Abstract
Long noncoding RNAs (lncRNAs) play key roles in various malignancy pathogenesis. However, the mechanisms remain poorly understood in the development and progression of colorectal cancer (CRC). Here, we focused on the specific role of human leukocyte antigen (HLA) Complex P5 (HCP5) in CRC. Quantitative real-time PCR (qRT-PCR) analysis and western blot were used to assess the expression of HCP5 in CRC tissues. The association between the expressions of HCP5 and miR-139-5p was assessed by Pearson’s correlation analysis. The prognosis of CRC patients was analyzed by Kaplan-Meier survival analysis. Specific siRNAs were stably transfected into CRC cells with lentivirus approaches. The proliferative, migrative and invasive capacities of CRC cells were detected by Transwell, MTT and scratch assay, respectively. Dual-luciferase assay was performed to measure miR-139-5p-targeted relationship with lncRNA HCP5. HCP5 overexpression and of miR-139-5p downregulation were dramatically correlated with low TNM stage, poor differentiation, low tumor depth invasion in CRC patients (P < 0.05). Besides, HCP5 overexpression or ZEB1 knockdown repressed Snail family transcriptional repressor (SNAI) and vimentin expressions, upregulated E-cadherin expression, and inhibited cell proliferation and metastasis (P < 0.05). Moreover, luciferase reporter assay demonstrated that miR-139-5p was a directly target of HCP5 (P < 0.05). Overall, the present study indicated that HCP5 played a key regulator in CRC development and progression by targeting HCP5/miR-139-5p/ZEB1 axis, which may serve as a novel therapeutic target for CRC therapy.
Keywords: Colorectal cancer (CRC), long noncoding RNAs (lncRNA), human leukocyte antigen (HLA) complex P5 (HCP5), miR-139-5p, proliferation, metastasis
Introduction
Colorectal cancer (CRC) is one of the most frequently diagnosed malignancies and the third cause of cancer-related deaths in the world [1]. In the past decades, despite the survival of CRC patients has to some extent been improved by earlier diagnosis and novel therapy strategies, over 50% of CRC patients still encounter local or distant recurrence risk after conventional therapy [2,3]. Epithelial-mesenchymal transition (EMT) is the process by which epithelial cells are transformed into mesenchymal cells under specific pathological conditions [4]. The epithelial cells lose their typical intercellular junction, remodel the cytoskeleton, and become fibroblasts during EMT, resulting in apoptosis resistance and enhanced motor ability. Therefore, suppression of EMT can effectively inhibit metastasis of tumor cells [5,6]. Recent studies have shown that not only coding RNA, but non-coding RNA also regulates EMT [7,8].
Long non-coding RNAs (lncRNAs) are functional non-coding RNAs (length ~200 nt) without the ability to encode proteins [9]. Present studies have shown that a variety of lncRNAs are involved in transcriptional regulation, nuclear structure organization and post-transcriptional processing [10-12]. LncRNAs have been identified as pivotal regulators in many cell biological processes, including cell growth, proliferation and differentiation. Dysregulation of lncRNAs has been preliminarily demonstrated to promote the development, invasion and metastasis of many cancers. Recent studies have shown that aberrant expressions of lncRNAs, such as MAPKAPK5-AS1, BANCR and DLEU1, are associated with CRC, which were regarded as new therapeutic targets for CRC [13-15]. LncRNA human leukocyte antigen (HLA) Complex P5 (HCP5) was regarded as a new gene locus of thyroid diseases [16]. Down-regulation of HCP5 was shown in patients with ovarian cancer, which was considered to be a susceptible gene locus of HCV-related HCC [17,18].
EMT refers to the process of transforming epithelial cells into mesenchymal cells. EMT can be physiologically embryonic development, or pathologically the development of cancer, which is the key initial event of the metastatic cascade [19]. EMT plays a significant role on invasion, proliferation and migration of tumor cells, and is the main cause of drug resistance in tumors. Researchers found that the expression alterations of key molecules were observed during the EMT phenotype acquisition [20]. Nevertheless, the potential mechanism of Wnt signaling pathway to alter EMT process in CRC is unclear. Hence this study was purposed to investigate whether HCP5 promotes EMT process in CRC through regulating downstream miR-139-5p and ZEB1. In addition, we also evaluated the carcinogenic effect of HCP5 in CRC cells.
Materials and methods
CRC tissue samples
CRC species and para-carcinoma control tissues (n = 135) were collected from patients who underwent resection, and were examined as primary CRC by two independent pathologists between January 2014 and December 2015 at Henan University of Science and Technology First Affiliated Hospital. The patients who had chemotherapy history prior to surgery were excluded. The average follow-up period was 5 years. All patients signed the written informed consent. This study was approved by the Ethics Committee of Henan University of Science and Technology First Affiliated Hospital.
