Abstract
STOML2 (Stomatin-like protein 2) is up-regulated and acts as an oncogenic protein in multiple cancers. However, the role and regulatory mechanism of STOML2 in head and neck squamous cell carcinoma remain unclear. Here, we found that STOML2 is overexpressed and indicates poor outcomes in HNSCC. In addition, the expression of STOML2 correlates positively with T stage, lymph node metastasis and recurrence. Reduced STOML2 dramatically inhibits cell proliferation, colony formation and motility of HNSCC cells in vitro. Furthermore, the sensitivity of HNSCC cells towards cisplatin is obviously improved in STOML2-silencing cells. Subsequent studies suggest that STOML2 could regulate the expression of IL6 transcriptionally and then further induce the phosphorylation of Tyr705 residue of Stat3, whose activation plays a critical role in HNSCC. Taken together, these results for the first time demonstrate that STOML2 promotes HNSCC progression through activating IL6-Stat3 pathway and provide a promise for diagnosis and treatment for HNSCC.
Keywords: HNSCC, STOML2, Stat3, proliferation, invasion, chemo-sensitivity
Introduction
Head and neck squamous cell carcinoma (HNSCC) is among the most common types of human malignancies, with an annual worldwide incidence rate of 600,000 [1]. Despite developed systemic therapies have improved outcomes, the overall 5-year survival rate remains only about 50% [2]. Malignant proliferation, invasion-metastasis and chemo-resistance are the main causes of relapse and poor prognosis [3], but our understanding about them still remains poor. Therefore, based on above, exploring novel molecules and underlying mechanisms involved in HNSCC progression are urgently needed to improve patient survival.
As a mitochondrial protein, STOML2 is an unusual member of the stomatin family, lacking an NH2-terminal hydrophobic domain, which distinguishes it from other members [4]. Functionally, STOML2 is a regulator of mitochondrial membrane potential and ATP production [5]. An increasing number of studies have found that STOML2 is implicated in tumor progression and development. STOML2 is aberrantly expressed and predicted poor prognosis in multiple cancers, including esophageal squamous cell carcinoma [6], rectal cancer [7], epithelial ovarian cancer [8], cervical cancer [9], breast cancer [10] and gastric cancer [11]. Moreover, STOML2 overexpression is associated with cell growth, motility and chemo-sensitivity [6,12,13]. Rebecca et al. demonstrated that in progressing oral premalignant lesions, DNA copy number is increased at chromosome 9p13, where STOML2 located in [14]. It is suggested that STOML2 is likely to act as an oncogene candidate involved in the progression from normal tissue to oral cancer. However, the function and molecular mechanism of STOML2 in HNSCC still remains to be explored.
Signal transducer and activator of transcription 3 (Stat3) is a critical transcriptional factor, implicated in cell proliferation, apoptosis and metastasis of multiple cancers, including HNSCC [15]. Once treated with interleukin (IL)-6, the JAK/Stat3 signaling pathway is activated [16]. Then, Stat3 undergoes phosphorylation, homodimerization, nuclear translocation and induces gene transcription. Furthermore, previous studies revealed that phosphorylation at the Tyr705 residue of Stat3 is significantly increased in HNSCC [17]. On this basis, regulation of the IL-6/Stat3 signaling pathway could be a promising therapy for HNSCC.
Here, we demonstrated that STOML2 was overexpressed in HNSCC and correlated positively with T stage, lymph node metastasis, recurrence and unfavorable prognosis. Moreover, STOML2 knockdown impaired the capacity of cell proliferation and colony formation, impeded migration and invasion, but increased chemo-sensitivity of HNSCC cells. Mechanically, STOML2 depletion suppressed the activation of Stat3 in an IL6-dependent manner. Together, these results indicate that STOML2 acts as a crucial regulator in HNSCC progression through activating IL6-Stat3 pathway.
Materials and methods
Cell culture and reagents
Human HNSCC cell lines SCC25 and SCC15 were obtained from ATCC (Manassas, VA, USA), while CAL27, TSCC1 and Tca-8113 were purchased from the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. All HNSCC cells above were authorized by STR analysis, which were maintained in DMEM/Ham’s F12 (1:1), DMEM, MEM or RPMI1640 (Invitrogen, Camarillo, CA, USA) supplemented with 10% FBS and penicillin (100 U/ml)/streptomycin (100 mg/ml), respectively. The cells were cultured under a humidified atmosphere (37°C, 5% CO2).
IL6 was acquired from R&D Systems (Minnesota, USA) for activating Stat3 signaling pathway.
Database analysis
The online software cBioPortal (www.cbioportal.org) [18,19] was applied to analyze TCGA (The Cancer Genome Atlas) database. HNSCC GEO datasets GSE25099 and GSE37991 were provided to compare the expression of STOML2 and MMP9 between cancerous tissues and the adjacent normal tissues as well as determine the correlation between them.
Immunohistochemistry (IHC)
For immunohistochemical staining, the paraffin-embedded HNSCC tissues were deparaffinized in xylene, rehydrated and washed in PBS. After antigen retrieval, the samples were blocked and incubated with primary antibody against STOML2 (1:200, Proteintech, Rosemont, USA) at 4°C overnight. Then, the visualization was performed with the PV-9000 Polymer Detection kit and DAB chromagen (Beijing Golden Bridge Biotechnology Company, Beijing, China). The Kaplan-Meier estimates and the log-rank test were done in the OS (Overall Survival) analysis by using GraphPad Prism 6.
Real-time reverse transcription PCR
Total RNA was extracted by using TRIzol reagent (Invitrogen) according to manufacturer’s instruction, and reversely transcribed to cDNA with Quantscript RT kit produced by Tiangen (Beijing, China). Then, quantitative PCR was performed by using SYBR Premix Ex TaqTM II (TaKaRa, Japan) on ABI PRISM 7300 sequence detection system (Applied Biosystems, Foster City, CA). The primer pairs used were as follows: STOML2 forward primer, 5’-GTGACTCTCGACAATGTAAC-3’; STOML2 reverse primer, 5’-TGATCTCATAACGGAGG-CAG-3’; IL6 forward primer, 5-ACTCACCTCTTCAGAACGAATTG-3’; IL6 reverse primer, 5’-CCATCTTTGGAAGGTTCAGGTTG-3’. GAPDH was used as an internal control.
