Abstract
Background: Gastric cancer (GC) is one of the most common digestive tract tumors, and a serious threat to human health. Long non-coding RNA (lncRNA) are involved in many cancers. However, the biological functions of most lncRNAs are unclear. In this study, we investigated the mechanisms by which FLVCR1-AS1 regulated GC progression. Methods: FLVCR1-AS1 expression in GC tissues and 3 GC cell lines were measured by quantitative real-time PCR (qRT-PCR). Invasion, proliferation, and apoptosis profiles were analyzed by commercial assays to determine the biological functions of FLVCR1-AS1 in GC cells. The binding sites of micro RNA-155 (miR-155) on FLVCR1-AS1 were predicted using the miRDB program. Luciferase reporter assay was used to validate direct targeting of FLVCR1-AS1 by miR-155. The effects of FLVCR1-AS1 on expressions of c-Myc and p21 were assessed by western blotting. In vivo experiments were performed to analyze the effects of FLVCR1-AS1 on GC tumor growth. Results: High expression of FLVCR1-AS1 correlated with poor clinical outcomes and prognosis in patients with GC. FLVCR1-AS1 promoted proliferation and invasion of GC cells by acting as a ceRNA to sponge miR-155. Conclusion: FLVCR1-AS1 acted as an oncogene in GC via FLVCR1-AS1-miR-155-c-Myc signaling and may serve as a novel therapeutic target for treatment of patients with GC.
Keywords: LncRNA, FLVCR1-AS1, miR-155, c-Myc, gastric cancer
Introduction
Gastric cancer (GC) is one of the most common digestive tract tumors, and is a serious threat to human health. In recent decades, with improvements in human living conditions, establishment of good eating habits, and eradication of Helicobacter pylori (Hp), the incidence of gastric cancer has generally declined. However, global incidence of GC is fourth-highest among male cancers, mortality rate ranks third, and the male incidence rate is twice that of females [1,2]. Although recent studies have evaluated the molecular mechanisms regulating GC, the underlying mechanisms have not been fully elucidated and existing therapies are not sufficient. Therefore, it is necessary to characterize the molecular mechanisms of GC to identify new therapeutic targets.
Long non-coding RNAs (LncRNAs) are a class of RNAs that are > 200 nucleotides in length but do not encode proteins and have been considered the “dark matter” of gene transcription [3]. Recent studies have shown that abnormal expression of lncRNA is associated with human diseases and participates in tumor growth, invasion, metastasis, recurrence, and drug resistance [4]. Therefore, lncRNA may become a new target for tumor gene therapy. With continuous development of high-throughput transcriptome sequencing technology, increasing numbers of dysregulated lncRNAs in gastric cancer species have been discovered [5]. For example, gastric cancer high expression transcript 1 (GHET1) and colorectal cancer-related transcript 1 (CCAT1) can interact with c-Myc to promote gastric cancer progression [6,7], while maternally expressed gene 3 (MEG3) can inhibit the occurrence of gastric cancer [8]. However, the exact biological functions of most lncRNAs remain unclear.
Previous reports showed that the lncRNA FLVCR1-AS1 was aberrantly expressed in some tumor types, such as lung cancer and hepatoma [9,10]. However, the role of FLVCR1-AS1 in tumorigenesis of GC is unclear. Thus, we investigated the mechanisms by which FLVCR1-AS1 regulates GC progression.
Materials and methods
Patient samples
Thirty patients with a clear histological diagnosis of gastric cancer underwent surgery in our department from January 2015 to October 2017. None of the patients had received preoperative radiotherapy, chemotherapy, or any other cancer treatment. Gastric cancer tissues and paired adjacent normal tissues (with > 2 cm distance from the edge of the tumor) were collected and snap-frozen in liquid nitrogen immediately after surgical resection. Clinical data is summarized in Table 1, including gender, age, histological grade, invasion depth, lymph node metastasis, distant metastasis and TNM stage. Histological grade was assigned according to American Joint Committee on Cancer (AJCC) standards. Informed written consent was obtained for all patients and the study was approved by The Fifth Subsidiary Sun Yat-sen University Hospital Animal Experimental Ethics Committee. Experiments involving human subjects were in accordance with the Helsinki Declaration.
Table 1.
The expression of FLVCR1-AS1 in gastric cancer patients’ tissues
| Factors | No. | FLVCR1-AS1 | P value |
|---|---|---|---|
| Gender | 0.331 | ||
| Male | 18 | 2.255±1.193 | |
| Female | 12 | 2.651±0.995 | |
| Age (years) | 0.728 | ||
| < 50 | 9 | 2.365±1.262 | |
| ≥ 50 | 21 | 2.512±1.009 | |
| Histological grade | 0.118 | ||
| Well-intermediately differentiation | 17 | 2.178±1.142 | |
| Poor differentiation | 13 | 2.813±0.964 | |
| Invasion depth | 0.123 | ||
| T1-T2 | 18 | 2.200±1.111 | |
| T3-T4 | 12 | 2.833±1.004 | |
| Lymph node metastasis | 0.18 | ||
| N0-N1 | 18 | 2.261±1.070 | |
| N2-N3 | 12 | 2.837±1.106 | |
| Distant metastasis | 0.034* | ||
| M0 | 22 | 2.043±0.831 | |
| M1 | 8 | 3.075±1.231 | |
| TNM stage | 0.032* | ||
| I, II | 17 | 1.921±1.113 | |
| III, IV | 13 | 2.987±1.068 |
P < 0.05, Student’s t-test.
Cell lines
The human gastric epithelial cell line GES-1 and three human gastric cancer cell lines (AGS, MNK-45, and MGC-803) were obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, USA) in 5% CO2 atmosphere at 37°C.
