Abstract
Aim: To investigate the expression of lncRNA SNHG6 in esophageal squamous cell carcinoma (ESCC), the biological function of SNHG6 in ESCCs, and evaluate its diagnostic value in ESCC. Methods: SNHG6 expression in tissues and cells was detected by quantitative real-time polymerase chain reaction (qRT-PCR). The effect of SNHG6 on the cell proliferation, migration and invasion was detected by CCK-8 assay and transwell assay, respectively. Results: SNHG6 expression was upregulated in ESCC tissues and ESCCs. Upregulated SNHG6 expression was correlated with prognosis, lymph node metastasis, distant metastasis and TNM stage of ESCC patients. SNHG6 promoted the abilities of cell proliferation, migration and invasion in ESCCs, knockdown of SNHG6 reversed these effects. SNHG6 might serve as an independently diagnostic biomarker for lymph node metastasis, distant metastasis and TNM stage. Conclusion: SNHG6 may exert oncogenic function in ESCC and may be a potential diagnostic marker for this cancer.
Keywords: Long non-coding RNAs, SNHG6, esophageal squamous cell carcinoma, diagnostic
Introduction
Esophageal cancer (EC) is one of the most common aggressive malignancies and which is the eighth most common cancer and sixth leading cause of mortality in the world [1,2]. EC mainly consists of squamous cell carcinoma and adenocarcinoma according to histopathological types. China is the highest-risk areas, and more than 90% of esophageal cancer are esophageal squamous cell carcinoma (ESCC) [3,4]. Despite continuous advances in diagnostic techniques and therapeutic modalities, the prognosis of ESCC is still unsatisfactory [5]. Recently, targeted cancer therapy such as trastuzumab and ramucirumab bring a new hope to patients with esophageal cancer, however, which is only applicable to a small number of patients. Thus, the discovery of new molecular disease markers is still urgently needed, which will contribute to the diagnosis and prognosis of complex ESCC.
Whole genome sequencing found that only 2% of the genome encoded proteins, remaining genome is transcribed into non-coding RNAs (ncRNAs). Long non-coding RNAs (lncRNAs) are a class RNA with more than 200 nucleotides and which is more than 76% of ncRNAs [6,7]. Increasing evidence indicates that lncRNAs play a crucial role in various cellular processes, such as development, differentiation and metabolism, and these effects may occur in various link such as epigenetic regulation, transcription and post-transcriptional regulation [8-11]. Recently, lncRNAs have been demonstrated to associated with the tumorigenesis and Progression in various tumors and to function as oncogenes or tumor suppressors in multiple signaling pathways [12,13]. Increasing evidence has shown that some lncRNAs are high expression in esophageal cancer and function as oncogenes, such as lncRNA POU3F3, HOTAIR, MALAT1 [14-17]. LncRNA POU3F3 promotes the methylation level of POU3F3 gene via interaction with histone methylase EZH2 mRNA, which leads to abnormal expression of the POU3F3 and promotes the occurrence and development of ESCC [14]. LncRNA HOTAIR inhibits the expression of WIF-1 in ESCC by binding to PRC2 complexes and then activates the histone of H3K27 promoter region and Wnt/β-catenin pathways [15]. In addition, some lncRNAs are down-regulated and function as tumor suppressors in esophageal cancer, such as lncRNA 91H [18]. LncRNA 91H expression may be related to H19 ICR methylation regulation, and up-regulation of 91H and inhibition of IGF2 expression may inhibit the disease progression of ESCC.
Recent researches found that some snoRNAs exhibit an important biological role in various human cancers. SnoRNA host gene 6 (SNHG6) was up-regulated and identified as an oncogene in colorectal cancer, stomach cancer, hepatocellular carcinoma, osteosarcoma, et al. [19-22]. However, studies of SNHG6 in ESCC are still scarce. In this study, we investigated the expression of SNHG6 and its functional role in ESCC and ESCCs, and observed the effect of knockdown of SNHG6 on the cells proliferation, migration and invasion in ESCCs. In addition, we evaluated the diagnostic value of lncRNA SNHG6 for clinicopathologic characteristics of ESCC patients.
