Graphical abstract
Keywords: Lipid accumulation, Metabolic disorder, siRNA, Non-alcoholic fatty liver disease, Fibrosis, Liver tumour
Highlights
-
•
Repin1 is a potential target gene for the treatment of obesity.
-
•
The effects of hepatic Repin1 deletion were evaluated in a mouse model of progressive NAFLD.
-
•
Liver-specific Repin1 knockout alleviated systemic and hepatic lipid accumulation.
-
•
Liver injury, lw/bw index and tumour load were reduced in LRep1−/− STZ/HFD mice.
-
•
A therapeutic approach using Repin1 siRNA had beneficial effects on early NAFLD.
Abstract
There is an increasing prevalence of obesity and metabolic syndrome, which promote the development of non–alcoholic fatty liver disease (NAFLD), a disease that can evolve into cirrhosis and hepatocellular carcinoma. Repin1 loss was previously shown to have beneficial effects on lipid and glucose metabolism and obesity regulation. Herein, we characterized NAFLD in mice with hepatic deletion of Repin1 (LRep1−/−). For this purpose, liver disease was analysed in male LRep1−/− and wild-type mice treated with streptozotocin/high fat diet or a control diet over a period of 20 wks. Streptozotocin/high fat diet treated LRep1−/− mice showed a significant decrease in systemic and hepatic lipid accumulation, accompanied by diminished chronic inflammation and a subsequent reduction in liver injury. Remarkably, Repin1-deficient mice exhibited a lower tumour prevalence and tumour frequency, as well as a reduced liver weight/body weight index. A therapeutic approach using Repin1 siRNA in the early phase of NAFLD verified the observed beneficial effects of Repin1 deficiency. This study provides evidence that loss of Repin1 in the liver attenuates NAFLD progression, most likely by reducing fat accumulation and alleviating chronic tissue inflammation. Thus, modulating Repin1 expression may become a novel strategy and potential tool to inhibit NAFLD progression.
Introduction
The global problem of obesity and consequent chronic hepatic fat deposition is one of the main contributors to the increasing development of non-alcoholic fatty liver disease (NAFLD). With a worldwide prevalence of approximately 25% [1], [2], NAFLD is becoming an increasingly important risk factor for the development of more severe forms of chronic liver disease, such as cirrhosis and hepatocellular carcinoma (HCC) [1], [2]. Within this complex and multifactorial disease, excessive hepatic fat accumulation is the most striking feature [3]. Exploring specific pathways and factors that regulate and modulate metabolic dysfunction can lead to the early diagnosis of NAFLD and the development of novel therapies to reduce its progression. In this context, Repin1 was identified as a candidate gene in the development of metabolic disorders because of its crucial effects on lipid and glucose metabolism [4], [5], [6]. In human subjects, increased Repin1 mRNA expression in visceral and subcutaneous adipose tissue was confirmed to significantly correlate with total body fat mass and adipocyte size [7]. In line with these studies, Repin1 expression was also related to determinants of human obesity and insulin resistance [7], [8], [9]. Repin1 is a ubiquitously expressed zinc finger protein, with the highest levels found in adipose and liver tissue [5]. In addition to its important roles in adipose tissue development, fat mass, adipocyte size and lipid storage [7], [8], [10], [11], Repin1 was recently shown to be involved in hepatic lipid transport and storage [10]. There is evidence of beneficial metabolic effects of Repin1 deficiency in the liver due to the regulated expression of key glucose and lipid metabolism genes [10]. Thus, among other findings, it was observed that hepatic Repin1 deletion leads to lower body weight, improved insulin sensitivity and reduced body and liver fat [10].
Understanding the features of Repin1 in liver disease may offer new therapeutic approaches to metabolic-related disorders. Given the observed positive effects on metabolism of liver-specific Repin1 deficiency, particularly with respect to lipid metabolism, we analysed the consequences of hepatic Repin1 deletion on NAFLD progression in mice. Therapeutic strategies to reduce Repin1 action in liver tissue may have potential in tackling NAFLD and its progression.