LncRNA in situ hybridization
Biotin-labeled specific FLVCR1-AS1 probes were used to analyze expression of FLVCR1-AS1. Samples were fixed with 4% formaldehyde and embedded in paraffin at a melting point of 57°C. After dewaxing, biotin-labeled probes were incubated overnight with the sections at 55°C and then DAB substrate was used for colorimetric detection of FLVCR1-AS1. Finally, the sections were co-stained with hematoxylin. FLVCR1-AS1 probe sequences were as follows: #1: 5’-CAGGAAAATGTCAGCCAGCG-3’; #2: 5’-GCCTCTAAGTAGTGACACTA-3’; #3: 5’-GATTAATCACAGCTCCTCAC-3’.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Invitrogen, USA). The standards for RNA purity and concentration were RIN ≥ 7.0 and 28S/18S > 0.7, respectively. cNDA was synthesized using PrimeScript RT reagent Kit with gDNA Eraser (Takara, Dalian, China). Quantitative analysis of RNA expression by qRT-PCR using SYBR green. The housekeeping gene RNU6-2 was used as an internal standard for miRNA. GADPH was used as an internal standard for FLVCR1-AS1 and c-Myc. qRT-PCR amplification conditions were as follows: 95°C for 14 min, followed by 40 cycles each at 95°C for 10 s, 60°C for 30 s, 72°C for 30 s. The 2-ΔΔCT formula was used to calculate relative expression levels.
Gene knockdown and overexpression
For FLVCR1-AS1 knockdown, FLVCR1-AS1 siRNA (siFLVCR1-AS1 5’-GGTAAGCAGTGGCTCCTCTAA-3’) were purchased from Genepharma (Shanghai, China). MGC803 cells were seeded onto 6-well plates and transfected with siFLVCR1-AS1. For FLVCR1-AS1 overexpression, lentiviral expressing vector pITA was used to construct a pITA-FLVCR1-AS1 lentiviral plasmid. After sequencing to verify the correctness of the sequence, pITA-FLVCR1-AS1 was transfected into MKN45 cells.
For miR-155 knockdown and overexpression, miR-155 inhibitor and miR-155 mimics were purchased from Genepharma (Shanghai, China). MGC803 and MKN45 cells were transfected with the above constructs using Lipofectamine 2000 (Invitrogen; USA) according to standard protocols. MiR-155 mimic sequences were as follows: sense: 5’-UUAAUGCUAAUUGUGAUA GGGGU-3; antisense: 5’-CCCUAUCACAAUUAGCAUUAAUU-3’. MiR-155 inhibitor sequence was as follows: 5’-ACCCCUAUCACAAUUAGCAUUAA-3’.
Cell proliferation and apoptosis assay
Cell proliferation assays were performed using the Cell Counting Kit-8 (CCK-8, Dojindo, Japan) according to the manufacturer’s instructions. Cells were seeded onto 96-well plates (density: 8 × 103 cells/well, volume: 100 μL) and cultured at 37°C for 24 h. Ten microliters of CCK-8 reagent was added to each well, the plate was incubated at 37°C. Absorbance at 450 nm (A450) was measured at 0, 24, 48 and 72 h. All experiments were performed in triplicate.
To measure apoptosis, cells were centrifuged at 1000 rpm for 5 min and then stained using Annexin V-FITC and propidium iodide (PI) kits (KeyGen Biotech, Nanjing, China) according to the manufacturer’s protocol. Cell apoptosis was quantified by flow cytometry on a Beckman Coulter flow cytometer (Becton Dickinson; USA).
Cell Ki67 immunofluorescence staining
Cells were fixed with 4% formaldehyde solution at room temperature for 1 h and then washed twice with PBS buffer. Cells were lysed with 0.5% Triton-X for 0.5 h and incubated with 5% BSA at room temperature for 5 min to block non-specific background staining. Cells were then incubated with the primary antibody anti-Ki67 (Thermo Fisher Scientific, Invitrogen, USA, 1:100) at 37°C for 2 h. After incubation of the secondary antibody for 1 h, the cells were treated with DAPI (5 μg/mL) at room temperature for 10 min. Visualization was performed using a fluorescence microscope.
Cell invasion assay
Transwell assay was used to evaluate cell invasion ability. Two hundred microliters of serum-free medium and 500 μL of medium containing 10% FBS were added to the upper chamber and the lower chamber, respectively. Cells were seeded into the upper chamber at a final density of 1 × 105 cells/mL and incubated at 37°C for 24 h. The upper chamber was removed and the cells were wiped on the chamber filter with a cotton swab. After washing with PBS, the chamber was fixed in 95% ethanol for 10 min and then stained with 1% crystal violet for 30 min. Cells that invaded to the lower chamber were counted using a Nikon TE2000 microscope (Nikon, Tokyo, Japan).
Flow cytometric analysis of the cell cycle
To analyze cell cycle, cells were centrifuged at 1000 rpm for 5 min then stained using a cell cycle kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. Cell cycle was analyzed by flow cytometry on a Beckman Coulter flow cytometer (Becton Dickinson; USA).
Luciferase assay
Bioinformatics analysis was performed to predict FLVCR1-AS1-targeted miRNAs using miRDB (http://mirdb.org/miRDB/). miR-155 binding sites on FLVCR1-AS1 were predicted by RNAhybrid 2.2 (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). Wild-type binding sites (5’-UUUUCAGACCUUUAGCAUUAACCUUAUAGG-3’; position: 882-890) and mutant binding sites (5’-UUUUCAGACCUUAAUCAGGAACCUUAUAGG-3’; position: 882-890) were synthesized and cloned into psiCHECK2 vectors to produce FLVCR1-AS1-WT and FLVCR1-AS1-MT vectors, respectively.
Cells were seeded onto 96-well plates at a final density of 1 × 105 cells/well. The recombinant vectors were co-transfected into MGC803 and MKN45 cells with miR-155 mimic or miR-ctrl. Dual-luciferase reporter assay system (E1910, Promega) was used to measure luciferase activity according to the manufacturer’s protocol.
Western blotting
Total protein was extracted using RIPA buffer (Beyotime, China) and separated on a 10% SDS-PAGE gel. The separated proteins were transferred onto PVDF membranes (Bio-Rad, USA). Membranes were blocked with TBST buffer containing 5% non-fat milk for 1 h at room temperature. Membranes were then incubated with primary antibodies (1:1000) at 4°C for 12 h. After washing with TBST buffer, membranes were incubated with secondary antibodies (1:10000) at room temperature for 1 h. Protein expression levels were measured using an enhanced chemiluminescence detection kit (Thermal Scientific, USA). All experiments were repeated in triplicate.
