Abstract
Bariatric surgery (BS) success rates vary in the long-time. A better understanding of weight-loss response may help improve the outcomes of BS. Gut microbiome could be implicated in the successful rate of BS. The aim of the study is to analyze the role of gut microbiome in the successful rate of BS. This is a cross-sectional study of a prospective cohort of 24 patients who underwent gastric bypass. Patients were classified based on excess weight loss (EWL) as: Success (EWL50% at nadir weight and throughout follow-up), Primary Failure (EWL<50% at nadir weight and thereafter), and Weight Regain (EWL>50% at nadir weight, but <50% at last follow-up visit). Gut microbiome analysis was assessed by High Throughput Sequencing. Cholesterol metabolism was shown as the most affected parameter among groups. Studied groups registered minor changes between their gut microbiome abundances, with Butyrivibrio, Lachnospira and Sarcina among them. However, Success group shared a more diverse core microbiome than the other groups. We showed evidence of a possible role of gut microbiome in the cholesterol metabolism, possibly through bile acids, relative to the success or failure of BS outcomes. Acinetobacter and Serratia, from Primary Failure core microbiome, could have implications in its successful rate. Sarcina abundance was presented as the best genera related to the body mass index (BMI) post-surgery. Gut microbiota could mediate, at least partially, the success rate of BS through their interaction with the bile acids milieu. Further studies are necessary to validate this probe of concept.
Keywords: Bariatric surgery, gut microbiota, RYGB, cholesterol, bile acids
Introduction
Obesity rates are increasing worldwide. Although different interventions have been tried, mainly in the lifestyle (diet, physical activity), bariatric surgery (BS) continues being one of the most effective and durable method against morbid obesity and its complications. Despite its effectiveness, a significant number of patients experience poor weight-loss outcomes, and long-term weight regain.
BS success generally consists of three primary outcomes measures including sustained weight loss, improvement/resolution of associated comorbidities and quality of life [1]. However, the most widely used outcome is the amount of excess weight loss [1,2]. An EWL of ≥50% has been shown to be a good predictor of clinically significant change [3]. Recently, several studies have focused on success rate for BS, although the factors that contribute to the final outcome remain unknown [4,5].
Gut microbiota has emerged as an important factor underlying changes in the metabolic processes of the host. Recent works have indicated that gut microbiota may mediate some of the beneficial effects of BS [6,7], although the extent of its contribution is difficult to measure. Changes in the diversity and composition of the gut microbiota have been registered after BS [8]. These changes could be mediated through the changes suffered in the host environment [6]. In this manner, metabolomics studies have revealed profound changes in gut microbiome-host interactions after surgery [7]. However, little is known about the mid- and long-term effects and the gut microbiota contribution on the BS success. Thus, the aim of our study was to analyze the role of the gut microbiota in the successful rates of the BS in the mid-term. In this manner, we characterized the gut microbiome of patients who underwent BS but differed in their successful outcomes in the mid-term.
Material and methods
Participants in our study underwent a standardized laparoscopic RYGB [9] at Hospital Clinic, Barcelona, Spain, between 2005 and 2009. RYGB was performed by the same surgical team using a laparoscopic approach. In brief, laparoscopic RYGB included the creation of a small proximal gastric pouch of about 20 ml along the lesser curvature of the stomach, the division of the jejunum 40 cm distal to the ligament of Treitz, and end-to-side gastrojejunostomy of about 1.5 cm in diameter using a circular stapler, and a side-to-side jejunojejunostomy 150 cm distal to the gastrojejunostomy.
Eligibility criteria included age of at least 18 years, RYGB procedure performed more 24 months prior to inclusion and weight stability (±3 kg) for at least 3 months before examination. Every patient followed a diet according to the recommendations for Post-gastric bypass patients [10]. Primary Failure (n=6) was selected based on a %EWL<50% from nadir weight after RYGB up to the end of follow-up. Weight Regain (n=12) response was adjudicated based on a %EWL≥50% at the time of postsurgical nadir weight, but a %EWL<50% at the time of inclusion. Success (n=6) was selected based on a %EWL>50% from nadir weight onwards after RYGB up to the end of follow-up and to match the failure groups for gender, age, presurgical body mass index (BMI), and follow-up duration. %EWL was calculated as previously reported [11]. The study was approved by the Regional Ethical Committee CEIm (Comité d’ética d’Investigació).
Anthropometrical and biochemical measures
Blood samples were obtained at preSurgery and studyTime, while fecal samples were obtained at studyTime (8.3±1.7 years after undergoing BS). Anthropometrical measurements were collected prior to surgery and at multiple time-points after the intervention.