Materials and methods
Patients and tissue samples
Seventy-five human ESCC tissues and their corresponding noncancerous esophageal tissues were collected from Zhengzhou central hospital affiliated Zhengzhou university between October 2012 and February 2013. All tissue samples were stored in liquid nitrogen immediately after surgical resection and were transferred to -80°C freezer before use. The diagnosis of ESCC for all patients was confirmed by three professional pathologists. Written informed consent was obtained from all patients. This study was approved by the ethics committee of Zhengzhou central hospital affiliated Zhengzhou University. Survival time was calculated from the date of diagnosis to death, the last follow-up time was June 31, 2018.
Cell culture and transfection
The human ESCC cell lines (ESCCs) EC9706, EC109, EC1 and an immortalized normal esophagus epithelial cell line HET-1A were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI 1640 medium (GIBCO, HyClone, USA) containing 10% fetal bovine serum and 5% CO2 at 37°C. To ensure the efficiency of knockdown, three different siRNAs were designed, two of which were considered appropriate for SNHG6 knockdown, the knockdown efficiency above of 70%, si-SNHG6-1 (sense: 5’-GCAGUUUACUGAGUCAUUACU-3’), si-SNHG6-2 (sense: 5’-UCGAAUAUGUUCAAAACAGGU-3’), si-NC (sense: 5’-UUCUCCGAACGUGUCACGUTT-3’) (Genechem, China). Si-SNHG6 or si-NC was transfected into EC109 and EC1 cells with Lipofectamine 2000 (Invitrogen, USA).
Cell proliferation, migration and invasion assays
Cell proliferation were measured by Cell Counting Kit-8 (CCK-8) (Beyotime, Jiangsu, China). In total, 5 × 103 cells were seeded into 96-well plates in 100 μL of medium, followed by incubation with 10 μL of CCK-8 per well at 24, 48, 72 and 96 hours for 1 hour. For transwell migration assays, the cells were seeded in the upper chamber with the non-coated membrane. For invasion assays, pre-coated with matrigel solution and polymerized in transwell inserts. The top chamber with FBS-free medium and the lower chamber filled with 10% FBS. Cells were incubated for 48 h at 37°C and the cells that had migrated or invaded the lower surface of the membrane were fixed in paraformaldehyde and stained with 0.1% crystal violet (Sigma), and the cells were counted from five independent visual fields.
qRT-PCR
Total RNA from tissues specimens and cells were extracted by Trizol reagent (Thermo Scientific, USA) and which were treated with DNase I to eliminate the genomic DNA contamination. Total RNA reverse-transcribed into cDNA by PrimeScriptTM RT reagent Kit (Takara Bio Inc, Japan). Real-time PCR analysis at Roche 480 II real-time PCR detection system with 20 μL reaction mixtures. The PCR reactions at 95°C for 5 minutes, followed by 40 cycles of 95°C for 15 s, 60°C for 34 s and 72°C for 30 s. The relative expression levels were calculated by the 2-ΔΔCt method, with all experiments repeated three times. The gene GAPDH was amplified as an endogenous control. The primers were as follows: SNHG6 forward primer: 5’-ATACTTCTGCTTCGTTACCT-3’, reverse primer: 5’-CTCATTTTCATCATTTGCT-3’; GAPDH forward primer: 5’-GGGAGCCAAAAGGGTCAT-3’, reverse primer: 5’-GAGTCCTTCCACGATACCAA-3’.
Statistical analysis
SPSS 19.0 software (IBM, Chicago, IL, USA) and GraphPad Prism (GraphPad Software 5.0, La Jolla, CA) was used to perform statistical analysis. Values were represented as mean ± standard difference (SD). Student’s t-test or one-way ANOVA were used for comparisons between groups and correlation analysis was performed by chi-square (χ2) test. P < 0.05 was considered as statistically significant.
