
Samples from the external auditory canal were collected with a sterile swab at the baseline and at the end (6 months) of the study and submitted for a bacterial analysis following the guidelines recommended by the Spanish Society of Infectious Diseases and Clinical Microbiology [53]. Microbial isolates were identified by a MALDI-TOF analysis as previously reported.
2.13. Statistical Analysis
All the quantitative assays included in this study were performed at least in triplicate. The quantitative data were expressed as the mean and standard deviation (SD). When not normally distributed, the data were presented as the median and interquartile range (IQR). For the murine assays, the data were expressed as the means ± standard error of the mean (SEM) of the determinations. The differences between the control and treated groups were evaluated with a one-way analysis of variance (ANOVA) followed by Dunnett’s test. A Wilcoxon signed rank test was used to compare paired microbiological data before and after the probiotic intake and the χ2 test was used to find differences in the detection frequencies of the bacterial species in the external auditory canal samples. For all the comparisons, differences were considered significant at p < 0.05. The statistical analyses were conducted using R (version 3.0.2, R-project) software [54].
3. Results
3.1. Identification and In Vitro Characterization of the Strain
Strain PS7 was identified as a member of the L. salivarius species (from now, L. salivarius PS7). A RAPD genotyping analysis showed that the profile of this strain was different to those of other L. salivarius strains from our own collection. Subsequently, the strain was deposited at the Spanish Type Culture Collection (CECT; Burjassot, Spain) under accession number CECT 9422.
In relation to the survival of L. salivarius PS7 after exposition to conditions resembling those found in the human digestive tract, an exposure to a saliva-like solution had no deleterious effect on the strain while the survival rate after a transit through the stomach- and small intestine-like compartments was at approximately 52% of the population initially inoculated.
The strain produced neither folates nor cyanocobalamin under the assayed conditions. In contrast, it was able to produce riboflavin at a total concentration of ~200 ng/mL (intracellular riboflavin: 165.00 ± 0.52 ng/mL; extracellular riboflavin: 34.74 ± 3.06 ng/mL).
L. salivarius PS7 showed a high ability to adhere to both Caco-2 and HT-29 cells. The mean ± SD of the number of adhered lactobacilli in 20 random microscopic fields was 697.1 ± 297.6 and 251.7 ± 82.3, respectively.
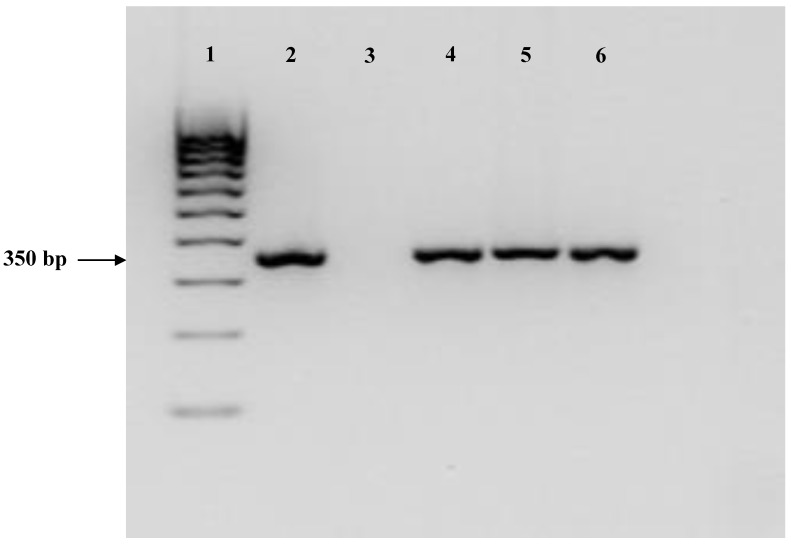
L. salivarius PS7 showed a clear antimicrobial activity (inhibition zone >2 mm around the streak) against most of the otitis-related indicator organisms used in this study (Table 1). Subsequently, and to try to elucidate the compound(s) responsible for the antimicrobial activity, the strain was screened for production of bacteriocins, organic acids (D-and L-lactic acid, acetic acid) and hydrogen peroxide. The strain displayed bacteriocin activity against the Gram-positive strains included as indicator organisms in this study (Table 1). A PCR analysis for structural genes of known salivaricins revealed that this strain produced the bacteriocin Abp-118 (both structural genes were identical) (Figure 1). L. salivarius PS7 produced a high concentration of L-lactic acid in the MRS broth while it did not produce the D-lactic acid isomer (Table 2). A significant concentration of acetic acid could also be detected in the culture supernatants of the strain (0.68 ± 0.17 mg/mL). This strain did not produce hydrogen peroxide.
Table 1.
Antimicrobial activity of L. salivarius PS7 against bacterial strains isolated from clinical cases of AOM, as assessed by different assays.
| Indicator Strain | Overlay Method (cm) | Well Diffusion Assay (cm) | Co-Cultures (CFU/mL) | ||
|---|---|---|---|---|---|
| Non-Adjusted pH | pH 6.2 | Initial Load | Final Load | ||
| S. pneumoniae MP07 | 3.6 | 1.1 | 1.1 | 7.40 | Nd a |
| S. pyogenes MP03 | 2.0 | 1.2 | 0.7 | 7.74 | Nd a |
| S. aureus MP29 | 1.6 | 1.3 | 1.2 | 7.52 | 4.81 a |
| S. epidermidis MP33 | 2.3 | 1.1 | 1.0 | 7.53 | 5.90 a |
| A. otitidis MP02 | 1.0 | 1.2 | 1.2 | – | – |
| E. faecalis MP64 | 0.5 | 0.4 | 0.4 | 8.02 | 8.78 b |
| H. influenzae MP04 | 2.2 | – | – | – | – |
| M. catarrhalis MP08 | 2.1 | – | – | – | – |
| P. aeruginosa MP24 | 1.5 | – | – | – | – |
| E. coli MP69 | 1.4 | – | – | 7.70 | 5.74 a |
ap < 0.01. b p = 0.29; Nd, no growth detected; – not assayed. AOM: Acute otitis media; CFU: colony-forming unit.
