Abstract
In this study, we investigated the role of eukaryotic translation initiation factor 3 subunit G (EIF3G) in colorectal cancer. Immunohistochemical analysis showed higher EIF3G expression in stage IV human colorectal cancer tissues than in adjacent normal tissues (P<0.01). EIF3G short hairpin RNA (shRNA) knockdown in HCT116 colon cancer cells reduced proliferation and increased apoptosis as compared to control. EIF3G knockdown also increased autophagy and reduced mTOR signaling, as evidenced by low phospho-AKT, phospho-S6K and phospho-4EBP1 levels. Functional experiments indicated that overexpression of EIF3G promoted HCT-116 cells proliferation, migration and xenograft tumor growth. Finally, we observed lower xenograft tumor weights and volumes with EIF3G-silenced HCT116 cells than with control cells. These findings demonstrate that EIF3G promotes colon cancer growth and is a potential therapeutic target.
Keywords: EIF3G, colorectal cancer, proliferation, apoptosis, autophagy
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the fourth leading cause of cancer mortality in the worldwide [1,2]. The survival rate for patients with CRC depends on the stage at diagnosis. Although localized and regional stage patients with CRC have 5-year survival rates of 90% and 70%, respectively, patients with regional and distant metastatic stage CRC have very low survival rates of 13% [3]. Liver is the most common site of CRC metastasis since majority of the intestinal mesenteric drainage enters the hepatic portal venous system. More than 50% of patients with CRC develop liver metastatic disease, which leads to death of more than two thirds of these patients [4,5]. Therefore, novel effective therapies for CRC are urgently needed. Previous studies indicated that somatic mutations in KRAS, BRAF, PIK3CA, AKT1, PTEN, NRAS, and TGFBR2 genes are common in CRC and have been used as prognostic biomarkers or therapeutic targets [6-12]. However, the exact mechanisms underlying the onset of colorectal cancer are still unknown.
Eukaryotic translation initiation factors (EIFs) regulate cellular protein synthesis at the level of mRNA translation initiation [13]. EIF3 is the most complex translation initiation factor that mediates mRNA binding to the 40S ribosomal subunit [14]. In most multicellular eukaryotes, EIF3 is composed of 13 subunits, named eIF3a-eIF3m [15-17]. Differential expression of EIF3 subunits are frequently observed in many human cancers [18,19]. Moreover, overexpression of some EIF subunits drives de novo holo-complex formation, which results in modest increase in protein synthesis along with cell transformation [20]. However, the specific mechanisms related to transformation are unknown.
The eukaryotic translation initiation factor 3 subunit G (EIF3G) binds to the eIF3 holo-complex via EIF3E. The presence of EIF3G and EIF3E distinguishes a sub-complex that translates a restricted set of mRNAs from another complex, which contains the full complement of EIF3 subunits and promotes the translation of all mRNAs [21,22]. Recently, EIF3G has been implicated in the development and progression of several tumors. It is involved in caspase-mediated apoptosis in breast cancer [23], ovarian cancer [24], and prostate cancer cells [25], thereby suggesting an oncogenic role for EIF3G in cancer. However, the mechanisms by which EIF3G regulated the apoptosis and autophagy in CRC remain unclear. Therefore, in this study, we explored the role of EIF3G in colorectal cells and pathways involved in its regulation.
Materials and methods
Cell culture
Human colon cancer HCT116, SW480 and human embryonic kidney 293T cell lines were purchased from American type culture collection (ATCC, Manassas, VA, USA). HCT116 cells were grown in McCoy’s 5A medium (Sigma Aldrich, St. Louis, MO, USA) with 10% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific, MA, USA) at 37°C and 5% CO2. HEK293T cells were grown in Dulbecco’s modified eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% heat-inactivated FBS.
Immunohistochemistry
Immunohistochemical staining of human colon cancer tissue specimens collected from Liaocheng People’s Hospital were carried out as previously described (25). In brief, the specimens were first deparaffinized and subjected to heat-induced antigen retrieval with citrate buffer. Then, the samples were incubated with primary anti human EIF3G antibody overnight at 4°C (1:1000; Bethyl Laboratories, Inc., Montgomery, TX, USA; cat. no. A301-757A). After subsequent washes, the samples were treated with biotinylated anti-rabbit secondary antibody followed by avidin-conjugated horseradish peroxidase and developed with diaminobenzidine as the substrate (1:1000; Santa Cruz, CA, USA; cat. no. SC-2379). The samples were counterstained with hematoxylin and the images were visualized and scored under microscope.
The scoring criteria for staining intensity were as follows: score 0, negative staining; score 1, weak staining; score 2, moderate staining; and score 3, strong staining. The scoring for positive staining was as follows: score 0, no staining; score 1, 0-20% staining; score 2, 21-60% staining; and score 3, 61-100% staining. Total score was the sum of scores for staining intensity and the proportion of positive staining. A total score of 0 indicated negative; score 1, weak; score 2-4, moderate and score 5-6, strong.
EIF3G silencing and lentiviral infection
The 4 EIF3G shRNA designed by Invitrogen Block-iT RNAi Designer (http://rnaidesigner.thermofisher.com/rnaiexpress/) were as follows: EIF3G shRNA1, 5’-GCGGAATCGAATGAGATTTGC-3’; EIF3G shRNA2, 5’-GGAACATGTTGCAGTTCAACC-3’; EIF3G shRNA3, 5’-GCCCTAGAATACTACGACAAA-3’; and EIF3G shRNA4, 5’-GGGATAAATGCTGTCTGATAG-3’. The control scramble shRNA 5’-GCGGAGGGTTTGAAAGAATTA-3’ was used as a negative control. The shRNAs were cloned in lentiviruses. High-titer lentiviruses were generated by transient transfection of HEK293T cells with Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific, MA, US). Stably transfected HCT116 cells were generated with the relevant lentiviruses along with polybrene. The stable control and EIF3G transfectants were selected in fresh growth medium supplemented with puromycin at 16 h post-infection. After subsequent culturing in puromycin select medium, the knockdown efficiency was evaluated by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western blot.