Experimental
Mice
Male mice with hepatocyte-restricted, Cre-loxP-mediated Repin1 deletion (LRep1AlbCre; LRep1−/−) and wild-type (Wt) littermates (background C57BL/6N) [10] were housed on a 12 h/12 h day/night cycle with ad libitum access to water and standard laboratory chow or high fat diet (HFD). Experiments were approved by the local Landesamt für Landwirtschaft, Lebensmittelsicherheit und Fischerei Mecklenburg-Vorpommern (LALLF M-V/TSD/7221.3-1.1-039/14) and conducted in accordance with the German legislation on the protection of animals and EU-directive 2010/63/EU.
Mouse model and experimental groups
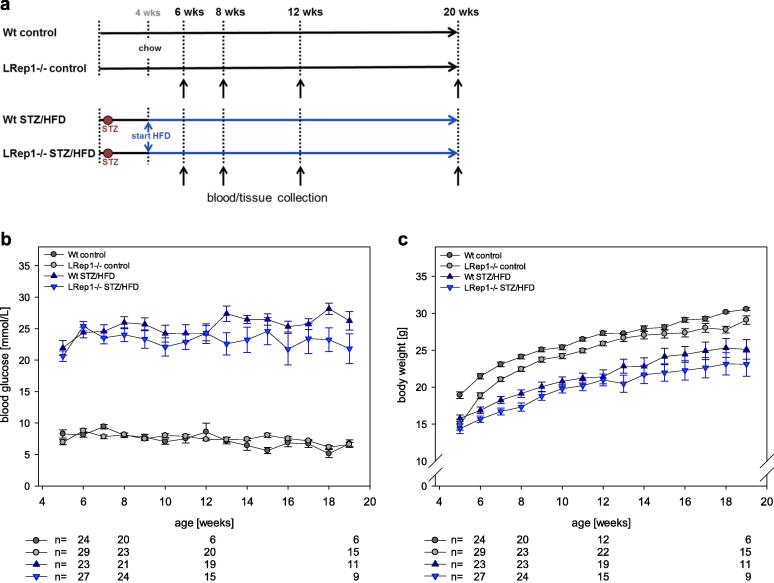
For the induction of progressive NAFLD in LRep1−/− and Wt mice by streptozotocin (STZ) and a HFD (LRep1−/− STZ/HFD and Wt STZ/HFD) (Fig. 1a), we used the non-alcoholic steatohepatitis (NASH)-fibrosis-tumour model [12] as described previously by our group [13]. Over the course of 20 wks, the mice developed progressive NAFLD with steatosis (6 wks), steatohepatitis (8 wks), fibrosis (12 wks) and liver tumours (20 wks). Standard chow-fed LRep1−/− and Wt mice without STZ application served as healthy controls (LRep1−/− control and Wt control) (Fig. 1a). General health was monitored daily, and blood glucose levels and body weight were measured weekly. STZ/HFD animals exhibiting normal blood glucose levels were excluded from the experiment. Under ketamine/xylazine anaesthesia (90/7 mg/kg bw, ip), mice were sacrificed at the final observation time points of 6, 8, 12, and 20 wks (7–15 animals per time point and group). Blood was collected, and the livers were excised, weighed and processed for subsequent analysis.
Fig. 1.
(a) Experimental design of Repin1-deficient and wild-type (Wt) mice treated with or without STZ/HFD. (b) Blood glucose levels and (c) body weight of Wt (dark grey) and LRep1−/− control mice (light grey) and of STZ/HFD-treated Wt (dark blue) and LRep1−/− mice (light blue) at 5 to 19 wks of age (mean ± SEM). The sample sizes at 6, 8, 12 and 19 wks are indicated below the graph. Differences between STZ/HFD and control groups in blood glucose and body weight (Kruskal-Wallis and pairwise Mann-Whitney tests) were statistically significant in Wt mice and LRep1−/− mice, as expected (P < 0.001). Further significant differences were observed between both control groups (P = 0.006, P = 0.002) and between Wt STZ/HFD and LRep1−/− STZ/HFD mice in body weight (P = 0.001) but not in blood glucose (P = 0.014) (all with αadj = 0.008). Blood glucose remained stable over time (P = 0.449), but body weight increased over time (P < 0.001).