Results
SNHG6 is upregulated in ESCC and is associated with ESCC progression
To investigate SNHG6 expression in 75 pairs ESCC tissues, qRT-PCR analysis was performed. Compared to the corresponding non-cancerous tissues, SNHG6 expression was significantly upregulated in ESCC tissues (Figure 1A, P < 0.01). Then, the expression of SNHG6 in ESCCs and HET-1A was detected, the result demonstrated that SNHG6 expression was higher in the ESCCs compared to immortalized normal esophagus epithelial cell line HET-1A (Figure 1C). Meanwhile, we evaluate the prognostic value of SNHG6, the survival analysis indicated that ESCC patients with high SNHG6 expression showed poor overall survival than those with low SNHG6 expression (log rank = 5.812, P = 0.013, Figure 1B). The overall survival time of high and low SNHG6 expression group was 36.78±2.71 and 47.08±2.84 months in ESCC patients, respectively.
Figure 1.
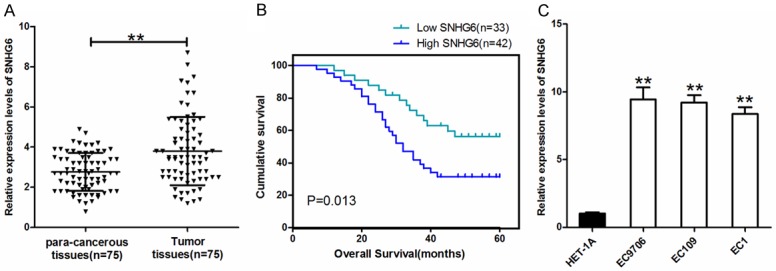
Relative expression of lncRNA SNHG6 in ESCC. A. SNHG6 expression in ESCC and para-cancerous tissues, **P < 0.01; B. Kaplan-Meier survival analysis revealed that high-level of SNHG6 was associated with poor prognosis of ESCC patients; C. The expression of SNHG6 in normal esophagus cell line (HET-1A) and ESCC cell lines. **P < 0.01 vs HET-1A group.
LncRNA SNHG6 expression and clinicopathological features
In order to investigate the relationship between SNHG6 expression and clinicopathological features, the 75 ESCC patients were divided into two groups according to SNHG6 median expression: the high expression group (n = 42) and the low expression group (n = 33). Significant correlation were observed between SNHG6 expression and lymph node metastasis, distant metastasis and TNM stage (P = 0.003, P = 0.021, P = 0.003, respectively, Table 1). There were no significantly difference between the expression of SNHG6 and other clinical parameters such as gender, age, smoking, drinking and tumor size. In order to avoid bias caused by univariate analysis, multivariate analysis was performed to explore the relationship between various parameters and SNHG6 expression. Multivariate linear regression analysis revealed that lymph node metastases and TNM stage independently predicted the MALAT1 expression, the total R2 = 0.619 (Table 2).
Table 1.
The correlations between SNHG6 expression and clinicopathological characteristics in ESCC patients
| Variables | Cases | SNHG6 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Gender | 0.300 | |||
| Male | 51 | 24 | 27 | |
| Female | 24 | 9 | 15 | |
| Age (years) | 0.115 | |||
| < 60 | 23 | 13 | 10 | |
| ≥ 60 | 52 | 20 | 32 | |
| Smoking | 0.359 | |||
| Yes | 38 | 18 | 20 | |
| No | 37 | 15 | 22 | |
| Drinking | 0.137 | |||
| Yes | 45 | 17 | 28 | |
| No | 30 | 16 | 14 | |
| Tumor size | 0.081 | |||
| ≥ 4 cm | 42 | 15 | 27 | |
| < 4 cm | 33 | 18 | 15 | |
| Lymph node metastasis | 0.003 | |||
| Yes | 44 | 13 | 31 | |
| No | 31 | 20 | 11 | |
| Distant metastasis | 0.021 | |||
| Yes | 13 | 2 | 11 | |
| No | 62 | 31 | 31 | |
| TNM stage | 0.003 | |||
| I/II | 23 | 16 | 7 | |
| III/IV | 52 | 17 | 35 | |
Table 2.