EIF3G stably overexpressed HCT116 cell line construction
In order to construct EIF3G stably overexpressed CRC cell line, the over-expression vector of EIF3G was constructed in pcDNA3.1 by Genscript (Nanjing, China). The plasmid was transfected into HCT116 cells, and selected using neomycin (1000 µg/ml) for four weeks.
Quantitative RT-PCR (qRT-PCR)
Total RNA was isolated from human tissues or cells with Trizol (Invitrogen, Thermo Fisher Scientific, MA, USA) according to the manufacturer’s instructions. Then, cDNA was synthesized with 2 μg total RNA using SuperScript II RNase H Reverse Transcriptase Kit (Life Technologies). Real-time PCR was performed with SYBR Green PCR mix (Thermo Fisher) with the following EIF3G primers: 5’-CTGGAGGAGGGCAAATACCT-3’ (sense) and 5’-CTCGGTGGAAGGACAAACTC-3’ (antisense). As control, the following GAPDH primers were used: 5‘-ACACTCACTCTTCTACCTTC-3’ (sense) and 5’-TTGCTGTAGCCAAATTCATT-3’ (anti-sense). The qPCR procedure was performed in a Bio-Rad CFX Real-time System (Bio-Rad, USA) as follows: initial denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 15 s and annealing extension at 60°C for 30 s. The relative expression of EIF3G was determined by the 2-ΔΔCt method where Ct refers to cycle threshold.
Western blot
Total protein was extracted from HCT116 cells with Sodium dodecyl sulfate (SDS) sample buffer (50 mM Tris-HCl pH 6.8, 5 mM EDTA, 2% SDS and 5% glycine). Protein concentration was determined by BCA Protein Assay Kit (Thermo Fisher, MA, USA). Equal amounts (30 μg) of protein from each sample were seperated in a 10% polyacrylamide SDS gel at 80V for 2 h. The separated proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, MA, USA) at 200 mA for 3 h. The membranes were blocked for 1 h with 5% non-fat milk in 1X Tris-buffered Saline with Tween 20 (TBST, 20 mM Tris pH 7.6, 150 mM NaCl and 0.05% Tween-20) followed by primary antibodies at 4°C overnight. Then, the membranes were washed thrice with 1XTBST and incubated for 2 h at room temperature with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (Santa Cruz, dilution 1:5000). The primary antibodies included rabbit anti human EIF3G (1:10,000; Bethyl Laboratories, Inc., Montgomery, TX, USA; cat. no. A301-757A), mouse anti human β-actin (1:1000; cat. no. 4970), phospho-Akt (Ser 473; 1:1000; cat. no. 4058), phospho-p70 S6K (Thr389; 1:1000; cat. no. 9430), phospho-4E-BP1 (Ser65; 1:1000; cat. no. 9456) and cleaved caspase-3 (1:1000; cat. no. 4970) from Cell Signaling Technology (Danvers, MA, USA).
Cell proliferation assay
Cell proliferation was measured by cell counting kit 8 (#96992, Sigma Aldrich, USA) according to manufacturer’s protocol. Briefly, experimental and control HCT116 cells were seeded in 96-well plate at a density of 3000 cells per well and cultured for 3 days. Then, the cells were further incubated for another 2 hrs with 1/10th volume of 2-(2-methoxy-4-nitroph-enyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt solution. The plate was read in a microplate reader at 450 nm. Cell proliferation was also detected using Ethynyl deoxyuridine (EdU) Kit (Thermo Fisher, MA, USA), according to the manufacturer’s protocol. All experiments were performed in triplicates.
Flow cytometry
Apoptosis was measured in control and EIF3G shRNA transfected HCT116 cells with Annexin V-FITC and propidium iodide (PI) using the Dead Cell Apoptosis Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions and then assessed by flow cytometry using the FACS Calibur (BD Biosciences). In brief, cells were harvested at 80% confluence after trypsinization and washed with ice-cold PBS. Then, the cells were fixed in 70% cold ethanol, centrifuged, washed with 1X PBS washes and stained with Annexin V/PI solution for 30 min at room temperature. The cells were analyzed by flow cytometry. FACS plots were analyzed to determine percent early apoptotic (AnnexinV+ PI-), late apoptotic/secondary necrotic (AnnexinV+ PI+), and necrotic (AnnexinV- PI+) cells.
Monodansylcadaverine (MDC) staining
The auto fluorescent agent, monodansylcadaverine (MDC), was used to analyze the autophagic process. After culturing for three days, autophagy was induced in HCT116 cells by amino acid starvation for 24 h. Then, the cells were incubated with 0.05 mM MDC in PBS at 37°C for 10 minutes. After incubation, the cells were washed thrice with PBS and immediately observed under a fluorescence microscope (Olympus, Tokyo, Japan). The cells were assessed by flow cytometry and MDC stained cells were quantified.
Cell migration assay
Cell migration was measured by using 24-well poly-carbonate transwell inserts (Millipore, Bedford, MA, USA) according to manufacturer’s protocol. Briefly, indicated CRC cells suspended in FBS free McCoy’s 5A medium plated in the upper well of transwell inserts. Medium supplemented with 10% FBS was added to the lower well. The cells on the upper surface of inserts were scraped off, and the cells on the lower surface were fixed and stained with crystal violet after 48 hrs incubation.