Blood samples were obtained from the antecubital vein and placed in vacutainer tubes (BDvacutainerTM, London, UK) after an overnight fast on the day of the surgery and on the moment of the study. The serum was separated and immediately frozen at -80 until analysis. Serum glucose, cholesterol, triglycerides and high density lipoprotein cholesterol (HDL cholesterol), were measured in a Dimension autoanalyzer (Dade Behring Inc., Deerfield, IL) by enzymatic methods (RandoxLabo- ratories Ltd., UK and WakoBioproducts, Richmond, VA). Low-density lipoprotein cholesterol (LDL cholesterol) was calculated using the Friedewald formula.
DNA extraction from fecal samples
Fecal samples were collected and immediately stored at -80°C until analysis. DNA extraction from stools was done using the QIAamp DNA stool Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA concentration and purity were estimated with a Nanodrop spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Gut microbiome analysis
Libraries from stool samples were built with the 16S Metagenomics kit (Thermofisher), consisting of primer pools to amplify multiple variable regions (V2, 3, 4, 6-7, 8 and 9) of the 16S rRNA. After generating amplicons, the Ion PlusTM Fragment Library Kit (Thermofisher) was used to ligate barcoded adapters and synthesize libraries. Barcoded libraries from all the samples were pooled and templated on the automated Ion Chef system (Thermofisher) followed by a 400 bp sequencing on the Ion S5 (Thermofisher).
Base calling and run demultiplexing were performed by using Torrent SuiteTM Server software (Thermofisher), version 5.4.0, with default parameters for the 16S Target Sequencing (bead loading ≤30, key signal ≤30 and usable sequences ≤30). Quality sequences were analyzed using QIIME 1.9.1 software [12]. Briefly, the workflow was the following: Operational taxonomic units (OTUs) were calculated by clustering sequences at a similarity of 97% with a closed-reference OTU picking approach. The representative sequences were submitted to the UCLUST to obtain the taxonomy assignment and the relative abundance of each OTU using the Greengenes 16S rRNA gene database. To correct for differences in sequencing depth, CSS normalization method [13] was used before the group significance tests (group_significance.py), while a randomly sub-sampled for each sample at the same number of sequences were used to evaluate alpha and beta diversity through QIIME, aligning the OTUs with PyNAST to build a phylogenetic tree. Core microbiomes were assessed with the compute_core_microbiome.py script in QIIME. Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to assess the associations of the core microbiomes from the different groups.
PICRUSt 1.1.1 (Investigation of Communities by Reconstruction of Unobserved States) was used to calculate the functional profiles of the microbial communities [14]. Bile salt hydrolase (BSH) gene content was predicted through the metagenome_contributions.py script of the KEGG orthology K01442, cholylglycine hydrolase.
Statistical analysis
Operational taxonomic units (OTUs) (at different taxonomic levels, from phylum to genus level) differing between groups were identified in QIIME with the nonparametric Kruskal-Wallis test (group_significance.py). The Spearman correlation coefficients were calculated to estimate the correlations between variables through QIIME (observation_metadata_correlation.py). OTUs differences reported in QIIME, were further analyzed with the statistical software package SPSS version 22.0 (SPSS Inc., IL, USA). Multiple linear regression models were also used to determine the associations between variables. Values were considered to be statistically significant when P<0.05.
Results
Primary failure patients showed the worst biochemical and anthropometric values at the time of the study
Anthropometric and biochemical characteristics of the patients are depicted in Table 1. Patients at the preSurgery time presented no differences between the groups. At study Time, BMI, weight, waist circumference, total cholesterol and LDL-cholesterol differed among groups. Success patients showed the best values of these metabolic variables, differing from those of Primary Failure patients, without finding statistical differences in relation to the Weight Regain patients.
Table 1.
Anthropometric and biochemical variables of the study groups at the preSurgery and StudyTime points
| Success (n=6) | Primary Failure (n=6) | Weight Regain (n=12) | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| preSurgery | StudyTime | preSurgery | StudyTime | preSurgery | StudyTime | |
| Age (years) | 43.33±9.97 | 44.67±7.63 | 48.83±9.81 | |||
| Sex (M/F) | 2/4 | 0/6 | 1/11 | |||
| Weight* (Kg) | 130.57±21.34 | 82.52±14.85$ | 128.75±17.47 | 108.53±14.99$ | 126.62±18.73 | 103.39±16.56$ |
| BMI* (Kg/m2) | 47.03±6.01 | 29.72±4.70$ | 50.21±5.72 | 42.33±4.97$ | 48.44±5.60 | 39.49±4.53$ |
| Waist* (cm) | 133±22.06 | 87.4±13.18$ | 136.2±9.98 | 121.67±17.90$ | 129.23±14.19 | 111.75±13.29$ |
| EWL (%) | 81.5±18.4 | 31.7±8.0 | 37.7±13.8 | |||
| Glucose (mmol/L) | 86.5±8.53 | 90.17±5.71 | 102±10.28 | 95.17±6.55 | 100.58±16.39 | 97.75±8.33 |
| HbA1c | 5.02±0.19 | 5.35±0.27 | 5.45±0.28 | 5.72±0.27 | 5.34±0.54 | 5.67±0.39 |
| Cholesterol* (mmol/L) | 186.83±41.34 | 171.67±23.99$ | 201.83±39.68 | 211.83±13.86$ | 205±33.29 | 188.25±17.64 |
| HDL-Chol (mmol/L) | 44±11.24 | 58.5±13.19 | 47.5±17.74 | 60±16.36 | 49.42±12.60 | 61.17±11.26 |
| LDL-Chol* (mmol/L) | 115.17±22.47 | 100.17±17.45$ | 129.67±36.74 | 134±11.84$ | 132.25±25.64 | 110.25±20.99 |
| TCG (mmol/L) | 138.5±94.26 | 73.83±21.98 | 124±73.12 | 89.5±35.02 | 116.42±41.18 | 84.5±24.87 |
Data are expressed as mean ± SD. Statistical differences: P<0.05.