Multivariate analysis of different clinicopathologic factors for lncRNA SNHG6 expression
| Variable | Standardised | t | P value |
|---|---|---|---|
| β-coefficient | |||
| Gender | 0.060 | 0.526 | 0.601 |
| Age | 0.030 | 0.339 | 0.736 |
| Smoking | 0.047 | 0.419 | 0.676 |
| Drinking | 0.019 | 0.158 | 0.875 |
| Tumor size | 0.001 | 0.012 | 0.991 |
| Lymph node metastases | 0.224 | 2.433 | 0.018 |
| Distant metastasis | 0.139 | 1.385 | 0.171 |
| TNM stage | 0.549 | 4.099 | 0.000 |
Total R2 = 0.619.
Knockdown of SNHG6 inhibited proliferation, migration and invasion in ESCCs
To investigate the biological function of SNHG6 in ESCCs, si-SNHG6 was transfected into EC109 and EC1 cells to knocked down SNHG6 expression. First, to investigate the knockout efficiency, we analyzed SNHG6 expression in ESCCs and HET-1A cell by qRT-PCR. The results revealed that SNHG6 expression was significantly decreased after transfected with si-SNHG6-1 and si-SNHG6-2, the silence efficiency was 75.4 and 77.3 in EC109 and the silence efficiency was 80.8 and 79.4 in EC1, respectively (Figure 2A).
Figure 2.
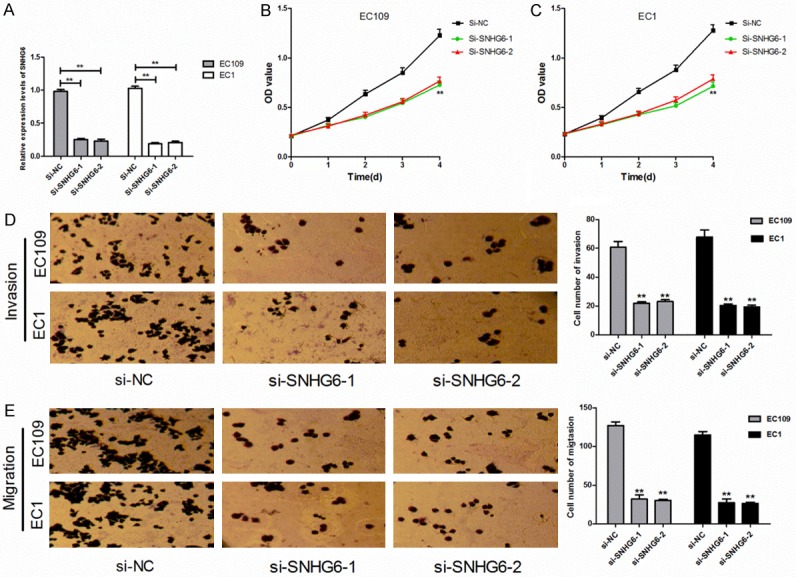
Knockdown of SNHG6 inhibited proliferation, migration and invasion in ESCCs. A. The expression of SNHG6 in EC109 and EC1 cells after cells transfected with si-SNHG6 or si-NC. B, C. The effect of SNHG6 knockdown on the proliferation of EC109 and EC1 cells detected by CCK-8 assay. D, E. The effect of SNHG6 knockdown on invasion and migration abilities of EC109 and EC1 cells detected by Transwell assays. **P < 0.01 vs si-NC group.
We further analyzed the effect of knocked down SNHG6 on the cell proliferation, migration and invasion of EC109 and EC1 cells. The CCK-8 assay revealed that knocked down SNHG6 decreased the OD value at every interval examined both in EC109 and EC1 cells compared with the si-NC group (Figure 2B, 2C), which means that knocked down SNHG6 significantly inhibited the proliferation abilities of ESCCs. The transwell assays showed that SNHG6 enhance the migration abilities of EC109 and EC1 cells, the number of migrated cells in si-SNHG6-1 and si-SNHG6-2 groups were significantly decreased compared to the si-NC group (Figure 2E). Meanwhile, the invasion abilities were significantly inhibited in si-SNHG6-1 and si-SNHG6-2 groups compared to the si-NC group (Figure 2D). The above results indicated that SNHG6 may exerts oncogenic function in ESCCs.