Repin1 siRNA experiments
Repin1 siRNA (siRep1) and non-targeting control siLuciferase (siLuci) were formulated in lipid nanoparticles (LNPs) prepared by Silence Therapeutics GmbH essentially as described [14], [15]. siRNA-LNPs were synthesized by mixing an siRNA-containing aqueous phase with a lipid-containing ethanol phase at a 3:1 vol ratio in a microfluidic chip device using syringe pumps as previously described [16] to a final concentration of 0.12 mg/ml. The resultant siRNA-LNPs were dialysed overnight in a 10,000 molecular weight cut-off (MWCO) cassette against 270 mmol/l sucrose at 4 °C and stored at −80 °C until use. The siRNA-LNPs had a mean diameter of ∼120–150 nm, with a polydispersity index between 0.1 and 0.3 as measured by dynamic light scattering (ZetaSizer, Malvern Instruments, Malvern, Worcestershire, UK). The siRNA encapsulation efficiency of greater than 90% was determined using a Quant-iT Ribogreen Assay (Invitrogen, ThermoFisher Scientific, Waltham, Massachusetts, USA). Simultaneous with the start of HFD feeding (4 wks of age), Wt mice were injected intravenously under inhalation anaesthesia (1.5 vol% isoflurane) via the jugular vein with siRep1 (2.063 mg/kg bw; 0.05 ml/10 g bw; n = 7) every 72 h for 14 days. Control animals received equivalent volumes of siLuci (n = 7) or sucrose buffer (n = 6, buffer). Mice were sacrificed after 14 days of siRNA and HFD treatment at an age of 6 wks. In the siRNA experiments, the following parameters were assessed as described in the appropriate methods section: plasma glutamate dehydrogenase (GLDH); mRNA expression of Repin1, F4/80 and collagen 1α; CAE-positive cells; and F4/80- and Sirius Red-positive area.
Haematological measurements and plasma analyses
Blood samples were collected biweekly and at the final time points by retrobulbar sinus puncture. Red blood cell, white blood cell and blood platelet counts, as well as haemoglobin and haematocrit, were assessed with an automated cell counter (Sysmex KX-21, Sysmex Deutschland GmbH, Norderstedt, Germany). The activities of alanine aminotransferase (ALT) and GLDH in ethylenediaminetetraacetic acid (EDTA)-treated plasma were measured spectrophotometrically as indicators of hepatocellular disintegration and necrosis using a cobas c 111 analyser (Roche Diagnostics, Rotkreuz, Switzerland) according to the manufacturer’s instructions. Plasma triglycerides were measured using a triglyceride assay kit (10010303, Cayman Chemical Company, Ann Arbor, Michigan, USA) according to the manufacturer’s instructions.
Histopathology/cell staining
Lipid accumulation was visualized and quantified by Oil Red O staining in 8 µm thick frozen liver sections as described previously [13]. Additionally, liver tissue was fixed in 4% phosphate-buffered formaldehyde for two to three days, embedded in paraffin, and cut into 5 µm thick sections. Haematoxylin and eosin (H&E) staining was used to determine the NAFLD activity score (NAS) as described previously by our group [13]. All scores were determined by three to four independent observers in a blinded manner. For the tumour analysis, photomicrographs of one H&E-stained liver section per animal were taken using a 1.25× objective. The micrographs were then combined with a picture of the whole liver section. The number of neoplastic foci was counted, and the areas of neoplastic foci and the whole section were selected and measured using ImageJ 1.47v (Wayne Rasband, National Institutes of Health, USA). All histological staining was assessed in a blinded manner. For the assessment of granulocyte infiltration of tissues, paraffin-embedded liver tissue sections were stained for chloroacetate esterase (CAE) with naphthol AS-D chloroacetate (Sigma-Aldrich, St. Louis, Missouri, USA) and quantified as described by Liebig et al. 2018 [13]. Sirius Red staining was used to quantify collagen deposition. Using a 20× objective and a polarizing filter, 20 consecutive photomicrographs per liver section (one section per animal) were taken, and the Sirius Red-stained area was quantified using Adobe Photoshop CS5 12.0.4 (Adobe, San José, California, USA). Digital images were taken with a Color View II FW camera (Color 10 View, Munich, Germany). Immunohistochemical staining for F4/80 was conducted as described previously by our group [13]. For the staining analysis, at least 30 consecutive photomicrographs were taken per section (one section per animal) using a 40× objective. The red F4/80-positive area was quantified using the colour threshold tool in ImageJ 1.47v.