Western blot
Immunoblotting assay was performed according to standard procedure. The membranes were incubated with specific primary antibodies at 4°C overnight: STOML2 (1:1000, Proteintech), p-Stat3 (Y705, 1:1000), p-Stat3 (S727, 1:1000), Stat3 (1:1000), cleaved Caspase 3 (1:1000, all purchased from Cell Signaling Technology, Danvers, MA, USA), MMP9 (1:1000, Abcam, Cambridge, UK), and GAPDH (1:5000, Sigma-Aldrich, Missouri, USA). All images were obtained by ImageQuant LAS-4000 System (GE, Fairfield, Connecticut, USA).
Transfection
HNSCC cells transfected with siRNAs against STOML2, Stat3 or negative control oligos (Ribobio, Guangzhou, China) were labeled as si-STOML2, si-Stat3 or si-NC, respectively. Lipofectamine 2000 (Invitrogen) was used for Oligonucleotide transfection according to the manufacturers’ recommendations.
Immunofluorescence staining
HNSCC cells were plated on 18-mm cover glasses after 48 hours transfection. Immunofluorescence staining was conducted with primary antibodies against p-Stat3 (1:100, Cell Signaling Technology) at 4°C overnight. Then, the cells were incubated with Alexa Fluor 488 secondary antibodies (1:500, Cell Signaling Technology). All images were obtained by Iamger.Z2 (Zeiss, Oberkochen, Germany).
Cell growth assay
HNSCC cells (2,000 cells/well) were seeded into 96-well plates and cultured for 4 days. MTT assay was performed to measure the level of cell growth at 0 day, 2 day and 4 day. The absorbance of each well was quantified by measuring at 490 nm.
Clonogenic assay
HNSCC cells were added into a 6-well plate (1000 cells/well) and cultured for nearly two weeks. Then, colonies (>50 cells) were washed, fixed, stained (with 0.1% crystal violet) and counted.
Wound healing assay
The transfected cells and the control group were added into 6-well dish with equal amount. The scratches were made by using a 10 μl pipette tip when cells grew nearly to 100% confluence. Images of gap from random fields were captured at 0 hour and 24 hour of experiment using an inverted microscope (DMI6000B, Leica, German).
Transwell assay
In invasion or migration assay, SCC25 cells (1×105 cells/chamber) or SCC15 (6×104 cells/chamber) were plated in the upper chambers coated with Matrigel (BD) or uncoated. After 24 hours, the penetrated cells were fixed, stained and counted from at least three random fields.
Flow cytometry
For detecting cell cycle, the harvested cells were washed and fixed by 75% ethanol at 4°C. Before detection, cells were incubated with PBS containing propidium iodide and RNase at 37°C for 30 min in the dark. Apoptosis assay was performed with Annexin V/PI Apoptosis Detection kit (BD, Franklin Lakes, NJ, USA) according to the manufacturer instruction on the same FACS Canto II (BD).
Statistical analysis
All experiments other than IHC assay were repeated at least three times. The results presented as mean ± SD were analyzed with a double-sided Student’s t-test using GraphPad Prism 6. In the graphs, *, **, *** and **** indicated P<0.05, 0.01, 0.001 and 0.0001, respectively. P<0.05 was regarded as statistically significant.
Results
STOML2 is aberrantly expressed and indicates poor prognosis of HNSCC patients
Abundant evidence shows that STOML2 is upregulated in various cancerous specimens, which suggests poor prognosis. Therefore, we firstly verified the frequency of STOML2 gene alteration across multiple cancers by using cBioPortal. The graph suggested that copy number of STOML2 gene was significantly amplified, which based on TCGA database in various cancers including HNSCC (Figure 1A). Then, mining two accessible datasets of gene expression profiling (GSE25099, GSE37991) in Gene Expression Omnibus (GEO) confirmed that the mRNA expression of STOML2 was obviously increased in HNSCC (Figure 1B). Moreover, the result of IHC staining showed that in the majority of HNSCC specimens, STOML2 protein was overexpressed at the invasive margin (Figure 1C, Case 1) and the invasive front (Figure 1C, Case 2) of cancer nests. In the meanwhile, we found that the level of STOML2 in poorly differentiated tumors was higher than that in moderately and well differentiated tumors (Figure 1D). Analysis of the relationship between STOML2 expression and clinicopathological features of HNSCC indicated that enhanced STOML2 correlated positively with T stage, lymph node metastasis as well as recurrence (Table 1, P<0.05). Kaplan-Meier survival curve revealed that higher STOML2 expression indicated worse outcome of HNSCC patients (Figure 1D, P=0.0379).
Figure 1.