Diagnostic value of lncRNA SNHG6
To investigate the diagnostic value of lncRNA SNHG6 for predict clinicopathological features, three receiver operating characteristic curves (ROCs) were generated. The results showed that SNHG6 might serve as a diagnostic biomarker for lymph node metastasis, distant metastasis and TNM stage. For lymph node metastasis, the area under the ROC curve (AUC) was 0.8326 (95% CI 0.7352-0.9300, P < 0.001), when the cutoff value was 3.300, the sensitivity and specificity was 82.2 and 80.0% (Figure 3A), respectively. For distant metastasis, the AUC was 0.8759 (95% CI 0.7717-0.9802, P < 0.001), when the cutoff value was 4.800, the sensitivity and specificity was 84.6 and 87.1% (Figure 3B), respectively. Finally, for TNM stage, the AUC was 0.8838 (95% CI 0.8108-0.9568, P < 0.001), when the cutoff value was 3.45, the sensitivity and specificity was 73.1 and 91.3% (Figure 3C), respectively.
Figure 3.
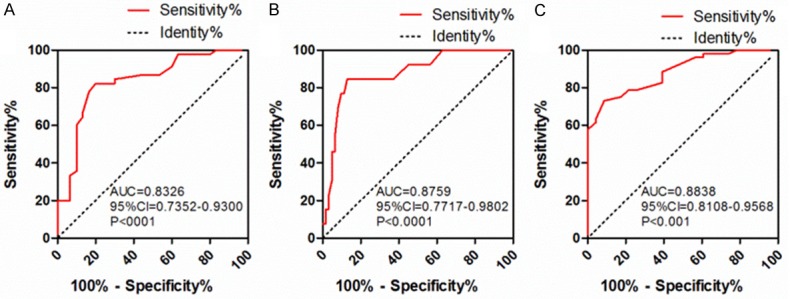
ROC curve analysis of SNHG6 as a diagnostic marker for ESCC. A-C. Predictive value of SNHG6 for lymph node metastasis, distant metastasis, TNM stage in ESCC patients, respectively.
Discussion
The emergence of new surgical methods and adjuvant therapies for ESCC has significant improved the 5-year survival rate of ESCC patients. However, the 5-year survival rate of malignant esophageal cancer patients is still less than 5% [23,24]. Thus, the discovery of new molecular markers for early diagnosis and prognosis of complex esophageal cancer is still urgently needed. Recent study found that linc-POU3F3 was detected in the tissues and plasma of ESCC patients, and its expression level in plasma of ESCC patients was significantly higher than that of normal control group. Therefore, lncRNA may be a potential biomarker for early diagnosis and prognosis of ESCC [25].
Emerging study has shown that SNHG6 can be influence on the occurrence and development of various tumor by multiple signaling pathways. SNHG6 functions as a competing endogenous RNA, regulating ZEB1 expression by binding miR-101-3p and which then inducing epithelial to mesenchymal transition in hepatocellular carcinoma [21]. SNHG6 regulates p21 expression via activation of the JNK pathway and suppression of EZH2 expression in gastric cancer cells, which inhibits gastric cancer development [26]. SNHG6 suppressed cell proliferation by regulating p21 and KLF2, and inducing cell apoptosis [22]. In addition, SNHG6 may influence on tumor therapy by regulating the levels of genomic methylation. SNHG6 dysregulation can lead to genome-wide hypomethylation via binding two miR-1297-mediated SAMe-dependent positive feedback loops in hepatocellular carcinoma cell and that exogenous SAMe may be beneficial in the treatment of hepatocellular carcinoma [27]. In this study, we first observed the expression of SNHG6 in ESCC tissues and ESCCs, the results showed that SNHG6 expression were significantly up-regulated both in ESCC tissues and ESCCs. Then, the SNHG6 expression were divided into two groups, we found that SNHG6 expression was closely related to prognosis of ESCC patients. In addition, we analyzed the relationship between SNHG6 expression and clinicopathologic characteristics via univariate analysis and multivariate analysis. The results showed that SNHG6 expression was significant correlation with lymph node metastasis, distant metastasis and TNM stage. Study by Zhang et al. also found that SNHG6 was significantly up-regulated in ESCC tissues and ESCCs, and SNHG6 expression associated with tumor size and tumor stage, these results was consistent with our research [28]. However, the study did not evaluate the diagnostic value of SNHG6 as a biomarker in ESCC. The above results indicated that SNHG6 may exerts oncogenic function in ESCC.
