Abstract
Esophageal squamous cell carcinoma (ESCC) is one of the most prevalent types of upper gastrointestinal malignancy. Here, we used 1H nuclear magnetic resonance spectroscopy (1H-NMR) to identify potential pre- and post-operative serum biomarkers in patients with early stage ESCC using metabolomic fingerprint spectrum. Serum samples from preoperative patients with ESCC (ESCC, n = 25), postoperative patients with ESCC (PO, n = 24), and controls (n = 40) were analysed using 1H-NMR spectroscopy. Using orthogonal partial least squares-discriminant analysis, 31 altered serum metabolites were successfully identified among the three groups. These metabolites are indicative of the changes that occur with glycometabolism, the metabolism of fatty acids, amino acids, choline, ketone bodies, nucleotides, and lipids. Based on receiver operating characteristic (ROC) curve analysis and a biomarker panel with an area under the curve (AUC) of 0.969, six serum metabolites (α-glucose, choline, glutamine, glutamate, valine, and dihydrothymine) were selected as potential diagnostic biomarkers for early stage ESCC. Additionally, four potential PO biomarkers (α-glucose, pyruvate, glutamate, and valine) with an AUC of 0.985 were selected to distinguish ESCC and PO. Many metabolites trended towards normalisation in PO patients, with only choline remaining high with an AUC of 0.858, suggesting that it may be a valuable potential biomarker for neoplasm progression, recurrence, chemoradiotherapy, and prognosis. 1H-NMR spectroscopy may be a useful tumour detection approach in the early diagnosis of ESCC. These results also indicate that it is useful to differentiate pre- and post-operative ESCC, evaluate surgery therapeutic responses, and monitor postoperative chemoradiotherapy.
Keywords: Esophageal squamous cell carcinoma, metabolomics, biomarker, 1H-NMR spectroscopy
Introduction
Esophageal squamous cell carcinoma (ESCC), a major histologic type of oesophageal cancer, is a prevalent upper gastrointestinal malignancy that affects major populations in China [1]. Patients diagnosed during the early stages of ESCC have significantly greater long-term survival rates (at least 5 years or more) than do those diagnosed at middle or later stages [2]. Most ESCC patients exhibit metastasis or locally advanced ESCC at the time of diagnosis and have a five-year survival rate of only 5-15% [3]. Current techniques, including computed tomography scanning, upper gastrointestinal radiography, endoscopic ultrasonography, and chromoendoscopy with iodine staining have limitations or low specificities and sensitivities [4,5]. These limitations highlight the need for accurate non-invasive screening tools to facilitate early ESCC detection. Thus, there is a need for the development of a diagnostic tool and for reliable biomarkers with high sensitivity and specificity at an early curative stage.
Metabolomics is a powerful approach for surveying endogenous small molecule metabolites (less than 1000 Da) through the non-invasive analysis of cells, tissues, or biofluids [6,7]. It focuses on the unique metabolomic fingerprint spectrum generated by metabolic processes in a biological system through targeted or non-targeted strategies [8,9]. Hence, metabolomics represents a broad field for the detection of useful biomarkers for disease diagnosis, therapy, and prognosis, and for insights into the pathophysiologic mechanisms of oncogenesis and tumour staging. 1H nuclear magnetic resonance (1H-NMR) spectroscopy is a non-destructive and non-invasive technique that requires a small quantity of sample to screen cancer-associated perturbations in cellular metabolism. Currently, many studies are applying metabolomics technology to tissue, plasma, serum, and urine samples to reveal variation in the tricarboxylic acid (TCA) cycle and in the metabolism of choline, amino acids, fatty acids, and urea to identify metabolite biomarkers in patients with esophageal cancer [10-12]. However, some questions remain unanswered: What are the critical metabolite changes that occur in the early stages of ESCC? What levels of serum metabolomics sensitivity and specificity are required to distinguish patients with early ESCC from healthy groups? For assessment of operative results and to monitor postoperative chemoradiotherapy, what metabolite changes occur after tumour removal? Answering these questions may provide a means of improving early diagnosis, therapy, and prognoses for ESCC.
Therefore, we applied non-targeted (principal component analysis, PCA) and targeted (partial least squares-discriminant analysis, PLS-DA) methods based on 1H-NMR spectroscopy to identify global changes in serum metabolic profiles. We analysed and compared the serum metabolic profiles of control groups (C) and of patients with early stage ESCC before and after operation, respectively. Furthermore, orthogonal partial least squares-discriminant analysis (OPLS-DA) was applied to visualise the metabolic variation among the three serum samples. The general objectives of this study were to: 1) identify potential diagnostic serum biomarkers for early stage pre- and post-operative ESCC by metabolomic fingerprint spectrum; 2) identify potential biomarkers of operative effects and monitor postoperative chemoradiotherapy with early ESCC; and 3) increase our understanding of the underlying mechanisms of ESCC.
Materials and methods
Study subjects and sample collection
Serum samples (89) were collected from the Department of Cardiothoracic Surgery and the Medical Examination Center in the Second Affiliated Hospital of Shantou University Medical College. These samples included those from 40 controls (C), 25 pre-operative patients with ESCC (ESCC), and 24 post-operative patients with ESCC (PO) without chemotherapy, radiotherapy, or chemoradiotherapy. Patient information and clinical characteristics are summarised in Table 1. The clinical stages of ESCC patients (stage I/II) were diagnosed by esophagoscopy examination with biopsy, X-ray barium radiography, and chest computed tomography. Tumour staging was based on the American Joint Committee on Cancer (AJCC)7th staging system. Healthy controls were matched with patients with ESCC based on age, gender, BMI, and place of residence. Fasting blood was collected from patients and healthy controls between 7 and 8 am and was centrifuged at 3000 rpm for 10 min. Serum samples were isolated and immediately stored at -80°C for further analysis. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethical Committee of the Second Affiliated Hospital of Shantou University Medical College (Registration No. 2016-32). The approval date was 21 November 2016. Informed consent was obtained from each subject before participation in the study.
Table 1.