RNA pull-down
FLVCR1-AS1 were biotinylated by GenePharma Company (Shanghai, China) and processed using a Pierce Magnetic RNA Pull-Down Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. MiR-155 expression levels were measured by qRT-PCR.
Animal study
BALB/c nude mice (6 weeks old) were obtained from the Experimental Animal Center of Southern Medical University. 5 × 106 MKN45 cells with stable expression of pITA-FLVCR1-AS1 lentiviral plasmid and control cells were injected into the nude mice. Tumor volumes were measured and calculated weekly using the formula (length × width2)/2 for four weeks. Mice were euthanized by inhalation of diethyl ether, and tumor volume and weight were measured. Tumor section slides were analyzed by immunohistochemistry assay with Ki67.
Statistical analysis
SPSS software (SPSS standard version 19.0; SPSS Inc.) was used for all statistical analyses. Survival curves were calculated using the Kaplan-Meier method and were analyzed using the log-rank test. Values were expressed as mean ± SD. Two groups of experiments were conducted using Student’s t-test. Statistical significance was determined by one-way analysis of variance (ANOVA) followed by Dunnett’s test among multiple groups. P < 0.05 was considered significant.
Results
Up-regulation of FLVCR1-AS1 correlated with clinical indices and prognosis in patients with gastric cancer
To investigate regulation of FLVCR1-AS1 expression in gastric cancer, thirty patients with gastric cancer were evaluated in this study. qRT-PCR was performed to measure mRNA expression levels in gastric cancer tissues and corresponding normal tissues. As shown in Figure 1A, mRNA expression levels of FLVCR1-AS1 in gastric cancer tissues were significantly higher than those in normal tissues (P < 0.01). Patients were divided into two groups according to expression levels of FLVCR1-AS1. Kaplan-Meier survival analysis was used to compare overall survival rates of gastric cancer patients with different levels of FLVCR1-AS1. The results showed that overall survival rates of patients with high FLVCR1-AS1 expression were significantly lower than those of patients with low FLVCR1-AS1 expression level (Figure 1B). Subsequently, we analyzed expression levels of FLVCR1-AS1 in both normal and tumor tissues by in situ hybridization. As shown in Figure 1C, FLVCR1-AS1 had higher expression levels in tumor tissues compared with normal tissues. This result was consistent with the results of qRT-PCR analyses. In summary, FLVCR1-AS1 was abnormally enriched in gastric cancer tissues and was associated with poor GC prognosis.
Figure 1.
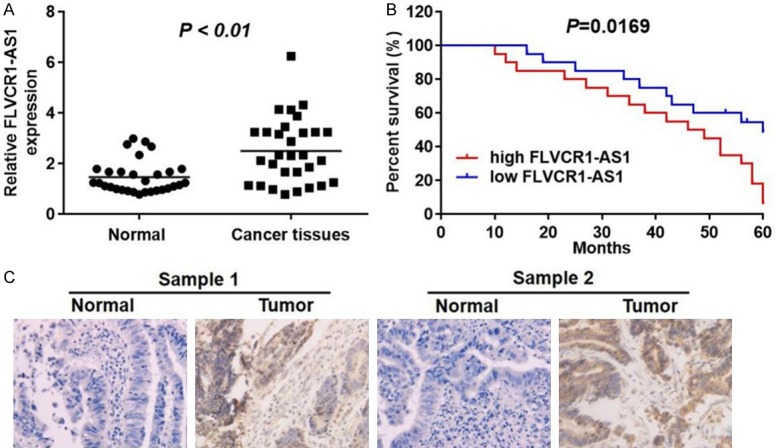
FLVCR1-AS1 was upregulated in GC and was correlated with clinical and prognosis in GC patients. A. qRT-PCR analysis was used to detect the relative expression levels of FLVCR1-AS1 in normal tissues (adjacent tissues of GC patients) and tumor tissues of GC patients (n=30). B. GC patients with higher expression of FLVCR1-AS1 showed lower overall survival rate and the correlation between FLVCR1-AS1 and overall survival of osteosarcoma patients was analyzed by Kaplan Meier method analysis (log rank test). C. Histologic examinations were performed after H&E staining to observe the morphology of GC tissues in normal tissues and tumor tissues. FLVCR1-AS1 had higher expression levels in GC tissues compared with the normal tissues. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. *P < 0.05.
FLVCR1-AS1 knockdown inhibited proliferation and invasion, and enhanced cell apoptosis in gastric cancer cells
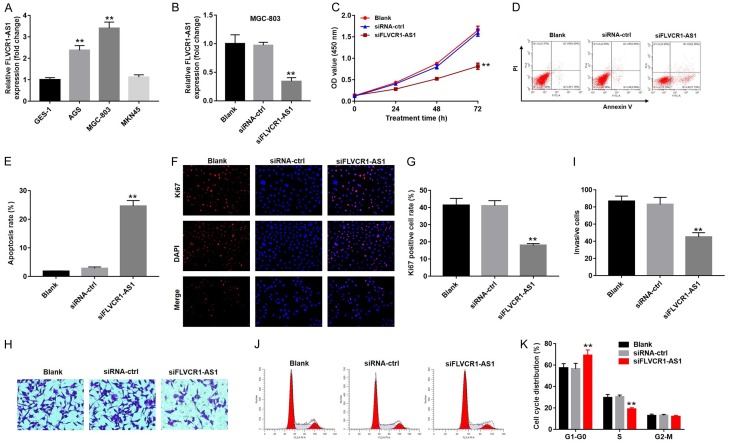
To characterize the role of FLVCR1-AS1 in gastric cancer, we measured mRNA expression levels GES-1 cells and three human gastric cancer cell lines (AGS, MGC-803, and MNK-45). As shown in Figure 2A, expression levels of FLVCR1-AS1 in AGS and MGC-803 cells were significantly higher than those in GES-1 cells. However, there was no significant difference in FLVCR1-AS1 expression between MNK-45 and GES-1 cells.
