Abstract
Radiation-induced lung toxicity, including radiation pneumonitis and pulmonary fibrosis, often occurs in patients receiving radiation therapy. Epithelial-mesenchymal transition (EMT) of alveolar epithelial cells (AECs) plays critical roles in radiation-induced lung toxicity. In the present study, RNA sequencing was applied to examine the whole transcriptomes of human pulmonary AEC cells (HPAEpiC) with or without radiation treatment. We found that cytokine, chemokine and cell adhesion signaling pathways were enriched in radiation-treated cells. CCL2 (C-C Motif Chemokine Ligand 2), CCL5 and CCR4 (C-C Motif Chemokine Receptor 4) were among the top enriched genes in chemokine signaling pathway. The upregulation of CCL2, CCL5 and CCR4 in response to irradiation was confirmed at both mRNA and protein levels by real-time PCR, western blotting and enzyme-linked immunosorbent assay analyses. Ophiopogonin B, a bioactive ingredient of Radix Ophiopogon japonicas, was found to attenuate radiation-induced EMT in HPAEpiC cells as demonstrated by the alteration in cell morphology, and the expression of E-cadherin and Vimentin. Ophiopogonin B could also reduce radiation-induced expression of CCL2, CCL5, CCR4 and phosphorylated ERK (p-ERK). Moreover, CCR4 knockdown, U0126 (a MEK/ERK inhibitor) or ophiopogonin B also partially blocked the EMT promoting effects of CCL2 and CCL5. Our data suggested CCL2, CCL5 and CCR4 may be potential therapeutic targets for radiation-induced lung toxicity. Ophiopogonin B, which could down-regulate CCL2, CCL5 and CCR4, may be a useful radioprotective agent.
Keywords: Chemokines, EMT, alveolar epithelial cells, ophiopogonin B
Introduction
The lung is highly sensitive to radiation and represents the major organ targeted for irradiation-induced damage [1]. Radiation-induced lung toxicity, including radiation pneumonitis and pulmonary fibrosis, is common in patients receiving radiotherapy [2]. Radiation pneumonitis usually occurs within a few weeks to several months after the start of radiation therapy, whose symptoms include cough, shortness of breath and fever. Pulmonary fibrosis is a permanent scarring of lung tissue that responds more slowly to initial tissue damage (lasting months to years) [3-5]. The inflammation and fibrosis processes are initiated and maintained by a complex network of cytokines and chemokines [1,6]. Alveolar epithelial cells (AECs), as a rich cellular source of chemokines, play important roles in radiation-induced lung toxicity. By utilizing a co-culture system, Hong et al. found that irridiation significantly increased the levels of several cytokines (CSF2 [Colony Stimulating Factor 2], CSF3, IL-6 [interleukin-6] and CCL2 [C-C Motif Chemokine Ligand 2]) in a dose-dependent manner [7]. During the processes of radiation-induced lung toxicity, AECs convert into myofibroblasts by epithelial-mesenchymal transition (EMT) [8], in which TGF-β (transforming growth factor-β)/Smad [9] and ERK (extracellular signal-regulated kinase) [10] signaling pathways are involved. The myofibroblasts produce matrix molecules, thus contributing to the radiation-induced lung toxicity [11,12].
Increasing evidence has been presented that traditional Chinese medicine [13], such as ginseng [14], astragalus [15] and turmeric [16], have potential radioprotective effects. Ophiopogonin B is a bioactive ingredient of Radix Ophiopogon japonicas, a traditional Chinese herbal drug which has been reported to exert cardioprotective, antibacterial, anti-cancer, anti-inflammation and radiation protective activities [17,18]. Ophiopogonin B has been reported to exert anticancer effects in cervical cancer, gastric cancer and lung cancer via regulating JNK (c-Jun N-terminal kinase), ERK or PI3K (phosphatidylinositol 3-kinase)/AKT signaling [19-23]. However, little is known about the role of ophiopogonin B in radiation-induced lung toxicity.
In the present study, whole transcriptomes of human pulmonary AECs (HPAEpiC) in response to radiation was sequenced by next-generation RNA sequencing. Significantly changed genes were identified. Functional analysis suggested that radiation was positively correlated with multiple genes in cytokine, chemokine, cell adhesion signaling pathways. CCL2, CCL5 and CCR4 (C-C Motif Chemokine Receptor 4) were enriched genes in chemokine signaling pathway. Real-time PCR, western blot and ELISA (enzyme-linked immunosorbent assay) analyses confirmed the upregulation of CCL2, CCL5 and CCR4 in response to irradiation. We also investigate the effects of ophiopogonin B on radiation-induced EMT and the expression of CCL2, CCL5, CCR4 and phosphorylated ERK (p-ERK) in HPAEpiC cells.
Materials and methods
Cell culture
Human pulmonary alveolar epithelial cells (HPAEpiC) obtained from ScienCell Research Laboratories (Carlsbad, CA, USA) were maintained in Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) in under 5% CO2 at 37°C.
RNA extraction and RNA sequencing
HPAEpiC cells were exposed to 8 Gray of 60Co γ-rays (Hangzhou Cancer Hospital) and cultured for 24 h. Then, total RNA was extracted from 3 samples with radiation (Radiation group) and 3 samples without radiation (Control group) using a Trizol kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The isolated RNA was quantified using a Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and the integrity of isolated RNA was checked by denaturing formaldehyde gel electrophoresis. Complementary DNA (cDNA) libraries were constructed with TruSeq® RNA LT Sample Prep Kit v2 (Illumina, San Diego, CA, USA) following the manufacturer’s protocol. Sequencing was performed using Illumina Hiseq 2500 instruments (Illumina). RNA sequencing data was analyzed as previously described [24]. Differentially expressed genes (DEG) were chosen according to the p value ≤ 0.05 and the Fold Change (FC) value ≥ 2. Gene set enrichment analysis (GSEA) was performed as describe previously [24] to identify KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways enriched in the Radiation group and the Control group.