Conventional RT-PCR analysis
Total RNA was isolated from snap frozen liver tissue lysates or cell lysates using an RNeasy Mini Kit including on-column genomic DNA digestion with an RNase-free DNase Set (Qiagen, Hilden, Germany). Two micrograms of RNA was reverse transcribed to cDNA using oligo(dT)18 primers (New England Biolabs GmbH, Ipswich, Massachusetts, USA) and Superscript II RNase H- Reverse Transcriptase (Invitrogen, ThermoFisher Scientific, Waltham, Massachusetts, USA). Real-time PCR gene expression analysis was performed using SYBR Green I (Roche, Mannheim, Germany) for the detection of amplified dsDNA strands on the LightCycler 1.5 system (Roche, Basel, Switzerland) with the following primers: Cd36: for 5′-GGTGATGTTTGTTGCTTTTATGATTTC-3′, rev 5′-GTAGATCGGCTTTACCAAAGATG-3′; collagen 1α: for 5′-TGGACCTCCGGCTCCTGCTC-3′, rev 5′-TCGCACACAGCCGTGCCATT-3′, Rps18: for 5′-AGGATGTGAAGGATGGGAAG-3′, rev 5′-TTGGATACACCCACAGTTCG-3′; Repin1: for 5′-GCCTTCTGTTGTGCCATCTGT-3′, rev 5′-TCTCAGGCATCGTGCTTCTTCC-3′; Gapdh: 5′-GAATTTGCCGTGAGTGGAGT-3′, rev 5′-CGTCCCGTAGACAAAATGGT-3′; and F4/80: for 5′-CTTTGGCTATGGGCTTCCAGTC-3′, rev 5′-GCAAGGAGGACAGAGTTTATCGTG-3′. All data were calculated by using the comparative ddCt method, and expression values were normalized to those of the housekeeping gene Rps18 or Gapdh. Target gene expression was compared to a Wt C57BL/6N liver tissue pool.
Electron microscopy
For fixation, small blocks of liver tissue were immersed in a fixative containing 2% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.3. Prior to resin embedding for transmission electron microscopy (TEM), samples were washed three times with 0.1 M sodium phosphate buffer and then postfixed with a solution of 1% aqueous osmium tetroxide for 1 h. After being washed in distilled water, the tissue blocks were dehydrated in a graded series of acetone, finishing with two steps in pure acetone. Next, the specimens were infiltrated overnight with epoxy resin (Epon 812, Serva, Heidelberg, Germany), starting with a 1:1 mixture of acetone and resin and continuing with pure resin for 4 h, and then were transferred to rubber moulds on the following day. After curing the resin at 60 °C for 2 days, semithin sections (0.5 µm) and thin sections (50–70 nm) were cut on an ultramicrotome (Ultracut E, Reichert&Jung, Wien, Austria) using diamond knifes (Diatome, Biel, Switzerland). Semithin sections were stained with toluidine blue to visualize the tissue structure for further morphometric measurements (see below). Thin sections for ultrastructural inspection were cut from these areas, transferred to 300 mesh copper grids and stained with lead citrate and uranyl acetate. The grids were examined using a Zeiss EM902 electron microscope (Carl Zeiss, Oberkochen, Germany) operated at 80 kV. Digital images were acquired with a side-mounted 1 × 2k FT-CCD Camera (Proscan, Scheuring, Germany) using iTEM camera control and imaging software (Olympus-SIS, Münster, Germany) with calibrated morphometric measurement tools to determine lipid droplet diameter, area and number. For the morphometric analysis, approximately 15 consecutive photomicrographs per section (one section per animal) were taken and analysed in a blinded manner.