Figure 1.
PCR assay for the detection of the bacteriocin Abp118 structural gene. Lane 1: marker (HyperladderTM 100 bp, BIOLINE, London, UK). Lane 2: positive control. Lane 3: negative control. Lanes 4, 5 and 6: L. salivarius PS7.
Table 2.
Final pH and production of organic acids (mg/mL; mean ± SD) in an MRS broth by L. salivarius PS7.
| pH | L–Lactic Acid | D-Lactic Acid | Acetic Acid |
|---|---|---|---|
| 4.01± 0.04 | 10.29 ± 0.70 | Nd | 0.68 ± 0.17 |
Nd, not detectable.
L. salivarius PS7 showed a high potential for co-aggregating with bacterial strains involved in AOM cases, particularly with those belonging to the genera Streptococcus, Alloiococcus, Enterococcus and Staphylococcus. In addition, most of the otitis-related Gram-positive pathogens could not be detected, or their concentrations decreased notably after their overnight co-culture with the strain PS7 in the BHI broth (Table 1).
The antibiotic sensitivity assays showed that all the MIC values were below the cut-off values recommended by EFSA for all antibiotics, with the exception of kanamycin (MIC in this study: 128 µg/mL; EFSA cut-off value: 64 µg/mL) (Table 3). However, recent reports suggest that L. salivarius may be intrinsically resistant to kanamycin, and this aspect will be discussed below. Finally, L. salivarius PS7 was able neither to degrade gastric mucin in vitro nor to produce biogenic amines.
Table 3.
Minimum inhibitory concentrations (MICs) and cut-off values (μg/mL) of a variety of antibiotics for L. salivarius PS7.
| Antibiotics | Cut-Off Values * | MIC (L. salivarius PS7) |
|---|---|---|
| Ampicillin | 4 | 0.5 |
| Clindamycin | 4 | 0.5 |
| Chloramphenicol | 4 | 2 |
| Erythromycin | 1 | 0.25 |
| Streptomycin | 64 | 32 |
| Gentamicin | 16 | 2 |
| Kanamycin | 64 | 128 |
| Tetracyclin | 8 | 2 |
| Vancomycin | n.r. | >128 |
| Linezolid | 2 | 1 |
| Penicillin | 1 | 0.25 |
* EFSA (2018), except for linezolid and penicillin (Klare et al., 2007). n.r.: not required.
3.2. Acute and Repeated Dose (4-Weeks) oral Toxicity Studies in a Rat Model
All animals survived both oral toxicity trials. The development of the treated animals during the experimental periods corresponded to their species and age. At no time point of the experimental period were there any significant differences in body weight or body weight gain among the groups treated with L. salivarius PS7 (including the satellite ones) in comparison to the control groups. No abnormal clinical signs, behavioural changes, body weight changes, haematological and clinical chemistry parameters, macroscopic or histological findings, or organ weight changes were observed. There were no statistical differences in body weights among the groups. Similarly, no statistically significant differences in body weight gain, or in food and water consumption were observed between the groups.
No significant differences in the liver GSH concentration were observed between the control and treated groups (9.67 ± 1.42 vs. 9.71 ± 1.56 mmol/g, p > 0.1), and lactobacilli could not be isolated from the blood, liver or spleen of the treated rats.
3.3. Pilot Clinical Trial: Prevention of AOM in Children
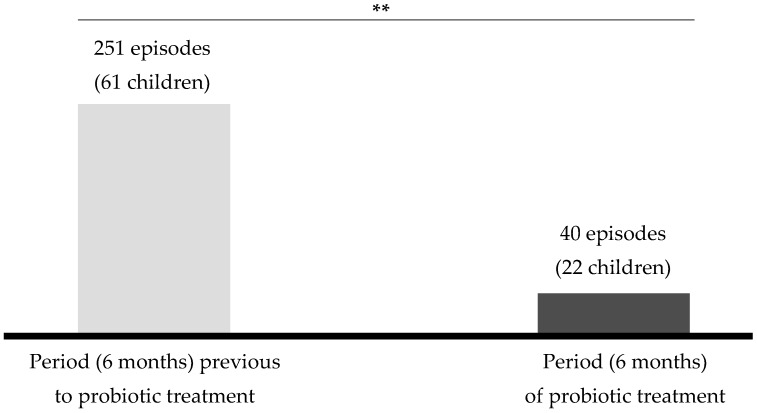
A total of 64 children who fulfilled the inclusion criteria were enrolled and received the probiotic treatment (Table 4). Three children (~4.6%) dropped out (one due to antibiotic intake; one due to tympanostomy; one due to a previously unadvertized allergy to cow’s milk protein); thus, 61 children completed the study. Compliance during the study was very high (≥ 96%). Twenty-two out of the 61 recruited children (36%) suffered at least one episode of AOM (median [25Q–75Q]), with a median duration of 4 days (Table 4). In contrast, ~70% of the children with rAOM who were attended by the same pediatrician in the same period but did not receive the probiotic strain suffered at least one AOM episode with a median duration of 6 days. The number of AOM episodes during the 6-month intervention period decreased by 84% when compared to the number registered in the 6 months that preceded the intervention (Figure 2). Although any child suffering an AOM episode received an antibiotic treatment, the number of antimicrobial treatments decreased by ~60% in the children receiving the probiotic strain with respect to the other otitis-prone children treated by the same pediatrician.