Increasing evidence has shown that the change of SNHG6 expression may be related with the proliferative, migratory and invasive abilities of cancer cells [22,29]. Study by Meng et al. found that SNHG6 promoted glioma cell proliferation, migration and increased apoptosis by sponging miR-101-3p, knockdown of SNHG6 reversed the effects of SNHG6 on cell malignancy [29]. To investigate the effect of knocked down SNHG6 on the cell proliferation, migration and invasion, two si-SNHG6 were transfected into ESCCs. Coincidentally, we identified that SNHG6 regulated the malignant abilities of ESCCs. Knockdown of SNHG6 significantly attenuated the ability of cell proliferation, migration and invasion of EC109 and EC1 cells.
Recent study found that SNHG6 expression is highly cancer type specific, SNHG6 as a potential biomarker for hepatocellular carcinoma [30]. Since SNHG6 expression could affect the prognosis of ESCC patients and promoted ESCC cells malignancy, whether SNHG6 expression could independently predict the clinicopathological features of ESCC patients. We further evaluated the diagnostic value of SNHG6 as a biomarker of clinicopathological features by ROCs. We confirmed that SNHG6 can be serves as a diagnostic biomarker for lymph node metastasis, distant metastasis and TNM stage, the AUC more than 0.8. However, our results need further big sample research for verification and further exploration of the mechanisms of SNHG6 affect on biological characteristics of ESCC is needed.
In conclusion, lncRNA SNHG6 was upregulated in ESCC tissues and ESCCs, and its expression level was closely related to prognosis, lymph node metastasis, distant metastasis and TNM stage. In addition, SNHG6 could regulated the malignant abilities of ESCCs, which as a potential diagnostic marker for ESCC.
Acknowledgements
This work was supported by the Medical Science Research Project of Henan Province (Grants 2018020765, 201601030).
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Cente MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Rice TW, Gress DM, Patil DT, Hofstetter WL, Kelsen DP, Blackstone EH. Cancer of the esophagus and esophagogastric junction - major changes in the American joint committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:304–317. doi: 10.3322/caac.21399. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet. 2017;390:2383–2396. doi: 10.1016/S0140-6736(17)31462-9. [DOI] [PubMed] [Google Scholar]
- 6.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein LD. Human genome: end of the beginning. Nature. 2014;431:915–916. doi: 10.1038/431915a. [DOI] [PubMed] [Google Scholar]
- 8.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 9.Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16:71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-kappaB signalling circuit. Immunology. 2017;150:379–388. doi: 10.1111/imm.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 12.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Zheng J, Deng J, You Y, Wu H, Li N, Lu J, Zhou Y. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714–1726. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 15.Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY, Xue WQ, Chen YB, Zhang Y, Jia WH. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104:1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y, Yang K. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:1–13. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CM, Wu QQ, Li SQ, Chen FJ, Tuo L, Xie HW, Tong YS, Ji L, Zhou GZ, Cao G, Wu M, Lv J, Shi WH, Cao XF. Upregulation of the long noncoding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591–597. doi: 10.1007/s10620-013-2956-7. [DOI] [PubMed] [Google Scholar]