Figure 2.
FLVCR1-AS1 knockdown inhibited cell proliferation and invasion, and enhanced cell apoptosis. (A) qRT-PCR analysis was used to detect the relative expression levels of FLVCR1-AS1 in GES-1, AGS, MGC-803 or MKN45 cell lines. (B) qRT-PCR analysis was used to detect the relative expression levels of FLVCR1-AS1 in MGC-803 cells following transfected with FLVCR1-AS1 siRNA (siFLVCR1-AS1) or a non-target siRNA control (siRNA-ctrl). (C) Cell viability was determined using CCK-8 assay in MGC-803 cells following transfected with siFLVCR1-AS1 or siRNA-ctrl for 0, 24, 48 and 72 h. (D) Cell apoptosis of MGC-803 cells after transfecting with siFLVCR1-AS1 or siRNA-ctrl was detected with flow cytometry. (E) Apoptosis rate of MGC-803 cells after transfecting with siFLVCR1-AS1 or siRNA-ctrl. (F) MGC-803 cells proliferation after transfecting with siFLVCR1-AS1 or siRNA-ctrl was observed with Ki67 and DAPI staining. (G) Ki67 positive cell rate of MGC-803 cells after transfected with siFLVCR1-AS1 or siRNA-ctrl. (H) The transwell invasion assay and (I) the invasion rate of MGC-803 cells following siFLVCR1-AS1 or siRNA-ctrl were measured. (J) The cell cycle assay and (K) the cell cycle distribution rate of MGC-803 cells following siFLVCR1-AS1 or siRNA-ctrl were measured. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. **P < 0.01.
Expression of FLVCR1-AS1 was knocked down in MGC-803 cells following transfection with siFLVCR1-AS1 (P < 0.01) (Figure 2B). The effect of FLVCR1-AS1 expression on cell proliferation was evaluated using a CCK-8 kit. The results indicated that knockdown of FLVCR1-AS1 significantly reduced cell proliferation ability after transfection with siFLVCR1-AS1 within 72 h (59% decrease, P < 0.01) (Figure 2C). FACS analysis and AnnexinV/PI staining were used to measure cell apoptosis. As shown in Figure 2D and 2E, FLVCR1-AS1 knockdown significantly enhanced apoptosis (5.0-fold increase, P < 0.01). Ki67 immunofluorescence staining was also performed to measure the effects of FLVCR1-AS1 expression on cell proliferation. After transfection with siFLVCR1-AS1, proliferation ability of MGC-803 cells decreased significantly (55% decrease, P < 0.01) (Figure 2F and 2G). This result was consistent with that of CCK-8 assay, which indicated that knockdown of FLVCR1-AS1 inhibited cell proliferation.
In addition, we performed transwell assay to analyze the effect of FLVCR1-AS1 expression on cell invasion ability. The results showed that knockdown of FLVCR1-AS1 expression significantly reduced the invasive ability of MGC-803 cells (45% decrease, P < 0.01) (Figure 2H and 2I). Moreover, the results of cell cycle assays indicated that knockdown of FLVCR1-AS1 expression significantly elevated the proportion of cells in G0/G1 phase, and reduced the proportion of cells in S phase (Figure 2J and 2K). Collectively, FLVCR1-AS1 knockdown inhibited proliferation and invasion ability of gastric cancer cells, enhanced cell apoptosis, and induced cell cycle arrest.
FLVCR1-AS1 overexpression enhanced cell proliferation and invasion
To further explore the role of FLVCR1-AS1 in gastric cancer cells, FLVCR1-AS1 was overexpressed in MNK-45 cells (P < 0.01) (Figure 3A). CCK-8 and Ki67 immunofluorescence staining experiments were performed to evaluate the effects of FLVCR1-AS1 expression on cell proliferation. As shown in Figure 3B, proliferation of FLVCR1-AS1-overexpressing cells was significantly increased compared with cells transfected with control vectors at 72 h (1.6-fold increase, P < 0.01). In contrast, the Ki67 positive cell rate of FLVCR1-AS1-overexpressing cells was significantly higher than that of the control group (2.1-fold increase, P < 0.01) (Figure 3C and 3D). Transwell assay was also used to analyze the effect of FLVCR1-AS1 expression on invasion ability of MNK-45 cells. Overexpression of FLVCR1-AS1 significantly increased cell invasion ability (1.5-fold increase, P < 0.01) (Figure 3E and 3F). In summary, FLVCR1-AS1 overexpression enhanced cell proliferation and invasion. These results were consistent with those of FLVCR1-AS1 knockdown in MGC-803 cells.
Figure 3.