Cell culture
Human CRC cell lines (GEO, HCT116, SW620 and LOVO) were obtained from ATCC (Manassas, VA, USA) in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (GIBCO, Invitrogen Inc., Carlsbad, CA, USA). The normal colonic epithelial cell line CCD-18Co was obtained from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI1640 medium (Gibco). The cells were cultured in a humidified atmosphere of 5% CO2 at 37°C.
qRT-PCR
Total RNA was extracted from the cancer cells using TRIzol reagent and cDNA was generated using M-MLV Reverse Transcriptase (Takara Bio Inc., Kusatsu, Japan). qRT-PCR was performed using LightCycler480 (Roche, San Francisco, CA, USA) under the following steps: pre-denaturation at 95 for 40 s; denaturation with 40 cycles of 95 for 5 s and 60 for 40 s; and termination at 95 for 10 min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the endogenous control. Relative expression levels were calculated using the 2-ΔΔCT method.
Transwell assay
Transwell filter (BD Biosciences) was employed to perform Transwell migration and invasion assay as previous described [21].
Scratch wound-healing assay
The cells were seeded in 6-well culture plates at 95%-100% confluence. Cells were harvested 48 hours after transfection. The scratch was created using a 200 µL pipette tip. PBS was used to wash the plates for three times to remove cellular debris. Images for the wound closure were photographed at different times (12, 24, 36 and 48 h) by microscope (Nikon).
Western blot
Protein concentration was determined by BCA kit (Dingguo Company). Proteins were separated by 10% SDS-PAGE and then transferred to polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). The membrane was then blocked with PBST solution containing 5% nonfat dry milk for 1 hour. Rabbit polyclonal primary antibodies against ZEB1 (1:800, Abcam, Cambridge, MA, USA), Snail (1:800, Abcam), β-catenin (1:250, Santa Cruz Biotech, Santa Cruz, CA), E-cadherin (1:1,000, Proteintech, Chicago, USA), E-cadherin (1:100, Bioss), and vimentin (1:200, Spring Bioscience, Pleasanton, CA, USA) were used. After incubating the membranes at 4°C overnight, the membranes were incubated with HRP-conjugated secondary antibodies at room temperature for 2 hours. (1:1,000; Proteintech, Rosemont, IL, USA). The protein expression level was normalized by GAPDH (1:1000, Santa Cruz Biotech). Blots were visualized by ECL Western blotting kit (Amersham Biosciences, Buckinghamshire, UK).
Cell transfection
MiR-139-5p mimics, miR-139-5p inhibitors, negative control (NC), pcDNA3.1-HCP5, pcDNA3.1-ZEB1, pcDNA3.1, HCP5 siRNA (si-HCP5) and si-NC were synthesized by Shanghai GenePharma Co., Ltd. Cells were placed on 12-well plate (Corning, USA) and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were harvested 48 hours after transfection.
MTT
5 mg/ml MTT (20 μl) was added when the bottom of the well was covered with single-layer cells. After incubation for 4 h, 150 μl DMSO was added to each well. The mixture vibrated for 10 min at low speed to fully dissolve the crystal. The absorbance value of samples at 490 nm was detected by enzyme linked immunosorbent assay (ELISA).
Dual luciferase reporter assay
TCF4/LEF reporter plasmids (pTOP-Luc) were transfected into cells by Lipofectamine 2000. After 12 h, the cells were digested and inoculated in the 24-well plates. After 24 h, the cells were lysed and the luciferase activity was detected according to the instructions of the luciferase reporter detection kit (NEB Cor.). Luciferase reporter genes of HCP5 Mut and ZEB1 Mut containing mutation sequence at the binding site of microRNA-139-5p were constructed. The above plasmids and reporter gene vectors were synthesized by Shanghai Sango Corporation (China). Next, microRNA-139-5p mimics were transfected into CRC cells that were transfected HCP5 Wt, HCP5 Mut, ZEB1 Wt, ZEB1 Mut or psiCHECK2, respectively. After 6 h transfection, the luciferase reporter gene assay (Promega, USA) of lysed cells was detected, and Renilla Luciferase Reporter was as internal reference.
Statistical analysis
GraphPad Prism (GraphPad Software, Inc., USA) was used for statistical analysis. All data are presented as the mean ± SD. Comparison between groups was performed by one-way ANOVA followed by student’s t-test. Differences in the survival curve were evaluated using a log-rank test and drawn using Kaplan-Meier method. P < 0.05 were considered as statistically significant difference.