Animal study
Six to eight weeks old athymic nu/nu nude mice were purchased from Vital River (China, Beijing) and housed under 12-hour light/dark cycles with autoclaved laboratory rodent diet and water. Aliquots of HCT116 cells (5×106 cells, in 100 μL of PBS) were injected subcutaneously into the right armpit area of the mice. When the tumor volume reached 180 mm3, 50 nM EIF3G shRNA2, control shRNA, pcDNA3.1 or pcDNA3.1-EIF3G were injected into tumors twice a week. Tumor volume and body weight of tumor bearing mice were measured twice a week. Tumor volume was calculated by the following formula: Tumor volume (mm3) = length (mm) × width (mm) × width (mm) × 1/2.
Statistical analysis
Statistical analysis was performed with the SPSS 11.0 software (SPSS Inc., Chicago, IL, USA). The immunohistochemical data was analyzed by the Kruskal-Wallis nonparametric one-way ANOVA followed by the Nemenyi pairwise multiple tests. Data were expressed as mean ± SD and were analyzed by one-way analysis of variance followed by the Student-Neuman-Keul’s test. P<0.05 was considered statistically significant.
Ethics statement
Informed consent was obtained from all patients, and experiments were performed in accordance with the Declaration of Helsinki rules. The animal study was conducted as approved by the local ethics committee of Liaocheng People’s Hospital.
Results
EIF3G is highly expressed in colorectal cancer tissues
Immunohistochemical analysis of EIF3G staining in colorectal cancer tissue specimens showed no correlation between EIF3G expression and different cancer stages (Table 1, P=0.286). However, EIF3G staining gradually increased from stages I to IV and was the strongest in stage IV, which is highly metastatic. Furthermore, as shown in Figure 1A, EIF3G was mainly localized to the cytoplasm, and its expression was higher in tumor tissues than in adjacent normal tissues of stages III and IV. Quantitative RT-PCR and western blot analysis demonstrated high EIF3G mRNA and protein expression in tumor tissues than in adjacent tissues, respectively (Figure 1B and 1C).
Table 1.
The expression of EIF3G in CRC patients’ tissues (IHC)
| Cancer staging AJCC/UICC | n | EIF3G expression | P-Value# | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Negative | Weak | Moderate | Strong | ||||
| 0 | 31 | 18 | 10 | 2 | 1 | ||
| I | A | 21 | 10 | 7 | 4 | 0 | |
| B | 18 | 7 | 7 | 1 | 3 | ||
| C | 27 | 11 | 7 | 7 | 2 | ||
| II | A | 17 | 4 | 12 | 0 | 1 | |
| B | 16 | 10 | 1 | 3 | 2 | 0.286 | |
| C | 27 | 5 | 10 | 6 | 6 | ||
| III | A | 18 | 10 | 0 | 3 | 5 | |
| B | 19 | 6 | 4 | 5 | 4 | ||
| C | 28 | 9 | 4 | 7 | 8 | ||
| IV | A | 23 | 4 | 2 | 7 | 10 | |
| B | 19 | 5 | 3 | 8 | 3 | ||
The data was analyzed by the Kruskal-Wallis nonparametric one-way ANOVA followed by the Nemenyi pairwise multiple test.
P<0.05 was considered a significant different.
Figure 1.
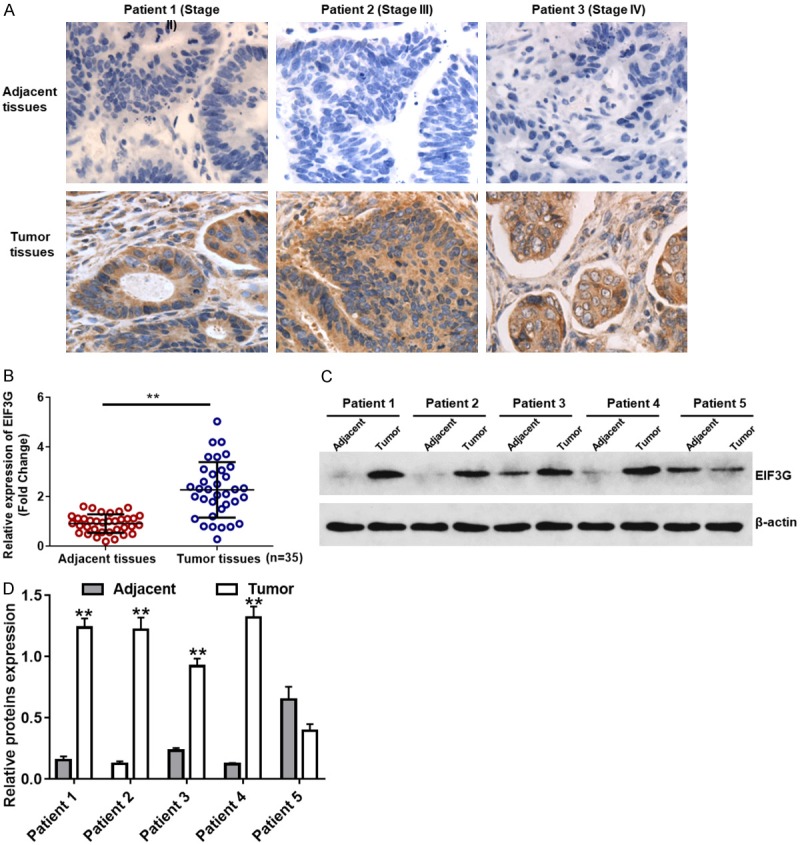
Immunochemistry of EIF3G in stage IV colorectal cancer. A. Three representative images showing immunohistochemical staining of EIF3G in stage IV colorectal cancer patient tumor and adjacent normal tissues (magnification, ×200). The brown dots represent EIF3G staining and the blue dots represent nuclear staining by hematoxylin. B. Quantitative RT-PCR analysis showing EIF3G mRNA expression in tumor and adjacent normal tissues from CRC patients (n=35). C. Western blot analysis showing EIF3G protein expression in tumor and adjacent normal tissues from CRC patients. D. Quantitative analysis of relative EIF3G protein expression in tumor and adjacent normal tissues from CRC patients. **P<0.01 compared with the adjacent group.