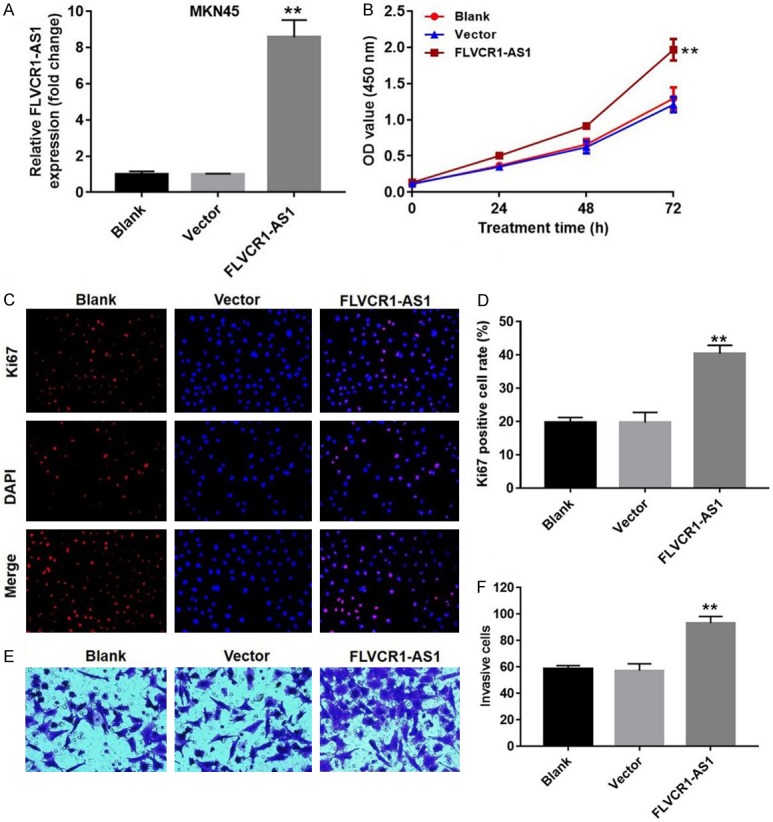
FLVCR1-AS1 overexpression enhanced cell proliferation and invasion. A. qRT-PCR analysis of the relative expression levels of FLVCR1-AS1 in MKN45 cells that transfected with FLVCR1-AS1 overexpression plasmid or an empty vector. B. The effect of FLVCR1-AS1 overexpression on proliferation of MKN45 cells was detected with CCK8. C. Ki67 immunofluorescence staining were performed to evaluate the effect of FLVCR1-AS1 overexpression on proliferative activity of MKN45 cells. D. Ki67 positive MKN45 cells were quantified. E. The effect of FLVCR1-AS1 overexpression on invasion ability of MKN45 cells was detected with transwell assay. F. The invasion rate of MKN45 cells following FLVCR1-AS1 were measured. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. **P < 0.01.
FLVCR1-AS1 sponges miR-155 in gastric cancer cells
Bioinformatics analysis was performed to predict target miRNAs for FLVCR1-AS1. MiR-155, miR-513c, miR-514b, and miR-670 were the top four predicted targets. Subsequently, levels of these four miRNAs in MGC-803 and MNK-45 cells were measured by qRT-PCR. Knockdown of FLVCR1-AS1 expression in MGC-803 cells resulted in significantly increased miR-155 (3.9-fold increase, P < 0.01), while there were no significant differences in miR-513c, miR-514b, and miR-670 levels (Figure 4A). Consistent with the above results, overexpression of FLVCR1-AS1 in MNK-45 cells resulted in significantly decreased miR-155 levels (64% decrease, P < 0.01), while there were no significant differences in miR-513c, miR-514b, and miR-670 levels (Figure 4B). According to the above results, miR-155 may directly target FLVCR1-AS1.
Figure 4.
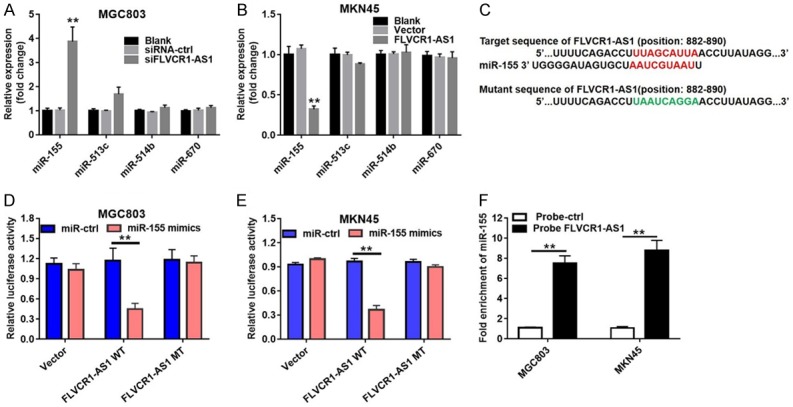
FLVCR1-AS1 functioned as a sponge for miR-155 in GC. A. qRT-PCR analysis of top four predicted miRNAs in MGC803 cells transfected with siFLVCR1-AS1 or si-NC. B. qRT-PCR analysis of top four predicted miRNAs in MKN45 cells transfected with FLVCR1-AS1 overexpression plasmid or an empty vector. C. Schematic description of miR-155 binding sites in the FLVCR1-AS1. D. FLVCR1-AS1-mut as well as a FLVCR1-AS1-wt luciferase reporter plasmid was cloned by mutating the predicted miR-155 binding site in FLVCR1-AS. FLVCR1-AS1-wt or FLVCR1-AS1-mut was co-transfected into MGC803 cells with si-NC or miR-155 mimics. E. FLVCR1-AS1-mut as well as a FLVCR1-AS1-wt luciferase reporter plasmid was cloned by mutating the predicted miR-155 binding site in FLVCR1-AS. FLVCR1-AS1-wt or FLVCR1-AS1-mut was co-transfected into MKN45 cells with si-NC or miR-155 mimics. F. The binding relation between FLVCR1-AS1 and miR-155 was verified by RNA pull-down assay. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. **P < 0.01.
To confirm this prediction, luciferase reporter assay was used in this study to further confirm whether miR-155 directly targets FLVCR1-AS1 in MGC-803 and MNK-45 cells. The targeted sequence and mutant sequence of FLVCR1-AS1 are shown in Figure 4C. We constructed FLVCR1-AS1-WT and FLVCR1-AS1-MT luciferase reporter plasmids by cloning the wild-type and mutant sequences downstream of the luciferase gene. Then, FLVCR1-AS1-WT and FLVCR1-AS1-MT were transfected into MGC-803 and MNK-45 cells with miR-155 mimics or miR-ctrl. Co-transfection with FLVCR1-AS1-WT and miR-155 mimics resulted in significantly decreased luciferase activity in both MGC-803 (58%, P < 0.01) and MNK-45 (53%, P < 0.01) cells (Figure 4D and 4E). The interaction between FLVCR1-AS1 and miR-155 was further examined by RNA pull-down. We found significant enrichment of miR-155 by using biotin-labeled FLVCR1-AS1 (Figure 4F).