Table 4.
Main demographic characteristics of the infants and children recruited in the pilot trial (n = 61), and main outcomes of the study.
| Characteristic | Sex | n | Mean ± SD or Median * | p-Value |
|---|---|---|---|---|
| Age (Years) and Gender | 3.31 ± 1.7 | |||
| <3 years (n = 30) | Males | 16 | ||
| Females | 14 | |||
| ≥3 years (n = 31) | Males | 15 | ||
| Females | 16 | |||
| Inclusion months: | ||||
| September | 7 | |||
| October | 5 | |||
| November | 6 | |||
| December | 7 | |||
| January | 6 | |||
| February | 10 | |||
| March | 9 | |||
| April | 6 | |||
| May | 3 | |||
| June | 2 | |||
| History of AOM episodes/child | ||||
| Preceding 6 months | 4 (3–4) * | <0.001 ¥ | ||
| Preceding 12 months | 5 (5–6) * | <0.001 ¥ | ||
| During the study | 0 (0–1) * |
n, number of children; * Median (25Q–75Q); ¥ Wilcoxon rank sum test paired comparison.
Figure 2.
Comparison between the number of AOM episodes reported during the 6 months of the probiotic treatment and those reported during the previous 6 months. **, statistically significant change (p < 0.05; χ2 test).
Globally, the microbial density in the external auditory canal decreased notably along the intervention period, from ≥3.5 log10 CFU at the baseline (all the cultures were positive at this sampling time) to ≤ 2 log10 CFU at the end of the intervention (with 22% of the cultures being negative). At time 0, M. catharralis, coagulase-negative staphylococci and A. otitidis were the dominant bacteria in swabs taken from the external auditory canal of the recruited children, while coagulase-negative staphylococci and viridans streptococci dominated after 6 months of the probiotic treatment (Table 5). A. otitidis, H. haemolyticus, H. influenzae, M. catharralis, Neisseria spp, P. aeruginosa, S. aureus, S. pneumoniae and S. pyogenes, which were relatively common in the samples obtained at time 0, were not detected or their frequency of detection decreased notably at the end of the intervention (Table 5).
Table 5.
Results of the microbiological analysis of the samples from the external auditory canal taken at day 0 before the probiotic intervention and at the end of the 6-month treatment period.
| Time 0 | After 6 Months | p-Value | |
|---|---|---|---|
| Number of positive samples (bacterial growth) | 61 | 17 | < 0.001 ¥ |
| Median log10 CFU (per swab) | 4 (3.5–5) | 2 (1.5–2) | 0.012 * |
| Number of samples with the presence of: | |||
| Actinomyces europaeus | 2 | 0 | |
| Alloiococcus otitidis | 12 | 2 | |
| Coagulase-negative staphylococci | 14 | 15 | |
| Haemophilus haemolyticus | 11 | 1 | |
| Haemophilus influenza | 7 | 1 | |
| Moraxella catharralis | 14 | 3 | |
| Neisseria spp. | 6 | 0 | |
| Pseudomonas aeruginosa | 4 | 0 | |
| Rhodococcus ruber | 2 | 0 | |
| Staphylococcus aureus | 11 | 3 | |
| Streptococcus pneumoniae | 9 | 3 | |
| Streptococcus pyogenes | 10 | 6 | |
| Group viridans streptococci | 9 | 11 |
¥ Chi-squared test. * Wilcoxon rank sum test.
4. Discussion
In the present study, the probiotic potential of L. salivarius PS7 was characterized by using a wide variety of in vitro and in vivo assays, including its ability to antagonize otopathogens, its toxicological assessment in rats, and a pilot clinical trial for the initial evaluation of its efficacy for preventing rAOM in infants and children. The characterization scheme of this strain followed the guidelines for the evaluation of probiotics provided by FAO and WHO [55].
Both the survival rate (52%) of L. salivarius PS7 when exposed to conditions similar to those found in the digestive tract and its capacity for adhesion to intestinal cells were similar or higher than those found for commercial probiotic strains in previous studies using the same in vitro models [31,56]. These properties indicate that a high concentration of the strain can reach the pharynx and the gut mucosal surfaces and adhere to them. The adhesion ability of this strain may allow the competitive exclusion of pathogenic bacteria [57]. While its presence in the pharynx may facilitate its direct interaction with otopathogens, its presence in the gut might also have positive consequences. It has been shown that exposure (after swallowing) of the gut to upper respiratory pathogens involved in otitis media provides a potent modulation of the immune responses in the middle ear [58]. The potential ear-related immunomodulatory properties of L. salivarius PS7 were not addressed in this work but deserve future research.