Quantitative real-time PCR
Total RNA was reversed transcribed into cDNA with the cDNA Reverse Transcription Kit (Thermo fisher, Rockford, IL, USA). SYBR green PCR mix (Thermo fisher) was used for quantitative real-time PCR on an ABI 7300 series PCR machine (Applied Biosystems, Foster City, CA, USA). The mRNA expression levels were quantified using the 2-ΔΔCT method, with GAPDH as the internal control. The primers for real-time PCR analysis are as follows: CCL2, 5’-AACCGAGAGGCTGAGACTAAC-3’ (forward) and 5’-TGCCAACCCAGAGAAGAAATG-3’ (reverse); CCL5, 5’-AACCGAGAGGCTGAGACTAAC-3’ (forward) and 5’-AGGACAAGAGCAAGCAGAAAC-3’ (reverse); CCR4, 5’-CCTTCCTGGCTTTCTGTTC-3’ (forward) and 5’-CATCTTCACCGCCTTGTTC-3’ (reverse); and GAPDH, 5’-AATCCCATCACCATCTTC-3’ (forward) and 5’-AGGCTGTTGTCATACTTC-3’ (reverse).
Western blotting analysis
The cells were washed twice with PBS and lysed in ice-cold radioimmunoprecipitation buffer (Solarbio, Beijing, China) following the manufacturer’s protocols. Equal amount of protein from each sample was separated by electrophoresis and transferred onto nitrocellulose membranes (Millipore, Bredford, USA). The membranes were blocked and incubation with the primary antibodies at 4°C overnight. Following incubation with horseradish peroxidase (HRP)-labeled secondary antibody (Beyotime, Shanghai, China; dilution 1:1000) at room temperature for 1 h, protein expression was detected with an enhanced chemiluminescence kit (Millipore). The sources of primary antibodies were as follows: anti-CCL2 (dilution 1:2000), anti-CCL5 (dilution 1:500), anti-CCR4 (dilution 1:500) were from Abcam (Cambridge, MA, USA), while anti-phosphor-ERK (dilution 1:1000) anti-ERK (dilution 1:1000), anti-Vimentin (dilution 1:1000), anti-E-cadherin(dilution 1:1000) and anti-GAPDH (dilution 1:2000) were obtained from Cell Signaling Technology (Danvers, MA, USA). Experiments were repeated at least for three times and representative blots are shown.
Enzyme-linked immunosorbent assay (ELISA)
The concentrations of CCL2 and CCL5 in the culture medium were measured with ELISA kits (R&D Systems, Minneapolis, MN, USA) following the manufacturer’s instructions.
Statistical analysis
All data were analyzed with Graphpad Prism 6.0 program (GraphPad, San Diego, CA, USA) and presented as the mean ± standard deviation (SD). The comparison among different groups was made by one-way analysis of variance (ANOVA). Values of P<0.05 were considered statistically significant.
Results
Whole transcriptional profile of the response to radiation in HPAEpic cells by RNA sequencing analysis
We examined the changes of the whole transcriptional profile of HPAEpic cells in response to radiation by using RNA-seq method. The heat map (Figure 1) clearly showed a noteworthy difference in gene expression pattern between the Radiation group and the Control group. The volcano analysis was conducted with a minimum of a 2-fold change and P ≤ 0.05 and showed more up-regulated genes than downregulated genes in response to radiation (Figure 2A). A total of 1,385 genes was found significantly changed after radiation treatment, of which 1,028 genes were up-regulated (Supplementary Table 1) and 357 genes were down-regulated (Supplementary Table 2). Base on GSEA analysis, 40 and 6 pathways were enriched in radiation-treated cells (Table 1) and control cells (Table 2), respectively. It is worth noting that radiation exposure was positively correlated with cytokine, chemokine (Figure 2B) and cell adhesion signaling pathways (Table 1), while negatively correlated with DNA replication and cell cycle processes (Table 2).
Figure 1.
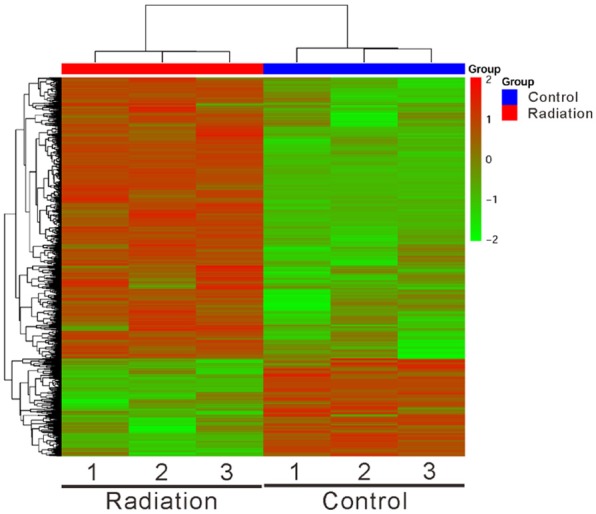
Cluster analysis and heatmap of RNA sequencing data from radiation-treated and control HPAEpic cells. The color bar across the top of the heat map indicates the Radiation group (red) and the Control group (blue). Color from red to green indicates high to low expression.
Figure 2.