Summary of clinical and demographic characteristics of preoperative ESCC patients (ESCC), postoperative ESCC patients (PO), and controls (C)
| Variable | C | ESCC | PO |
|---|---|---|---|
| Number of subjects | 40 | 25 | 24 |
| Male/Female | 31/9 | 19/6 | 19/5 |
| Age (year, range) | 60.3, 46-79 | 63.2, 49-81 | 63.5, 52-81 |
| BMI (mean, range) | 21.5, 16.2-29.9 | 21.7, 16.5-30.2 | 21.4, 16.8-30.7 |
| Differentiation degree | - | Well: 23 | Well: 22 |
| Middle: 2 | Middle: 2 | ||
| TNM classification | - | I/II: 25 | I/II: 24 |
Sample preparation and 1H-NMR spectroscopy
Frozen serum samples were thawed immediately and vortexed for 10 s at room temperature. Then, 200 μL of phosphate buffer solution (90 mM NaH2PO4/Na2HPO4, pH = 7.4) was added to 400 μL serum for NMR detection. After centrifugation at 10,000 rpm for 10 min at 4°C, approximately 550 μL of the clear supernatant was transferred into 5-mm NMR tubes (ST500, NORELL, Inc., Morganton, North Carolina, USA) for sampling. An NMR spectrometer (600.13 MHz, Bruker Avance III, Bruker Corporation, Kalsruhe, Germany) was used to obtain 1H-NMR spectra under CPMG (Carr-Purcell-Meboom-Gill) pulse sequence with a total spin relaxation delay (2 nτ) of 70 ms to weaken broad resonances from high molecular weight compounds and retain low molecular weight compounds and some lipids. All serum samples were analysed in random order at 298 K. One-dimensional spectrum was used to obtain CPMG spin echo pulse sequence (RD-90°-(τ-180°-τ)n-ACQ) to suppress water signal with a relaxation delay of 5 s. The acquisition parameters were: spectral width, SW = 20 ppm; recycle delay, RD = 4.0 s; t1 = 350 μs; mixing time, tm = 100 ms; number of scans, NS = 32; number of points, TD = 32768; and acquisition time, AQ = 2.73 s.
1H-NMR spectroscopy analysis
The raw data (free induction decays, FIDs) were input into MestReNova Version 9.0.1 (Mestrelab Research, Santiago de Compostela, Galicia, Spain, 9.0.1) for processing and complexity reduction to facilitate pattern recognition. To enhance the signal-to-noise ratio, all 1H-NMR spectra were multiplied by a 1.0 Hz exponential line broadening prior to Fourier transformation. The chemical shifts of serum spectra were referenced to the methyl doublet signal of lactate at δ 1.33 ppm. Both phase adjustment and baseline correction were performed manually. Each spectrum (9.0-0.5 ppm) was divided into approximately 419 segments with equal bin width (0.02 ppm) excluding the residual water (5.18-4.67 ppm) and urea (6.40-5.40 ppm) regions. To remove the dilution effect or bulk mass differences among samples due differing serum weights, the remaining spectra were internally normalised to a total spectral area of unity prior to pattern recognition analysis. Total detailed NMR rawdata is shown in Supplementary Material.
Pattern recognition (PR) analysis
To establish a global profile of the differential features of patients with ESCC and healthy controls, we used multivariate analysis to identify consistent variations between 1H-NMR data sets. Serum spectra data were input into the SIMCA-P+ version 14.1 software package (Umetrics Inc., Umea, Sweden, V 14.1) for PR analysis. First, PCA was performed using the Parato-scaled normalised 1H-NMR spectra to discover the intrinsic trends and outliers among the three serum sample groups. Then, PLS-DA and OPLS-DA were performed to prevent over-fitting of the statistical model and to select the potential biomarkers, respectively.
Model quality and reliability were assessed by R2X, R2Y, and Q2 values, which reflect the explained variance and model predictability. R2X represents the variation explained by the models and R2Y indicates the ‘goodness of fit’ in the data. Q2, calculated by a cross-validation procedure, indicates the predictability of the model. To avoid model overfitting, a default seven-round cross-validation procedure was performed in SIMCA-P+ 14.1 to determine the optimal number of principal components. Reliability of the models was further rigorously validated by a permutation analysis (n = 300 times). The variable importance in the projection (VIP) from OPLS-DA models was identified as a coefficient for peak selection. These variables were considered potential biomarker candidates as class discriminating information, the higher the value, the greater the discriminatory power of the metabolite. VIPs larger than 1.0 usually represent those metabolites with significant group discrimination.
Data pre-processing and statistical analysis
CV-ANOVA (analysis of variance testing of cross-validated predictive residuals) was performed to identify significantly different features between groups in OPLS-DA models. Univariate statistical significance of P < 0.05 was considered to distinguish metabolites. Student’s t (normal distribution) or Mann-Whitney U (if abnormal distribution) tests were performed to analyse the metabolic profiles among controls, ESCC, and PO patients. The metabolites were recognised according to the Human Metabolome Database (http://www.hmdb.ca/). Hierarchical clustering analysis (HCA) of these biomarkers was conducted using R-3.5.0 (www.r-project.org) software. To further evaluate the diagnostic power of the potential biomarkers whose levels differed significantly among the three groups, receiver operating characteristic (ROC) analysis was conducted by SPSS 20.0 (SPSS Inc., Chicago, IL, USA) to verify potential biomarkers, and the area under the ROC curve (AUROC), specificity, sensitivity were calculated, where AUROC > 0.80 indicated excellent diagnostic ability.
Results
Metabonomic profiling of serum samples for ESCC, PO, and controls
The one-dimensional 1H-NMR spectra of serum samples provide an overview of all metabolites from the control, ESCC, and PO groups (Figure 1). Approximately 47 metabolites were tagged in the spectra in the three groups, including amino acids, organic acids, energy metabolism molecules, methylguanidine, myo-inositol, trimethylamine N-oxide, glucose components, lipids, carbohydrates, and nucleotides.
Figure 1.