- 18.Gao T, He B, Pan Y, Xu Y, Li R, Deng Q, Sun H, Wang S. Long non-coding RNA 91H contributes to the occurrence and progression of esophageal squamous cell carcinoma by inhibiting IGF2 expression. Mol Carcinog. 2015;54:359–367. doi: 10.1002/mc.22106. [DOI] [PubMed] [Google Scholar]
- 19.Xue WN, Li JW, Wang F, Han P, Liu Y, Cui B. A long non-coding RNA expression signature to predict survival of patients with colon adenocarcinoma. Oncotarget. 2017;8:101298–101308. doi: 10.18632/oncotarget.21064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan K, Tian J, Shi WZ, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999–1012. doi: 10.1159/000478682. [DOI] [PubMed] [Google Scholar]
- 21.Chang L, Yuan YF, Li CC, Guo T, Qi H, Xiao Y, Dong X, Liu Z, Liu Q. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR-101-3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383:183–194. doi: 10.1016/j.canlet.2016.09.034. [DOI] [PubMed] [Google Scholar]
- 22.Zheng LL, Hu N, Guan GY, Guan G, Chen J, Zhou X, Li M. Long noncoding RNA SNHG6 promotes osteosarcoma cell proliferation through regulating p21 and KLF2. Arch Biochem Biophys. 2018;646:128–136. doi: 10.1016/j.abb.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Lordick F, Hölscher AH, Haustermans K, Wittekind C. Multimodal treatment of esophageal cancer. Langenbecks Arch Surg. 2013;398:177–87. doi: 10.1007/s00423-012-1001-1. [DOI] [PubMed] [Google Scholar]
- 24.Domper Arnal MJ, Ferrández Arenas A, Lanas Arbeloa A. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and eastern countries. World J Gastroenterol. 2015;21:7933–7943. doi: 10.3748/wjg.v21.i26.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong YS, Zhou XL, Wang XW, Wu QQ, Yang TX, Lv J, Yang JS, Zhu B, Cao XF. Association of decreased expression of long non-coding RNA LOC285194 with chemoradiotherapy resistance and poor prognosis in esophageal squamous cell carcinoma. J Transl Med. 2014;12:233–241. doi: 10.1186/s12967-014-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Li D, Zhao M, Huang S, Zhang Q, Lin H, Wang W, Li K, Li Z, Huang W, Che Y, Huang C. Long noncoding RNA SNHG6 regulates p21 expression via activation of the JNK pathway and regulation of EZH2 in gastric cancer cells. Life Sci. 2018;18:1–7. doi: 10.1016/j.lfs.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 27.Guo T, Wang H, Liu P, Xiao Y, Wu P, Wang Y, Chen B, Zhao Q, Liu Z, Liu Q. SNHG6 acts as a genome-wide hypomethylation trigger via coupling of miR-1297-mediated S-adenosylmethionine-dependent positive feedback loops. Cancer Res. 2018;78:3849–3864. doi: 10.1158/0008-5472.CAN-17-3833. [DOI] [PubMed] [Google Scholar]
- 28.Fan RH, Guo JN, Yan W, Huang MD, Zhu CL, Yin YM, Chen XF. Small nucleolar host gene 6 promotes esophageal squamous cell carcinoma cellproliferation and inhibits cell apoptosis. Oncol Lett. 2018;15:6497–6502. doi: 10.3892/ol.2018.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng Q, Yang BY, Liu B, Yang JX, Sun Y. Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p. Int J Biol Markers. 2018;33:148–155. doi: 10.1177/1724600817747524. [DOI] [PubMed] [Google Scholar]
- 30.Fan RH, Guo JN, Yan W, Khoshnevisan A, Shoja Z, Motahari P, Farhangi B. Long non-coding RNA SNHG6 as a potential biomarker for hepatocellular carcinoma. Oncol Lett. 2018;15:6497–650. doi: 10.1007/s12253-017-0241-3. [DOI] [PubMed] [Google Scholar]