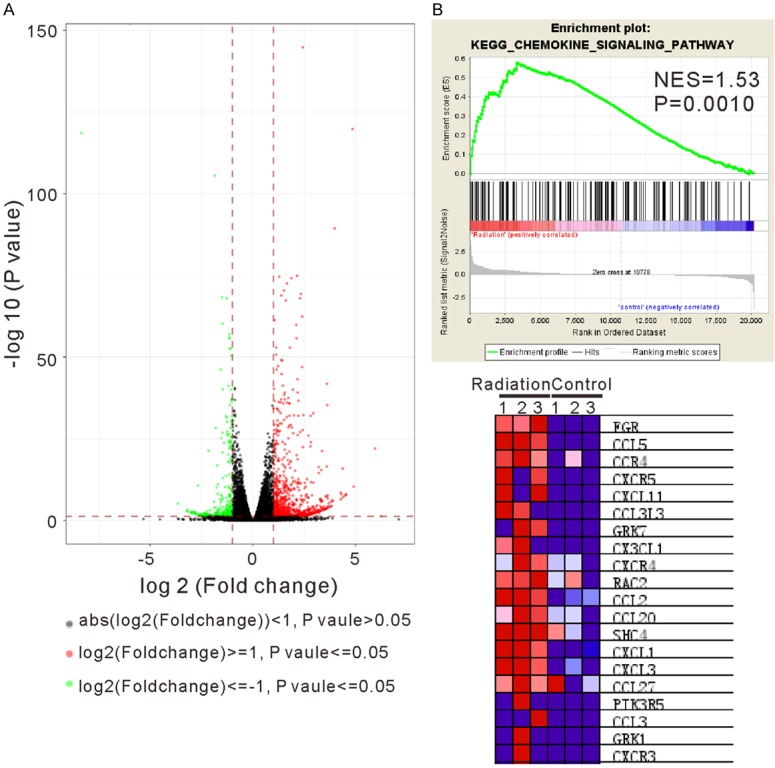
Analysis of significantly changed genes in response to radiation. A. Volcano analysis was performed with a minimum of a 2-fold change and P<0.05 between the Radiation group and the Control group. The log2 (fold change) is plotted on the x-axis and the negative log10 (P-value) is plotted on the y-axis. Red and green dots represented up-regulated and down-regulated genes, respectively. B. Gene set enrichment analysis (GSEA) showed that KEGG chemokine signaling pathway was correlated with radiation response. The enrichment plot (upper panel) and the top 20 enriched genes (bottom panel) are shown.
Table 1.
Statistically significant KEGG classifications of enrichment in radiation-treated HPAEpiC cells
| Name | Size | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| AUTOIMMUNE_THYROID_DISEASE | 27 | 1.7672 | 0.0000 | 0.0092 |
| HEMATOPOIETIC_CELL_LINEAGE | 61 | 1.7366 | 0.0000 | 0.0112 |
| GRAFT_VERSUS_HOST_DISEASE | 23 | 1.7120 | 0.0000 | 0.0144 |
| CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 186 | 1.6692 | 0.0000 | 0.0211 |
| B_CELL_RECEPTOR_SIGNALING_PATHWAY | 68 | 1.6452 | 0.0000 | 0.0223 |
| CHEMOKINE_SIGNALING_PATHWAY | 145 | 1.5332 | 0.0010 | 0.0594 |
| CELL_ADHESION_MOLECULES_CAMS | 107 | 1.5561 | 0.0011 | 0.0572 |
| TOLL_LIKE_RECEPTOR_SIGNALING_PATHWAY | 85 | 1.5568 | 0.0011 | 0.0608 |
| DRUG_METABOLISM_OTHER_ENZYMES | 39 | 1.6249 | 0.0012 | 0.0251 |
| TYPE_I_DIABETES_MELLITUS | 30 | 1.7054 | 0.0013 | 0.0124 |
| NOTCH_SIGNALING_PATHWAY | 42 | 1.6262 | 0.0024 | 0.0269 |
| PROXIMAL_TUBULE_BICARBONATE_RECLAMATION | 21 | 1.6600 | 0.0026 | 0.0194 |
| ALLOGRAFT_REJECTION | 24 | 1.6688 | 0.0027 | 0.0176 |
| LEISHMANIA_INFECTION | 56 | 1.5425 | 0.0035 | 0.0621 |
| VIRAL_MYOCARDITIS | 56 | 1.5742 | 0.0036 | 0.0514 |
| LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 100 | 1.4851 | 0.0043 | 0.0798 |
| VASCULAR_SMOOTH_MUSCLE_CONTRACTION | 99 | 1.4880 | 0.0064 | 0.0792 |
| CYTOSOLIC_DNA_SENSING_PATHWAY | 40 | 1.5406 | 0.0073 | 0.0599 |
| NEUROACTIVE_LIGAND_RECEPTOR_INTERACTION | 181 | 1.3489 | 0.0082 | 0.2307 |
| NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 53 | 1.5378 | 0.0091 | 0.0588 |
| PRION_DISEASES | 23 | 1.5768 | 0.0115 | 0.0532 |
| ONE_CARBON_POOL_BY_FOLATE | 16 | 1.5469 | 0.0139 | 0.0623 |
| ABC_TRANSPORTERS | 41 | 1.5293 | 0.0143 | 0.0596 |
| SYSTEMIC_LUPUS_ERYTHEMATOSUS | 38 | 1.4924 | 0.0157 | 0.0784 |
| PANTOTHENATE_AND_COA_BIOSYNTHESIS | 15 | 1.5206 | 0.0160 | 0.0641 |
| FOCAL_ADHESION | 187 | 1.3368 | 0.0164 | 0.2281 |
| INTESTINAL_IMMUNE_NETWORK_FOR_IGA_PRODUCTION | 31 | 1.5124 | 0.0172 | 0.0656 |
| AXON_GUIDANCE | 118 | 1.3750 | 0.0212 | 0.1975 |
| ADHERENS_JUNCTION | 68 | 1.4183 | 0.0245 | 0.1542 |
| STARCH_AND_SUCROSE_METABOLISM | 34 | 1.5007 | 0.0245 | 0.0736 |
| PENTOSE_AND_GLUCURONATE_INTERCONVERSIONS | 16 | 1.5189 | 0.0259 | 0.0625 |
| FC_EPSILON_RI_SIGNALING_PATHWAY | 66 | 1.4097 | 0.0264 | 0.1587 |
| EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION | 64 | 1.4159 | 0.0268 | 0.1532 |
| LONG_TERM_DEPRESSION | 59 | 1.4194 | 0.0272 | 0.1578 |
| JAK_STAT_SIGNALING_PATHWAY | 119 | 1.3342 | 0.0342 | 0.2230 |
| NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 97 | 1.3437 | 0.0380 | 0.2246 |
| ECM_RECEPTOR_INTERACTION | 80 | 1.3443 | 0.0403 | 0.2287 |
| OLFACTORY_TRANSDUCTION | 26 | 1.4469 | 0.0413 | 0.1238 |
| MELANOMA | 62 | 1.3664 | 0.0450 | 0.2083 |
| ASCORBATE_AND_ALDARATE_METABOLISM | 15 | 1.4317 | 0.0492 | 0.1431 |
Table 2.