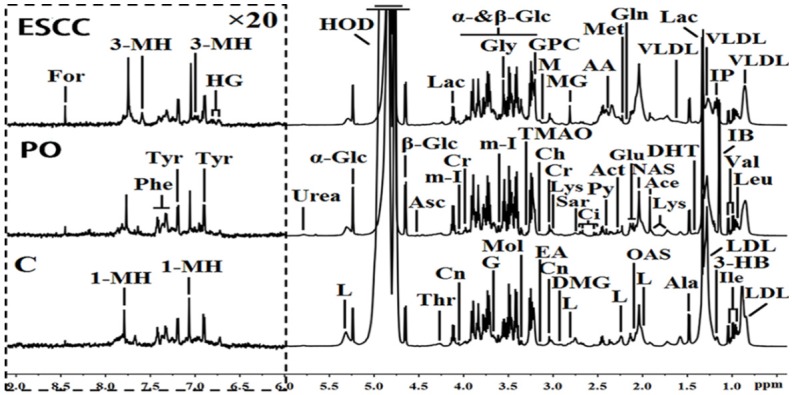
1H-NMR spectra (δ 0.5-9.0 ppm) of serum obtained from the controls (C), preoperative esophageal squamous cell carcinoma (ESCC), and postoperative ESCC patients (PO). The region of δ6.0-9.0 ppm (in the dashed box) was magnified 20 times compared with corresponding region of δ0.5-6.0 ppm for the purpose of clarity. Keys: 1-MH: 1-methylhistidene; 3-HB: 3-hydroxybutyrate; 3-MH: 3-methylhistidene; AA: acetoacetate; Ace: acetate; Ace: acetone; Ala: alanine; Asc: ascorbate; Cho: choline; Ci: citrate; Cn: creatinine; Cr: creatine; DHT: dihydrothymine; DMG: N, N-dimethylglycine; EA: ethanolamine; For: formate; G: glycerol; Gln: glutamine; Glu: glutamate; Gly: glycine; GPC: glycerolphosphocholine; HG: homogentisate; HOD: the residual water resonance; IB: isobutyrate; Ile: isoleucine; IP: isopropanol; L: lipid; Lac: lactate; LDL: low density lipoprotein; Leu: leucine; Lys: lysine; M: malonate; Met: methionine; MG: methylguanidine; m-I: myo-inositol; Mol: methanol; NAS: N-acetyl glycoprotein signals; OAS: O-acetyl glycoprotein signals; Phe: phenylalanine; Py: pyruvate; Sar: sarcosine; Thr: threonine; TMAO: trimethylamine N-oxide; Tyr: tyrosine; Val: valine; VLDL: very low density lipoprotein; α-Glc: α-glucose; β-Glc: β-glucose.
PR (PCA, PLS-DA and OPLS-DA) analysis of serum metabolomic profiling for ESCC, PO, and controls
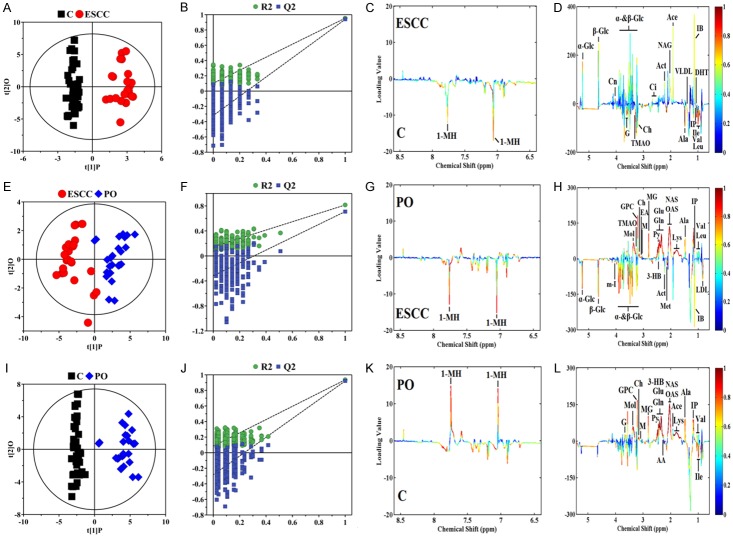
To obtain useful metabolomics profiles, unsupervised PCA analysis of 1H-NMR results showed the difference between the ESCC, PO, and control groups. The three-dimensional (3D) PCA score plots (Figure 2A) revealed separation trends and group clustering based on 1H-NMR spectra of the three groups (R2X = 69.0%, Q2 = 0.658). However, we were unable to identify a clear difference between them via the PCA scores plot. We performed supervised 3D PLS-DA score analysis, and the resultant plot showed obvious metabolic perturbations among the three groups (R2X = 68.6%, R2Y = 0.837, Q2 = 0.814), especially between the ESCC and PO patients (Figure 2B). To maximise the group separation and to visualise the metabolic distinctions, the supervised OPLS-DA classification model was used to investigate metabolomic alterations. We used three separate supervised OPLS-DA models to distinguish between ESCC patients and controls (Figure 3A-D; R2X = 72.3%, R2Y = 0.936, Q2 = 0.921, P = 2.06×10-26), ESCC and PO patients (Figure 3E-H, R2X = 64.2%, R2Y = 0.815, Q2 = 0.810, P = 2.27×10-11), and PO patients and controls (Figure 3I-L, R2X = 61.9%, R2Y = 0.954, Q2 = 0.940, P = 2.23×10-35). Taken together, these results suggest that the models were robust, and the random permutation tests indicated that the models were not over-fitted. Our results indicate that 1H-NMR-based serum metabolomics has potential applications for identifying early ESCC. Moreover, our results showed that changes in some endogenous metabolites were related to response to operative treatment intervention.
Figure 2.
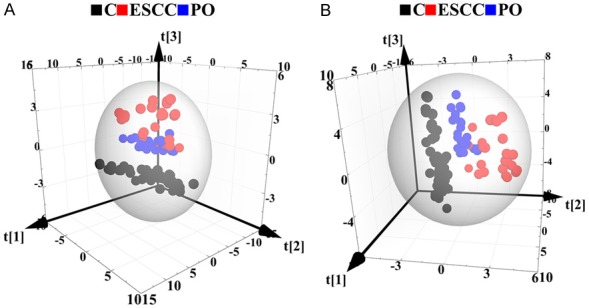
A. 3D PCA score plots based on 1H CPMG NMR spectra of serum obtained from controls (C), ESCC, and PO groups. PCA score plots revealed separation trends and group clustering based on 1H-NMR spectra of the three groups (R2X = 69.0%, Q2 = 0.658). B. 3D PLS-DA score plots based on 1H CPMG NMR spectra of serum obtained from controls (C), ESCC, and PO groups (R2X = 68.6%, R2Y = 0.837, Q2 = 0.814).
Figure 3.