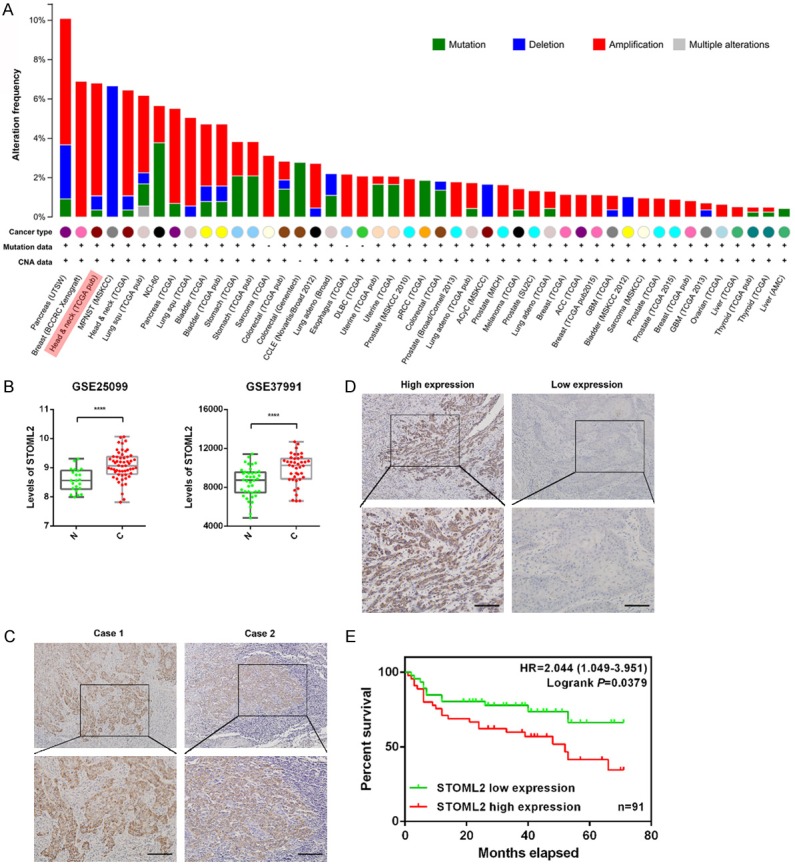
STOML2 expression level is increased in HNSCC and indicates poor prognosis. (A) The frequency of STOML2 somatic alteration across various cancers including HNSCC. (B) The analysis of GEO database (GSE25099 and GSE37991) indicated that the mRNA level of STOML2 was enhanced in HNSCC tissues. N, adjacent normal tissues, C, HNSCC tissues. (C) Representative photographs of IHC results of STOML2 at the invasive margin and the invasive front of HNSCC tissues. (D) Representative photographs of IHC results of STOML2 in HNSCC with different stages of differentiation. Scale bar in (C and D), 100 μm. (E) Kaplan-Meier survival curve showed that HNSCC patients (n=91) with high STOML2 expression level had an unfavorable overall survival (P=0.0379).
Table 1.
STOML2 expression and clinicopathological features of HNSCC
| Clinicopathological Features | PSMD14 expression | Total | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Overall | 46 (50.5%) | 45 (49.5%) | 91 | |
| Gender | ||||
| Male | 24 (42.9%) | 32 (57.1%) | 56 | 0.063 |
| Female | 22 (62.9%) | 13 (37.1%) | 35 | |
| Age | ||||
| ≤45 | 9 (64.3%) | 5 (35.7%) | 14 | 0.264 |
| >45 | 37 (48.1%) | 40 (51.9%) | 77 | |
| T stage | ||||
| T1-T2 | 31 (66%) | 16 (34%) | 47 | 0.003* |
| T3-T4 | 15 (34.9%) | 28 (65.1%) | 43 | |
| LN metastasis | ||||
| N0 | 29 (64.4%) | 16 (35.6%) | 45 | 0.009* |
| N1+N2+N3 | 17 (37%) | 29 (63%) | 46 | |
| Recurrence | ||||
| No | 37 (57.8%) | 27 (42.2%) | 64 | 0.033* |
| Yes | 9 (33.3%) | 18 (66.7%) | 27 | |
NOTE: The result was analyzed by the Pearson X2 test. P values with significance were shown with an asterisk.
P<0.05.
STOML2 knockdown suppresses cell growth of HNSCC in vitro
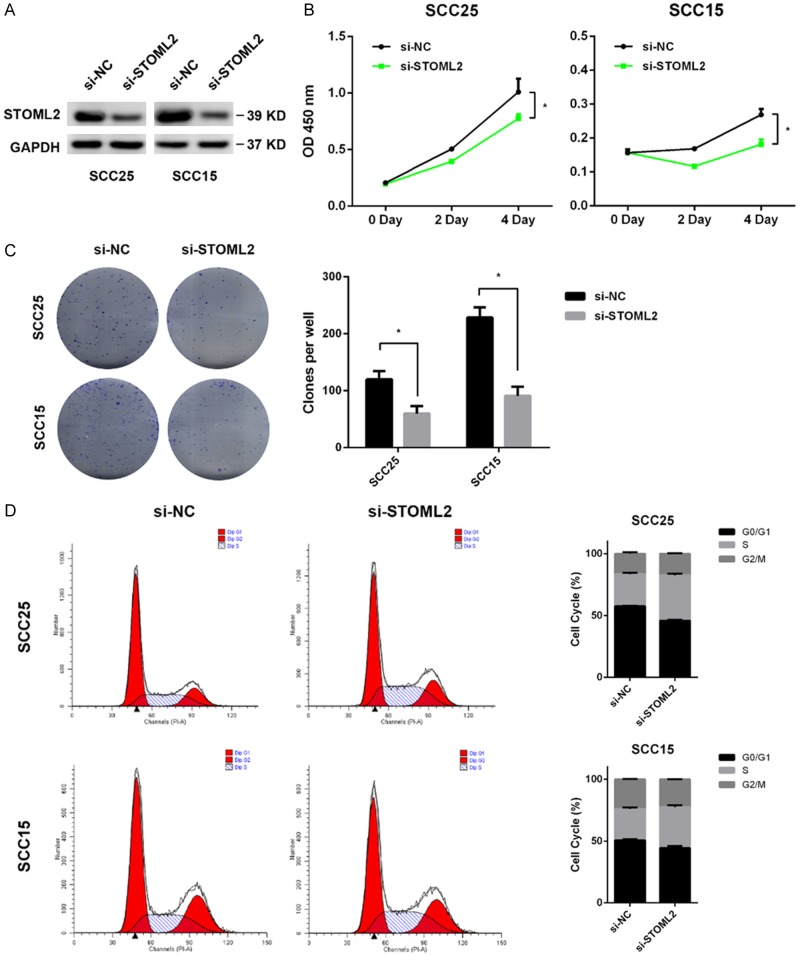
Since STOML2 expression correlated positively with T stage in HNSCC, we explored whether STOML2 played a role in cell growth of HNSCC. Real-time PCR and immunoblotting assay were performed to detect the level of STOML2 in a panel of HNSCC cell lines. The results showed that the mRNA and protein expression of STOML2 in SCC25 and SCC15 were higher than other cell lines (Figure 2A, 2B). Therefore, STOML2 was antagonized by siRNAs in vitro in these two cell lines. The inhibitory effect of three specific siRNAs towards STOML2 were evaluated (Figure 2C). Then, reduced STOML2 impaired the cell proliferation of SCC25 and SCC15 cells in vitro by using a pool of three siRNAs above against STOML2 (Figure 3A, 3B). In comparison to the negative control, both the size and number of colonies were decreased in the STOML2-silenced cells (Figure 3C). As shown in Figure 3D, STOML2 knockdown resulted in a cell cycle arrest at S phase, which may be the main cause of decreased STOML2-mediated inhibition of cell growth.