Indicates statistical differences among the groups.
Indicates statistical differences between the preSurgery and StudyTime within the group.
Figure 1 shows the progression of BMI from the preSurgery point until 60 months post-surgery. Primary Failure patients presented the worst post-surgery evolution, even at the very beginning (4 months). However, Weight Regain and Success patients showed a very similar trend until 36 months. At 48 months, Weight Regain patients statistically differed from success patients.
Figure 1.
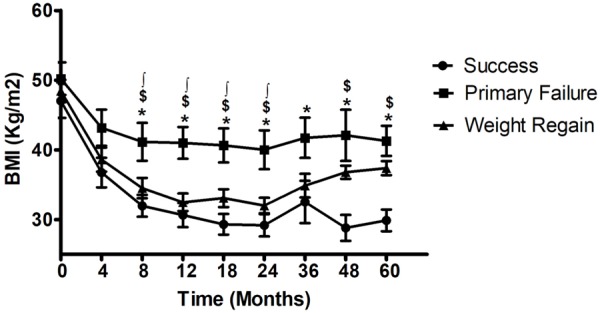
Retrospective BMI record according to the study groups. Circle: Success, square: Primary Failure, triangle: Weight Regain. *Indicates statistical differences between Success and Primary Failure groups. $ Indicates statistical differences between Success and Weight Regain groups. ∫Indicates statistical differences between Primary Failure and Weight Regain groups.
Gut microbiota diversity
To investigate the mid-term effects of BS on the gut microbiota, we analyzed fecal samples of the study population. After the quality assessment, total of 387,882 quality sequences were used for the posterior analysis of the gut microbiome, with 6,852 OTUs identified. For diversity analysis, samples were rarefied to 1,645 sequences, corresponding to the lowest number of quality reads obtained from any patient sample in the data set.
A dimensional Principal Coordinates Analysis plot of unweighted UniFrac distance was used for the visualization of complex relationships among groups. No differences were observed (Anosim test, P>0.05), suggesting a high similarity among the groups (Figure 2A).
Figure 2.
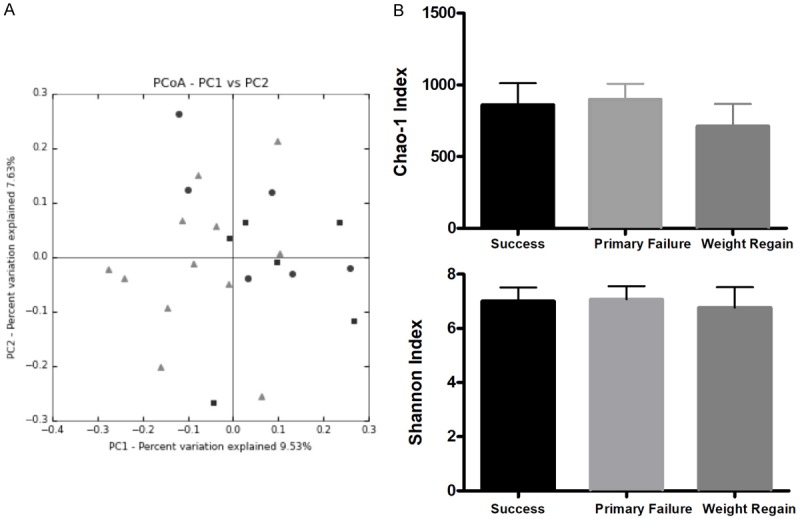
A. Clustering of fecal bacterial communities according to the different study groups by Principal Coordinates Analysis (PCoA) using Unweighted UniFrac distances. B. Chao1 Richness estimator and Shannon Diversity indexes of the study groups at the StudyTime. Black: Success, grey: Primary Failure, dark grey: Weight Regain.
Alpha diversity assessment using rarefaction curves revealed no significant differences between study groups, estimated by the indexes of Chao1 (Richness) and Shannon (Diversity). Weight Regain group registered a lower richness with respect to the Primary Failure group nearly reaching the statistical difference (P=0.06: Figure 2B).