Orthogonal partial least squares-discriminant analysis score plots (A, E, I) derived from 1H CPMG NMR spectra of serum and corresponding coefficient loading plots (C, D, G, H, K, L) obtained from control (C), ESCC, and PO groups and cross validation (B, F, J) by permutation test (n = 300). The colour map shows the significance of metabolite variations between the two classes. Peaks in the positive direction indicate metabolites that are more abundant in the groups in the positive direction of the first principal component. Consequently, metabolites that are more abundant in the groups in the negative direction of the first primary component are presented as peaks in the negative direction.
Discovery, description, and identification of potential biomarkers and biomarker panel
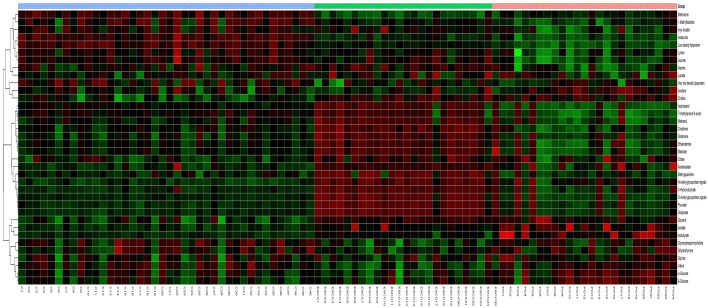
A total of 31 differential metabolites were identified as characteristic metabolites (Table 2). We constructed a heat map to visualise the discriminatory power of biomarkers among the three groups (Figure 4). Compared to controls, 29 significant biomarkers were identified in ESCC patients. Based on the sensitivity and specificity of this approach, ROC analyses were performed for further prediction of potential biomarkers (Table 3). As neoplastic diseases involve systematic disturbance of metabolic biochemical pathways, a biomarker panel including multiple biomarkers, rather than a single biomarker, could better distinguish the different groups and supply useful information for clinicians. Therefore, we identified a panel of six biomarkers (glucose, choline, glutamine, glutamate, valine, and dihydrothymine) that were combined together using binary logistic regression to give a high AUC value of 0.969 (Figure 5A).
Table 2.
Summary of metabolite statistical data from controls (C), preoperative ESCC (ESCC), and postoperative ESCC (PO) groups
| Metabolites | ESCC vs C | ESCC vs PO | PO vs C | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| VIP | P Value | Trend | VIP | P Value | Trend | VIP | P Value | Trend | |
| 1-Methylhistidine | 1.097 | 2.54×10-8 | ↓ | 1.006 | 0.0014 | ↓ | 1.003 | 1.89×10-5 | ↓ |
| 3-Hydroxybutyrate | 2.431 | 2.21×10-26 | ↑ | 2.462 | 3.71×10-7 | ↑ | - | - | - |
| Acetate | 2.177 | 8.06×10-6 | ↑ | 1.062 | 0.0014 | ↑ | - | - | - |
| Acetoacetate | - | - | - | - | - | - | 1.320 | 6.98×10-9 | ↑ |
| Acetone | 2.148 | 0.0005 | ↑ | 1.429 | 0.003 | ↑ | - | - | - |
| Alanine | 2.510 | 0.0004 | ↓ | 2.430 | 8.42×10-7 | ↓ | 2.273 | 0.024 | ↓ |
| Choline | 3.319 | 0.0008 | ↑ | 3.064 | 1.22×10-14 | ↑ | 2.401 | 1.78×10-15 | ↑ |
| Citrate | 1.134 | 0.031 | ↑ | - | - | - | - | - | - |
| Creatinine | 1.028 | 2.60×10-7 | ↑ | - | - | - | - | - | - |
| Dihydrothymine | 1.055 | 3.25×10-22 | ↓ | 1.813 | 0.018 | ↓ | - | - | - |
| Glutamate | 2.837 | 1.33×10-10 | ↑ | 2.705 | 7.26×10-26 | ↑ | - | - | - |
| Glutamine | 2.335 | 1.48×10-8 | ↑ | 2.532 | 7.88×10-15 | ↑ | - | - | - |
| Glycerol | 1.735 | 2.78×10-5 | ↑ | - | - | - | - | - | - |
| Glycerophosphorylcholine | - | - | - | 2.665 | 0.04 | ↓ | - | - | - |
| Isobutyrate | 2.542 | 1.24×10-5 | ↑ | 1.728 | 0.002 | ↑ | - | - | - |
| Isoleucine | 1.898 | 6.73×10-19 | ↓ | 1.275 | 6.99×10-15 | ↓ | - | - | - |
| Isopropanol | 1.988 | 0.0007 | ↓ | 2.129 | 8.81×10-10 | ↓ | 1.403 | 1.34×10-13 | ↓ |
| Leucine | 1.954 | 1.38×10-10 | ↓ | 1.190 | 1.91×10-6 | ↓ | - | - | - |
| Low density lipoprotein | 2.091 | 0.017 | ↑ | 1.375 | 0.044 | ↑ | - | - | - |
| Lysine | 2.436 | 7.38×10-7 | ↑ | 2.254 | 2.10×10-5 | ↑ | - | - | - |
| Malonate | 1.197 | 2.48×10-7 | ↓ | 1.072 | 2.53×10-12 | ↓ | - | - | - |
| Methanol | 2.271 | 1.36×10-8 | ↓ | 2.016 | 1.20×10-13 | ↓ | - | - | - |
| Methionine | 1.318 | 1.69×10-5 | ↓ | 1.526 | 2.80×10-15 | ↓ | - | - | - |
| Methylguanidine | 2.643 | 1.94×10-10 | ↓ | 2.563 | 1.09×10-14 | ↓ | - | - | - |
| myo-Inositol | 1.578 | 0.003 | ↓ | - | - | - | - | - | - |
| Pyruvate | 2.154 | 1.54×10-9 | ↓ | 2.029 | 4.67×10-27 | ↓ | - | - | - |
| Trimethylamine N-oxide | 1.123 | 6.45×10-6 | ↓ | 1.518 | 4.44×10-8 | ↓ | - | - | - |
| Valine | 2.594 | 6.45×10-6 | ↓ | 1.289 | 0.003 | ↓ | 2.033 | 0.039 | ↓ |
| Very low density lipoprotein | 1.944 | 0.014 | ↓ | - | - | - | - | - | - |
| α-Glucose | 4.672 | 0.0003 | ↓ | 3.661 | 2.33×10-5 | ↓ | - | - | - |
| β-Glucose | 4.650 | 3.05×10-6 | ↓ | 3.449 | 0.0001 | ↓ | - | - | - |
VIP values > 1.000 were used to indicate statistical significance. Univariate statistical significance of P < 0.05 was used to distinguish metabolites. “-” indicates VIP value is less than 1.000, or that the P-value is greater than 0.05. ‘↑’: up-regulated. ‘↓’: down-regulated.