Statistical analysis
The results are presented as the mean ± standard error of the mean (SEM.) or as box plots of the distribution, indicating the median, the interquartile range (box), and the minimum and maximum (whiskers). The influence of time in dependent samples was assessed by the Friedman test, and group differences were assessed by Kruskal-Wallis test as a non-parametric analysis of variance, followed by the pairwise Mann-Whitney comparison with alpha adjustment (Bonferroni αadj; Fig. 1). Differences among independent samples in the four groups and at different time points (Repin1 knockout experiment) were assessed by two-way ANOVA, followed by the Holm-Sidak post hoc comparison. For reasons of clarity, only statistically significant differences between groups at each time point are indicated in the figures. Group differences were assessed using the t-test for electron microscopy data (Fig. 3b–e) and the Mann-Whitney rank sum test for tumour data (Fig. 5b and c). The distribution of tumour prevalence among groups was tested by the chi square test. Differences among groups concerning siRNA data (Fig. 6) were analysed using one-way ANOVA (followed by the Holm-Sidak method) or, in cases of non-normality, using one-way ANOVA on ranks (followed by Dunn’s method). Kaplan-Meier survival curves were compared using the log-rank test (αadj is given). The hazard ratio (HR) of group comparisons and the respective 95% confidence interval (CI) are reported. The data are presented as the mean ± SEM (LRep1−/− experiment) or the minimum, quartiles and maximum (categorical data; siRNA experiment) for each parameter and group, along with the sample size, in Table 1 (Repin1 knockout experiment) and Table 2 (siRNA experiment).
Fig. 3.
Quantitative analysis of the (a) proportion (grouped according to diameter), (b) diameter, (c) size, (d) frequency and (e) total area of lipid droplets in hepatocytes of STZ/HFD-treated wild-type (Wt) and LRep1−/− mice at 8 and 12 wks by means of TEM. Values are presented as the mean ± SEM. Differences between groups (b–e) were evaluated by t-tests. (f) Representative TEM images (scale bar, 5 µm; lipid droplets are marked by arrowheads). (g) Hepatic Cd36 mRNA expression in 6-, 8-, 12-, and 20-wk-old control and STZ/HFD-treated Wt and LRep1−/− mice. Values are presented as the mean ± SEM. Group differences were evaluated by two-way ANOVA with the Holm-Sidak post hoc test.
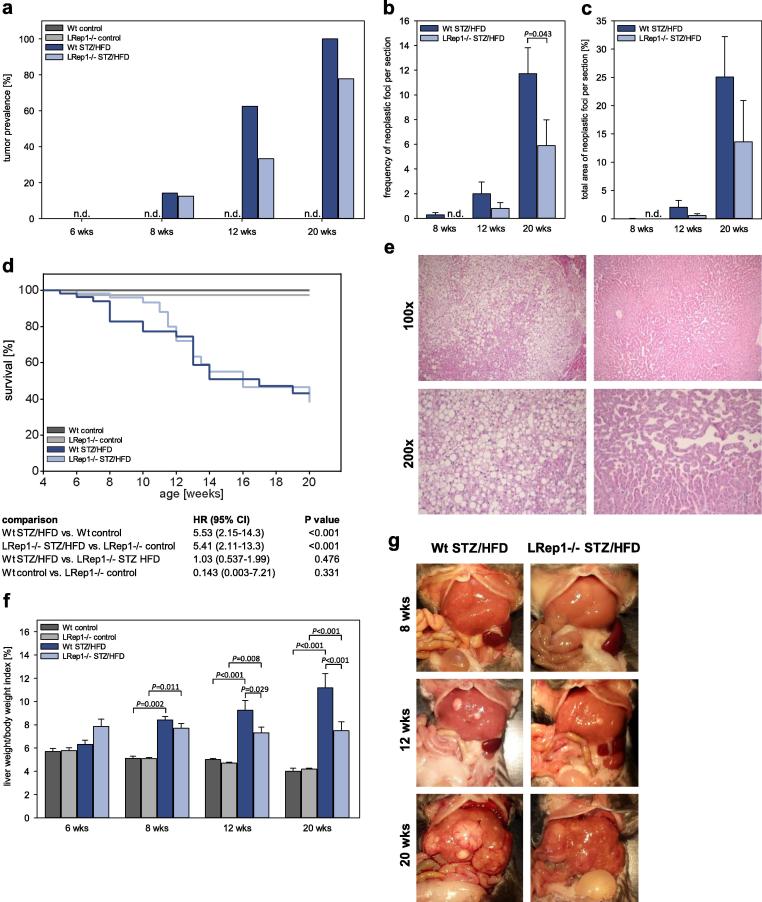
Fig. 5.