Results
Expression of lncRNA HCP5 and miR-139-5p in CRC tissues and cells
The expression level of HCP5 in CRC tissues was obviously higher compared to para-carcinoma tissues (P < 0.05). However, the expression level of miR-139-5p decreased in CRC tissues compared to para-carcinoma tissues (P < 0.05, Figure 1A). Moreover, overexpression HCP5 was found in CRC cell lines (GEO, HCT116, LOVO and SW620) compared to normal colon cell line (CCD-18CO) (P < 0.05). However, miR-139-5p expression levels in CRC cell lines were obviously lower compared to CCD-18Co cells (P < 0.05, Figure 1B). Spearman’s correlation analysis suggested a significant negative correlation between HCP5 and miR-139-5p in CRC tissues (r = -0.765, P < 0.05, Figure 1C).
Figure 1.
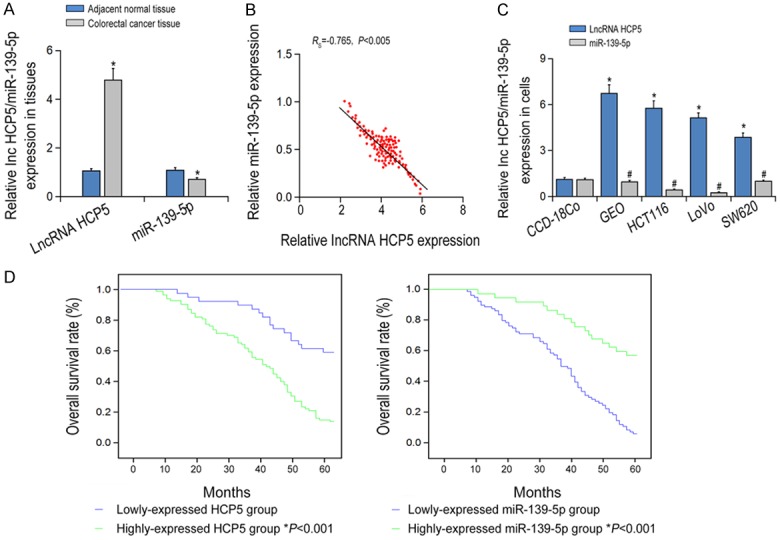
The expression levels of lncRNA HCP5 and miR-139-5p in colorectal cancer (CRC) tissues and cell lines. A: LncRNA HCP5 and miR-139-5p expressions were compared between CRC tissues and adjacent normal tissues. *P < 0.05 vs adjacent normal tissues. B: LncRNA HCP5 and miR-139-5p expression were compared among CRC cells. *P < 0.05 vs HCP5 expression in CCD-18Co cell line; #P < 0.05 vs miR-139-5p expression in CCD-18CO cell line. C: LncRNA HCP5 expression was negatively correlated with miR-139-5p expression within CRC tissues. D: The Kaplan-Meier analysis of correlation between the level of LncRNA HCP5 and miR-139-5p with OS.
Correlation between HCP5/miR-139-5p expressions and clinicopathological parameters of CRC patients
Correlation analysis revealed that overexpression of HCP5 and down-regulation of miR-139-5p were significantly correlated with clinical stage, differentiation and distant metastasis in CRC patients (P < 0.05, Table 1). Furthermore, regression analysis showed that over-expressed HCP5 and down-regulated miR-139-5p predicted poor differentiation, higher TNM stage and distant metastasis in CRC patients (P < 0.05, Data not shown). As shown in Figure 1D, Kaplan-Meier survival analysis showed that the overall survival (OS) of patients with low HCP5 and high miR-139-5p expression was much higher than those with high HCP5 and low expression of miR-139-5p (P < 0.05).
Table 1.