Figure 2.
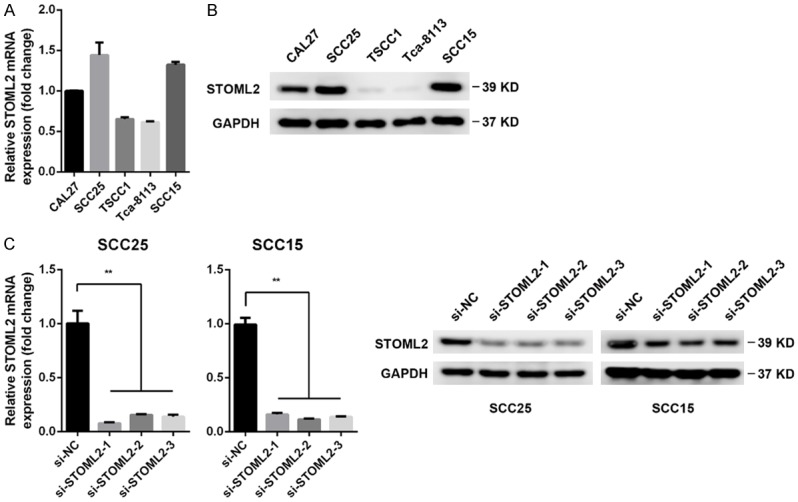
STOML2 expression in HNSCC cell lines. A. The mRNA level of STOML2 was measured in a panel of HNSCC cells by real-time PCR. B. The protein expression of STOML2 was detected in a panel of HNSCC cells by immunoblots. C. Three distinct siRNAs were introduced into both SCC25 and SCC15 cells, respectively. The STOML2 expressions in these cells were measured via real-time PCR and western blotting. Data, mean ± SD, **P<0.01.
Figure 3.
STOML2 knockdown suppresses proliferation of HNSCC cells. A. The expression of STOML2 in SCC25 and SCC15 cells transfected with a pool of siRNAs against STOML2. B. Growth curve suggested that reduced STOML2 significantly inhibited cell proliferation. Data, mean ± SD, *P<0.05. C. Reduction of colony formation capacity in STOML2-silenced SCC25 and SCC15 cells. Data, mean ± SD, *P<0.05. D. STOML2 knockdown arrested cell cycle at S phase in SCC25 and SCC15 cells.
Depletion of STOML2 inhibits migration and invasion of HNSCC cells in vitro
Owing to the overexpression of STOML2 at the invasion front of HNSCC sample, we supposed whether STOML2 could regulate motility of HNSCC cells in vitro. Upon delivery of STOML2 siRNAs into SCC25 and SCC15 cells, the healing velocity of these recipient cells was dramatically retarded within 24 hours (Figure 4A). Additionally, STOML2 depletion impeded migration and invasion of HNSCC cells in vitro (Figure 4B). As an important member of matrix metalloproteinase family, MMP9 plays a crucial role in cell invasion. Therefore, we analyzed GEO data and found that the expression of MMP9 was positively correlated with that of STOML2 in HNSCC (Figure 4C). The result of western blot reconfirmed that STOML2 could regulate the expression of MMP9 (Figure 4D), which was in line with the conclusion in glioma [20]. Taken together, the above results demonstrated that STOML2 could modulate cell motility in vitro, which may be partially MMP9-dependent.
Figure 4.