EIF3G knockdown efficiency in HCT116 cells by shRNAs
Next, we compared EIF3G knock down efficiency in HCT116 cells transfected with 4 different EIF3G shRNAs (1-4) and 1 control scramble shRNA. QRT-PCR and western blot analysis revealed that EIF3G mRNA and protein levels were significantly decreased by shRNAs 2 and 4 than shRNAs 1, 3 and scramble, respectively (Figure 2A and 2B).
Figure 2.
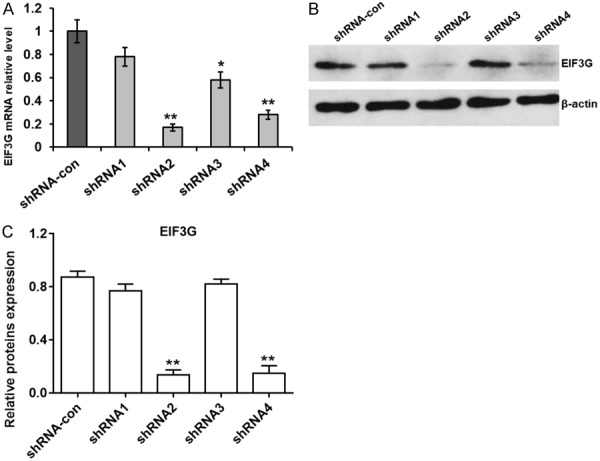
EIF3G shRNA knockdown in HCT116 cells. A. QRT-PCR analysis of EIF3G mRNA levels in control and EIF3G shRNA (1-4) transfected HCT116 cells with β-actin as internal control (n=3). B. Representative western blot showing EIF3G protein levels in control and EIF3G shRNA (1-4) transfected HCT116 cells with β-actin as internal control. C, Quantitative analysis of relative EIF3G protein expression in control and EIF3G shRNA (1-4) transfected HCT116 cells (n=3). **P<0.01, *P<0.05 compared with the shRNA-con group.
EIF3G silencing suppresses HCT116 cell proliferation
Next, we analyzed the consequences of EIF3G knockdown in HCT116 cells. As shown in Figure 3A, there were no changes in cell morphology due to EIF3G knockdown. However, EIF3G shRNA2 reduced cell density after 72 h. CCK-8 cell proliferation assay demonstrated that EIF3G silencing decreased HCT116 growth rate (Figure 3B). This suggested that EIF3G promoted colorectal cell proliferation.
Figure 3.
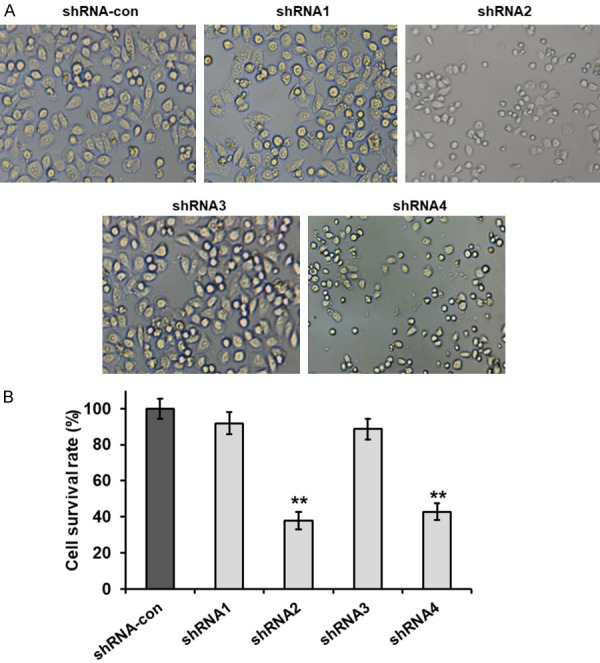
EIF3G knockdown inhibits HCT116 cell proliferation. A. Representative images showing cell numbers and morphology of control and EIF3G shRNA transfected HCT116 cells (200×). B. CCK8 cell proliferation analysis of control and EIF3G shRNA (1-4) transfected HCT116 cells at 72 h. **P<0.01 compared with the shRNA-con group.
EIF3G silencing promotes HCT116 cell apoptosis
We analyzed apoptosis by flow cytometry of AnnexinV and PI double stained control and EIF3G silenced HCT116 cells. EIF3G silenced HCT116 cells showed increased early and late apoptosis than controls (Figure 4A and 4B). This was further confirmed by increased cleaved caspase-3 in EIF3G silenced HCT116 cells than in controls (Figure 4C and 4D). We have repeated these experiments using another CRC SW480 cell lines. The results were consistent with data using HCT-116 (Figure S1A-D).
Figure 4.