Statistically significant KEGG classifications of enrichment in control cells
| Name | Size | NES | NOM p-val | FDR q-val |
|---|---|---|---|---|
| SPLICEOSOME | 116 | -1.741 | 0.0000 | 0.108 |
| DNA_REPLICATION | 36 | -1.519 | 0.0109 | 0.285 |
| RNA_DEGRADATION | 54 | -1.464 | 0.0175 | 0.272 |
| AMINOACYL_TRNA_BIOSYNTHESIS | 41 | -1.404 | 0.0444 | 0.300 |
| CELL_CYCLE | 124 | -1.261 | 0.0196 | 0.395 |
| PARKINSONS_DISEASE | 108 | -1.252 | 0.0492 | 0.364 |
Radiation induced CCL2, CCL5 and CCR4 in HPAEpic cells
CCL5, CCR4 and CCL2, which were among the top 20 enriched genes in chemokine signaling pathway (Figure 2B), were selected for further study. As shown in Figure 3A, radiation time-dependently increased the mRNA expression of CCL2, CCL5 and CCR4 comparing with Control cells. Furthermore, a significant increase in protein level of CCL2, CCL5 and CCR4 were observed in radiation-treated cells as measured by western blot (Figures 3B, S1). ELISA assay confirmed the elevated release of CCL2 and CCL5 to the medium in response to radiation at a time-dependent manner (Figure 3C).
Figure 3.
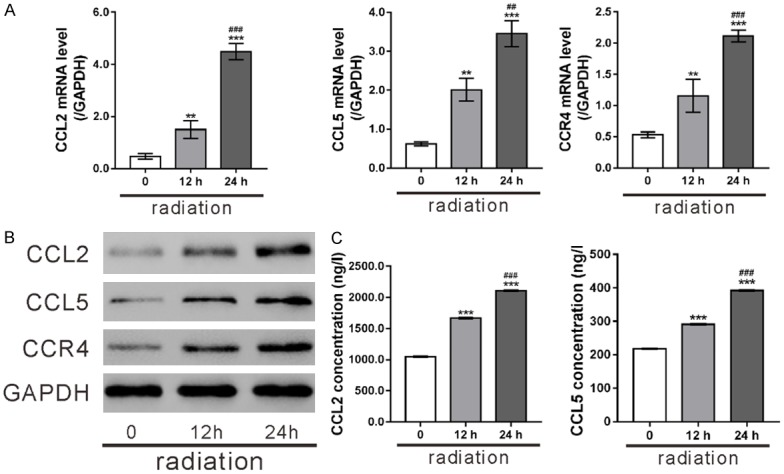
Radiation induced CCL2, CCL5 and CCR4 in HPAEpic cells. HPAEpic cells subjected to radiation (8 Gray) and then cultured for 0, 12 and 24 h. (A, B) The mRNA and protein expression of CCL2, CCL5 and CCR4 was assessed by real-time PCR analysis (A) and western blotting (B), respectively. (C) The release of CCL2 and CCL5 in the culture medium was detected by ELISA assays. **P<0.01, ***P<0.001 versus 0 h; ##P<0.01, ###P<0.001 versus 12 h.
Ophiopogonin B attenuated radiation-induced EMT of HPAEpic cells
Upon radiation, AECs undergo EMT, which plays a critical role in the pathogenesis of radiation-induced lung toxicity. To study the radioprotective effects of ophiopogonin B, HPAEpic cells were pre-treated with ophiopogonin B (OP-B) and then subjected to radiation. As illustrated in Figure 4A, Control cells without radiation treatment displayed classical epithelial morphology, while radiation-treated cells had elongated spindle-like shape. Pre-treatment with ophiopogonin B improved the morphological alteration caused by radiation.
Figure 4.
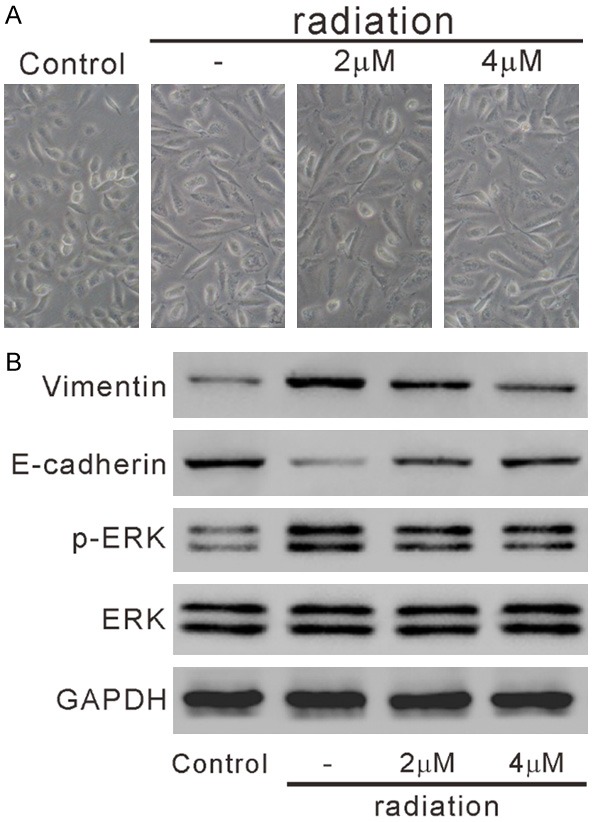
Ophiopogonin B (OP-B) affected the morphology changes and the levels of EMT-related proteins in radiation-treated HPAEpic cells. HPAEpic cells were pre-treated with 0, 2 or 4 mM OP-B for 2 h, then subjected to radiation (8 Gray) and cultured for another 24 h. Cells without any treatment were served as negative control. A. Cell morphology was observed under a microscope. The representative phase contrast images from three independent experiments are shown (magnification 200×). B. The levels of E-cadherin, Vimentin, p-ERK and ERK was detected by western blotting.