Figure 4.
Hierarchical cluster analysis of serum metabolic profile for distinguishing ESCC and PO from controls (C). Each column represents one serum sample, and each row represents a single metabolite. The expression values are represented by the color scale. The intensity increases from green (relatively down-regulated) to red (relatively up-regulated).
Table 3.
Sensitivity, specificity, and AUC value of the metabolites for discrimination of controls (C), ESCC, and postoperative ESCC (PO) groups
| Metabolites | ESCC vs C | PO vs ESCC | PO vs C | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Sensitivity | Specificity | AUC | Sensitivity | Specificity | AUC | Sensitivity | Specificity | AUC | |
| 1-Methylhistidine | 0.694 | 0.783 | 0.754 | 0.744 | 0.645 | 0.756 | 0.823 | 0.659 | 0.749 |
| 3-Hydroxybutyrate | 0.712 | 0.754 | 0.712 | 0.738 | 0.670 | 0.717 | 0.520 | 0.855 | 0.686 |
| Acetate | 0.649 | 0.525 | 0.637 | 0.699 | 0.562 | 0.663 | 0.789 | 0.467 | 0.586 |
| Acetoacetate | 0.794 | 0.583 | 0.701 | 0.632 | 0.535 | 0.574 | 0.885 | 0.566 | 0.748 |
| Acetone | 0.432 | 0.708 | 0.584 | 0.496 | 0.665 | 0.546 | 0.734 | 0.513 | 0.614 |
| Alanine | 0.753 | 0.822 | 0.789 | 0.687 | 0.795 | 0.752 | 0.624 | 0.837 | 0.777 |
| Choline | 0.825 | 0.856 | 0.839 | 0.807 | 0.861 | 0.843 | 0.811 | 0.867 | 0.858 |
| Citrate | 0.587 | 0.682 | 0.633 | 0.625 | 0.489 | 0.587 | 0.784 | 0.651 | 0.763 |
| Creatinine | 0.691 | 0.535 | 0.579 | 0.587 | 0.731 | 0.663 | 0.486 | 0.645 | 0.542 |
| Dihydrothymine | 0.836 | 0.821 | 0.827 | 0.779 | 0.650 | 0.711 | 0.775 | 0.672 | 0.733 |
| Glutamate | 0.829 | 0.835 | 0.829 | 0.809 | 0.855 | 0.827 | 0.636 | 0.828 | 0.738 |
| Glutamine | 0.815 | 0.841 | 0.830 | 0.736 | 0.613 | 0.658 | 0.695 | 0.879 | 0.769 |
| Glycerol | 0.662 | 0.713 | 0.678 | 0.537 | 0.718 | 0.629 | 0.664 | 0.845 | 0.765 |
| Glycerophosphorylcholine | 0.654 | 0.451 | 0.575 | 0.715 | 0.623 | 0.661 | 0.541 | 0.743 | 0.663 |
| Isobutyrate | 0.630 | 0.512 | 0.594 | 0.635 | 0.579 | 0.607 | 0.432 | 0.801 | 0.587 |
| Isoleucine | 0.610 | 0.675 | 0.625 | 0.794 | 0.765 | 0.773 | 0.504 | 0.779 | 0.661 |
| Isopropanol | 0.723 | 0.641 | 0.662 | 0.791 | 0.621 | 0.775 | 0.653 | 0.792 | 0.725 |
| Leucine | 0.749 | 0.810 | 0.786 | 0.590 | 0.767 | 0.662 | 0.863 | 0.545 | 0.721 |
| Low density lipoprotein | 0.520 | 0.812 | 0.642 | 0.503 | 0.827 | 0.744 | 0.529 | 0.842 | 0.757 |
| Lysine | 0.707 | 0.556 | 0.634 | 0.834 | 0.587 | 0.785 | 0.837 | 0.628 | 0.778 |
| Malonate | 0.498 | 0.674 | 0.589 | 0.622 | 0.833 | 0.757 | 0.785 | 0.711 | 0.736 |
| Methanol | 0.476 | 0.683 | 0.529 | 0.603 | 0.881 | 0.762 | 0.673 | 0.804 | 0.753 |
| Methionine | 0.588 | 0.786 | 0.671 | 0.724 | 0.785 | 0.746 | 0.874 | 0.523 | 0.729 |
| Methylguanidine | 0.801 | 0.721 | 0.742 | 0.658 | 0.814 | 0.772 | 0.574 | 0.783 | 0.684 |
| myo-Inositol | 0.719 | 0.843 | 0.785/ | 0.439 | 0.824 | 0.648 | 0.688 | 0.456 | 0.587 |
| Pyruvate | 0.723 | 0.810 | 0.773 | 0.816 | 0.857 | 0.835 | 0.653 | 0.828 | 0.782 |
| Trimethylamine N-oxide | 0.675 | 0.497 | 0.576 | 0.826 | 0.685 | 0.782 | 0.650 | 0.803 | 0.723 |
| Valine | 0.838 | 0.803 | 0.821 | 0.548 | 0.843 | 0.705 | 0.599 | 0.773 | 0.715 |
| Very low-density lipoprotein | 0.611 | 0.720 | 0.651 | 0.432 | 0.675 | 0.554 | 0.570 | 0.775 | 0.622 |
| α-Glucose | 0.840 | 0.881 | 0.862 | 0.827 | 0.868 | 0.855 | 0.762 | 0.654 | 0.705 |
| β-Glucose | 0.853 | 0.769 | 0.799 | 0.811 | 0.749 | 0.792 | 0.547 | 0.601 | 0.588 |
Numbers in bold italics are potential biomarkers for distinguishing between ESCC and C, ESCC and PO, and PO and C.
Figure 5.