Gut microbiota profile
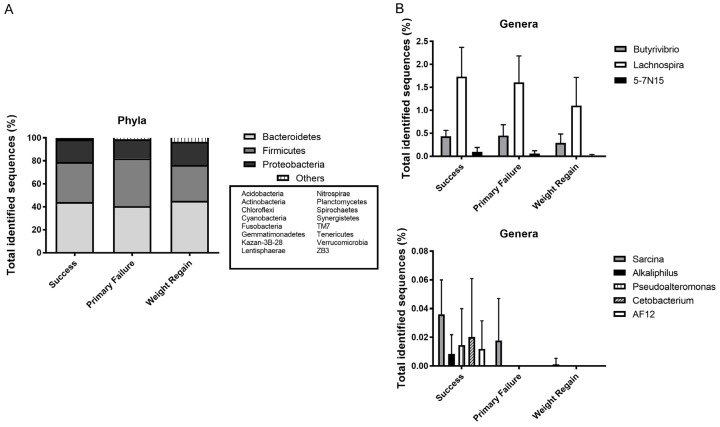
According to the abundance of each identified OTU, the dominant bacteria were, as expected, the phyla Bacteroidetes and Firmicutes, followed by Proteobacteria and Actinobacteria (Figure 3A), although 15 other phyla were also identified but with minor abundances (less than 1%).
Figure 3.
A. Phylum level distribution of bacteria in fecal samples from the study groups. B. Genera statistically significant among the groups of study.
Different changes were registered in the OTU abundances among the groups. Family abundances did not show any difference among the groups. However, some genera presented statistically significant differences (P<0.05) among groups for success, primary failure and weight regain groups: within the Firmicutes genera Sarcina (0.06, 0.03 and 0.00%, respectively), Butyrivibrio (0.79, 0.84 and 0.61%), Alkaliphilus (Success group abundance of 0.02%) as well as Lachnospira (2.85, 2.63 and 2.11%, respectively); Pseudoalteromonas (Success group abundance of 0.14%) from Proteobacteria phylum, and from Fusobacteria, the genus Cetobacterium (Success group abundance of 2.33%); while within Bacteroidetes only two minor genera 5-7N15 (0.31, 0.23 and 0.05%, respectively) and AF12 (Success group abundance of 0.05%) registered differences. However, the representation of most of these bacteria was scarce. Figure 3B shows these differences: Success group showed the highest abundance of these bacteria in relation to Weight Regain group, who showed the lowest abundance.
Patients from the success group shared a more diverse core microbiome than the other groups
After the observation of few differences among the study groups, we wondered if each group was characterized by a concrete core gut microbiome profile. For this purpose, we investigated the core microbiome of each group, meaning those OTUs that were shared among the 100% of the samples of the study group according to the Greengenes dataset at a 97% of identity. In this manner, we discovered that the Success group shared a more diverse core microbiome than the other groups, with the Weight Regain group as the one with the least number of OTUs shared among the whole samples (110 OTUs in the Success group, versus 54 and 9 in Primary Failure and Weight Regain, respectively). Only 9 OTUs were shared by the 100% of the patients analyzed in this study (Figure S1). When these OTUs were translated into taxa data at genus level, more informative results were obtained. Thus, core microbiomes of Success patients contained exclusively 16 different taxa, while Primary Failure patients showed 5 and Weight Regain only 1 different taxa from the other groups (Figure 4).
Figure 4.
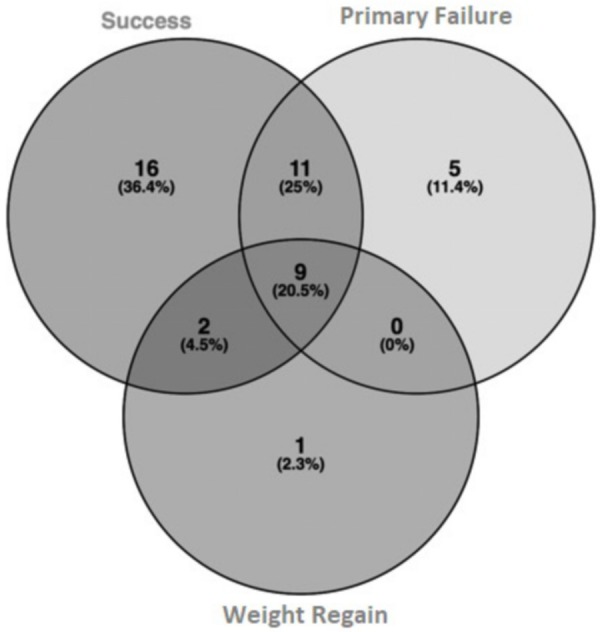
Venn diagram of the core microbiomes from the study groups at taxa level.