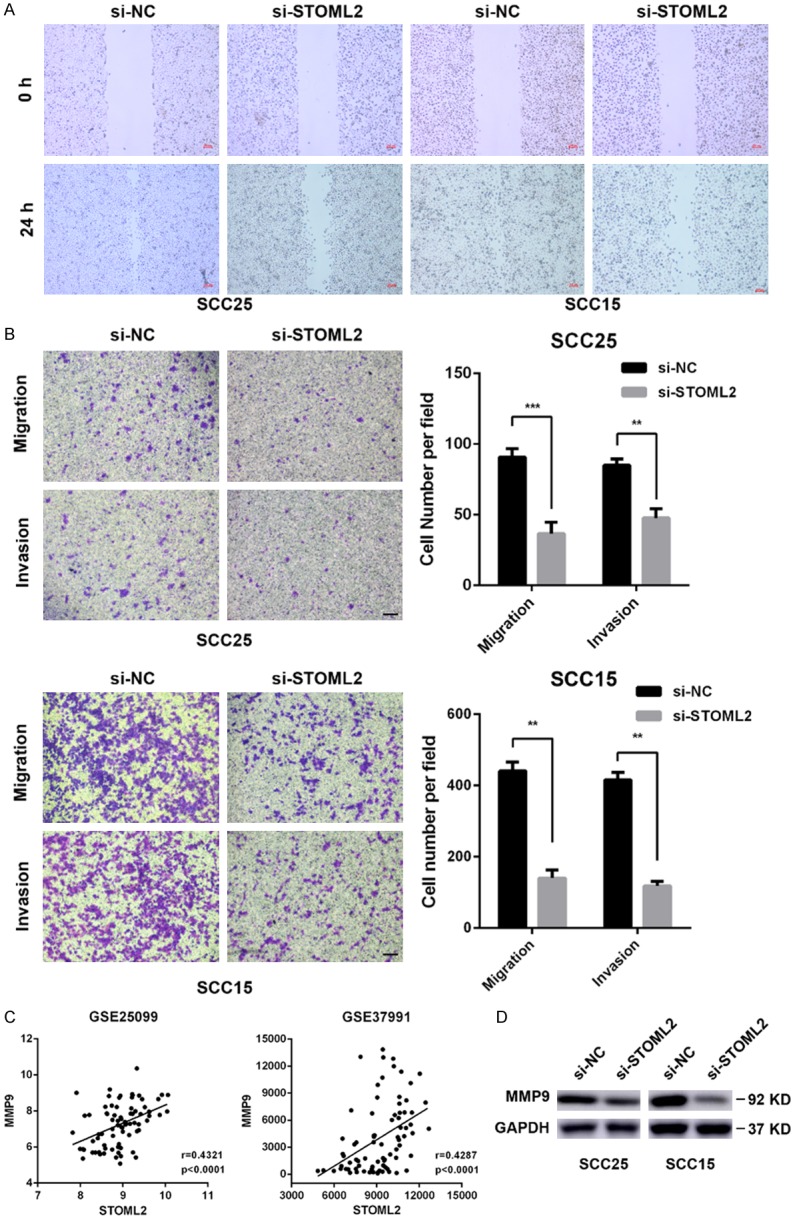
STOML2 knockdown inhibits motility of HNSCC cells in vitro. A. Representative photographs of gap at 0 hr and 24 hr of wound healing assay performed in HNSCC cells transfected with siRNAs against STOML2 or negative control. B. Transwell assay showed that depletion of STOML2 dramatically attenuated the abilities of migration and invasion of SCC25 and SCC15 cells. Scale bar, 100 μm. Data, mean ± SD, *P<0.05, **P<0.01. C. GEO database revealed that MMP9 correlated positively with STOML2. D. Western blotting results showed that STOML2 depletion reduced expression of MMP9.
STOML2 knockdown enhances chemo-sensitivity of HNSCC in vitro
Besides the malignant proliferation and invasion, chemo-resistance is another vital cause of poor prognosis in HNSCC. Hence, we also explored the role of STOML2 in it. As shown, Knockdown of STOML2 dramatically reduced the cisplatin IC50 in SCC25 and SCC15 cells (Figure 5A). With the treatment of CDDP (cisplatin), the apoptosis rate of STOML2 silencing cells was obviously increased and the number of survival decreased significantly (Figure 5B, 5C). In addition, the expression of cleaved Caspase 3 was elevated once STOML2 was antagonized (Figure 5D), which indicated that STOML2 weakened CDDP-mediated apoptosis in vitro.
Figure 5.
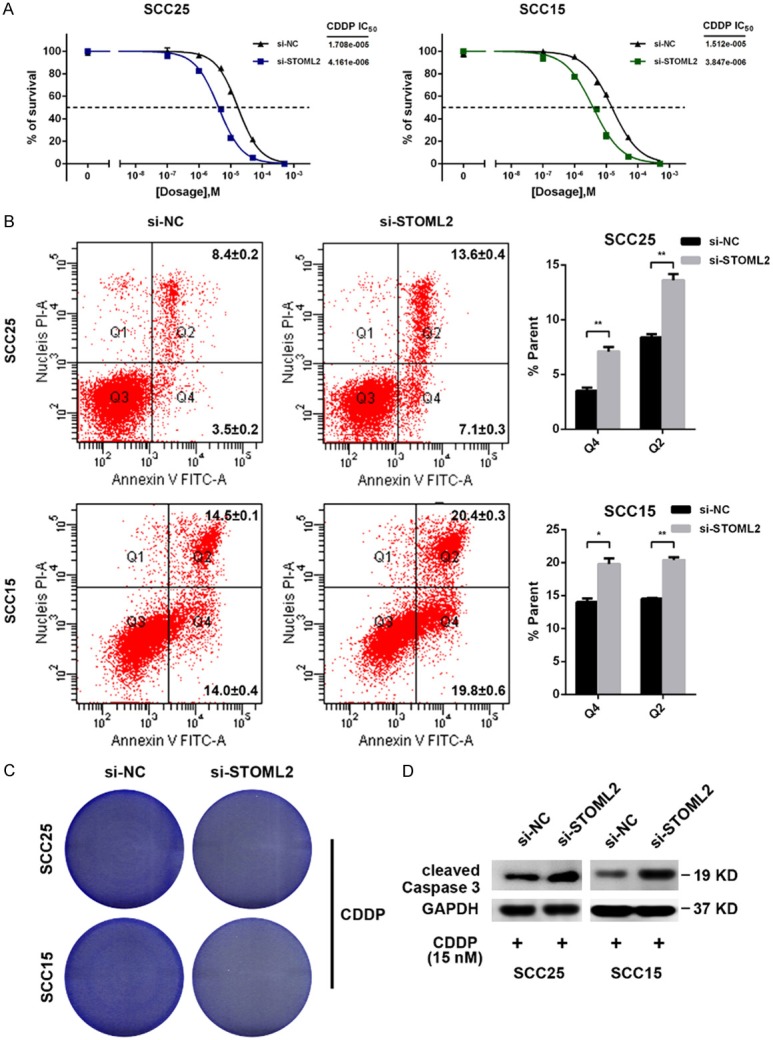
Reduced STOML2 enhances sensitivity of HNSCC cells to cisplatin in vitro. (A) STOML2 deletion decreased cisplatin IC50 in SCC25 (from 17.08 to 4.161 μM) and SCC15 cells (from 15.12 to 3.847 μM). (B and C) Silenced STOML2 sensitized HNSCC cells to cisplatin treatment and increased apoptosis using flow cytometry (B) and colony formation assay (C). CDDP, cisplatin. Data, mean ± SD, *P<0.05, **P<0.01. (D) Depletion of STOML2 enhanced the level of cleaved Caspase 3 in the case of cisplatin dosing.