In order to select a strain for the prevention of otitis, its ability to exert antagonism against otopathogens seems an essential property. The results of the in vitro assays to evaluate the antimicrobial activity of L. salivarius PS7 against a wide variety of strains and species previously isolated from clinical rAOM cases confirmed that it was a suitable candidate for such a target. All the otitis-related indicator strains were inhibited when the overlay method was used and, among them, all the Gram-positive strains were sensitive to the bacteriocin-like compound produced by L. salivarius PS7. The PCR analyses revealed that L. salivarius PS7 produces Abp-118. This is a potent broad-spectrum class II bacteriocin, which was originally described in L. salivarius UCC118 [59]. A previous study demonstrated that Abp-118 is produced in vivo and that it was the primary mediator conferring protection against infection by Listeria monocytogenes in mice [60]. The lack of inhibition of a few otopathogens (E. faecalis MP64) in the co-culture assay may be due to the fact that Abp-118 is not expressed when the bacteriocinogenic strain grows in a BHI medium. In addition, L. salivarius PS7 produced high amounts of L-lacic acid in the MRS cultures, and this is in agreement with the results obtained with other L. salivarius strains in previous studies [31,56]. Smaller concentrations of acetic acid were also present in the MRS cultures. The fact that L. salivarius PS7 cells were able to co-aggregate with most of the otopathogenic strains tested in this work may facilitate the exposure of otopathogens to the antimicrobial substances produced by L. salivarius PS7 [44]. Recently, it was shown that, to exert antimicrobial activity in mucosal surfaces, physical contact between lactobacilli and pathogens may be required [61]. Interestingly, the presence of lactobacilli in the nasopharynx has been associated with a reduced risk of respiratory conditions in children [62]. Unfortunately, the presence of Lactobacillus in ear or nasopharynx samples was not tested in this study.
Although L. salivarius is a species included among those with the “qualified presumption of safety” (QPS) status [63], L. salivarius PS7 was submitted to a thorough safety assessment. In relation to its pattern of antibiotic sensitivity, all the MIC values were below the cut-off values recommended by EFSA [47], with the exception of kanamycin. A resistance of lactobacilli to kanamycin is a common finding among L. salivarius strains [64,65,66] due to the lack of a transport system for this antibiotic. This type of intrinsic resistance does not represent a human health risk [47]. In a previous study, the MIC value of L. salivarius CECT 5713 (a commercial probiotic strain) for kanamycin (128 µg/mL) [46] was the same as the one obtained with L. salivarius PS7 in this study. The analysis of the genome of L. salivarius CECT 5713 revealed the absence of transmissible genes involved in kanamycin resistance. Moreover, L. salivarius PS7 did not produce biogenic amines and did not show any ability for mucin degradation. These results are also similar to those reported for other L. salivarius strains using the same assays [31,56]. An acute and repeated-dose oral toxicity assessment in rats showed that L. salivarius PS7 was completely safe in this animal model when administrated in doses ranging from 9 to 10 log10 CFU daily. No significant differences in the liver GSH concentration were observed between the control and treated groups, indicating that the treatment did not cause oxidative stress to rats. This result is consistent with the absence of bacteremia since no lactobacilli could be isolated from the blood, liver or spleen of the rats, indicating that the tested strains do not cause either a local or a systemic infection in rats. These findings are in agreement with those previously reported for another L. salivarius strain also isolated from human milk [51].
Because of the wide antimicrobial activity of L. salivarius PS7 against otopathogens and its safety, a pilot clinical trial was carried out to assess its efficacy in preventing rAOM in infants and children. Oral intakes of L. salivarius PS7 over 6 months led to a statistically significant reduction (84%) in the number of episodes of AOM in comparison to those observed in the same population during the 6 months preceding the probiotic intervention. Such a percentage is higher than that observed in previous studies after oral or nasal administration of other probiotic strains, such as Lactobacillus rhamnosus GG and LC705, Bifidobacterium breve 99 and Propionibacterium freudenreichii JS [67,68], and Streptococcus salivarius 24SMB or Streptococcus oralis 89a [69,70]. When AOM occurred, the median duration of AOM episodes was 4 days, which is lower than the value obtained in previous probiotic trials (5.6 days) [68].
The external ear canal may act as a bacterial reservoir for the same otopathogens usually found in the middle ear during AOM [17,25,71]. Therefore, ear canal samples were analyzed before and after the probiotic intake period. The probiotic treatment led to a statistically significant decrease in the number of ear canal samples that were positive for bacterial growth and in the bacterial density of the positive samples (Table 5). The frequency of detection of potential otopathogens (A.otitidis, Haemophilus haemolyticus, H. influenzae, M. catharralis, Neisseria spp., P. aeruginosa, S. aureus, S. pneumoniae and S. pyogenes) in the samples also decreased notably after the trial (Table 5). The only bacterial groups whose detection frequency remained similar before and after the probiotic treatment were coagulase-negative staphylococci and viridans streptococci. This change in the composition of the ear canal microbiota seems to be closely associated with the decrease in the number of AOM episodes. In previous studies, otopathogens were more frequently detected in children with otitis than in healthy controls [10,72,73]. In fact, a failure to erradicate otopathogens from the ear and/or nasoparhyngeal surfaces has been viewed as an explanation for rAOM episodes after treatments with antibiotics [4] or with generic non-otitis-targeted probiotics [68,74]. A lack of specificity may explain the conflicting evidence on the effectiveness of probiotics in preventing AOM [75,76]. In contrast, the presence of coagulase-negative staphylococci and viridans streptococci seems to be a feature of a healthy ear and nasopharynx [13,77].
We acknowledge that the clinical trial performed in this study must be considered as a preliminary “proof of concept” trial since it faces some design limitations (e.g., lack of a placebo group and randomization). However, the results have shown that, while antibiotics may still play a relevant role in treatment of AOM, a target-specific probiotic strain may have a preventive role, significantly reducing the number of episodes in children with rAOM and the number of antibiotic doses required to treat them. This is a relevant finding within the context both of the present “antibiotic resistance crisis” and of the steady rise in autoimmune diseases associated with host microbiota disturbances. Additional work is in progress in order to initiate a well-designed multicenter clinical trial to further confirm the results obtained in this work.