The relationship between lncRNA HCP5/miR-139-5p expression and the colorectal cancer patients’ clinical characteristics
| Characteristics | LncRNA HCP5 expression | miR-139-5p expression | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Low | High | P | Low | High | P | |
| N = 135 | 43 | 92 | 82 | 53 | ||
| Age | ||||||
| ≤ 60 | 24 | 48 | 42 | 30 | ||
| > 60 | 19 | 44 | 0.651 | 40 | 23 | 0.502 |
| Gender | ||||||
| Male | 20 | 56 | 48 | 28 | ||
| Female | 23 | 36 | 0.093 | 34 | 25 | 0.364 |
| Smoking History | ||||||
| Yes | 10 | 15 | 13 | 13 | ||
| No | 33 | 77 | 0.276 | 69 | 40 | 0.213 |
| Tumor size | ||||||
| ≥ 5 cm | 16 | 49 | 44 | 20 | ||
| < 5 cm | 27 | 43 | 0.028 | 38 | 32 | 0.026 |
| Tumor Differentiation Statue | ||||||
| Poor/Moderate | 14 | 52 | 49 | 19 | ||
| Well | 29 | 40 | 0.004 | 33 | 34 | 0.008 |
| Lymphatic metastasis | ||||||
| Yes | 25 | 33 | 28 | 31 | ||
| No | 18 | 59 | 0.003 | 54 | 22 | 0.001 |
| Tumor Depth Metastasis | ||||||
| pT3 + pT4 | 17 | 54 | 49 | 22 | ||
| pT1 + pT2 | 26 | 38 | 0.017 | 33 | 31 | 0.008 |
| Histological Grade | ||||||
| III | 23 | 36 | 36 | 23 | ||
| I + II | 20 | 56 | 0.038 | 46 | 30 | 0.875 |
| TNM Stage | ||||||
| III + IV | 18 | 53 | 48 | 23 | ||
| 0 + I + II | 25 | 39 | 0.034 | 34 | 30 | 0.041 |
HCP5 and miR-139-5p regulate EMT, invasion and migration of CRC
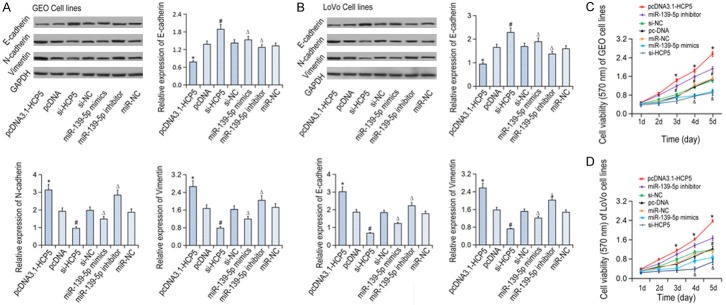
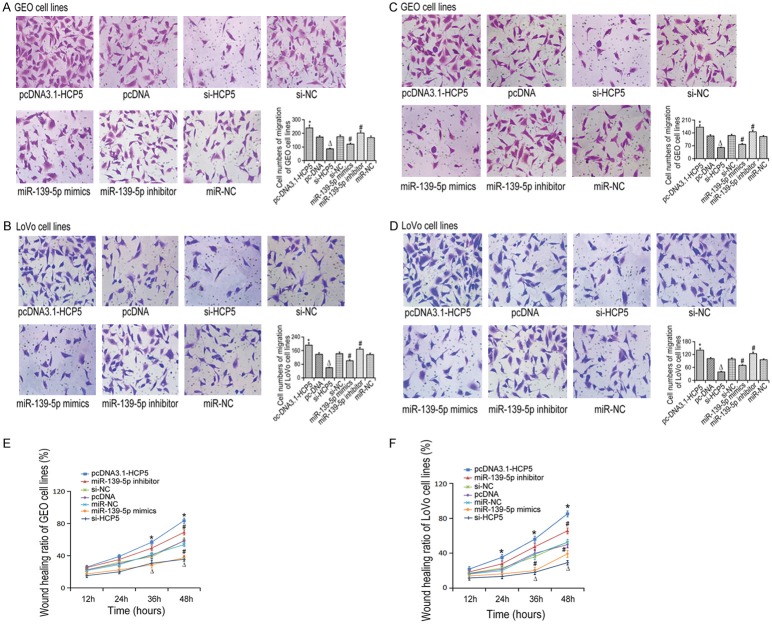
In order to analyze whether HCP5 and miR-139-5p regulate cell invasive and migrative capabilities by inducing EMT, the expression of EMT related gene expressions we detected in CRC cell lines (Figure 2A, 2B). The expression of E-cadherin was up-regulated in the si-HCP5 group as compared to si-NC group, while the expressions of Snail and vimentin decreased significantly (P < 0.05). In addition, microRNA-139-5p inhibitors could significantly suppressed E-cadherin expression and increase Snail and vimentin expressions (P < 0.05). Furthermore, MTT results showed that HCP5 knockdown or the mi-139-5p overexpression led to decreased cell viability compared with the control group (P < 0.05). However, cell viability increased significantly in the miR-139-5p inhibitor group (P < 0.05, Figure 2C, 2D). Meanwhile, Transwell assay results showed that cells transfected with si-HCP5 or mir-139-5p mimic underwent a decreased closing trend of scratch wound (P < 0.05). The number of transmembrane cells in the mir-139-5p inhibitor group increased significantly compared with cells transfected miR-NC (P < 0.05, Figure 3A-D). Finally, scratch assay showed that the migration was significantly reduced in CRC cells transfected with si-HCP5 or the miR-139-5p mimic (P < 0.05). Nevertheless, the cell migration was significantly increased following downregulation of miR-139-5p expression (P < 0.05, Figure 3E, 3F).
Figure 2.
The expression levels of EMT related gene expression and the viability of in CRC cell lines in various groups. A: The expression levels of EMT related gene expression in GEO cells; B: The expression levels of EMT related gene expression in LOVO cells; C: The viability of GEO cell lines; D: The viability of LOVO cell lines; *P < 0.05 vs pcDNA; ΔP < 0.05 vs si-NC; #P < 0.05 vs miR-NC.