The reduced expression of epithelial molecular markers (such as E-cadherin) and elevated expression of mesenchymal molecular markers (such as Vimentin) are main characteristics of EMT [8]. Western blot analysis showed that radiation increased Vimentin expression and decreased E-cadherin expression, which was partially reversed by ophiopogonin B treatment (Figures 4B, S2). Additionally, the ERK signaling pathway is an important regulatory factor in radiation-induced EMT [10,25]. Ophiopogonin B treatment obviously decreased radiation-elevated ERK phosphorylation (p-ERK) (Figure 4B). These data suggested that ophiopogonin B attenuated radiation-induced EMT of HPAEpic cells.
Ophiopogonin B inhibited CCL2, CCL5 and CCR4 in radiation-exposed HPAEpic cells
The effects of ophiopogonin B on the expression of CCL2, CCL5 and CCR4 were also studied. As shown in Figures 5A, 5B and S3, ophiopogonin B dose-dependently suppressed the expression of CCL2, CCL5 and CCR4 in radiation-treated HPAEpic cells at both mRNA and protein levels. Moreover, ophiopogonin B treatment reduced the release of CCL2 and CCL5 in radiation-exposed HPAEpic cells at a dose-dependent fashion (Figure 5C). The above findings strongly indicated that ophiopogonin B regulated CCL2, CCL5 and CCR4 expression in AECs cells.
Figure 5.
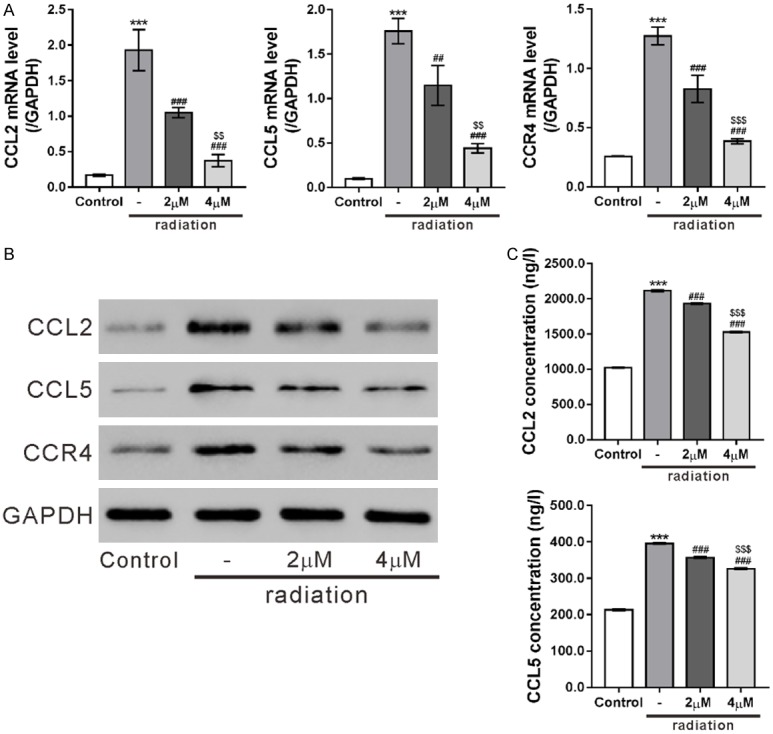
Ophiopogonin B (OP-B) inhibited CCL2, CCL5 and CCR4 in HPAEpic cells. HPAEpic cells were pre-treated with 0, 2 or 4 mM OP-B for 2 h, then subjected to radiation (8 Gray) and cultured for another 24 h. Cells without any treatment were served as negative control. The mRNA and protein expression of CCL2, CCL5 and CCR4 was assessed by real-time PCR analysis (A) and western blotting (B), respectively. (C) The release of CCL2 and CCL5 in the culture medium was detected by ELISA assays. ***P<0.001 versus Control; ##P<0.01, ###P<0.001 versus radiation; $$P<0.01, $$$P<0.001 versus 2 mM OP-B + radiation.
CCR4 knockdown and ophiopogonin B affected the levels of EMT-related proteins in CCL2 or CCL5 treated HPAEpic cells
To explore whether CCR4 mediated the functions of CC2 or CCL5, CCR4 expression was knocked down in HPAEpic cells. HPAEpic cells were transduced with CCR4 shRNAs (sh#1, sh#2 and sh#3) or control shRNA (shCon). Real-time PCR (Figure 6A) and western blotting analysis (Figures 6B, S4A) showed that CCR4 was efficiently knocked down by all CCR4 shRNAs, of which sh#3 displayed the best knock-down efficiency. Next, HPAEpic cells were transduced with sh#3 or shCon in the present of CCL2 or CCL5. Western blot analysis (Figures 6C, S4B) showed that CCL2 and CCL5 increased p-ERK and Vimentin, but decreased E-cadherin. CCR4 knockdown partially blocked the effects of CCL2 and CCL5. These data suggested that the effect of CCL2 and CCL5 on the EMT-related proteins was dependent on CCR4.
Figure 6.
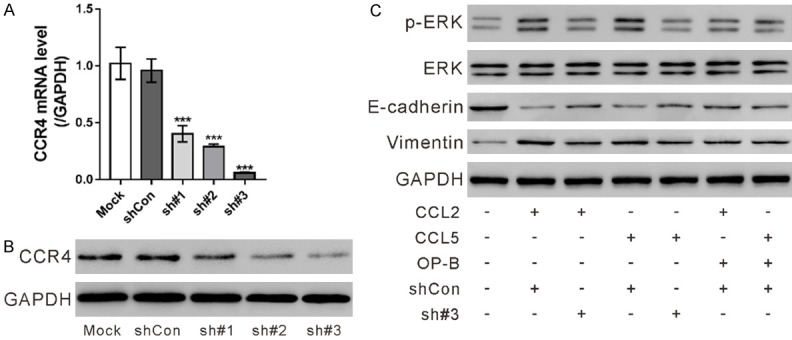
CCR4 knockdown and ophiopogonin B (OP-B) affected the levels of EMT-related proteins in CCL2 or CCL5 treated HPAEpic cells. (A, B) HPAEpic cells were transduced with CCR4 shRNAs (sh#1, sh#2 and sh#3) or control shRNA (shCon). The mRNA and protein expression of CCR4 was assessed by real-time PCR (A) and western blotting analysis (B), respectively, at 48 h post transduction. Cells without any treatment (Mock) were used as negative control. ***P<0.001 versus shCon. (C) HPAEpic cells were divided into 7 groups and pre-treated with shCon or sh#3 for 24 h, or with 4 mM OP-B for 2 h, and then exposed to 100 ng/ml CCL2 (R&D Systems, Minneapolis, MN, USA) or CCL5 (R&D Systems) or not as indicated for 24 h. The levels of E-cadherin, Vimentin, p-ERK and ERK was detected by western blotting.