FLVCR1-AS1 promoted c-Myc expression by sponging miR-155
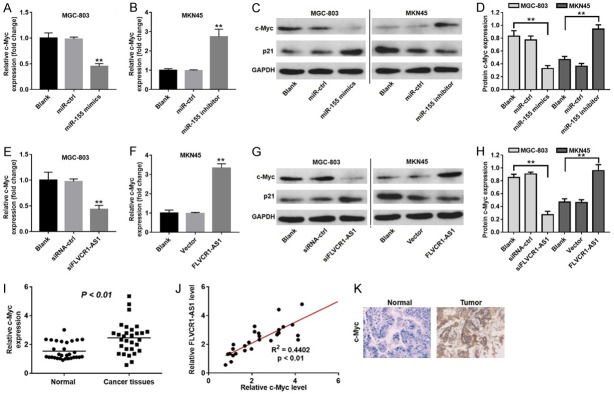
Previous studies showed that c-Myc is an important miR-155 target gene [11]. However, interactions between c-Myc and FLVCR1-AS1 are not well-understood. To investigate whether FLVCR1-AS1 regulated c-Myc expression through the sponge mechanism, we examined the relationship between miR-155 land c-Myc expression levels. As shown in Figure 5A, overexpression of miR-155 resulted in a significant decrease in the relative mRNA expression of c-Myc (55% decrease, P < 0.01). Furthermore, transfection with a miR-155 inhibitor significantly increased the relative mRNA expression of c-Myc (2.9-fold, P < 0.01) (Figure 5B). Western blot was used to measure protein levels of c-Myc and p21. c-Myc protein was decreased and p21 was increased in MGC-803 cells transfected with miR-155 mimic, while miR-155 inhibitors reduced c-Myc and p21 expression (P < 0.01) (Figures 5C, 5D and S1). In addition, FLVCR1-AS1 knockdown in MGC-803 cells resulted in a significant decrease in c-Myc mRNA levels (52% decrease, P < 0.01) (Figure 5E). Overexpression of FLVCR1-AS1 in MKN-45 cells resulted in a significant increase in c-Myc mRNA levels (52% increase, P < 0.01) (Figure 5F). Consistent with the above results, c-Myc protein levels were decreased and p21 protein levels were increased by siFLVCR1-AS1 in MGC-803 cells, while overexpression of FLVCR1-AS1 resulted in the opposite effect (P < 0.01) (Figure 5G and 5H).
Figure 5.
C-Myc was a target gene of miR-155 and was suppressed by siFLVCR1-AS1. A. qRT-PCR analysis was used to detect the relative expression of c-Myc in MGC803 cells following transfected with miR-155 mimics or NC. B. qRT-PCR analysis was used to detect the relative level of c-Myc in MKN45 cells following transfected with miR-155 inhibitors or NC. C. Relative protein levels of c-Myc and p21 in MGC803 or MKN45 cells following transfected with miR-155 mimics, miR-155 inhibitors or corresponding ncRNA were detected with Western blot. D. Relative protein levels of c-Myc and p21 in MGC803 or MKN45 cells were quantified. GAPDH was used as the internal standard. E. qRT-PCR analysis was used to detect the relative gene levels of c-Myc and p21 in MGC803 cells following transfected with si-NC or siFLVCR1-AS1. F. qRT-PCR analysis was used to detect the relative gene levels of c-Myc and p21 in MKN45 cells following transfected with FLVCR1-AS1 overexpression plasmid or an empty vector. G. Relative protein levels of c-Myc in MGC803 or MKN45 cells following transfected with siFLVCR1-AS1, FLVCR1-AS1 overexpression plasmid or corresponding NC were deteced. H. Relative protein levels of c-Myc in MGC803 or MKN45 cells were quantified. I. qRT-PCR analysis was used to detect the relative gene level of c-Myc in GC tissues. J. Pearson’s correlation scatter plot of the fold change of c-Myc and FLVCR1-AS1 in GC tissues. K. IHC staining was used to detect the expressions of c-Myc in peri-tumor and tumor tissues. C-Myc had higher expression levels in GC tissues compared with the adjacent tissues. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. **P < 0.01.
Next, we measured relative mRNA expression levels of c-Myc in the thirty patients with gastric cancer by qRT-PCR. The results showed that mRNA levels of c-Myc in gastric cancer tissues were significantly higher than those in normal tissues (Figure 5I) (P < 0.01). In addition, c-Myc expression levels positively correlated with those of FLVCR1-AS1 (R2=0.4402, P < 0.01) (Figure 5J). In addition, as shown in Figure 5K, c-Myc expression levels were higher in tumor tissues compared with those in normal tissues. These findings indicated that FLVCR1-AS1 promoted c-Myc expression by sponging miR-155.
Overexpression of FLVCR1-AS1 promoted GC growth in vivo
To further characterize the influence of FLVCR1-AS1 on growth of GC xenografts in mice, MKN45 cells transfected with a FLVCR1-AS1 overexpression lentivirus were subcutaneously implanted into nude mice. As shown in Figure 6A and 6B, tumors derived from FLVCR1-AS1 overexpression cells grew more quickly than those derived from the vector control, and subsequent tumor weight was also significantly higher than that of the vector control (Figure 6C). Furthermore, qRT-PCR analysis showed that FLVCR1-AS1 overexpression significantly increased FLVCR1-AS1 and c-Myc mRNA expression and downregulated miR-155 expression (Figure 6D-F). Western blot analysis showed that FLVCR1-AS1 overexpression promoted expression of c-Myc protein (Figure 6G and 6H). Finally, proliferative activity of tumor cells was assessed via Ki-67 immunohistochemical staining. Ki-67 staining was increased in the group that was implanted with FLVCR1-AS1 overexpression cells (Figure 6I and 6J). These results suggested that FLVCR1-AS1 overexpression promoted expression of c-Myc through competitive binding with miR-155, resulting in tumorigenesis.
Figure 6.