Acknowledgments
We acknowledge all the families that participated in this study. We are grateful to Leónides Fernández for the critical reading of the manuscript.
Author Contributions
J.M.R. and C.B. designed the study; N.C., V.M., R.A. and E.J. characterized the strain and performed the microbial analyses. M.L. and M.C. were the pediatricians involved in this study. J.M.R. and E.J. wrote, edited and revised the manuscript. All the authors approved the final version of the manuscript.
Funding
This work was supported by Laboratorios Casen Recordati SL. The funding agency had no role in study design, data collection and analysis.
Conflicts of Interest
E.J. and N.C. are employees of Probisearch S.L.U. C.B. is employee of Casen Recordati SL. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Rovers M.M. The burden of otitis media. Vaccine. 2008;26:G2–G4. doi: 10.1016/j.vaccine.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Venekamp R.P., Sanders S.L., Glasziou P.P., Del Mar C.B., Rovers M.M. Antibiotics for acute otitis media in children. Cochrane Database Syst. Rev. 2015:CD000219. doi: 10.1002/14651858.CD000219.pub4. [DOI] [PubMed] [Google Scholar]
- 3.Klein J.O. The burden of otitis media. Vaccine. 2000;19:S2–S8. doi: 10.1016/S0264-410X(00)00271-1. [DOI] [PubMed] [Google Scholar]
- 4.Pelton S.I., Leibovitz E. Recent advances in otitis media. Pediatr. Infect. Dis. J. 2009;28:S133–S137. doi: 10.1097/INF.0b013e3181b6d81a. [DOI] [PubMed] [Google Scholar]
- 5.Pichichero M.E. Ten-year study of acute otitis media in Rochester, NY. Pediatr. Infect. Dis. J. 2016;35:1027–1032. doi: 10.1097/INF.0000000000001216. [DOI] [PubMed] [Google Scholar]
- 6.Jervis-Bardy J., Rogers G.B., Morris P.S., Smith-Vaughan H.C., Nosworthy E., Leong L.E., Smith R.J., Weyrich L.S., de Haan J., Carney A.S., et al. The microbiome of otitis media with effusion in Indigenous Australian children. Int. J. Pediatr. Otorhinolaryngol. 2015;79:1548–1555. doi: 10.1016/j.ijporl.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 7.Chan C.L., Wabnitz D., Bardy J.J., Bassiouni A., Wormald P.J., Vreugde S., Psaltis A.J. The microbiome of otitis media with effusion. Laryngoscope. 2016;126:2844–2851. doi: 10.1002/lary.26128. [DOI] [PubMed] [Google Scholar]
- 8.Ngo C.C., Massa H.M., Thornton R.B., Cripps A.W. Predominant bacteria detected from the middle ear fluid of children experiencing otitis media: A systematic review. PLoS ONE. 2016;11:e0150949. doi: 10.1371/journal.pone.0150949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granath A. Recurrent acute otitis media: What are the options for treatment and prevention? Curr. Otorrhinolatyngol. Rep. 2017;5:93–100. doi: 10.1007/s40136-017-0151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlidağ T., Demirdağ K., Kaygusuz I., Ozden M., Yalçin S., Oztürk L. Resistant bacteria in the adenoid tissues of children with otitis media with effusion. Int. J. Pediatr. Otorhinolaryngol. 2002;64:35–40. doi: 10.1016/S0165-5876(02)00038-1. [DOI] [PubMed] [Google Scholar]
- 11.Brook I., Gober A.E. In vitro bacterial interference in the nasopharynx of otitis media-prone and non-otitis media-prone children. Arch. Otolaryngol. Head Neck Surg. 2000;126:1011–1013. doi: 10.1001/archotol.126.8.1011. [DOI] [PubMed] [Google Scholar]
- 12.Leibovitz E., Greenberg D., Piglansky L., Raiz S., Porat N., Press J., Leiberman A., Dagan R. Recurrent acute otitis media occurring within one month from completion of antibiotictherapy: Relationship to the original pathogen. Pediatr. Infect. Dis. J. 2003;22:209–216. doi: 10.1097/01.inf.0000066798.69778.07. [DOI] [PubMed] [Google Scholar]
- 13.Marchisio P., Claut L., Rognoni A., Esposito S., Passali D., Bellussi L., Drago L., Pozzi G., Mannelli S., Schito G., et al. Differences in nasopharyngeal bacterial flora in children with nonsevere recurrent acute otitis media and chronic otitis media with effusion: Implications for management. Pediatr. Infect. Dis. J. 2003;22:262–268. doi: 10.1097/01.inf.0000055063.40314.da. [DOI] [PubMed] [Google Scholar]
- 14.Klein J.O. Evidence Based Otitis Media. BC Decker; Hamilton, ON, Canada: 2003. Bacterial resistance and antimicrobial drug selection; pp. 429–437. [Google Scholar]