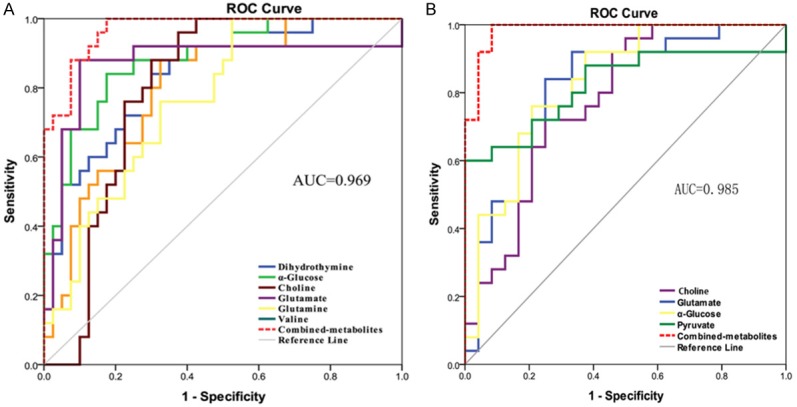
ROC curves of the discriminatory power of the combined potential biomarker panel for ESCC and controls (A, AUC = 0.969), ESCC and PO patients (B, AUC = 0.985).
To evaluate the effects of surgical treatment, 25 serum metabolites that differed between patients with ESCC and PO were identified. Four potential biomarkers (glucose, glutamate, pyruvate, and valine) with an AUC value of 0.985 for distinguishing the two groups, were further selected for the biomarker panel (Figure 5B). Changes in these metabolite biomarkers could be related to the surgical removal of tumour burden and could be used to further evaluate postoperative effects. Six metabolites were identified by comparing PO patients with controls. In PO patients, most of these metabolites tended to gradually return to levels observed in controls. Unfortunately, we could not identify a biomarker panel for differentiating between PO patients and controls, although the choline level decreased slowly with an AUC of 0.858. Therefore, choline may be a key potential biomarker for postoperative therapy and prognostics. According to the human metabolome database and the Kyoto Encyclopedia of Genes and Genomes (KEGG), a map of the metabolic pathways involved based on the metabonomic profiling of ESCC is shown in Figure 6.
Figure 6.
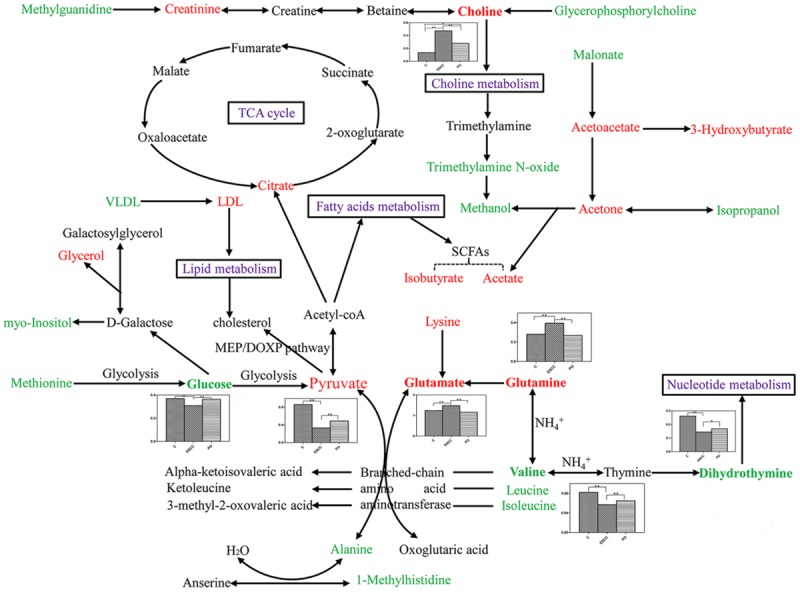
Metabolic pathways of the altered metabolites that include controls, preoperative and postoperative early stage ESCC metabolite biomarkers identified in this study. The bold in metabolites with histograms represents potential biomarkers among three groups. Red texts mean up-regulated with respect to controls, and green texts mean down-regulated with respect to controsl. ‘▲▲’ means P < 0.01, ‘▲’ means P < 0.05.
Discussion
Patients with ESCC have greater long-term survival when ESCC is treated in its early stages. Early detection may increase diagnostic accuracy, promote personalised treatment, and improve prognostic effects. However, the main causes of tumour occurrence remain unknown. Therefore, recognition of the characteristic metabolites of early stage ESCC would enable us to identify the disease and intervene earlier, which is more likely to prevent and/or delay the development of ESCC to medium-term or advanced stage and ultimately result in improved prognoses for patients. Previous studies found that variations in molecular and biochemical metabolism occur before histopathological and morphological changes [13,14]. Here, we used 1H-NMR spectroscopy to identify useful metabolic ESCC biomarkers for early diagnosis and determination of operative effects, influence therapeutic prognosis, and explore the molecular pathogenesis of ESCC.