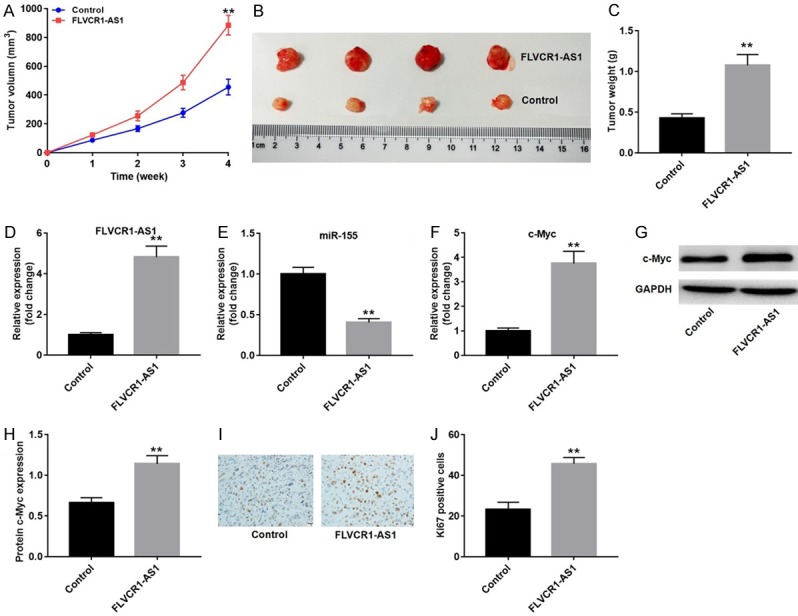
FLVCR1-AS1 promoted GC growth in vivo. A. FLVCR1-AS1-overexpression cells were injected into nude mice. Then tumor volumes were measured at different time points. B. Representative image of the excised tumors on day 28. C. Tumor weights were weighted and quantified, n=4. D. qRT-PCR analysis was used to detect the expression level of FLVCR1-AS1 in excised tumors. E. qRT-PCR analysis was used to detect the expression level of miR-155 in excised tumors. F. qRT-PCR analysis was used to detect the expression level of c-Myc in excised tumors. G. Relative protein level of c-Myc in excised tumors was detected with western blot. H. Relative protein level of c-Myc in tumors were quantified in each group. GAPDH was used as the internal standard in the western blot assay. I. Representative of immunohistochemical staining for Ki-67 in excised tumors. J. Quantification of Ki-67 positive cells in tumor tissues. Data were presented as mean ± standard deviation (SD). Each experiment was repeated three times. **P < 0.01.
Discussion
Increased study of lncRNA in gastric cancer has resulted in an important new direction in molecular biology and clinical medical research. LncRNA can regulate expression of coding genes through different molecular biological mechanisms, and play an important role in occurrence, development, and treatment of diseases. LncRNAs have the potential to serve as a new class of molecular markers with broad clinical application prospects. Abnormal expression of lncRNAs has been found in plas-ma, serum, gastric juice, and urine of patients with gastric cancer [12,13]. Up-regulation of lncRNAs such as HOTAIR, H19, HIF1A-AS2, GAPLINC, UCA1, LSINCT5, and PVT1 [14-16], and down-regulation of lncRNAs such as MEG3, AC1381281, AA174084, and FER1L4 [8,17,18] are thought to be associated with tumor size, tumor invasion, lymph node metastasis, and TNM staging of GC. GC patients with high or low levels of lncRNAs generally have shorter overall survival, shorter disease-free survival, worse prognosis, and poorer outcomes. These lncRNAs have the potential to become biomarker molecules for diagnosis, prognosis, and outcome of gastric cancer. In this study, we showed that FLVCR1-AS1 was upregulated in GC cells and tissues and correlated with clinical outcome and prognosis in GC patients. Therefore, FLVCR1-AS1 may serve as a novel GC diagnostic and prognostic marker.
In addition, signaling pathways mediated by lncRNAs in regulation of GC may provide new targets for drug-targeted therapy of GC. For example, Zhang et al. showed that the lncRNA ANRIL is highly expressed in GC tissues and cell lines. Correlation analysis between ANRIL and clinicopathological factors in GC patients showed that ANRIL expression level was closely related to tumor size, TNM stage, and survival prognosis of GC patients. In addition, the study found that ANRIL, miR-99a/miR-449a, and EZH2 have a mutual regulation relationship, which indirectly regulates proliferation of GC cells [19]. Many studies have shown that regulation of lncRNA expression by siRNA can significantly change the progression of GC. Yang et al. confirmed by qRT-PCR that tumor-associated lncRNA H19 is highly expressed in GC and found that it can promote proliferation of GC cells. Transfection of GC cells with H19 siRNA induced apoptosis of GC cells [20]. Metastasis-associated lung adenocarcinoma transcript1 (MALAT1) and HOX antisense intergenic RNA (HOTAIR) play important roles in GC metastasis. Okugawa et al. examined 300 specimens of GC and found that MALAT1 and HOTAIR were significantly increased in gastric cancer and closely related to peritoneal metastasis. Down-regulation of HOTAIR inhibits migration and invasion of GC cells. In vivo experiments demonstrated that HOTAIR siRNA significantly inhibited tumor growth and peritoneal metastasis [21]. Our findings demonstrated that FLVCR1-AS1 knockdown inhibited proliferation and invasion ability of GC cells, and enhanced cell apoptosis. FLVCR1-AS1 may also act as a target for GC treatment.
The ‘sponge’ theory of competing endogenous RNAs (ceRNA) regulatory networks suggests that lncRNA can directly participate in expression of target regulatory genes and also may contain some core seed sequences of miRNAs. In view of the long length of the IncRNA, these core sequences can affect the abundance of the target gene mRNA by adsorbing to the corresponding miRNA, thereby further affecting gene expression. The ceRNA network formed by lncRNA and miRNA is one of the foundations for lncRNA to play a role in tumors [22]. For example, Hu et al. found that the IncRNA GAPLINC is highly expressed in gastric cancer, which attenuates inhibition of CD44 by miR-311-3p by competitively binding to miR-311-3p. As such, the lncRNA GAPLINC can be used as a ceRNA to indirectly regulate migration and proliferation of gastric cancer cells [23]. A similar study indicated that ZEB1 acts as a target gene for miR-141 regulation and plays an important role in epithelial-mesenchymal transition. H19 specifically binds to miR-141, and expression of H19 and miR-141 are negatively correlated in GC tissues and cells. Knockdown of H19 significantly reduced ZEB1 expression, while overexpression of H19 significantly up-regulated ZEB1 expression. H19 can also act as a ceRNA to indirectly regulate expression of ZEB1 by inhibiting the degradation of target gene ZEB1 by miR-141 [24]. To demonstrate the mechanism of FLVCR1-AS1 function in GC, we predicted the target microRNA of FLVCR1-AS1 using an online database (http://www.mirdb.org). We identified miR-155 as a potential target of FLVCR1-AS1. Through luciferase activity reporter assay, we confirmed that FLVCR1-AS1 directly sponged miR-155. Therefore, FLVCR1-AS1 regulated GC progression via sponging miR-155. Our research expanded understanding of the regulatory mechanisms of GC progression.