Success patients were characterized by 10 genera from Firmicutes, 5 from Proteobacteria, and 1 from Bacteroidetes phyla. Core microbiomes of Primary Failure patients were characterized by 4 genera from Proteobacteria and 1 from Firmicutes phyla; and finally, Weight Regain patients only presented 1 genus from Bacteroidetes phylum that was not found in the other groups. These taxa are listed in the supplementary data (Table S1).
Different bacterial salt hydrolase activities, within a similar gut microbiome potentiality
To predict the abundance of gene families and related functional pathways of microbial communities in the fecal contents, PICRUSt analysis, a predictive metabolism approach, was performed with our 16S rRNA sequences. KEGG orthology (KO) counts from Kyoto Encyclopedia of Genes and Genomes (KEGG) database were used to estimate the gene richness of the groups through the Chao1 and Shannon indexes (Figure 5A). Membrane transport, carbohydrate and amino-acids metabolism, were the main activities represented in the gut microbiome of the patients (Figure S2).
Figure 5.
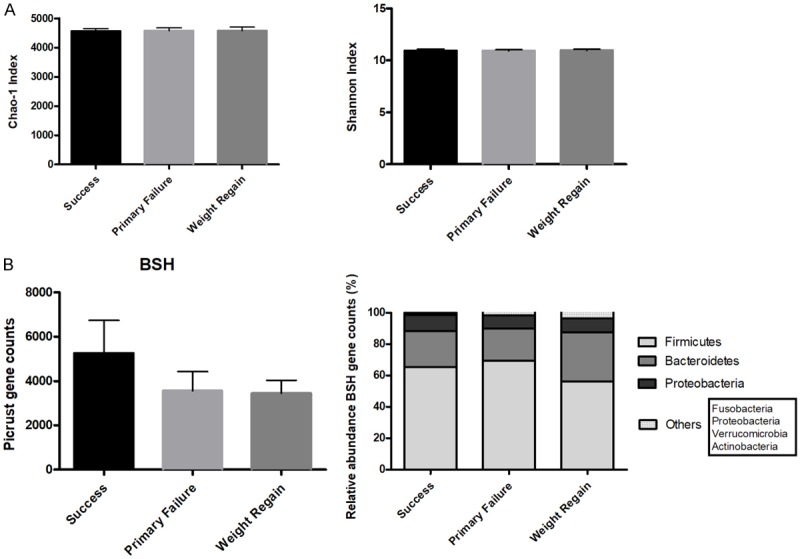
Predictive functionality of gut microbiome. A. Chao1 Richness estimator and Shannon Diversity indexes of the study groups at the StudyTime. B. Predictive values of the bacterial bile salt hydrolase (BSH) enzyme and its distribution among the different phyla. Black: Success, grey: Primary Failure, dark grey: Weight regain.
On the other hand, the levels of bacterial salt hydrolase (BSH) gene were predicted. Higher values of this enzyme were observed in the Success group (Figure 5B). The taxon with the highest rate of OTUs contributing to changes in BSH abundance was Firmicutes, followed by Bacteroidetes and Proteobacteria.
BMI and the cholesterol metabolism were related to the genus Sarcina
According to the differences found in the study groups profiles, we performed correlation analysis with the clinical variables. Three genera were related to BMI: Sarcina (R=-0.505, P=0.012), Alkaliphilus (R=-0.507, P=0.012) and AF12 (R=-0.566, P=0.004), while BSH levels were associated with Sarcina (R=0.518, P=0.010), Alkaliphilus (R=0.533, P=0.007) and Butyrivibrio (R=0.730, P=0.000). With these data, we performed multiple linear regression analysis to establish a model, in which Sarcina abundance was presented as the best genera related to BMI post-surgery (ß=-0.457, R2.adjusted =0.173, P=0.025).
Discussion
BS is positioned as an effective model to lose weight for morbid obesity. Although BS is a successful treatment for obesity, a high number of patients lose its benefits early in times or in the long-term, while others remain successfully over time. In the current study, we have characterized for the first time the gut microbiota of patients who underwent BS, but differed in their success rate. Clinical variables have indicated the worst metabolically condition of Primary Failure patients. This could indicate a role of the metabolism in the BS successful rate, especially lipid metabolism. On the other hand, gut microbiota has recently emerged as one of the mediators in the changes observed after BS [15]. Indeed, different core microbiomes were found among the groups. Our study shows that gut microbiota could be involved, at least partially, in the success or primary failure of BS.
BS induces changes in environmental factors and anatomy, which contribute to the weight loss and the amelioration of obesity comorbidities. These changes are procedural-related, as differences in the surgical interventions profoundly mark the digestive tract environment [16]. Our patients underwent RYGB surgery. RYGB is one of the most widely used interventions, which triggers in sharp changes because of its mixed, restrictive and malabsorptive, condition. RYGB provokes alterations in the bile acids levels, restriction of the stomach size, in the flow of nutrients, vagal manipulation and modulation of the enteric and adipose hormones [17].