STOML2 promotes malignant progression of HNSCC by activating IL6-Stat3 pathway
Since MMP9 can be transcriptionally up-regulated by the transcription factor Stat3 [21], which is implicated in malignant progression of multiple cancers, we asked if STOML2 regulated the transcriptional activity of Stat3. STOML2 silencing by siRNAs led to the attenuation of phosphorylated Stat3 (Y705) in SCC25 and SCC15 cells, but no difference in total expression of STAT3 was found (Figure 6A). Additionally, as shown in Figure 6A, STOML2 knockdown did not affect p-STAT3 (S727) levels in SCC25 and SCC15 cells. Through immunofluorescence staining, we found that STOML2 depletion suppressed the nuclear translocation of p-Stat3 (Figure 6B). On the contrary, Stat3 inhibition influenced little on the expression of STOML2 (Figure 6C). In glioma, knockdown of STOML2 transcriptionally down-regulated IL6 [20], which could activate Stat3. Therefore, we supposed whether STOML2 modulated IL6-Stat3 pathway. Real-time PCR results showed that IL6 mRNA level was reduced in STOML2-siRNA-transfected cells (Figure 6D). With the stimuli of IL6, Stat3 was activated and phosphorylated at Tyr705 (Figure 6E). Moreover, re-treatment of IL6 could rescue Stat3 activation in SCC25 and SCC15 cells pre-transfected STOML2 siRNAs (Figure 6F). Finally, we intended to verify the effect of Stat3 on HNSCC. Not only the capacities of cell proliferation and colony formation (Figure 7A, 7B), but the cell migration and invasion abilities (Figure 7C) of HNSCC cells were impaired with the inhibition of Stat3 by siRNAs. Besides, Stat3 knockdown sensitized HNSCC cells to cisplatin dramatically through Flow cytometry and colony formation assay (Figure 7D, 7E). Together, STOML2 could activate Stat3 in an IL6-dependent manner and then promote HNSCC progression.
Figure 6.
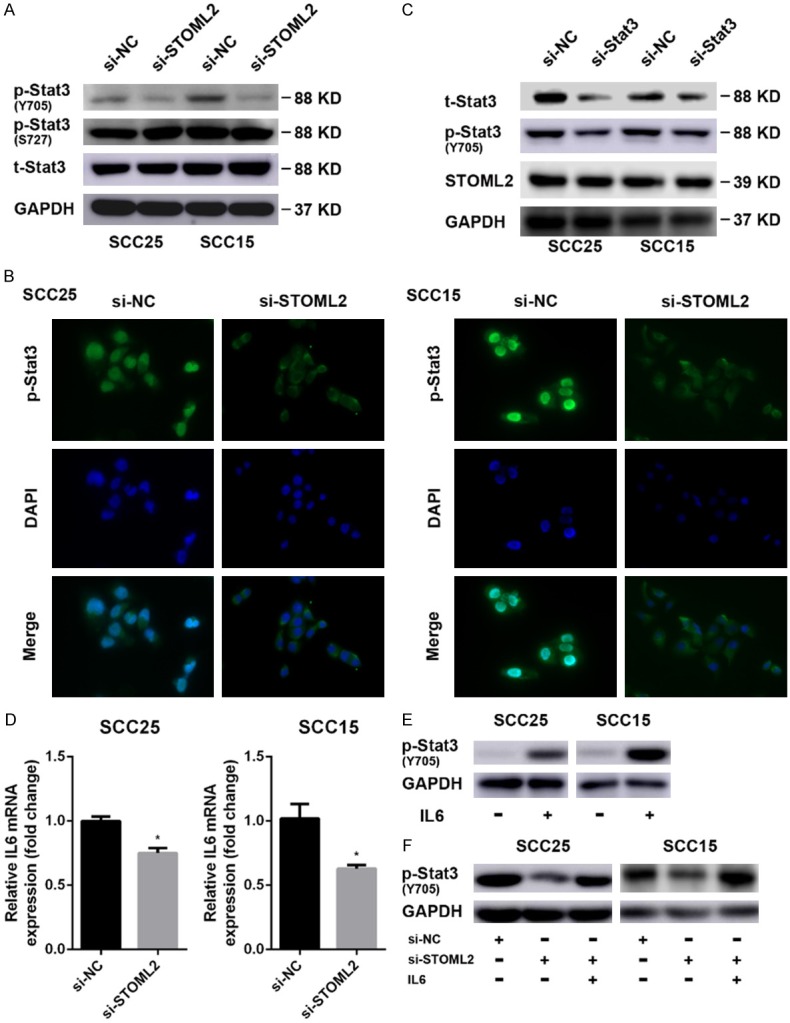
STOML2 regulates IL6-Stat3 pathway in HNSCC cells. (A) WB results showed that knockdown of STOML2 obviously reduced p-Stat3 (Y705), but affected little on the level of p-Stat3 (S727). (B) Immunofluorescence staining indicated that STOML2 knockdown impeded the accumulation of nuclear p-Stat3. (C) The expression of STOML2 was assessed by western blotting after blocking Stat3 expression. (D) Real-time PCR analysis indicated that STOML2 depletion decreased IL6 mRNA expression. Data, mean ± SD, *P<0.05. (E and F) Western blotting assay revealed that IL6 induced the phosphorylation of Stat3 at Y705 residue (E) and STOML2 regulated the activation of Stat3 in an IL-6 dependent manner (F).
Figure 7.