Figure 3.
The migration and invasion capabilities of CRC cell lines detected by Transwell assay. (A) Migration of GEO cells; (B) Migration of LOVO cells; (C) Invasion of GEO cells; (D) Invasion of LOVO cells. *P < 0.05 vs pcDNA; ΔP < 0.05 vs si-NC; #P < 0.05 vs miR-NC. The wound healing assay of GEO (E) and LOVO (F) cell lines in different groups. *P < 0.05 vs pcDNA; ΔP < 0.05 vs si-NC; #P < 0.05 vs miR-NC.
HCP5 can target miR-139-5p and inhibit the expression of miR-139-5p
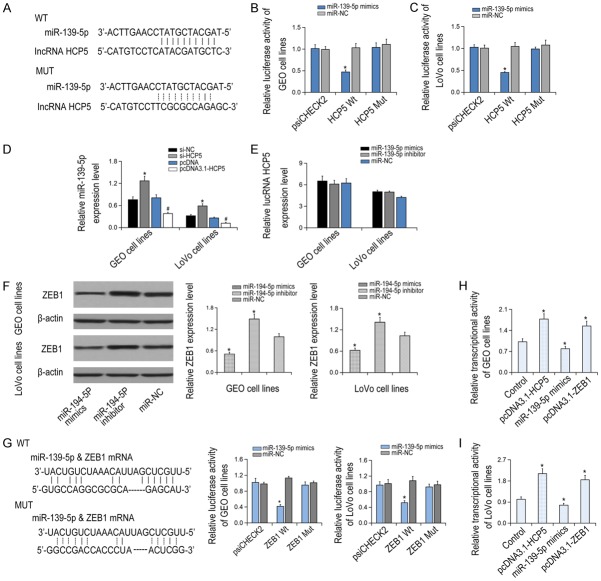
The potential binding sites of HCP5 and miR-139-5p were predicted by the bioinformatic database (Starbase software) as shown in Figure 4A. Moreover, compared with the NC group, the endogenous HCP5 was specifically enriched in the cells of miR-139-5p mimic transfected group, indicating that HCP5 directly targeted miR-139-5p. However, the luciferase activity between the HCP5 Mut + miR-139-5p mimic group and the psiCHECK2 group or the control group showed no statistical changes (P > 0.05, Figure 4B, 4C). qRT-PCR showed that miR-139-5p expression was notably increased in CRC cells after si-HCP5 transfection (P < 0.05, Figure 4D, 4E), suggesting that HCP5 functioned as a ceRNA by sponging miR-139-5p.
Figure 4.
Correlation between lncRNA HCP5 and miR-139-5p, as well as between miR-139-5p and ZEB1. A: LncRNA HCP5 can targeted binding to miR-139-5p. B, C: The luciferase activities in miR-139-5p mimic + HCP5 Wt and miR-139-5p mimic + HCP5 Mut groups, *P < 0.05 vs miR-NC. D: The miR-139-5p expression was detected after transfection of pcDNA3.1-HCP5. *P < 0.05 vs si-NC; #P < 0.05 vs pcDNA. E: HCP5 expressions in cells transfected with miR-139-5p inhibitor/mimics. F: ZEB1 expressions in LOVO and GEO cell lines after transfection of miR-194-5p mimic and miR-194-5p inhibitor. *P < 0.05 vs miR-NC. G: The luciferase activities of miR-139-5p mimic + ZEB1 Wt and miR-139-5p mimic + ZEB1 Mut were compared in LOVO and GEO cell lines. *P < 0.05 vs miR-NC. H, I: The transcriptional activities of GEO and LOVO cell lines were compared to control, pcDNA3.1-HCP5, miR-139-5p mimics and pcDNA3.1-ZEB1 groups. *P < 0.05 vs control.
MiR-139-5p regulates Wnt/β-catenin signaling pathway through targeting downstream ZEB1
Rescue experiments showed that suppression of miR-139-5p resulted in ZEB1 overexpression (P < 0.05), while promoting the expression of miR-139-5p caused down-regulation of ZEB1 (P < 0.05, Figure 4F). The luciferase activity in the mir-139-5p mimic + ZEB1 group was dramatically decreased than that in the NC group (P < 0.05, Figure 4G). In contrast, no significant difference was found between the miR-139-5p mut or ZEB1 group compared to the NC group (P > 0.05). Furthermore, the transcription level of β-catenin was significantly decreased in the miR-139-5p group and the si-ZEB1 group than that in the NC group (P < 0.05), while significantly increased in the HCP5 group (P < 0.05, Figure 4H, 4I). Similarly, the expression levels of Wnt/β-catenin signaling markers was inhibited in the miR-139-5p group and the si-ZEB1 group, but boosted significantly in the HCP5 group (P < 0.05, Figure 5).