(a) Tumour prevalence at 6, 8, 12, and 20 wks, (b) frequency of neoplastic foci per section and (c) total area of neoplastic foci per section in control and STZ/HFD-treated wild-type (Wt) and LRep1−/− mice; n.d., not detectable. (d) Kaplan-Meier survival curves of control and STZ/HFD-treated Wt and LRep1−/− mice over the period of 20 wks. (e) Representative photomicrographs of tumours in H&E-stained liver sections from Wt STZ/HFD mice (100× and 200× magnification). (f) Calculation of the liver to body weight ratio. No differences in tumour prevalence (a) at different time points (chi square test). Values (b, c, f) are presented as the mean ± SEM. Group differences were tested by the Mann-Whitney rank sum test (b, c) or by two-way ANOVA with the Holm-Sidak post hoc test (f). Log-rank tests for the similarity of survival curves with αadj = 0.008 (d). (g) Macroscopic images of livers from control and STZ/HFD-treated Wt and LRep1−/− mice at 8, 12, and 20 wks of age.
Fig. 6.
Therapeutic Repin1 siRNA treatment diminished liver injury and inflammation in 6-wk-old STZ/HFD mice. (a) Experimental design of buffer/siRNA-treated STZ/HFD wild-type (Wt) mice. Repeated injections with buffer or siRNA are marked with arrows (every 72 h). Hepatic mRNA expression of (b) Repin1, (c) F4/80 and (h) collagen 1α in 6-wk-old STZ/HFD Wt mice treated with buffer or siRNA specific for luciferase (siLuci) or Repin1 (siRep1) for 14 days. Histomorphometric quantification of liver sections stained for (d) F4/80, (f) CAE and (g) Sirius Red. (e) Representative images of F4/80-stained liver tissue from 6-wk-old siLuci- or siRep1-treated STZ/HFD Wt mice (macrophages, red; 400× magnification). (i) Analysis of glutamate dehydrogenase (GLDH) in the plasma of these mice. Data are presented as box plots (b–i, not e). Group differences were tested by one-way ANOVA (with the Holm-Sidak test; b, f and g) or one-way ANOVA on ranks (followed by Dunn’s method; c, d, h and i), depending on the data distribution.
Table 1.
Number of animals and data analysis for Repin1-deficient (LRep1−/−) and wild-type (Wt) mice treated with or without STZ/HFD.