Stat3 blocking attenuates proliferation, motility and chemo-resistance of HNSCC cells. (A and B) Transfection with si-Stat3 significantly impaired the capacities of cell proliferation (A) and colony formation (B) of SCC25 and SCC15 cells. (C) Stat3 depletion inhibited migration and invasion of HNSCC cells in vitro compared with negative control. Scale bar, 100 μm. (D and E) Stat3 knockdown increased the sensitivity of HNSCC cells to cisplatin and promoted apoptosis using flow cytometry (D) and colony formation assay (E). CDDP, cisplatin.
Discussion
Stomatin, firstly identified in human erythrocytes [22], is a plasma membrane protein found in various organisms ranging from mammals to bacteria. In mammals, Stomatin protein family consists of three members including STOML2, which is the only one lacking an NH2-terminal hydrophobic domain. Abundant evidence demonstrated that STOML2 is up-regulated in a variety of malignancies and predicts poor prognosis. Especially, previous study revealed that there is significant copy number amplification of STOML2 in progressing oral premalignant lesions [14]. We supposed that STOML2 may be fundamentally crucial in HNSCC tumorigenesis and be of great help for early detection of HNSCC. In addition, our finding showed that STOML2 expression was elevated in HNSCC tissues and correlated with T stage, lymph node metastasis and unfavorable outcome, which is consistent with previous studies. Therefore, STOML2 may serve as a novel prognostic biomarker in HNSCC, which provides a new promise to the strategy of diagnosis and therapy for HNSCC.
It has been shown that STOML2 knockdown could obviously reduce cell growth in endometrial adenocarcinoma [23] and lead to S phase arrest of ESCC cells [6]. In HNSCC, we reconfirmed that depletion of STOML2 resulted in cell cycle arrest at S phase and cell growth inhibition. For another, IHC analysis indicated that STOML2 expression was relative to lymph node metastasis in HNSCC, thereby we supposed whether STOML2 could play a role in motility of HNSCC cells. Subsequent experiments confirmed that reduced STOML2 hindered cell migration and invasion in HNSCC. It is worth noting that STOML2 level was significantly increased at the invasive front, which suggests that STOML2 may promote the survival of invasive cells and extracellular matrix (ECM) degradation. It is well known that Matrix Metalloproteinases (MMPs) play a critical role in ECM degradation and facilitate invasion-metastasis cascade. As one crucial member of MMPs family, MMP9 is up-regulated or super-activated in various cancer types [24,25]. According to database analysis and western blot results, we suggested that STOML2 silencing suppressed invasion of HNSCC cells through down-regulating the expression of MMP9. Besides, STOML2 correlated with high recurrence rate in HNSCC patients and a platinum-based regimen is the standard treatment for HNSCC [26]. So the influence of STOML2 on chemo-resistance of HNSCC cells was detected. Consequently, STOML2 deficiency increased sensitivity to cisplatin in HNSCC, in line with the conclusion reported in ESCC and cervical cancer. In our view, it is likely that silencing STOML2 reduced HNSCC malignancy through impairing not only mitochondrial function and ATP production, but activation of specific signaling pathway.
Recent studies has verified that abnormal activation of the Stat3 signaling pathway results in tumor growth, invasion, chemo-resistance, and recurrence [27,28]. In the CDDP-resistant HNSCC tissues and cells, Stat3 expression is dramatically elevated. While treated with specific inhibitor against Stat3, resistance to cisplatin is eliminated [29]. Moreover, Stat3 could be activated by IL6 and promote gene transcription including MMP9 [21,30]. In glioma, suppression of STOML2 reduces IL6 expression through inhibiting NF-κB transcription activity. Therefore, we assumed whether STOML2 could modulate Stat3 activation to regulate gene transcription and cancer progression. In this study, we confirmed that STOML2 depletion inhibited the activation and nuclear translocation of Stat3 in an IL6-dependent manner to attenuate tumor malignancy of HNSCC. On the contrary, there was no change in the expression of STOML2 under Stat3 inhibition, which suggested that STOML2 acted as an upstream regulator of Stat3. In the tumor microenvironment, IL6/Stat3 signaling also contributes to suppress the antitumor immune response [31]. Thus, targeting the STOML2-IL6-Stat3 pathway may provide therapeutic benefit by directly inhibiting tumor malignancy and by stimulating antitumor immunity.
In conclusion, our results for the first time demonstrate that STOML2 is up-regulated in HNSCC and modulate cell proliferation, migration, invasion and chemo-resistance via IL6-Stat3 signaling pathway. Furthermore, overexpression of STOML2 correlates positively with clinicopathologic characters, such as T stage, lymph node metastasis as well as recurrence, and indicates poor prognosis. These findings suggest the critical role of STOML2 in HNSCC malignant progression and provide STOML2 as an oncogene candidate for diagnosis and therapy of HNSCC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Posner M, Vermorken JB. Induction therapy in the modern era of combined-modality therapy for locally advanced head and neck cancer. Semin Oncol. 2008;35:221–8. doi: 10.1053/j.seminoncol.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Morrow JS. Identification and characterization of human SLP-2, a novel homologue of stomatin (band 7.2b) present in erythrocytes and other tissues. J Biol Chem. 2000;275:8062–71. doi: 10.1074/jbc.275.11.8062. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Cao W, Yu Z, Liu Z. Downregulation of a mitochondria associated protein SLP-2 inhibits tumor cell motility, proliferation and enhances cell sensitivity to chemotherapeutic reagents. Cancer Biol Ther. 2009;8:1651–8. doi: 10.4161/cbt.8.17.9283. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Ding F, Cao W, Liu Z, Liu W, Yu Z, Wu Y, Li W, Li Y, Liu Z. Stomatin-like protein 2 is overexpressed in cancer and involved in regulating cell growth and cell adhesion in human esophageal squamous cell carcinoma. Clin Cancer Res. 2006;12:1639–46. doi: 10.1158/1078-0432.CCR-05-1858. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Liu FJ. Expression of SLP-2 gene and CCBE1 are associated with prognosis of rectal cancer. Eur Rev Med Pharmacol Sci. 2017;21:1214–1218. [PubMed] [Google Scholar]