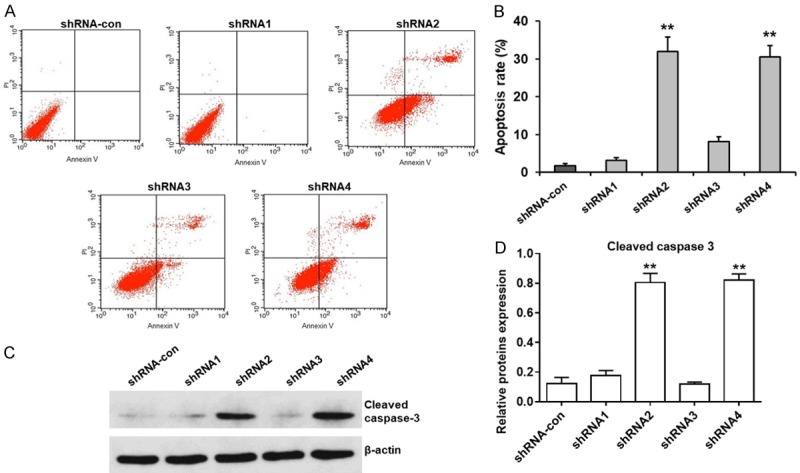
EIF3G knockdown promotes HCT116 cell apoptosis. A. Representative FACS plots show apoptosis in control and EIF3G shRNA (1-4) transfected HCT116 cells as determined by Annexin V-FITC/PI double staining. B. Quantitative analysis of percent early apoptotic (AnnexinV+ PI-) cells and late apoptotic (AnnexinV+ PI+) cells in control and EIF3G shRNA (1-4) transfected HCT116 cells (n=3). C. Representative western blot showing cleaved caspase-3 levels in control and EIF3G shRNA (2 and 4) transfected HCT116 cells at 72 h. D. Quantitative analysis of cleaved caspase 3 levels in control and EIF3G shRNA (2 and 4) transfected HCT116 cells at 72 h (n=3). **P<0.01, *P<0.05 compared with the shRNA-con group.
EIF3G silencing induces HCT116 cell autophagy
Next, we analyzed the effect of EIF3G silencing on autophagy in HCT116 cells by monodansylcadaverine (MDC) staining. EIF3G silenced HCT116 cells demonstrated autophagosome staining under fluorescence microscopy (Figure 5A). Flow cytometry analysis of MDC stained cells demonstrated increased autophagy in EIF3G silenced HCT116 cells than in controls (Figure 5B). Western blot analysis demonstrated that down-regulation of EIF3G reduced phosphorylated Akt, p70 S6K and 4E-BP1 levels (Figure 5C-F). These data demonstrated that down-regulation of EIF3G induced HCT116 cell autophagy by inhibiting the mTOR signaling pathway.
Figure 5.
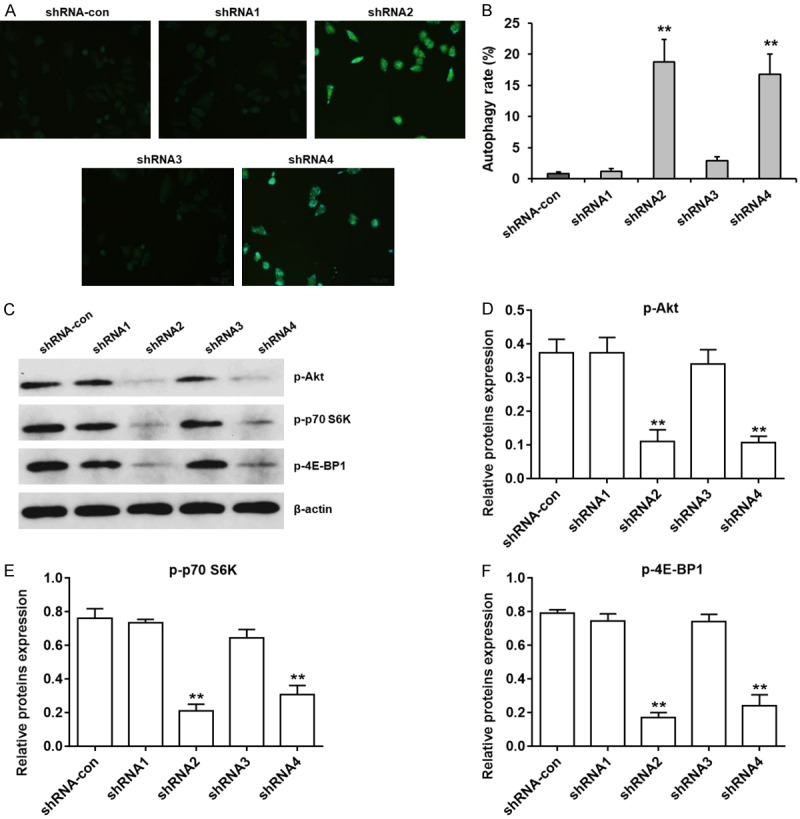
EIF3G knockdown induces autophagy in HCT116 cells. A. Representative fluorescence micrographs of monodansylcadaverine (MDC) stained control and EIF3G shRNA (2 and 4) transfected HCT116 cells at 72 h. B. Quantitative analysis of cell autophagy rate in control and EIF3G shRNA (2&4) transfected HCT116 cells at 72 h (n=3). C. Representative western blots showing levels of phosphorylated and total Akt, p70 S6K and 4-E-BP1 in control and EIF3G shRNA (2&4) transfected HCT116 cells at 72 h. D-F. Quantitative analysis of phospho-Akt, phospho-p70 S6K and phospho-4E-BP1 expressions in control and EIF3G shRNA (2 and 4) transfected HCT116 cells (n=3). **P<0.01 compared with the shRNA-con group.