- 15.Hall-Stoodley L., Hu F.Z., Gieseke A., Nistico L., Nguyen D., Hayes J. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton R.B., Rigby P.J., Wiertsema S.P., Filion P., Langlands J., Coates H.L., Vijayasekaran S., Keil A.D., Richmond P.C. Multi-species bacterial biofilm and intracellular infection in otitis media. BMC Pediatr. 2011;11:94. doi: 10.1186/1471-2431-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan C.L., Richter K., Wormald P.J., Psaltis A.J., Vreugde S. Alloiococcus otitidis forms multispecies biofilm with Haemophilus influenzae: Effects on antibiotic susceptibility and growth in adverse conditions. Front. Cell Infect. Microbiol. 2017;7:344. doi: 10.3389/fcimb.2017.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fergie N., Bayston R., Pearson J.P., Birchall J.P. Is otitis media with effusion a biofilm infection? Clin. Otolaryngol. Allied Sci. 2004;29:38–46. doi: 10.1111/j.1365-2273.2004.00767.x. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka S., Kobayashi T., Songjinda P., Tateyama A., Tsubouchi M., Kiyohara C., Shirakawa T., Sonomoto K., Nakayama J. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol. Med. Microbiol. 2009;56:80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 20.Blaser M.J., Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutten N.B., Rijkers G.T., Meijssen C.B., Crijns C.E., Oudshoorn J.H., van der Ent C.K., Vlieger A.M. Intestinal microbiota composition after antibiotic treatment in early life: The INCA study. BMC Pediatr. 2015;15:204. doi: 10.1186/s12887-015-0519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azad M.B., Konya T., Persaud R.R., Guttman D.S., Chari R.S., Field C.J., Sears M.R., Mandhane P.J., Turvey S.E., Subbarao P., et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: A prospective cohort study. BJOG. 2016;123:983–993. doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 23.Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeissig S., Blumberg R.S. Life at the beginning: Perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 2014;15:307–310. doi: 10.1038/ni.2847. [DOI] [PubMed] [Google Scholar]
- 25.Lappan R., Imbrogno K., Sikazwe C., Anderson D., Mok D., Coates H., Vijayasekaran S., Bumbak P., Blyth C.C., Jamieson S.E., et al. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 2018;18:13. doi: 10.1186/s12866-018-1154-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niittynen L., Pitkäranta A., Korpela R. Probiotics and otitis media in children. Int. J. Pediatr. Otorhinolaryngol. 2012;76:465–470. doi: 10.1016/j.ijporl.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Kullen M.J., Sanozky-Dawes R.B., Crowell D.C., Klaenhammer T.R. Use of DNA sequence of variable regions of the 16SrRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 2000;89:511–518. doi: 10.1046/j.1365-2672.2000.01146.x. [DOI] [PubMed] [Google Scholar]
- 28.Marín M., Arroyo R., Espinosa-Martos I., Fernández L., Rodríguez J.M. Identification of emerging human mastitis pathogens by MALDI-TOF and assessment of their antibiotic resistance patterns. Front. Microbiol. 2017;8:1258. doi: 10.3389/fmicb.2017.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martín V., Maldonado-Barragán A., Moles L., Rodriguez-Baños M., Campo R.D., Fernández L., Rodríguez J.M., Jiménez E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012;28:36–44. doi: 10.1177/0890334411424729. [DOI] [PubMed] [Google Scholar]
- 30.Marteau P., Minekus M., Havenaar R., Huis In’t Veld J.H.J. Survival of lactic acid bacteria in a dynamic model of the stomach and small intestine: Validation and the effects of bile. J. Dairy Sci. 1997;80:1031–1037. doi: 10.3168/jds.S0022-0302(97)76027-2. [DOI] [PubMed] [Google Scholar]
- 31.Martín R., Olivares M., Marín M.L., Fernández L., Xaus J., Rodríguez J.M. Probiotic potential of 3 Lactobacilli strains isolated from breast milk. J. Hum. Lact. 2005;21:8–17. doi: 10.1177/0890334404272393. [DOI] [PubMed] [Google Scholar]
- 32.Conway P.L., Gorbach S.L., Goldin B.R. Survival of lactic acid bacteria in the human stomach and adhesion to intestinal cells. J. Dairy Sci. 1987;70:1–12. doi: 10.3168/jds.S0022-0302(87)79974-3. [DOI] [PubMed] [Google Scholar]
- 33.Coconnier M.H., Klaenhammer T.R., Kernéis S., Bernet M.F., Servin A.L. Protein-mediated adhesion of Lactobacillus acidophilus BG2FO4 on human enterocyte and mucus-secreting cell lines in culture. Appl. Environ. Microbiol. 1992;58:2034–2039. doi: 10.1128/aem.58.6.2034-2039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juarez del Valle M., Laiño J., Savoy de Giori G., LeBlanc J.G. Riboflavin producing lactic acid bacteria as a biotechnological strategy to obtain bio-enriched soymilk. Food Res. Int. 2014;62:1015–1019. doi: 10.1016/j.foodres.2014.05.029. [DOI] [Google Scholar]
- 35.Laiño J.E., LeBlanc J.G., Savoy de Giori G. Production of natural folates by lactic acid bacteria starter cultures isolated from artisanal Argentinean yogurts. Can. J. Microbiol. 2012;58:581–588. doi: 10.1139/w2012-026. [DOI] [PubMed] [Google Scholar]