Further, HPAEpic cells were treated with 4 mM ophiopogonin B in the present of CCL2 or CCL5. Ophiopogonin B also partially blocked the effects of CCL2 and CCL5 on the EMT-related proteins.
U0126 suppressed CCL2 or CCL5-induced EMT
To investigate the involvement of ERK in CCL2 or CCL5-induced EMT, HPAEpic cells were pre-treated with a MEK/ERK inhibitor U0126 and then with CCL2/CCL5. As illustrated in Figure 7A, cells treated with CCL2 or CCL5 had elongated spindle-like shape, which was improved by the treatment with U0126. Western blot analysis showed that CCL2/CCL5 increased the levels of Vimentin and p-ERK, and decreased E-cadherin expression, which was partially reversed by U0126 treatment (Figures 7B, S5).
Figure 7.
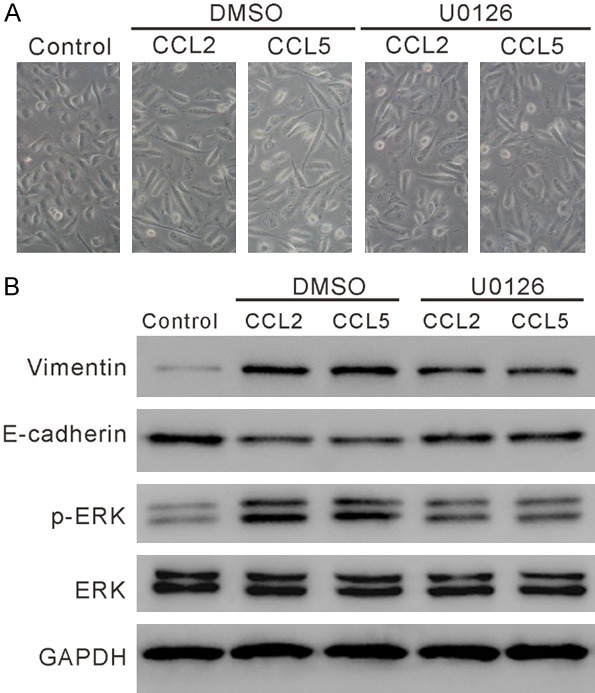
U0126 suppressed CCL2 or CCL5-induced EMT. HPAEpic cells were pre-treated with 10 mM U0126 (Selleck Chemicals, Houston, TX, USA) or DMSO for 2 h and then exposed to 100 ng/ml CCL2 (R&D Systems, Minneapolis, MN, USA) or CCL5 (R&D Systems) as indicated for 24 h. Cells without any treatment were served as negative control. A. Cell morphology was observed under a microscope. The representative phase contrast images from three independent experiments are shown (magnification 200×). B. The levels of E-cadherin, Vimentin, p-ERK and ERK was detected by western blotting.
Discussion
RNA sequencing has been applied to assess the global radiation responses at whole transcriptome level in non-small cell lung cancer (NSCLC) A549 cells [26]. In this study, we conducted RNA sequencing on normal alveolar epithelial cells (HPAEpic) with or without radiation treatment. We identified 1,385 significant changed genes (DEGs) in response to radiation (Supplementary Tables 1 and 2). Radiation-induced inflammation and fibrosis processes are initiated and maintained by a complex network of cytokines and chemokines [1,6]. It is not surprising that multiple genes in cytokine and chemokine signaling pathways were positively correlated with radiation treatment (Table 1).
Chemokines are a large family of small chemotactic cytokines and more than 40 members have been identified. The chemokines exert their biological functions by binding with the G protein-linked transmembrane receptors [27]. Here, CCL2 (also known as MCP1), CCL5 (also known as RANTES) and CCR4 were identified as up-regulated genes in response to radiation and enriched in chemokine signaling pathway (Figure 2B). It has been reported that metaplastic epithelial cells of idiopathic pulmonary fibrosis produce more CCL2 and the increased CCL2 was associated with disease activity [28]. CCL2 secretion was found significantly increased by radiation in type II AECs [7]. CCL2/CCL5 was upregulated in radiation-treated NSCLC cells [29]. CCR4, an important receptor for CCL2 and CCL5, can promote metastasis of lung cancer and colon cancer [30]. Thus, we then chose these three DEGs for subsequent experiments. The mRNA and protein expression of the above three DEGs were time-dependently increased by radiation exposure. In addition, the release of CCL2 and CCL5 was also elevated with time following radiation treatment (Figure 3). Further, CCL2 and CCL5 treatment could decrease E-cadherin expression and increased Vimentin expression, suggested the direct promotion effect of CCL2 and CCL5 on the EMT in HPAEpic cells (Figure 6C). Such effect was weakened by CCR4 knockdown, which suggesting that CCR4 mediated the EMT promoting effect of CCL2 and CCL5.