EIF3G silencing inhibits HCT116 xenograft tumor growth in nude mice
Next, we investigated the effect of EIF3G silencing by investigating the growth of HCT116 mouse xenografts. We observed decreased tumor volume in the EIF3G silenced HCT116 cells from days 14 to 21 than in controls (P<0.05; Figure 6A). We also observed decreased tumor weight in EIF3G silenced group (0.67±0.18 g) than in control group (1.26±0.31 g; P<0.01; Figure 6B and 6C) on day 21. The body weight of control and EIF3G silenced xenograft mice group was comparable (Figure 6D). Moreover, EIF3G silenced tumor xenografts showed low EIF3G protein levels on day 21 than in control tumor xenografts (Figure 6E and 6F).
Figure 6.
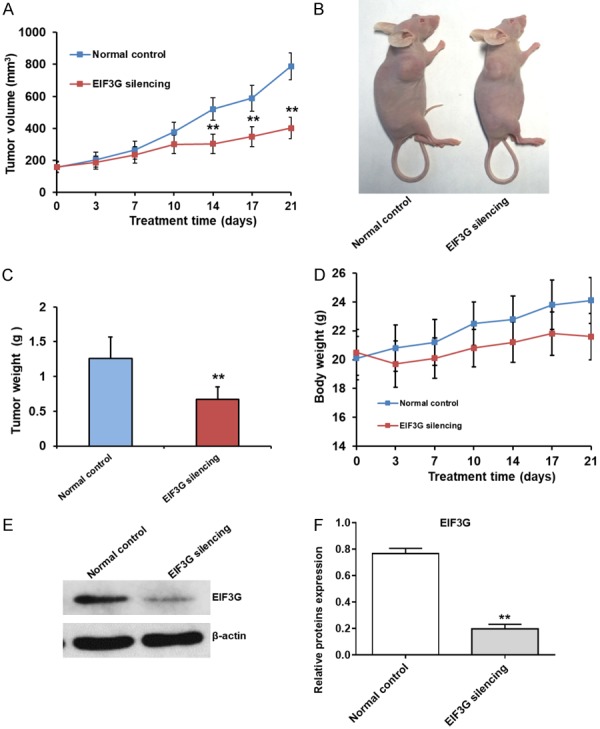
EIF3G silencing inhibits HCT16 xenograft growth in nude mice. A. Xenograft tumor volume from control and EIF3G transfected HCT116 cells at various time points (days 1-21). EIF3G silenced HCT116 cells show decreased tumor volume from days 14 to 21 than control shRNA transfected HCT116 cells (n=5). B. Photographs showing xenograft tumors from control and EIF3G shRNA transfected HCT116 cells on day 21 in nude mice. C. Bar graph shows mean xenograft tumor weight on day 21 from control and EIF3G shRNA transfected HCT116 cells (n=3). D. Analysis of body weight of nude mice xenografted with control and EIF3G shRNA transfected HCT116 cells at various time points (days 1-21; n=5). E. Representative EIF3G protein levels in xenograft tumor tissues from control and EIF3G shRNA transfected HCT116 cells with β-actin as internal control. F. Quantitative analysis of EIF3G proteins expression in xenograft tumors derived from control and EIF3G shRNA transfected HCT116 cells (n=3). **P<0.01 compared with the normal control group.
EIF3G overexpression promotes HCT116 cell proliferation and migration
In order to further investigate the biological functions of EIF3G in CRC, we stably overexpressed EIF3G in HCT116 cells by transfecting EIF3G overexpression plasmid (Figure 7A). The EIF3G protein levels in stably EIF3G overexpressed HCT116 or control cells were detected by western blotting (Figure 7B). The results of CCK-8 assay indicated that EIF3G overexpression drastically promoted HCT116 cell proliferation (Figure 7C). In addition, transwell migration assays showed that EIF3G overexpression significantly enhanced HCT116 cell migration (Figure 7D and 7E). Collectively, all these data indicated that EIF3G overexpression promoted HCT116 cell proliferation and migration.
Figure 7.
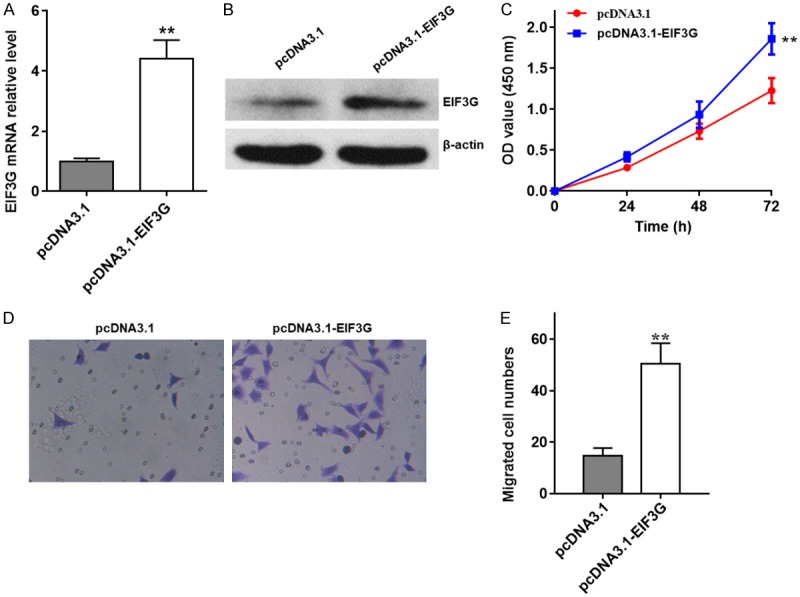
EIF3G overexpression promotes HCT116 cell proliferation and migration. A. EIF3G expression levels in EIF3G stably overexpressed and control HCT116 cells were analyzed by qRT-PCR (n=3). B. EIF3G protein levels in EIF3G stably overexpressed and control HCT116 cells were detected by western blotting. C. CCK8 cell proliferation analysis of EIF3G stably overexpressed and control HCT116 cells (n=3). D and E. Transwell migration assays analysis of the migrated cell numbers of EIF3G stably expressed and control HCT116 cells per field after 48 h incubation (n=3). **P<0.01 compared with the pcDNA3.1 group.