MiRNAs regulate gene expression at the post-transcriptional level. They play important roles in tumor development, organogenesis, viral defense, epigenetic regulation, and metabolism [25]. Previous studies showed that c-Myc is an important miR-155 target gene [11]. C-Myc is a proto-oncogene that can be over-amplified and highly expressed in tumor tissues by regulating physiological processes such as cell proliferation, differentiation, and apoptosis, and is closely related to the degree of malignancy of tumors [26]. Studies have shown that about 40% of patients with gastric cancer exhibit abnormally high expression of the c-Myc gene, which often leads to an increase in the recurrence rate of GC and poorer prognosis [27]. However, the regulatory mechanism of c-Myc expression remains unclear. Our findings indicated that overexpression of miR-155 decreased levels of c-Myc and FLVCR1-AS1 promoted c-Myc expression by sponging miR-155.
Taken together, up-regulation of FLVCR1-AS1 correlated with worse clinical outcomes and prognosis in GC patients. FLVCR1-AS1 promoted proliferation and invasion ability of GC cells by acting as a ceRNA to sponge miR-155. Thus, we identified a novel FLVCR1-AS1-miR-155-c-Myc signaling axis that promotes GC progression and provides a new therapeutic possibility for GC.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Tani H. Short-lived non-coding transcripts (SLiTs): clues to regulatory long non-coding RNA. Drug Discov Ther. 2017;11:20–24. doi: 10.5582/ddt.2017.01002. [DOI] [PubMed] [Google Scholar]
- 4.Hahne JC, Valeri N. Non-coding RNAs and resistance to anticancer drugs in gastrointestinal tumors. Front Oncol. 2018;8:226. doi: 10.3389/fonc.2018.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song YX, Sun JX, Zhao JH, Yang YC, Shi JX, Wu ZH, Chen XW, Gao P, Miao ZF, Wang ZN. Non-coding RNAs participate in the regulatory network of CLDN4 via ceRNA mediated miRNA evasion. Nat Commun. 2017;8:289. doi: 10.1038/s41467-017-00304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 7.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 9.Gao X, Zhao S, Yang X, Zang S, Yuan X. Long non-coding RNA FLVCR1-AS1 contributes to the proliferation and invasion of lung cancer by sponging miR-573 to upregulate the expression of E2F transcription factor 3. Biochem Biophys Res Commun. 2018;505:931–938. doi: 10.1016/j.bbrc.2018.09.057. [DOI] [PubMed] [Google Scholar]
- 10.Zhang K, Zhao Z, Yu J, Chen W, Xu Q, Chen L. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119:6045–6056. doi: 10.1002/jcb.26802. [DOI] [PubMed] [Google Scholar]
- 11.Sun S, Sun P, Wang C, Sun T. Downregulation of microRNA-155 accelerates cell growth and invasion by targeting c-myc in human gastric carcinoma cells. Oncol Rep. 2014;32:951–956. doi: 10.3892/or.2014.3288. [DOI] [PubMed] [Google Scholar]
- 12.Chen JS, Wang YF, Zhang XQ, Lv JM, Li Y, Liu XX, Xu TP. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63:223–230. doi: 10.4149/207_150821N454. [DOI] [PubMed] [Google Scholar]
- 13.Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8:81572–81582. doi: 10.18632/oncotarget.19197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu FT, Qiu C, Luo HL, Zhang Y, Xia GF, Hao TF, Zhu PQ. The association of HOTAIR expression with clinicopathological features and prognosis in gastric cancer patients. Panminerva Med. 2016;58:167–174. [PubMed] [Google Scholar]
- 15.Yang T, Zeng H, Chen W, Zheng R, Zhang Y, Li Z, Qi J, Wang M, Chen T, Lou J, Lu L, Zhou T, Dai S, Cai M, You W, Pan K. Helicobacter pylori infection, H19 and LINC00152 expression in serum and risk of gastric cancer in a Chinese population. Cancer Epidemiol. 2016;44:147–153. doi: 10.1016/j.canep.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Chen WM, Huang MD, Kong R, Xu TP, Zhang EB, Xia R, Sun M, De W, Shu YQ. Antisense long noncoding RNA HIF1A-AS2 is upregulated in gastric cancer and associated with poor prognosis. Dig Dis Sci. 2015;60:1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z, Guo X, Li G, Shi Y, Li L. Long noncoding RNAs as potential biomarkers in gastric cancer: opportunities and challenges. Cancer Lett. 2016;371:62–70. doi: 10.1016/j.canlet.2015.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 19.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 21.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The interaction between miR-141 and lncRNA-H19 in regulating cell proliferation and migration in gastric cancer. Cell Physiol Biochem. 2015;36:1440–1452. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 25.Figueroa-Bossi N, Bossi L. Sponges and predators in the small RNA world. Microbiol Spectr. 2018;6 doi: 10.1128/microbiolspec.rwr-0021-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trop-Steinberg S, Azar Y. Is Myc an important biomarker? Myc expression in immune disorders and cancer. Am J Med Sci. 2018;355:67–75. doi: 10.1016/j.amjms.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Calcagno DQ, Leal MF, Assumpcao PP, Smith MA, Burbano RR. MYC and gastric adenocarcinoma carcinogenesis. World J Gastroenterol. 2008;14:5962–5968. doi: 10.3748/wjg.14.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.