- 8.Sun F, Ding W, He JH, Wang XJ, Ma ZB, Li YF. Stomatin-like protein 2 is overexpressed in epithelial ovarian cancer and predicts poor patient survival. BMC Cancer. 2015;15:746. doi: 10.1186/s12885-015-1723-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao B, Xie Z, Guo L, Wu J, Zhang H. Stomatin-like protein 2 expression is associated with clinical survival in patients with cervical cancer. Int J Clin Exp Pathol. 2015;8:1804–9. [PMC free article] [PubMed] [Google Scholar]
- 10.Cao W, Zhang B, Li J, Liu Y, Liu Z, Sun B. SLP-2 overexpression could serve as a prognostic factor in node positive and HER2 negative breast cancer. Pathology. 2011;43:713–8. doi: 10.1097/PAT.0b013e32834c34ed. [DOI] [PubMed] [Google Scholar]
- 11.Liu D, Zhang L, Shen Z, Tan F, Hu Y, Yu J, Li G. Increased levels of SLP-2 correlate with poor prognosis in gastric cancer. Gastric Cancer. 2013;16:498–504. doi: 10.1007/s10120-013-0232-3. [DOI] [PubMed] [Google Scholar]
- 12.Cao W, Zhang B, Ding F, Zhang W, Sun B, Liu Z. Expression of SLP-2 was associated with invasion of esophageal squamous cell carcinoma. PLoS One. 2013;8:e63890. doi: 10.1371/journal.pone.0063890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu G, Zhang J, Xu F, Deng H, Zhang W, Kang S, Liang W. Stomatin-like protein 2 inhibits cisplatin-induced apoptosis through MEK/ERK signaling and the mitochondrial apoptosis pathway in cervical cancer cells. Cancer Sci. 2018;109:1357–1368. doi: 10.1111/cas.13563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towle R, Tsui IF, Zhu Y, MacLellan S, Poh CF, Garnis C. Recurring DNA copy number gain at chromosome 9p13 plays a role in the activation of multiple candidate oncogenes in progressing oral premalignant lesions. Cancer Med. 2014;3:1170–84. doi: 10.1002/cam4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan Z, Foster R, Bell DA, Mahoney J, Wolak K, Vaidya A, Hampel C, Lee H, Seiden MV. Signal transducers and activators of transcription 3 pathway activation in drug-resistant ovarian cancer. Clin Cancer Res. 2006;12:5055–63. doi: 10.1158/1078-0432.CCR-06-0861. [DOI] [PubMed] [Google Scholar]
- 16.Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review) Int J Oncol. 2014;44:1032–40. doi: 10.3892/ijo.2014.2259. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Ren Y, Liu A, Han L, Zhang K, Li S, Li P, Li P, Kang C, Wang X, Zhang L. STAT3 inhibitor WP1066 attenuates miRNA-21 to suppress human oral squamous cell carcinoma growth in vitro and in vivo. Oncol Rep. 2014;31:2173–80. doi: 10.3892/or.2014.3114. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song L, Liu L, Wu Z, Lin C, Dai T, Yu C, Wang X, Wu J, Li M, Li J. Knockdown of stomatin-like protein 2 (STOML2) reduces the invasive ability of glioma cells through inhibition of the NF-kappaB/MMP-9 pathway. J Pathol. 2012;226:534–43. doi: 10.1002/path.3008. [DOI] [PubMed] [Google Scholar]
- 21.Dechow TN, Pedranzini L, Leitch A, Leslie K, Gerald WL, Linkov I, Bromberg JF. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci U S A. 2004;101:10602–7. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart GW, Hepworth-Jones BE, Keen JN, Dash BC, Argent AC, Casimir CM. Isolation of cDNA coding for an ubiquitous membrane protein deficient in high Na+, low K+ stomatocytic erythrocytes. Blood. 1992;79:1593–601. [PubMed] [Google Scholar]
- 23.Cui Z, Zhang L, Hua Z, Cao W, Feng W, Liu Z. Stomatin-like protein 2 is overexpressed and related to cell growth in human endometrial adenocarcinoma. Oncol Rep. 2007;17:829–33. [PubMed] [Google Scholar]
- 24.Deryugina EI, Quigley JP. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006;25:9–34. doi: 10.1007/s10555-006-7886-9. [DOI] [PubMed] [Google Scholar]
- 25.Turpeenniemi-Hujanen T. Gelatinases (MMP-2 and -9) and their natural inhibitors as prognostic indicators in solid cancers. Biochimie. 2005;87:287–97. doi: 10.1016/j.biochi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, Peyrade F, Benasso M, Vynnychenko I, De Raucourt D, Bokemeyer C, Schueler A, Amellal N, Hitt R. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–27. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 27.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–60. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozovski U, Calin GA, Setoyama T, D’Abundo L, Harris DM, Li P, Liu Z, Grgurevic S, Ferrajoli A, Faderl S, Burger JA, O’Brien S, Wierda WG, Keating MJ, Estrov Z. Signal transducer and activator of transcription (STAT)-3 regulates microRNA gene expression in chronic lymphocytic leukemia cells. Mol Cancer. 2013;12:50. doi: 10.1186/1476-4598-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou X, Ren Y, Liu A, Jin R, Jiang Q, Huang Y, Kong L, Wang X, Zhang L. WP1066 sensitizes oral squamous cell carcinoma cells to cisplatin by targeting STAT3/miR-21 axis. Sci Rep. 2014;4:7461. doi: 10.1038/srep07461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawakita A, Yanamoto S, Yamada S, Naruse T, Takahashi H, Kawasaki G, Umeda M. MicroRNA-21 promotes oral cancer invasion via the Wnt/beta-catenin pathway by targeting DKK2. Pathol Oncol Res. 2014;20:253–61. doi: 10.1007/s12253-013-9689-y. [DOI] [PubMed] [Google Scholar]
- 31.Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15:234–248. doi: 10.1038/nrclinonc.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]