Figure 5.
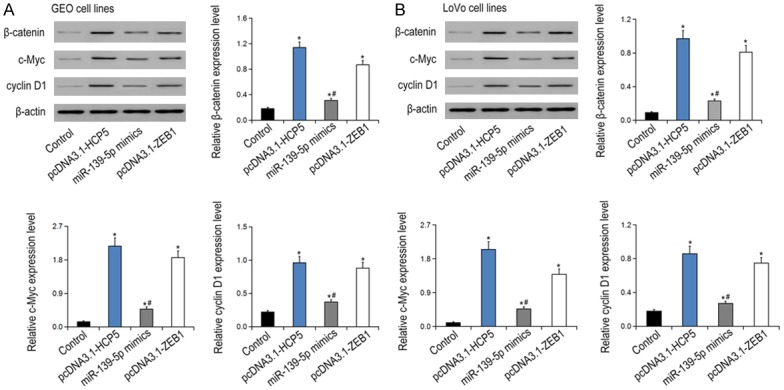
The EMT gene expressions in GEO (A) and LOVO (B) cell lines were among the groups of control, pcDNA3.1-HCP5, miR-139-5p mimics and pcDNA3.1-ZEB1. *P < 0.05 vs control; #P < 0.05 vs pcDNA3.1-ZEB1.
Discussion
Emerging evidences demonstrated that lncRNAs play a vital role in many human cancers, including CRC [22,23]. Aberrant expressions of lncRNAs were identified in many tumors [24]. It is reported that a variety of lncRNAs were used as the biomarkers for tumor diagnosis [25]. Finding key lncRNAs and understanding their mechanisms are essential for diagnosis, treatment and prognosis of tumors. However, the research on the role of lncRNA HCP5 in CRC has not been reported yet. At present, only a few CRC-related lncRNAs have been found, and further researches are needed to determine whether they can be used as predictive biomarkers. This study found that the expression of HCP5 was significantly ascended in CRC tissues and predicted poor prognosis. Functional investigation revealed the tumorigenic role of HCP5 in promoting the occurrence and metastasis of CRC in vivo and in vitro.
The results also showed that miR-29 was the target of HCP5, and their interaction mediated the progress of CRC. MiR-139-5p is the tumor suppressor in many kinds of cancers. Li et al found that the expression of miR-139-5p was specifically upregulated in breast cancer [26]. Wang et al reported that miR-139 could inhibit the development of glioma by targeting AMY-1, IGF-1R, and PGC-1β [27]. Previously, miR-139-5p strongly promoted the sensitivity of CRC cells to 5-fluorouracil by targeting NOTCH-1 [28]. In summary, miR-139-5p participated in inhibiting the risk of EMT, but the related mechanisms need further study.
In the current study, we observed that miR-139-5p could significantly regulate EMT process by directly targeting ZEB1 and altering downstream Wnt signal transduction. The relationship between ZEB1 and EMT in epithelial ovarian cancer (EOC) cells has been studied and found that ZEB1 regulated cell proliferation and invasion of EOC cells. The development of EMT is characterized by cytoskeleton recombination at first, then inhibition of cell adhesion, which ultimately enhances cell motility. Many studies have proved that EMT functions in tumor progression and metastasis [29]. Moreover, Wnt/β-catenin signaling pathway is involved in modulating EMT in tumor progression [30]. When the receptor of Wnt on the cell membrane is stimulated by signal, it can promote the translocation of β-catenin to the nucleus, resulting in the loss of E-cadherin and subsequent EMT [31,32]. Inhibition of Wnt/β-catenin pathway can block EMT transcription factors and promote epithelial differentiation [33]. Our previous studies showed that EphA2 regulated EMT through Wnt/β-catenin signaling pathway. In addition, suppressing Snail-induced EMT development can largely inhibit invasion and migration of rectal cancer. This further proved that EMT aggravated accompanying with invasion and migration. Collectively, it can be concluded that miR-139-5p/ZEB1/Wnt signaling pathway was involved in the pathogenesis of EMT process in CRC.
Conclusions
In conclusion, HCP5/miR-139-5p/ZEB1/Wnt signaling pathway may promote the occurrence of EMT in CRC cells. These results can help to develop diagnostic biomarkers and therapeutic targets for CRC.
Disclosure of conflict of interest
None.