| Parameter | 6 wks |
8 wks |
12 wks |
20 wks |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wt control | LRep1−/− control | Wt STZ/HFD | LRep1−/− STZ/HFD | Wt control | LRep1−/− control | Wt STZ/HFD | LRep1−/− STZ/HFD | Wt control | LRep1−/− control | Wt STZ/HFD | LRep1−/− STZ/HFD | Wt control | LRep1−/− control | Wt STZ/HFD | LRep1−/− STZ/HFD | ||
| Total number of mice | n | 7 | 7 | 8 | 8 | 7 | 7 | 7 | 8 | 12 | 7 | 8 | 10 | 9 | 15 | 11 | 9 |
| Plasma triglycerides [mg/dL] | Mean ± | 29.2± | 36.6± | 132 ± | 258± | 11.7± | 7 | 7 | 7 | 11 | 7 | 6 | 10 | 4 | 15 | 10 | 8 |
| SEM | 3.75 | 2.43 | 17.5 | 63.6 | 3.04 | 58.4± | 503 ± | 280± | 22.4± | 66.6± | 406± | 316± | 8.56± | 39.6± | 126 ± | 96.1± | |
| n | 6 | 7 | 7 | 8 | 5 | 4.72 | 94.6 | 85.9 | 4.23 | 8.02 | 155 | 75.8 | 2.07 | 3.18 | 17.4 | 28.8 | |
| Oil Red O-positive area [%] | Mean ± | 0 | 3.85± | 24.5± | 25.7± | 18.5± | 7 | 7 | 8 | 11 | 7 | 8 | 10 | 6 | 15 | 11 | 8 |
| SEM | 0 | 1.93 | 3.49 | 4.65 | 4.98 | 14.0± | 42.2± | 26.0± | 2.97± | 0.877± | 29.3± | 23.5± | 5.26± | 4.20± | 26.2± | 23.9± | |
| n | 7 | 7 | 8 | 8 | 7 | 6.20 | 5.99 | 3.21 | 1.50 | 0.651 | 5.03 | 5.60 | 2.90 | 1.30 | 4.34 | 4.71 | |
| Lipid droplet diameter [µm] | Mean ± | – | – | – | – | – | – | 4 | 4 | – | – | 3 | 3 | – | – | – | – |
| SEM | – | 2.44± | 2.57± | – | – | 2.48± | 2.62± | – | – | – | – | ||||||
| n | – | – | – | – | – | 0.0749 | 0.247 | 0.326 | 0.0847 | ||||||||
| Lipid droplet size [µm2] | Mean ± | – | – | – | – | – | – | 4 | 4 | – | – | 3 | 3 | – | – | – | – |
| SEM | – | 7.15± | 7.76± | – | – | 7.73± | 8.41± | – | – | – | – | ||||||
| n | – | – | – | – | – | 0.392 | 1.54 | 1.72 | 0.847 | ||||||||
| Number of lipid droplets/image | Mean ± | – | – | – | – | – | – | 4 | 4 | – | – | 3 | 3 | – | – | – | – |
| SEM | – | 44.1± | 35.1± | – | – | 68.1± | 24.8± | – | – | – | – | ||||||
| n | – | – | – | – | – | 1.46 | 5.09 | 14.8 | 1.74 | ||||||||
| Total lipid droplet area/image [µm2] | Mean ± | – | – | – | – | – | – | 4 | 4 | – | – | 3 | 3 | – | – | – | – |
| SEM | – | 279± | 249± | – | – | 447± | 193± | – | – | – | – | ||||||
| n | – | – | – | – | – | 32.0 | 28.8 | 34.5 | 11.3 | ||||||||
| mRNA expression [Cd36/Rps18] | Mean ± | 1.08± | 1.70± | 5.54± | 3.83± | 0.984± | 7 | 7 | 8 | 8 | 7 | 8 | 8 | 6 | 8 | 8 | 7 |
| SEM. | 0.186 | 0.398 | 0.917 | 0.622 | 0.144 | 1.66± | 11.9± | 4.80± | 1.48± | 0.923± | 7.84± | 5.59± | 2.39± | 1.37± | 15.8± | 4.23± | |