Gut microbiota profile is affected by RYGB, with an increase in the abundance of facultative anaerobic, bile-tolerant and acid-sensible microorganisms [15]. Our study shows a particular gut microbiota profile related to RYGB, remained even years after BS. Literature has established that remodeling of the microbial community occurred mainly within the first three months after the surgery, with minor changes afterwards [18]. Only few genera have differed among groups: Sarcina, Alkaliphilus, AF12 or Butyrivibrio, which although with a minor representation showed powerful relationships with BMI or cholesterol metabolism, especially Sarcina. However, Unifrac analysis could not discriminate between groups, as changes produced by BS predominate over any other situation [8]. Other studies reported Proteobacteria was the phylum most affected by RYGB [8]. However, due to the design of our experiment, we cannot assure this result, although Proteobacteria is our third most abundant phylum. The most relevant information has been extracted from the core microbiomes, with the potential to modulate the host metabolic function and interfere with the outcomes of the BS.
The host critically depends on a diverse array of microbial metabolites for normal development. This includes metabolites that are produced by bacteria from dietary components, metabolites that are produced by the host and biochemically modified by gut bacteria, and metabolites that are synthesized de novo by gut microbes [19]. Bile acids, which are cholesterol metabolites that facilitate the absorption of dietary fat and fat-soluble molecules, are found within these metabolites as bile acids are regulated by gut microbiota [20], performing a crucial role in the secondary metabolism of the primary bile acids. Different surgical techniques have different effects on cholesterol levels, independent of weight loss [21]. Thus, malabsorptive procedures are the most effective at decreasing the absorption of cholesterol and associated with a clear decrease of LDL-Cholesterol [21].
BS increases blood bile acid levels [22]. Increasing circulating levels of primary and secondary bile acids have been observed after RYGB [20]. We have predicted a higher level of BSH in the Success patients. Predicted BSH abundances have been significantly correlated with bile acid levels [23]. Success patients could present a higher level of bile acids than the other groups. Gut microbiota may promote bile acid deconjugation, dehydrogenetation and dehydoxylation, increasing the diversity of the systemic bile acids [24]. A higher amount of bile acids could increase the gut microbiome metabolism of the primary bile acids. Indeed, an increase in the microbial genes involved in the dehydroxylation (BSH enzyme) of primary to secondary bile acids in the RYGB patients was reported [8]. BSH enzyme is a conserved microbial adaptation that is unique to the gut associated microbiome and it is distributed among the major bacteria [25], with Firmicutes as the most represented phylum of the BSH gene in the present study.
The more diverse microbiomes are more resistant to perturbations associated to pathogenic situations [26]. Success patients have presented a more diverse core microbiome, what could represent a dysbiosis status of the other groups. Success patients could have been more prone to the environmental changes. Moreover, the low specificity of the Weight Regain patients could suggest a low implication of the gut microbiota in its successful rate.
Bile acids secreted by the liver also determine the composition of the gut microbiome, because bile acids can disturb bacterial membrane integrity [27]. BSH contributes to bile tolerance [26]. Success patients presented a more suitable microbiome than the other groups. But as environmental factors determine the gut microbiota profiles, failure patients need to adapt. Primary Failure patients present within their core microbiomes two interesting bacteria: Acinetobacter and Serratia. The correlation in their abundances suggests a possible interaction between them. Both bacteria are able to use carnitine in their metabolisms [28]. Carnitine is important for the dissemination and survival of the bacteria in the intestine, as enhances bile tolerance [29]. However, the metabolism of carnitine triggers the production of trimethylamine (TMA) and its subsequent oxidation to trimethylamine N-oxide (TMAO) by hepatic flavin-containing monooxygenases thanks to a two-component Rieske-type oxygenase/reductase (CntAB) and associated gene cluster [30]. Elevated levels of TMAO are related to an increased risk of atherosclerosis [31]. Primary Failure patients could present an altered profile of gut microbiota, namely dysbiosis, which could not be able to survive in a bile acid environment as the intestine, searching for other strategies as the metabolism of carnitine. However, this fact could develop a worst cardiometabolic prognostic. TMAO is able to promote the reduction in the expression levels of Cyp7a1, the major bile acid-producing enzyme and rate limiting step in the catabolism of cholesterol [31]. Indeed, Primary Failure patients could present a dysregulation in the conversion of the cholesterol to bile acids.
Moreover, we have also found that the genus Sarcina has a role in the BMI maintaining. TMA might also be involved in Sarcina’s action, because of its capacity of producing TMA from TMAO [32], removing TMAO from the environment. Moreover, Sarcina has been related to the BSH content and, consequently, to the cholesterol levels. In addition to the cholesterol removal via biliary sterol secretion, another pathway exists, the trans-intestinal cholesterol excretion (TICE). TICE is stimulated by the Farnesoid X Receptor (FXR) by the induction of its target gene FGF19, excreting up to 60% of the daily cholesterol [33]. Bile acids exert their actions in the energy metabolism through activation of the receptors FXR and TGR5 [34]. Thus, FXR controls fatty acid, triglyceride and BA metabolism, and it could be thanks to the activation of TICE pathway.