Ophiopogon decoction can prevent radiation pulmonary injury in rats [18], which prompted us to study the effects of ophiopogonin B, a bioactive ingredient of ophiopogon on irradiation-treated HPAEpic cells. In this study, ophiopogonin B attenuated radiation-induced EMT of HPAEpic cells as indicated by the changes of morphology and the levels of EMT-related proteins (Figure 4). More importantly, ophiopogonin B could reduce irradiation-induced expression of CCL2, CCL5 and CCR4, and attenuated the EMT promoting effect of CCL2 and CCL5. These data suggested that ophiopogonin B could protect alveolar epithelial cells from irradiation-induced EMT. Ophiopogonin B may be an effective radioprotective agent via downregulating CCL2 and CCL5, although further experiments with animal models are required.
The ERK signaling pathway is an important regulatory factor in radiation-induced EMT [10,25]. Here, Ophiopogonin B decreased radiation-induced phosphorylation of ERK (Figure 4B), which was in line was previous findings in human gastric cancer cells [22]. Evidence has been presented for the involvement of CCL2, CCL5 and CCR4 in the ERK signaling pathway. CCL2 [31] and CCL5 [32] could enhance the migration of chondrosarcoma and osteosarcoma cells respectively by activating the ERK signaling. CCR4 facilitated colon cancer cells metastasis via ERK/NF-κB/MMP13 pathway [33] and hepatocellular carcinoma cell metastases via ERK/AKT/MMP2 pathway [34]. Consistently with the previous findings, CCL2 and CCL5 treatment increased the phosphorylation of ERK, which was dependent on the expression of CCR4 (Figure 6C). U0126, a MEK/ERK inhibitor, attenuated the EMT promoting effect of CCL2 and CCL5 (Figure 7). Moreover, ophiopogonin B could also attenuate the promoting effect of CCL2 and CCL5 on ERK phosphorylation. Thus, we speculated that ophiopogonin B effectively improved radiation-induced EMT through CCL2-CCL5/CCR4/ERK signaling.
Collectively, we reported the expression profile of AECs in response to irradiation. We found that CCL2-CCL5/CCR4 was involved in radiation-induced AECs EMT via the ERK signaling. Our data suggested CCL2, CCL5 and CCR4 may be potential therapeutic targets for radiation-induced lung toxicity. Ophiopogonin B, which could down-regulate CCL2, CCL5 and CCR4, may be a useful radioprotective agent.
Acknowledgements
This study was supported by National Natural Science Foundation of China (81173246), the key project of the Science and Technology Planning Project of Hangzhou (2014Z08) and Chinese medicine technology project of Zhejiang Province (2017ZB078).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chen Y, Williams JP, Ding I, Hernady E, Liu W, Smudzin T, Finkelstein JN, Rubin P, Okunieff P. Radiation pneumonitis and early circulatory cytokine markers. Semin Radiat Oncol. 2002;12:26–33. doi: 10.1053/srao.2002.31360. [DOI] [PubMed] [Google Scholar]
- 2.Ataya S, Elwing J, Biddinger P, Panos RJ. Radiation-induced lung injury. Clin Pulm Med. 2006;13:232–242. [Google Scholar]
- 3.Timmerman R, Paulus R, Galvin J, Michalski J, Straube W, Bradley J, Fakiris A, Bezjak A, Videtic G, Johnstone D. Stereotactic body radiation therapy for inoperable early stage lung cancer. JAMA. 2010;303:1070–1076. doi: 10.1001/jama.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whelan TJ, Pignol JP, Levine MN, Julian JA, MacKenzie R, Parpia S, Shelley W, Grimard L, Bowen J, Lukka H, Perera F, Fyles A, Schneider K, Gulavita S, Freeman C. Long-term results of hypofractionated radiation therapy for breast cancer. N Engl J Med. 2010;362:513–520. doi: 10.1056/NEJMoa0906260. [DOI] [PubMed] [Google Scholar]
- 5.Yahalom J, Illidge T, Specht L, Hoppe RT, Li YX, Tsang R, Wirth A International Lymphoma Radiation Oncology Group. Modern radiation therapy for extranodal lymphomas: field and dose guidelines from the international lymphoma radiation oncology group. Int J Radiat Oncol Biol Phys. 2015;92:11–31. doi: 10.1016/j.ijrobp.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Ghafoori P, Marks LB, Vujaskovic Z, Kelsey CR. Radiation-induced lung injury. Assessment, management, and prevention. Oncology (Williston Park) 2008;22:37–47. [PubMed] [Google Scholar]
- 7.Hong ZY, Song KH, Yoon JH, Cho J, Story MD. An experimental model-based exploration of cytokines in ablative radiation-induced lung injury in vivo and in vitro. Lung. 2015;193:409–419. doi: 10.1007/s00408-015-9705-y. [DOI] [PubMed] [Google Scholar]
- 8.Almeida C, Nagarajan D, Tian J, Leal SW, Wheeler K, Munley M, Blackstock W, Zhao W. The role of alveolar epithelium in radiation-induced lung injury. PLoS One. 2013;8:e53628. doi: 10.1371/journal.pone.0053628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R. BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med. 2003;9:964–968. doi: 10.1038/nm888. [DOI] [PubMed] [Google Scholar]
- 10.Nagarajan D, Melo T, Deng Z, Almeida C, Zhao W. ERK/GSK3β/Snail signaling mediates radiation-induced alveolar epithelial-to-mesenchymal transition. Free Radic Biol Med. 2012;52:983–992. doi: 10.1016/j.freeradbiomed.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizuno S, Matsumoto K, Li MY, Nakamura T. HGF reduces advancing lung fibrosis in mice: a potential role for MMP-dependent myofibroblast apoptosis. FASEB J. 2005;19:580–582. doi: 10.1096/fj.04-1535fje. [DOI] [PubMed] [Google Scholar]