We found by OPLS-DA that the metabolic profiles of serum could differentiate patients with ESCC from controls. Altered metabolite levels could reflect disturbed glycometabolism (glycolysis and the TCA cycle), and fatty acid, amino acid, choline, ketone body, nucleotide, and lipid metabolism. The potential biomarker panel, which reflects metabolic pathways including glycolysis, and choline, amino acid, and nucleotide metabolism, could significantly differentiate ESCC patients from controls. One glucose molecule can generate 36 ATP molecules via the TCA cycle, while glycolysis produces only two. The use of glucose to generate energy under conditions of adequate oxygen supply by tumour cells is called the Warburg effect [15,16]. A significant decrease in glucose was found in our study, demonstrating a metabolic feature of ESCC with strong aerobic glycolysis, which is consistent with the results observed for many other rapidly proliferating cancers [17-19]. Accelerated glycolysis is a characteristic of all types of cancer and altered glycolysis has been previously examined in ESCC. The metabolomic pathway showed that energy metabolism was the dominant factor in the pathophysiologic mechanism of ESCC. It also identified triglycerides, glycoproteins, and acetone as important sources of energy. Lactate levels were disregarded because of the chance of glycolysis occurring in serum samples during the experiment. Choline, with higher VIP values in patients with ESCC, was the second most altered metabolite. Choline, phosphorylcholine, and glycerophosphorylcholine (GPC) are important for the phospholipid metabolism of cell membranes and have been previously identified as markers of cell proliferation and growth. The increased choline and decreased GPC identified in our research were probably membrane breakdown products because of accelerated tumour propagation. This result is consistent with those obtained for other tumour types [20-22], including in the high-resolution magic-angle spinning 1H-NMR spectroscopy study of squamous carcinoma tissues [23]. Elevated glutamine, glutamate, and glucogenic amino acid levels were also observed playing a distinct role in proliferating and cancer cells in early stage ESCC. To provide for continuous high-energy demands for fast cell proliferation, even under hypoxic conditions, glutamine is converted to glutamate and is further transformed into alpha-ketoglutarate for ATP synthesis through the TCA cycle [24], similar to the results of other studies [25-28]. However, our results are inconsistent with those previously reported for ESCC [29,30]. One reason for this is that these studies focused on the signatures of lymph node ESCC metastasis in which glutamine/glutamate could be consumed in the TCA cycle, whereas we only concentrated on the early stage of ESCC. Another plausible reason for the observed discrepancies is that they used a different platform and different parameters for serum/plasma metabolomic analysis. Compared with controls, we found that valine levels were significantly reduced in patients with ESCC. Valine is an essential branched-chain amino acid that serves as a nitrogen donor for nonessential amino acids and is important for energy consumption [29]. The decreased valine levels in ESCC patients indicate the need for glutamine biosynthesis, related to the TCA cycle, in response to higher energy requirements for tumour proliferation. We also observed that the dihydrothymine levels decreased more markedly in ESCC patients than in controls. Dihydrothymine is an intermediate decomposition product of thymine [31]. DNA replication in tumour cells rapidly exhausts thymine levels; hence, decreased dihydrothymine could be a feasible new biomarker associated with tumour initiation.
The observed differences in serum metabolites, including the potential biomarker panel, in ESCC and PO patients reflects the therapeutic effects of surgery. Comparison of the ESCC, PO, and controls showed that the PO serum metabolites tended toward normalisation, with the exception of choline, which tended to decline very slowly. Tumour burden could inhibit and lower the body’s immune function and further promote the development of cancer [32]. The results indicate that the removal of tumour burden also removes the tumour micro-environment causing the reprogramming of serum metabolites in PO patients. ESCC is characterised by rapid growth and spread and lymph node metastases in nearly 20-60% of TI/II stage cases, which could account for the slow recovery of choline levels following surgery. Therefore, choline may be a valuable potential biomarker for neoplasm progression, recurrence, chemoradiotherapy, or prognosis. We also speculate that these metabolites may be helpful in diagnosis, guiding effective surgical treatments, and forecasting outcomes. In the future, a large number of independent cohorts of patients with different varieties of cancer, as well as healthy controls, should be recruited to screen for potential biomarkers and to verify the findings reported here.
Taken together, the present study demonstrated the early stage serum metabolic alterations of pre- and post-operative ESCC by 1H-NMR-based metabolomics analysis. The results offer convincingly value in aiding diagnosis, monitoring treatment effects, and affording new insights with regard to the pathological mechanisms of ESCC. Furthermore, understanding the molecular pathogenesis of serum metabolic changes in ESCC could offer a new avenue for the individualisation of cancer therapy and prognostics [33].
Acknowledgements
This research was funded by the National Natural Science Foundation of China (Grant Number: 81672918), the Natural Science Foundation of Guangdong Province (Grant Number: 2018A030310154), and the Medical Scientific Research Foundation of Guangdong Province (Grant Number: A2017532).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Wang AH, Liu Y, Wang B, He YX, Fang YX, Yan YP. Epidemiological studies of esophageal cancer in the era of genome-wide association studies. World J Gastrointest Pathophysiol. 2014;5:335–343. doi: 10.4291/wjgp.v5.i3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert R, Hainaut P. The multidisciplinary management of gastrointestinal cancer. Epidemiology of oesophagogastric cancer. Best Pract Res Clin Gastroenterol. 2007;21:921–945. doi: 10.1016/j.bpg.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Deng W, Lin SH. Advances in radiotherapy for esophageal cancer. Ann Transl Med. 2018;6:79. doi: 10.21037/atm.2017.11.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuwahara A, Yamamori M, Nishiguchi K, Okuno T, Chayahara N, Miki I, Tamura T, Kadoyama K, Inokuma T, Takemoto Y, Nakamura T, Kataoka K, Sakaeda T. Effect of dose-escalation of 5-fluorouracil on circadian variability of its pharmacokinetics in Japanese patients with Stage III/IVa esophageal squamous cell carcinoma. Int J Med Sci. 2010;7:48–54. doi: 10.7150/ijms.7.