To our knowledge, this is the first attempt to relate the gut microbiota in the success rates of BS. Our study provides novel data in the implication of the gut microbiome in the maintenance of the BMI in the mid-term, after BS. We have shown evidence of the role of the cholesterol metabolism, possibly through the bile acids, in the success or failure of BS outcome, and that gut microbiota plays an important role in this process.
Although we have presented several interesting results, the present study shows some limitations. First of all, the unique moment of the sampling represents only a scheme in that point. Moreover, the interesting presented pathways should be demonstrated with further experimental procedures. However, we strongly believe that the novel results presented here open a new door to understand the mechanisms under the success or failure of BS, an aspect that could change the clinical aspects of the treatments against obesity. Thus, this probe of concept guarantees further large studies with a prospective design in which the exact moment when the benefits of the RYGB and other procedures of BS are lost or fading. Targeting the gut microbiota to modify cholesterol and bile acids metabolism might be a good candidate to improve, or at least ameliorate, the successful rates of BS.
Acknowledgements
The research groups belong to the “Centros de Investigación en Red” [CIBERobn and CIBERdem, of the “Instituto de Salud Carlos III”]. The authors thank the Metagenomic Plattform of Centro de Investigación Biomédica en Red de Fisiopatología de la Obesidad y la Nutrición, CIBERobn, Instituto de Salud Carlos III (ISCIII), Spain. CGR was supported by a “Juan de la Cierva, Formación” contract (FJCI-2015-24543), and IMI was supported by the “MS type I” program (CP16/00163). This work was supported in part by grants from the Instituto de Salud Carlos III co-founded by Fondo Europeo de Desarrollo Regional-FEDER, PI15/01114, CP16/00163, Madrid, Spain.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ariyarathenam AV, Pournaras DJ, Tham JC, Finlay I, Cota A. Need for standardization of the measurement of preoperative weight in bariatric surgical patients in the UK: a survey of british obesity and metabolic surgery society (BOMSS) members. Int J Surg. 2012;10:598–600. doi: 10.1016/j.ijsu.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 2.Sherf-Dagan S, Schechter L, Lapidus R, Sakran N, Goitein D, Raziel A. Perceptions of success in bariatric surgery: a nationwide survey among medical professionals. Obes Surg. 2018;28:135–141. doi: 10.1007/s11695-017-2800-9. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds CL, Byrne SM, Hamdorf JM. Treatment success: investigating clinically significant change in quality of life following bariatric surgery. Obes Surg. 2017;27:1842–1848. doi: 10.1007/s11695-017-2568-y. [DOI] [PubMed] [Google Scholar]
- 4.de Hollanda A, Jimenez A, Corcelles R, Lacy AM, Patrascioiu I, Vidal J. Gastrointestinal hormones and weight loss response after Roux-en-Y gastric bypass. Surg Obes Relat Dis. 2014;10:814–819. doi: 10.1016/j.soard.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 5.de Hollanda A, Casals G, Delgado S, Jiménez A, Viaplana J, Lacy AM, Vidal J. Gastrointestinal hormones and weight loss maintenance following Roux-en-Y gastric bypass. J Clin Endocrinol Metab. 2015;100:4677–4684. doi: 10.1210/jc.2015-3065. [DOI] [PubMed] [Google Scholar]
- 6.Aron-Wisnewsky J, Dore J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol Hepatol. 2012;9:590–598. doi: 10.1038/nrgastro.2012.161. [DOI] [PubMed] [Google Scholar]
- 7.Gralka E, Luchinat C, Tenori L, Ernst B, Thurnheer M, Schultes B. Metabolomic fingerprint of severe obesity is dynamically affected by bariatric surgery in a procedure-dependent manner. Am J Clin Nutr. 2015;102:1313–1322. doi: 10.3945/ajcn.115.110536. [DOI] [PubMed] [Google Scholar]
- 8.Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T, Fäncriks L, le Roux CW, Nielsen J, Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Hollanda A, Ruiz T, Jimenez A, Flores L, Lacy A, Vidal J. Patterns of weight loss response following gastric bypass and sleeve aastrectomy. Obes Surg. 2015;25:1177–1183. doi: 10.1007/s11695-014-1512-7. [DOI] [PubMed] [Google Scholar]
- 10.Moize VL, Pi-Sunyer X, Mochari H, Vidal J. Nutritional pyramid for post-gastric bypass patients. Obes Surg. 2010;20:1133–1141. doi: 10.1007/s11695-010-0160-9. [DOI] [PubMed] [Google Scholar]
- 11.Morínigo R, Moizé V, Musri M, Lacy AM, Navarro S, Marín JL, Delgado S, Casamitjana R, Vidal J. Glucagon-like peptide-1, peptide YY, hunger, and satiety after gastric bypass surgery in morbidly obese subjects. J Clin Endocrinol Metab. 2006;91:1735–1740. doi: 10.1210/jc.2005-0904. [DOI] [PubMed] [Google Scholar]