References
- 1.Park SY, Kim JY, Choi JH, Kim JH, Lee CJ, Singh P, Sarkar S, Baek JH, Nam JS. Inhibition of LEF1-mediated DCLK1 by niclosamide attenuates colorectal cancer stemness. Clin Cancer Res. 2019;25:1415–1429. doi: 10.1158/1078-0432.CCR-18-1232. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Fedewa SA, Anderson WF, Miller KD, Ma J, Rosenberg PS, Jemal A. Colorectal cancer incidence patterns in the united states, 1974-2013. J Natl Cancer Inst. 2017;109 doi: 10.1093/jnci/djw322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanwar SS, Poolla A, Majumdar AP. Regulation of colon cancer recurrence and development of therapeutic strategies. World J Gastrointest Pathophysiol. 2012;3:1–9. doi: 10.4291/wjgp.v3.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Wei X, Shen L, Zhu H, Zheng X. 20(S)-ginsenoside-Rg3 reverses temozolomide resistance and restrains epithelial-mesenchymal transition progression in glioblastoma. Cancer Sci. 2019;110:389–400. doi: 10.1111/cas.13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu GM, Li Q, Zhang PF, Shen SL, Xie WX, Chen B, Wu J, Hu WJ, Huang XY, Peng BG. Restoration of FBP1 suppressed Snail-induced epithelial to mesenchymal transition in hepatocellular carcinoma. Cell Death Dis. 2018;9:1132. doi: 10.1038/s41419-018-1165-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang JY, Kim ST, Kwon J, Lee J, Chun YO, Han JS, Han HS. Ultra-sensitive fluorescence monitoring and in vivo live imaging of circulating tumor cell-derived miRNAs using molecular beacon system. ACS Sens. 2018 doi: 10.1021/acssensors.8b01095. [DOI] [PubMed] [Google Scholar]
- 7.Kiran M, Chatrath A, Tang X, Keenan DM, Dutta A. A prognostic signature for lower grade gliomas based on expression of long non-coding RNAs. Mol Neurobiol. 2018 doi: 10.1007/s12035-018-1416-y. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Q, Yin J, Zeng A, Jin X, Zhang Z, Yan W, You Y. H19 Functions as a competing endogenous RNA to regulate EMT by sponging miR-130a-3p in glioma. Cell Physiol Biochem. 2018;50:233–245. doi: 10.1159/000494002. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Li H, Lu X, Wen C, Huo Z, Shi M, Tang X, Chen H, Peng C, Fang Y, Deng X, Shen B. Melittin-induced long non-coding RNA NONHSAT105177 inhibits proliferation and migration of pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9:940. doi: 10.1038/s41419-018-0965-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Guo D, Zhao Y, Ren M, Lu G, Wang Y, Zhang J, Mi C, He S, Lu X. Long non-coding RNA SNHG5 promotes human hepatocellular carcinoma progression by regulating miR-26a-5p/GSK3beta signal pathway. Cell Death Dis. 2018;9:888. doi: 10.1038/s41419-018-0882-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huo H, Tian J, Wang R, Li Y, Qu C, Wang N. Long non-coding RNA NORAD upregulate SIP1 expression to promote cell proliferation and invasion in cervical cancer. Biomed Pharmacother. 2018;106:1454–1460. doi: 10.1016/j.biopha.2018.07.101. [DOI] [PubMed] [Google Scholar]
- 12.Qin W, Kang P, Xu Y, Leng K, Li Z, Huang L, Gao J, Cui Y, Zhong X. Long non-coding RNA HOTAIR promotes tumorigenesis and forecasts a poor prognosis in cholangiocarcinoma. Sci Rep. 2018;8:12176. doi: 10.1038/s41598-018-29737-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ji H, Hui B, Wang J, Zhu Y, Tang L, Peng P, Wang T, Wang L, Xu S, Li J, Wang K. Long noncoding RNA MAPKAPK5-AS1 promotes colorectal cancer proliferation by partly silencing p21 expression. Cancer Sci. 2019;110:72–85. doi: 10.1111/cas.13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma S, Yang D, Liu Y, Wang Y, Lin T, Li Y, Yang S, Zhang W, Zhang R. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging (Albany NY) 2018;10:2062–2078. doi: 10.18632/aging.101530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu T, Han Z, Li H, Zhu Y, Sun Z, Zhu A. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17:118. doi: 10.1186/s12943-018-0873-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Lu B, Sun L, Yan X, Xu J. Identification of candidate genes or microRNAs associated with the lymph node metastasis of SCLC. Cancer Cell Int. 2018;18:161. doi: 10.1186/s12935-018-0653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu N, Zhang R, Zhao X, Su J, Bian X, Ni J, Yue Y, Cai Y, Jin J. A potential diagnostic marker for ovarian cancer: involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6:393–400. doi: 10.3892/ol.2013.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lange CM, Bibert S, Dufour JF, Cellerai C, Cerny A, Heim MH, Kaiser L, Malinverni R, Müllhaupt B, Negro F, Semela D, Moradpour D, Kutalik Z, Bochud PY Swiss Hepatitis C Cohort Study Group. Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma. J Hepatol. 2013;59:504–509. doi: 10.1016/j.jhep.2013.04.032. [DOI] [PubMed] [Google Scholar]