- 36.Horwitz W. Official Methods of Analysis of AOAC International. AOAC International; Gaithersburg, MD, USA: 2000. [Google Scholar]
- 37.Magnusson J., Schnürer J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001;67:1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodd H.M., Horn N., Zhang H., Gasson M.J. A lactococcal expression system for engineered nisins. Appl. Environ. Microbiol. 1992;58:3683–3693. doi: 10.1128/aem.58.11.3683-3693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Çataloluk O. Molecular characterization of the gene encoding for salivaricin B activity and its flanking sequences. Turk. J. Biol. 2001;25:379–386. [Google Scholar]
- 40.Flynn S., van Sinderen D., Thornton G.M., Holo H., Nes I.F., Collins J.K. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology. 2002;148:973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 41.Song Y.L., Kato N., Matsumiy Y., Lu C.X., Kato H., Watanabe K. Identification of an hydrogen peroxide production by fecal and vaginal lactobacilli isolated from Japanese women and newborn infants. J. Clin. Microbiol. 1999;37:3062–3064. doi: 10.1128/jcm.37.9.3062-3064.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap P.S., Gilliland S.E. Comparison of newly isolated strains of Lactobacillus delbrueckii susp. lactis for hydrogen peroxide production at 5 °C. J. Dairy Sci. 2000;83:628–632. doi: 10.3168/jds.S0022-0302(00)74922-8. [DOI] [PubMed] [Google Scholar]
- 43.Reid G., McGroarty J.A., Gil Domingue P.A., Chow A.W., Bruce A.W., Eisen A., Costerton J.W. Coaggregation of urogenital bacterial in vitro and in vivo. Curr. Microbiol. 1990;20:47–52. doi: 10.1007/BF02094024. [DOI] [Google Scholar]
- 44.Younes J.A., van der Mei H.C., van den Heuvel E., Busscher H.J., Reid G. Adhesion forces and coaggregation between vaginal staphylococci and lactobacilli. PLoS ONE. 2012;7:e36917. doi: 10.1371/journal.pone.0036917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klare I., Konstabel C., Müller-Bertling S., Reissbrodt R., Huys G., Vancanneyt M., Swings J., Goossens H., Witte W. Evaluation of new broth media for microdilution antibiotic susceptibility testing of lactobacilli, lactococci, pediococci, and bifidobacteria. Appl. Environ. Microbiol. 2005;71:8982–8986. doi: 10.1128/AEM.71.12.8982-8986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langa S., Maldonado-Barragán A., Delgado S., Martín R., Martín V., Jiménez E., Ruíz-Barba J.L., Mayo B., Connor R.I., Suárez J.E., et al. Characterization of Lactobacillus salivarius CECT 5713, a strain isolated from human milk: From genotype to phenotype. Appl. Microbiol. Biotechnol. 2012;94:1279–1287. doi: 10.1007/s00253-012-4032-1. [DOI] [PubMed] [Google Scholar]
- 47.EFSA Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 2018;16:e05206. doi: 10.2903/j.efsa.2018.5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bover-Cid S., Holzapfel W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999;53:33–41. doi: 10.1016/S0168-1605(99)00152-X. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J.S., Gopal P.K., Hill H.S. Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int. J. Food Microbiol. 2001;63:81–90. doi: 10.1016/S0168-1605(00)00398-6. [DOI] [PubMed] [Google Scholar]
- 50.Anadón A., Martínez M.A., Ares I., Ramos E., Martínez-Larrañaga M.R., Contreras M.M., Ramos M., Recio I. Acute and repeated dose (4 weeks) oral toxicity studies of two antihypertensive peptides, RYLGY and AYFYPEL, that correspond to fragments (90-94) and (143-149) from alpha(s1)-casein. Food Chem. Toxicol. 2010;48:1836–1845. doi: 10.1016/j.fct.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Lara-Villoslada F., Sierra S., Martín R., Delgado S., Rodríguez J.M., Olivares M., Xaus J. Safety assessment of two probiotic strains, Lactobacillus coryniformis CECT5711 and Lactobacillus gasseri CECT5714. J. Appl. Microbiol. 2007;103:175–184. doi: 10.1111/j.1365-2672.2006.03225.x. [DOI] [PubMed] [Google Scholar]
- 52.Del Castillo F., Baquero F., de la Calle T., López M.V., Ruiz-Canela J., Alfayate S., Llop F.M., Ortega M.J.C., Rey C.C. Documento de consenso sobre etiología, diagnóstico y tratamiento de la otitis media aguda. Rev. Pediatr. Aten. Primaria. 2012;14:195–205. doi: 10.4321/S1139-76322012000400002. [DOI] [Google Scholar]
- 53.Díez O., Batista N., Bordes A., Lecuona M., Lara M. Microbiological diagnosis of upper respiratory tract infections. Enferm. Infecc. Microbiol. Clin. 2007;25:387–393. doi: 10.1157/13106964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The R Project for Statistical Computing. [(accessed on 29 July 2016)]; Available online: http: www.R-project.org.