EIF3G overexpression enhances HCT116 xenograft tumor growth in nude mice
Finally, we analyzed the effect of EIF3G overexpression on HCT116 xenograft tumor growth in nude mice. We observed increased tumor volume in the EIF3G overexpressed HCT116 cells from days 10 to 21 than in controls (P<0.05; Figure 8A). We also observed enhanced tumor weight in EIF3G overexpressed group (2.25±0.25 g; P<0.01) than in control group (1.21±0.19 g; Figure 8B and 8C) on day 21. In addition, Edu staining indicated EIF3G overexpress promoted cell proliferation in tumor tissues (Figure 8D and 8E). Moreover, data in Figure 8F confirmed EIF3G stably high expressed in pcDNA3.1-EIF3G group. Inhibition or overexpression of EIF3G did not affect overall survival of colorectal-implanted mice.
Figure 8.
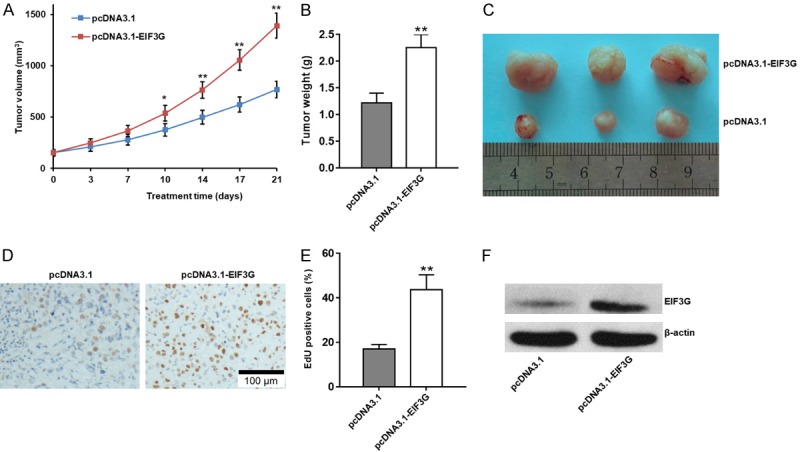
EIF3G overexpression enhances HCT116 xenograft tumor growth in nude mice. A. Xenograft tumor volume from pcDNA3.1 and pcDNA3.1-EIF3G transfected HCT116 cells at various time points (range from day 1 to day 21). EIF3G overexpressed HCT116 cells show increased tumor volume from days 10 to 21 than control cells (n=5). B. Photographs showing xenograft tumors from pcDNA3.1 and pcDNA3.1-EIF3G transfected HCT116 cells on day 21 in nude mice. C. Bar graph shows mean xenograft tumor weight on day 21 from pcDNA3.1 and pcDNA3.1-EIF3G transfected HCT116 cells (n=3). D. EdU staining analysis of EIF3G stably overexpressed in tumor tissues. E. Quantitative analysis of EdU staining (n=3). F. EIF3G protein levels in EIF3G stably overexpressed and control tumor tissues were detected by western blotting. **P<0.01, *P<0.05 compared with the pcDNA3.1 group.
Discussion
EIF3G belongs to the EIF3 family and is involved in transcriptional initiation. However, it has also been implicated in other roles. For example, it inhibits HIV replication and is cleaved by HIV-1 PR [26]. Recent studies have indicated that aberrant expression of EIF3G may be critical to many human diseases including cancers. In the present study, we demonstrated gradual increase in EIF3G expression with increasing colorectal cancer stages (I to IV). EIF3G was mainly localized to the cytoplasm, and its expression was higher in tumor tissues than in adjacent normal tissues of stages III and IV. Quantitative RT-PCR and western blot analysis demonstrated high EIF3G mRNA and protein expression in tumor tissues than in adjacent tissues, respectively.
Studies with the HCT116 colorectal cancer cell line revealed that EIF3G silencing resulted in decreased cell proliferation and increased apoptosis and autophagy via suppression of mTOR signaling pathway. This was further corroborated by in vivo mice xenograft study that showed decreased tumor volume and weight of mouse xenografts from EIF3G silenced HCT116 cells than in controls. Additionally, EIF3G overexpression promotes HCT116 cell proliferation and migration. Moreover, we also observed enhanced tumor weight of mouse xenografts in EIF3G overexpressed group. All these data demonstrate that EIF3G promotes colon cancer growth.
EIF3G expression regulated both HCT116 cell apoptosis and autophagy, which are activated during cellular stress [27]. Autophagy generally precedes apoptosis and is activated during cellular stress to maintain cellular homeostasis. Apoptosis or other types of cell death mechanisms are activated under prolonged stress [28]. Moreover, autophagy leads to autophagic cell death (ACD) [29]. Furthermore, the autophagic roles of caspases and apoptotic functions of autophagy-related proteins (ATGs) overlap. According to the present results, deregulation of EIF3G inhibited the phosphorylation of Akt, p70 S6K and 4E-BP1, which led to increased HCT116 cell apoptosis and autophagy. This suggests a role of EIF3G in the crosstalk between apoptosis and autophagy. These data also indicate that down-regulation of EIF3G induces HCT116 cell autophagy via inhibition of mTOR signaling pathway. However, the detailed interaction between apoptosis and autophagy needs to be investigated further.