- 19.Ouzounova M, Lee E, Piranlioglu R, El Andaloussi A, Kolhe R, Demirci MF, Marasco D, Asm I, Chadli A, Hassan KA, Thangaraju M, Zhou G, Arbab AS, Cowell JK, Korkaya H. Monocytic and granulocytic myeloid derived suppressor cells differentially regulate spatiotemporal tumour plasticity during metastatic cascade. Nat Commun. 2017;8:14979. doi: 10.1038/ncomms14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Smedt L, Palmans S, Andel D, Govaere O, Boeckx B, Smeets D, Galle E, Wouters J, Barras D, Suffiotti M, Dekervel J, Tousseyn T, De Hertogh G, Prenen H, Tejpar S, Lambrechts D, Sagaert X. Expression profiling of budding cells in colorectal cancer reveals an EMT-like phenotype and molecular subtype switching. Br J Cancer. 2017;116:58–65. doi: 10.1038/bjc.2016.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu G, Huang Q, Zheng W, Huang Y, Hua J, Yang S, Zhuang J, Wang J, Chang J, Xu J, Ye J. LPS upregulated VEGFR-3 expression promote migration and invasion in colorectal cancer via a mechanism of increased NF-kappaB binding to the promoter of VEGFR-3. Cell Physiol Biochem. 2016;39:1665–1678. doi: 10.1159/000447868. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Yu X, Xu Y, Shen H. Identification of dysregulated lncRNAs profiling and metastasis-associated lncRNAs in colorectal cancer by genome-wide analysis. Cancer Med. 2017;6:2321–2330. doi: 10.1002/cam4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, Chiao PJ, Huang P, Xie D, Li YH, Ju HQ, Xu RH. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu D, Xu B, Chen S, Yang Y, Zhang X, Liu J, Lu K, Zhang L, Liu C, Zhao Y, Jiang H, Liu N, Chen M. Long non-coding RNAs and prostate cancer. J Nanosci Nanotechnol. 2013;13:3186–3194. doi: 10.1166/jnn.2013.6870. [DOI] [PubMed] [Google Scholar]
- 26.Li HC, Chen YF, Feng W, Cai H, Mei Y, Jiang YM, Chen T, Xu K, Feng DX. Loss of the Opa interacting protein 5 inhibits breast cancer proliferation through miR-139-5p/NOTCH1 pathway. Gene. 2017;603:1–8. doi: 10.1016/j.gene.2016.11.046. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Yan X, Ji LY, Ji XT, Wang P, Guo SW, Li SZ. miR-139 Functions as an antioncomir to repress glioma progression through targeting IGF-1 R, AMY-1, and PGC-1beta. Technol Cancer Res Treat. 2017;16:497–511. doi: 10.1177/1533034616630866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Yin Y, Hu Y, Feng Y, Bian Z, Yao S, Li M, You Q, Huang Z. miR-139-5p sensitizes colorectal cancer cells to 5-fluorouracil by targeting NOTCH-1. Pathol Res Pract. 2016;212:643–649. doi: 10.1016/j.prp.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 29.Jiang C, Xu R, Li XX, Wang YY, Liang WQ, Zeng JD, Zhang SS, Xu XY, Yang Y, Zhang MY, Wang HY, Zheng XFS. p53R2 overexpression in cervical cancer promotes AKT signaling and EMT, and is correlated with tumor progression, metastasis and poor prognosis. Cell Cycle. 2017;16:1673–1682. doi: 10.1080/15384101.2017.1320629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan H, Yan Z, Chen W, Wu Y, Han J, Guo H, Qiao J. TET1 inhibits EMT of ovarian cancer cells through activating Wnt/beta-catenin signaling inhibitors DKK1 and SFRP2. Gynecol Oncol. 2017;147:408–417. doi: 10.1016/j.ygyno.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119:1623–1633. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 32.Stavrinou S, Clark A, Irving J, Lee CH, Oliva E, Young R, Sriraksa R, Magdy N, Van Noorden S, McCluggage WG, El-Bahrawy M. Differential expression of E-cadherin and catenins in ovarian sex cord stromal tumours. Histopathology. 2016;69:298–306. doi: 10.1111/his.12937. [DOI] [PubMed] [Google Scholar]
- 33.Xu HG, Zheng Q, Song JX, Li J, Wang H, Liu P, Wang J, Wang CD, Zhang XL. Intermittent cyclic mechanical tension promotes endplate cartilage degeneration via canonical Wnt signaling pathway and E-cadherin/beta-catenin complex cross-talk. Osteoarthritis Cartilage. 2016;24:158–168. doi: 10.1016/j.joca.2015.07.019. [DOI] [PubMed] [Google Scholar]