- 55.FAO. WHO . Guidelines for the Evaluation of Probiotics in Food. Joint FAO/WHO Working Group; London, ON, Canada: 2002. Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food. [Google Scholar]
- 56.Martín R., Jiménez E., Olivares M., Marín M., Fernández L., Xaus J. Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int. J. Food Microbiol. 2006;112:35–43. doi: 10.1016/j.ijfoodmicro.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 57.Lee Y.K., Puong K.Y., Ouwehand A.C., Salminen S. Displacement of bacterial pathogens from mucus and Caco-2 cell surface by lactobacilli. J. Med. Microbiol. 2003;52:925–930. doi: 10.1099/jmm.0.05009-0. [DOI] [PubMed] [Google Scholar]
- 58.Bresnick S.D., Ryan A.F. Modulation of middle ear immune response by gut immunization. Laryngoscope. 1989;99:1293–1298. doi: 10.1288/00005537-198912000-00015. [DOI] [PubMed] [Google Scholar]
- 59.Claesson M.J., Li Y., Leahy S., Canchaya C., van Pijkeren J.P., Cerdeño-Tárraga A.M., Parkhill J., Flynn S., O’Sullivan G.C., Collins J.K., et al. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA. 2006;103:6718–6723. doi: 10.1073/pnas.0511060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corr S.C., Li Y., Riedel C.U., O’Toole P.W., Hill C., Gahan C.G. Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA. 2007;104:7617–7621. doi: 10.1073/pnas.0700440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Younes J.A., Reid G., van der Mei H.C., Busscher H.J. Lactobacilli require physical contact to reduce staphylococcal TSST-1 secretion and vaginal epithelial inflammatory response. Pathog. Dis. 2016;74:ftw029. doi: 10.1093/femspd/ftw029. [DOI] [PubMed] [Google Scholar]
- 62.Rosas-Salazar C., Shilts M.H., Tovchigrechko A., Schobel S., Chappell J.D., Larkin E.K., Gebretsadik T., Halpin R.A., Nelson K.E., Moore M.L., et al. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J. Allergy Clin. Immunol. 2018;142:1447–1456. doi: 10.1016/j.jaci.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.EFSA Scientific opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA J. 2017;15:4664. doi: 10.2903/j.efsa.2017.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Danielsen M., Wind A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003;26:1–11. doi: 10.1016/S0168-1605(02)00254-4. [DOI] [PubMed] [Google Scholar]
- 65.Zhou J.S., Pillidge C.J., Gopal P.K., Gill H.S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005;98:211–217. doi: 10.1016/j.ijfoodmicro.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 66.Klare I., Konstabel C., Werner G., Huys G., Vankerckhoven V., Kahlmeter G., Hildebrandt B., Müller-Bertling S., Witte W., Goossens H. Antimicrobial susceptibilities of Lactobacillus; Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 2007;59:900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- 67.Tapiovaara L., Lehtoranta L., Swanljung E., Mäkivuokko H., Laakso S., Roivainen M., Korpela R., Pitkäranta A. Lactobacillus rhamnosus GG in the middle ear after randomized, double-blind, placebo-controlled oral administration. Int. J. Pediatr. Otorhinolaryngol. 2014;78:1637–1641. doi: 10.1016/j.ijporl.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Hatakka K., Blomgren K., Pohjavuori S., Kaijalainen T., Poussa T., Leinonen M., Korpela R., Pitkäranta A. Treatment of acute otitis media with probiotics in otitis-prone children-a double-blind, placebo-controlled randomised study. Clin. Nutr. 2007;26:314–321. doi: 10.1016/j.clnu.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Marchisio P., Santagati M., Scillato M., Baggi E., Fattizzo M., Rosazza C. Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:2377–2383. doi: 10.1007/s10096-015-2491-x. [DOI] [PubMed] [Google Scholar]
- 70.La Mantia I., Varricchio A., Ciprandi G. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for preventing recurrent acute otitis media in children: A real-life clinical experience. Int. J. Gen. Med. 2017;10:171–175. doi: 10.2147/IJGM.S137614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chan C.L., Wabnitz D., Bassiouni A., Wormald P.J., Vreugde S., Psaltis A.J. Identification of the bacterial reservoirs for the middle ear using phylogenic analysis. JAMA Otolaryngol. Head Neck Surg. 2017;143:155–161. doi: 10.1001/jamaoto.2016.3105. [DOI] [PubMed] [Google Scholar]
- 72.Torretta S., Marchisio P., Drago L., Baggi E., De Vecchi E., Garavello W., Nazzari E., Pignataro L., Esposito S. Nasopharyngeal biofilm-producing otopathogens in children with nonsevere recurrent acute otitis media. Otolaryngol. Head Neck Surg. 2012;146:991–996. doi: 10.1177/0194599812438169. [DOI] [PubMed] [Google Scholar]
- 73.Buzatto G.P., Tamashiro E., Proenca-Modena J.L., Saturno T.H., Prates M.C., Gagliardi T.B., Carenzi L.R., Massuda E.T., Hyppolito M.A., Valera F.C.P., et al. The pathogens profile in children with otitis media with effusion and adenoid hypertrophy. PLoS ONE. 2017;12:e0171049. doi: 10.1371/journal.pone.0171049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohen R., Martin E., de La Rocque F., Thollot F., Pecquet S., Werner A., Boucherat M., Varon E., Bingen E., Levy C. Probiotics and prebiotics in preventing episodes of acute otitis media in high-risk children: A randomized, double-blind, placebo-controlled study. Pediatr. Infect. Dis. J. 2013;32:810–814. doi: 10.1097/INF.0b013e31828df4f3. [DOI] [PubMed] [Google Scholar]
- 75.Popova M., Molimard P., Courau S., Crociani J., Dufour C., Le Vacon F., Carton T. Beneficial effects of probiotics in upperrespiratory tract infections and their mechanical actions to antagonize pathogens. J. Appl. Microbiol. 2012;113:1305–1318. doi: 10.1111/j.1365-2672.2012.05394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levi J.R., Brody R.M., McKee-Cole K., Pribitkin E., O’Reilly R. Complementary and alternative medicine for pediatric otitis media. Int. J. Pediatr. Otorhinolaryngol. 2013;77:926–931. doi: 10.1016/j.ijporl.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 77.Tano K., Grahn Håkansson E., Holm S.E., Hellström S. A nasal spray with alpha haemolytic streptococci as long term prophylaxis against recurrent otitis media. Int. J. Pediatr. Otorhinolaryngol. 2002;62:17–23. doi: 10.1016/S0165-5876(01)00581-X. [DOI] [PubMed] [Google Scholar]