In conclusion, our findings indicated that EI3FG was upregulated in colorectal cancer patient’s tumor tissues, promotes CRC cell proliferation, apoptosis, autophagy and migration in vitro, and CRC tumor growth in vivo. Our study demonstrates that EIF3G plays an important role in colon cancer progression and is a potential therapeutic target.
Acknowledgements
This study was supported by ZR2015HL082 Shandong Provincial Natural Science Foundation, China ZR2015HL082.
All patients provided written informed consent for the publication of all associated data in this study.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Society AC. Colorectal cancer facts & figures: 2014-2016. 2014 [Google Scholar]
- 4.Tomlinson JS, Jarnagin WR, DeMatteo RP, Fong Y, Kornprat P, Gonen M, Kemeny N, Brennan MF, Blumgart LH, D’Angelica M. Actual 10-year survival after resection of colorectal liver metastases defines cure. J. Clin. Oncol. 2007;25:4575–4580. doi: 10.1200/JCO.2007.11.0833. [DOI] [PubMed] [Google Scholar]
- 5.House MG, Kemeny NE, Gonen M, Fong Y, Allen PJ, Paty PB, DeMatteo RP, Blumgart LH, Jarnagin WR, D’Angelica MI. Comparison of adjuvant systemic chemotherapy with or without hepatic arterial infusional chemotherapy after hepatic resection for metastatic colorectal cancer. Ann Surg. 2011;254:851–856. doi: 10.1097/SLA.0b013e31822f4f88. [DOI] [PubMed] [Google Scholar]
- 6.Baba Y, Nosho K, Shima K, Hayashi M, Meyerhardt JA, Chan AT, Giovannucci E, Fuchs CS, Ogino S. Phosphorylated AKT expression is associated with PIK3CA mutation, low stage, and favorable outcome in 717 colorectal cancers. Cancer. 2011;117:1399–1408. doi: 10.1002/cncr.25630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 8.Dienstmann R, Vilar E, Tabernero J. Molecular predictors of response to chemotherapy in colorectal cancer. Cancer J. 2011;17:114–126. doi: 10.1097/PPO.0b013e318212f844. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Peralta AM, Nejda N, Oliart S, Medina V, Azcoita MM, Gonzalez-Aguilera JJ. Significance of mutations in TGFBR2 and BAX in neoplastic progression and patient outcome in sporadic colorectal tumors with high-frequency microsatellite instability. Cancer Genet Cytogenet. 2005;157:18–24. doi: 10.1016/j.cancergencyto.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, Settleman J, Giovannini M, Jacks T. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Negri FV, Bozzetti C, Lagrasta CA, Crafa P, Bonasoni MP, Camisa R, Pedrazzi G, Ardizzoni A. PTEN status in advanced colorectal cancer treated with cetuximab. Br J Cancer. 2010;102:162–164. doi: 10.1038/sj.bjc.6605471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 13.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spilka R, Ernst C, Mehta AK, Haybaeck J. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340:9–21. doi: 10.1016/j.canlet.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 15.Nesbit AD, Whippo C, Hangarter RP, Kehoe DM. Translation initiation factor 3 families: what are their roles in regulating cyanobacterial and chloroplast gene expression? Photosynth Res. 2015;126:147–159. doi: 10.1007/s11120-015-0074-4. [DOI] [PubMed] [Google Scholar]
- 16.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith MD, Arake-Tacca L, Nitido A, Montabana E, Park A, Cate JH. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure. 2016;24:886–896. doi: 10.1016/j.str.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershey JW. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Graf S, Provenzano E, Blows F, Pharoah P, Caldas C, Le Quesne J. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis. 2015;6:e1603. doi: 10.1038/cddis.2014.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- 21.Sha Z, Brill LM, Cabrera R, Kleifeld O, Scheliga JS, Glickman MH, Chang EC, Wolf DA. The eIF3 interactome reveals the translasome, a supercomplex linking protein synthesis and degradation machineries. Mol Cell. 2009;36:141–152. doi: 10.1016/j.molcel.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou C, Arslan F, Wee S, Krishnan S, Ivanov AR, Oliva A, Leatherwood J, Wolf DA. PCI proteins eIF3e and eIF3m define distinct translation initiation factor 3 complexes. BMC Biol. 2005;3:14. doi: 10.1186/1741-7007-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Q, Liu H, Ye J, Zhang H, Jia Z, Cao J. Nuclear distribution of eIF3g and its interacting nuclear proteins in breast cancer cells. Mol Med Rep. 2016;13:2973–2980. doi: 10.3892/mmr.2016.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, Zhang R, Zhang P. Eukaryotic translation initiation factor 3 subunit D overexpression is associated with the occurrence and development of ovarian cancer. FEBS Open Bio. 2016;6:1201–1210. doi: 10.1002/2211-5463.12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao Y, Teng J, Hong Y, Qu F, Ren J, Li L, Pan X, Chen L, Yin L, Xu D, Cui X. The oncogenic role of EIF3D is associated with increased cell cycle progression and motility in prostate cancer. Med Oncol. 2015;32:518. doi: 10.1007/s12032-015-0518-x. [DOI] [PubMed] [Google Scholar]
- 26.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ. Global landscape of HIV-human protein complexes. Nature. 2011;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang S, Yu ZM, Zhang CY, Zheng J, Li KL, Wu Y, Qian LL, Yang ZY, Li XR, Zhang Y, Wang RX. Autophagy promotes apoptosis of mesenchymal stem cells under inflammatory microenvironment. Stem Cell Res Ther. 2015;6:247. doi: 10.1186/s13287-015-0245-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