- 12.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koening JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotech. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ilhan ZE, DiBaise JK, Isern NG, Hoyt DW, Marcus AK, Kang DW, Crowell MD, Rittmann BE, Krajmalnik-Brown R. Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. ISME J. 2017;11:2047–2058. doi: 10.1038/ismej.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco JV, Ruiz PA, Palermo M, Gagner M. A review of studies comparing three laparoscopic procedures in bariatric surgery: sleeve gastrectomy, Roux-en-Y gastric bypass and adjustable gastric banding. Obes Surg. 2011;21:1458–1468. doi: 10.1007/s11695-011-0390-5. [DOI] [PubMed] [Google Scholar]
- 17.Laferrere B, Teixeira J, McGinty J, Tran H, Egger JR, Colarusso A, Kovack B, Bawa B, Koshy N, Lee H, Yapp K, Olivan B. Effect of weight loss by gastric bypass surgery versus hypocaloric diet on glucose and incretin levels in patients with type 2 diabetes. J Clin Endocrinol Metab. 2008;93:2479–2485. doi: 10.1210/jc.2007-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palleja A, Kashani A, Allin KH, Nielsen T, Zhang C, Li Y, Brach T, Liang S, Feng Q, Jorgensen NB, Bojsen-Moller KN, Dirksen C, Burgdorf KS, Holst JJ, Madsbad S, Wang J, Pedersen O, Hansen T, Arumugam M. Roux-en-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med. 2016;8:67. doi: 10.1186/s13073-016-0312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postler TS, Ghosh S. Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26:110–130. doi: 10.1016/j.cmet.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahmad NN, Pfalzer A, Kaplan LM. Roux-en-Y gastric bypass normalizes the blunted postprandial bile acid excursion associated with obesity. Int J Obes. 2013;37:1553–1559. doi: 10.1038/ijo.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benetti A, Del Puppo M, Crosignani A, Veronelli A, Masci E, Frigè F, Micheletto G, Panizzo V, Pontiroli AE. Cholesterol metabolism after bariatric surgery in grade 3 obesity: differences between malabsorptive and restrictive procedures. Diabetes Care. 2013;36:1443–1447. doi: 10.2337/dc12-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole AJ, Teigen LM, Jahansouz C, Earthman CP, Sibley SD. The influence of bariatric surgery on serum bile acids in humans and potential metabolic and hormonal implications: a systematic review. Curr Obes Rep. 2015;4:441–450. doi: 10.1007/s13679-015-0171-x. [DOI] [PubMed] [Google Scholar]
- 23.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, Korzenik JR. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016;43:1142–1153. doi: 10.1111/apt.13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gérard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens. 2014;3:14–24. doi: 10.3390/pathogens3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, Hill C, Gahan CG. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U S A. 2014;111:7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cani PD, Delzenne NM. Gut microflora as a target for energy and metabolic homeostasis. Curr Opin Clin Nutr Metab Care. 2007;10:729–734. doi: 10.1097/MCO.0b013e3282efdebb. [DOI] [PubMed] [Google Scholar]
- 27.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30:332–338. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meadows JA, Wargo MJ. Carnitine in bacterial physiology and metabolism. Microbiology. 2015;161:1161–1174. doi: 10.1099/mic.0.000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gahan CG, Hill C. Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol. 2014;4:9. doi: 10.3389/fcimb.2014.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Jameson E, Crosatti M, Schäfer H, Rajakumar K, Bugg TD, Chen Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A. 2014;111:4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fennema D, Phillips IR, Shephard EA. Trimethylamine and trimethylamine N-oxide, a flavin-containing monooxygenase 3 (FMO3)-mediated host-microbiome metabolic axis implicated in health and disease. Drug Metab Dispos. 2016;44:1839–1850. doi: 10.1124/dmd.116.070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Boer JF, Schonewille M, Boesjes M, Wolters H, Bloks VW, Bos T, van Dijk TH, Jurdzinski A, Boverhof R, Wolters JC, Kuivenhoven JA, van Deursen JM, Oude Elferink RPJ, Moschetta A, Kremoser C, Verkade HJ, Kuipers F, Groen AK. Intestinal farnesoid X receptor controls transintestinal cholesterol excretion in mice. Gastroenterology. 2017;152:1126–1138. e6. doi: 10.1053/j.gastro.2016.12.037. [DOI] [PubMed] [Google Scholar]
- 34.Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191–212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.