- 12.Biernacka A, Dobaczewski M, Frangogiannis NG. TGF-β signaling in fibrosis. Growth Factors. 2011;29:196–202. doi: 10.3109/08977194.2011.595714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi F, Li A, Inagaki Y, Gao J, Li J, Kokudo N, Li X, Tang W. Chinese herbal medicines as adjuvant treatment during chemo- or radio-therapy for cancer. Biosci Trends. 2010;4:297–307. [PubMed] [Google Scholar]
- 14.Lee TK, Johnke RM, Allison RR, O’brien KF, Dobbs LJ Jr. Radioprotective potential of ginseng. Mutagenesis. 2005;20:237–243. doi: 10.1093/mutage/gei041. [DOI] [PubMed] [Google Scholar]
- 15.Qi L, Liu CY, Wu WQ, Gu ZL, Guo CY. Protective effect of flavonoids from astragalus complanatus on radiation induced damages in mice. Fitoterapia. 2011;82:383–392. doi: 10.1016/j.fitote.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Bhartiya US, Raut YS, Joseph LJ, Hawaldar RW, Rao BS. Evaluation of the radioprotective effect of turmeric extract and vitamin E in mice exposed to therapeutic dose of radioiodine. Indian J Clin Biochem. 2008;23:382–386. doi: 10.1007/s12291-008-0084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen MH, Chen XJ, Wang M, Lin LG, Wang YT. Ophiopogon japonicus-A phytochemical, ethnomedicinal and pharmacological review. J Ethnopharmacol. 2016;181:193–213. doi: 10.1016/j.jep.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Kang G, Bai X. Prevention of Ophiopogon decoction on radiation pulmonary injury. Hebei Journal of Traditional Chinese Medicine. 2012;4:1596–1610. [Google Scholar]
- 19.Chen M, Du Y, Qui M, Wang M, Chen K, Huang Z, Jiang M, Xiong F, Chen J, Zhou J, Jiang F, Yin L, Tang Y, Ye L, Zhan Z, Duan J, Fu H, Zhang X. Ophiopogonin B-induced autophagy in non-small cell lung cancer cells via inhibition of the PI3K/Akt signaling pathway. Oncol Rep. 2013;29:430–436. doi: 10.3892/or.2012.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen M, Guo Y, Zhao R, Wang X, Jiang M, Fu H, Zhang X. Ophiopogonin B induces apoptosis, mitotic catastrophe and autophagy in A549 cells. Int J Oncol. 2016;49:316–324. doi: 10.3892/ijo.2016.3514. [DOI] [PubMed] [Google Scholar]
- 21.Nazim UM, Jeong JK, Park SY. Ophiopogonin B sensitizes TRAIL-induced apoptosis through activation of autophagy flux and downregulates cellular FLICE-like inhibitory protein. Oncotarget. 2018;9:4161–4172. doi: 10.18632/oncotarget.23647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W, Zhang Q, Jiang Y, Li F, Xin H. Effects of ophiopogonin B on the proliferation and apoptosis of SGC7901 human gastric cancer cells. Mol Med Rep. 2016;13:4981–4986. doi: 10.3892/mmr.2016.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu QJ, Hou LL, Hu GQ, Xie SQ. [Molecular mechanism of ophiopogonin B induced cellular autophagy of human cervical cancer HeLa cells] . Yao Xue Xue Bao. 2013;48:855–859. [PubMed] [Google Scholar]
- 24.He K, Wu G, Li WX, Guan D, Lv W, Gong M, Ye S, Lu A. A transcriptomic study of myogenic differentiation under the overexpression of PPARγ by RNA-Seq. Sci Rep. 2017;7:15308. doi: 10.1038/s41598-017-14275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding NH, Li JJ, Sun LQ. Molecular mechanisms and treatment of radiation-induced lung fibrosis. Curr Drug Targets. 2013;14:1347–1356. doi: 10.2174/13894501113149990198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang HJ, Kim N, Seong KM, Youn H, Youn B. Investigation of radiation-induced transcriptome profile of radioresistant non-small cell lung cancer A549 cells using RNA-seq. PLoS One. 2013;8:e59319. doi: 10.1371/journal.pone.0059319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinader V, Afarinkia K. A beginner’s guide to chemokines. Future Med Chem. 2012;4:845–852. doi: 10.4155/fmc.12.49. [DOI] [PubMed] [Google Scholar]
- 28.Iyonaga K, Takeya M, Saita N, Sakamoto O, Yoshimura T, Ando M, Takahashi K. Monocyte chemoattractant protein-1 in idiopathic pulmonary fibrosis and other interstitial lung diseases. Hum Pathol. 1994;25:455–463. doi: 10.1016/0046-8177(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Yang X, Tsai Y, Yang L, Chuang KH, Keng PC, Lee SO, Chen Y. IL-6 mediates macrophage infiltration after irradiation via up-regulation of CCL2/CCL5 in non-small cell lung cancer. Radiat Res. 2017;187:50–59. doi: 10.1667/RR14503.1. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura ES, Koizumi K, Kobayashi M, Saitoh Y, Arita Y, Nakayama T, Sakurai H, Yoshie O, Saiki I. RANKL-induced CCL22/macrophage-derived chemokine produced from osteoclasts potentially promotes the bone metastasis of lung cancer expressing its receptor CCR4. Clin Exp Metastasis. 2006;23:9–18. doi: 10.1007/s10585-006-9006-1. [DOI] [PubMed] [Google Scholar]
- 31.Tang CH, Tsai CC. CCL2 increases MMP-9 expression and cell motility in human chondrosarcoma cells via the Ras/Raf/MEK/ERK/NF-κB signaling pathway. Biochem Pharmacol. 2012;83:335–344. doi: 10.1016/j.bcp.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Wang SW, Wu HH, Liu SC, Wang PC, Ou WC, Chou WY, Shen YS, Tang CH. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PLoS One. 2012;7:e35101. doi: 10.1371/journal.pone.0035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ou B, Zhao J, Guan S, Feng H, Wangpu X, Zhu C, Zong Y, Ma J, Sun J, Shen X. CCR4 promotes metastasis via ERK/NF-κB/MMP13 pathway and acts downstream of TNF-α in colorectal cancer. Oncotarget. 2016;7:47637–47694. doi: 10.18632/oncotarget.10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng X, Wu H, Jin ZJ, Ma D, Yuen S, Jing XQ, Shi MM, Shen BY, Peng CH, Zhao R. Up-regulation of chemokine receptor CCR4 is associated with human hepatocellular carcinoma malignant behavior. Sci Rep. 2017;7:12362. doi: 10.1038/s41598-017-10267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