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang A, Sun H, Wang P, Wang X. Salivary proteomics in biomedical research. Clin Chim Acta. 2013;415:261–265. doi: 10.1016/j.cca.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Klupczynska A, Dereziński P, Garrett TJ, Rubio VY, Dyszkiewicz W, Kasprzyk M, Kokot ZJ. Study of early stage non-small-cell lung cancer using Orbitrap-based global serum metabolomics. J Cancer Res Clin Oncol. 2017;143:649–659. doi: 10.1007/s00432-017-2347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Idle JR, Gonzalez FJ. Metabolomics. Cell Metab. 2007;6:348–351. doi: 10.1016/j.cmet.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Espiridion B, Liang D, Ajani JA, Liang S, Ye Y, Hildebrandt MA, Gu J, Wu X. Identification of serum markers of esophageal adenocarcinoma by global and targeted metabolic profiling. Clin Gastroenterol Hepatol. 2015;13:1730–1737. doi: 10.1016/j.cgh.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Chen Y, Zhang R, He J, Song Y, Wang J, Wang H, Wang L, Zhan Q, Abliz Z. Global metabolomics reveals potential urinary biomarkers of esophageal squamous cell carcinoma for diagnosis and staging. Sci Rep. 2016;6:35010. doi: 10.1038/srep35010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YX, Wang LJ, Wang SM, Liang S, Chen A, Tang H, Chen L, Deng F. Study of metabolomic profiles of human esophageal carcinoma by use of high-resolution magic-angle spinning H-NMR spectroscopy and multivariate data analysis. Anal Bioanal Chem. 2013;405:3381–3389. doi: 10.1007/s00216-013-6774-8. [DOI] [PubMed] [Google Scholar]
- 11.Mir SA, Rajagopalan P, Jain AP, Khan AA, Datta KK, Mohan SV, Lateef SS, Sahasrabuddhe N, Somani BL, Keshava Prasad TS, Chatterjee A, Veerendra Kumar KV, VijayaKumar M, Kumar RV, Gundimeda S, Pandey A, Gowda H. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J Proteom. 2015;127:96–102. doi: 10.1016/j.jprot.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, Jin H, Hou X, Lv J, Gao X, Zheng G. Disturbed tryptophan metabolism correlating to progression and metastasis of esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2017;486:781–787. doi: 10.1016/j.bbrc.2017.03.120. [DOI] [PubMed] [Google Scholar]
- 13.Dawiskiba T, Deja S, Mulak A, Ząbek A, Jawień E, Pawełka D, Banasik M, Mastalerz-Migas A, Balcerzak W, Kaliszewski K, Skóra J, Barć P, Korta K, Pormańczuk K, Szyber P, Litarski A, Młynarz P. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J Gastroenterol. 2014;20:163–174. doi: 10.3748/wjg.v20.i1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei EK, Wolin KY, Colditz GA. Time course of risk factors in cancer etiology and progression. J. Clin. Oncol. 2010;28:4052–4057. doi: 10.1200/JCO.2009.26.9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 16.Gillies RJ, Robey I, Gatenby RA. Causes and consequences of increased glucose metabolism of cancers. J Nucl Med. 2008;49(Suppl 2):24S–42S. doi: 10.2967/jnumed.107.047258. [DOI] [PubMed] [Google Scholar]
- 17.Luan W, Zhou Z, Ni X, Xia Y, Wang J, Yan Y, Xu B. Long non-coding RNA H19 promotes glucose metabolism and cell growth in malignant melanoma via miR-106a-5p/E2F3 axis. J Cancer Res Clin Oncol. 2018;144:531–542. doi: 10.1007/s00432-018-2582-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 19.M’Koma AE, Blum DL, Norris JL, Koyama T, Billheimer D, Motley S, Ghiassi M, Ferdowsi N, Bhowmick I, Chang SS, Fowke JH, Caprioli RM, Bhowmick NA. Detection of pre-neoplastic and neoplastic prostate disease by MALDI profiling of urine. Biochem Biophys Res Commun. 2007;353:829–834. doi: 10.1016/j.bbrc.2006.12.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen W, Zu Y, Huang Q, Chen F, Wang G, Lan W, Bai C, Lu S, Yue Y, Deng F. Study on metabonomic characteristics of human lung cancer using high resolution magic-angle spinning 1H NMR spectroscopy and multivariate data analysis. Magn Reson Med. 2011;66:1531–1540. doi: 10.1002/mrm.22957. [DOI] [PubMed] [Google Scholar]
- 21.Monteggia E, Colombo I, Guerra A, Berra B. Phospholipid distribution in murine mammary adenocarcinomas induced by activated neu oncogene. Cancer Detect Prev. 2000;24:207–211. [PubMed] [Google Scholar]
- 22.Dobrzyńska I, Szachowicz-Petelska B, Sulkowski S, Figaszewski Z. Changes in electric charge and phospholipids composition in human colorectal cancer cells. Mol Cell Biochem. 2005;276:113–119. doi: 10.1007/s11010-005-3557-3. [DOI] [PubMed] [Google Scholar]
- 23.Newsholme P, Lima MM, Procopio J, Pithon-Curi TC, Doi SQ, Bazotte RB, Curi R. Glutamine and glutamate as vital metabolites. Braz J Med Biol Res. 2003;36:153–163. doi: 10.1590/s0100-879x2003000200002. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Xu L, Shen J, Cao B, Cheng T, Zhao T, Liu X, Zhang H. Metabolic signatures of esophageal cancer: NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochim Biophys Acta. 2013;1832:1207–1216. doi: 10.1016/j.bbadis.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Wu H, Xue R, Lu C, Deng C, Liu T, Zeng H, Wang Q, Shen X. Metabolomic study for diagnostic model of oesophageal cancer using gas chromatography/mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3111–3117. doi: 10.1016/j.jchromb.2009.07.039. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Liu L, Wei S, Nagana Gowda GA, Hammoud Z, Kesler KA, Raftery D. Metabolomics study of esophageal adenocarcinoma. J Thorac Cardiovasc Surg. 2011;141:469–475. doi: 10.1016/j.jtcvs.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Wang L, Hou Z, Ma H, Mamtimin B, Hasim A, Sheyhidin I. Metabolomic profiling reveals potential biomarkers in esophageal cancer progression using liquid chromatography-mass spectrometry platform. Biochem Biophys Res Commun. 2017;491:119–125. doi: 10.1016/j.bbrc.2017.07.060. [DOI] [PubMed] [Google Scholar]
- 28.Wang ZN, Lin Y, Liang JH, Huang Y, Ma C, Liu X, Yang J. NMR-based metabolomic techniques identify potential urinary biomarkers for early colorectal cancer detection. Oncotarget. 2017;8:105819–105831. doi: 10.18632/oncotarget.22402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin H, Qiao F, Chen L, Lu C, Xu L, Gao X. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J Proteome Res. 2014;13:4091–4103. doi: 10.1021/pr500483z. [DOI] [PubMed] [Google Scholar]
- 30.Ma H, Hasim A, Mamtimin B, Kong B, Zhang HP, Sheyhidin I. Plasma free amino acid profiling of esophageal cancer using high-performance liquid chromatography spectroscopy. World J Gastroenterol. 2014;20:8653–8659. doi: 10.3748/wjg.v20.i26.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pero RW, Johnson D, Olsson A. Catabolism of exogenously supplied thymidine to thymine and dihydrothymine by platelets in human peripheral blood. Cancer Res. 1984;44:4955–4961. [PubMed] [Google Scholar]
- 32.Zhou SB, Guo XW, Gu L, Ji SJ. Influential factors on radiotherapy efficacy and prognosis in patients with secondary lymph node metastasis after esophagectomy of thoracic esophageal squamous cell carcinoma. Cancer Manag Res. 2018;10:217–225. doi: 10.2147/CMAR.S147324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.