| n | 7 | 7 | 7 | 8 | 7 | 0.515 | 1.23 | 1.32 | 0.501 | 0.198 | 2.00 | 1.67 | 0.725 | 0.317 | 4.96 | 1.15 | |
| ALT [U/L] | Mean ± | 59.2± | 50. 7± | 107± | 118 ± | 40.7± | 6 | 6 | 8 | 10 | 6 | 6 | 9 | 9 | 15 | 11 | 8 |
| SEM | 5.26 | 11.9 | 29.8 | 16.6 | 2.69 | 56.3± | 103± | 95.6± | 38.1± | 50.7± | 176± | 55.9± | 97.1± | 87.8± | 169± | 100± | |
| n | 6 | 6 | 7 | 8 | 6 | 5.92 | 6.38 | 6.70 | 8.65 | 4.06 | 65.7 | 8.63 | 26.4 | 14.1 | 30.8 | 23.2 | |
| GLDH [U/l] | Mean ± | 14± | 32.3± | 88.6± | 149 ± | 7.17± | 7 | 7 | 7 | 10 | 7 | 5 | 9 | 9 | 14 | 11 | 8 |
| SEM | 1.66 | 17.0 | 33.9 | 46.8 | 0.543 | 40.0± | 95.3± | 65.7± | 15.1± | 14.0± | 83.8± | 28.6± | 45.9± | 18.9± | 60.6± | 37.6± | |
| n | 7 | 6 | 7 | 8 | 6 | 15.8 | 11.7 | 5.01 | 3.41 | 6.60 | 46.4 | 7.71 | 16.1 | 2.60 | 8.43 | 7.69 | |
| NAFLD activity score (NAS) | Mean ± | 1.33± | 2.48± | 4.50± | 3.75± | 1.57± | 7 | 7 | 8 | 12 | 7 | 8 | 10 | 8 | 15 | 10 | 9 |
| SEM. | 0.44 | 0.46 | 0.624 | 0.435 | 0.422 | 2.14± | 4.24± | 4.29± | 2.14± | 1.05± | 5.79± | 4.07± | 1.22± | 2.00± | 5.93± | 5.07± | |
| n | 7 | 7 | 8 | 8 | 7 | 0.453 | 0.339 | 0.222 | 0.409 | 0.321 | 0.735 | 0.391 | 0.698 | 0.272 | 0.139 | 0.247 | |
| NAS subscore steatosis | Mean ± | 0.333± | 0.476± | 1.46± | 1.00± | 0.571± | 7 | 7 | 8 | 12 | 7 | 8 | 10 | 8 | 15 | 11 | 9 |
| SEM | 0.126 | 0.143 | 0.274 | 0.167 | 0.095 | 0.429± | 1.10± | 1.17± | 0.361± | 0.095± | 2.13± | 1.37± | 0.258± | 0.711± | 2.03± | 1.63± | |
| n | 7 | 7 | 8 | 8 | 7 | 0.158 | 0.061 | 0.089 | 0.112 | 0.061 | 0.333 | 0.144 | 0.183 | 0.184 | 0.145 | 0.171 | |
| NAS subscore inflammation | Mean ± | 0.476± | 0.714± | 1.58± | 1.17± | 0.111± | 7 | 7 | 8 | 12 | 7 | 8 | 10 | 8 | 15 | 11 | 9 |
| SEM | 0.123 | 0.198 | 0.258 | 0.178 | 0.070 | 0.762± | 1.38± | 1.50± | 0.667± | 0.381± | 1.833± | 1.30± | 0.688± | 0.644± | 1.67± | 1.79± | |
| n | 7 | 7 | 8 | 8 | 7 | 0.158 | 0.184 | 0.089 | 0.149 | 0.135 | 0.252 | 0.136 | 0.353 | 0.110 | 0.127 | 0.061 | |
| NAS subscore ballooning | Mean ± | 0.524± | 1.24± | 1.46± | 1.58± | 0.714± | 7 | 7 | 8 | 12 | 7 | 8 | 10 | 8 | 15 | 10 | 9 |
| SEM. | 0.228 | 0.288 | 0.188 | 0.137 | 0.246 | 0.952± | 1.76± | 1.63± | 0.917± | 0.571± | 2.05± | 1.40± | 0.333± | 0.644± | 2.07± | 1.78± | |
| n | 7 | 7 | 8 | 8 | 7 | 0.223 | 0.174 | 0.117 | 0.170 | 0.215 | 0.135 | 0.178 | 0.227 | 0.124 | 0.067 | 0.056 | |
| CAE-positive cells [n/HPF] | Mean ± | 5.52± | 1.74± | 5.04± | 4.097± | 2.25± | 7 | 7 | 7 | 12 | 7 | 8 | 10 | 9 | 15 | 11 | 9 |
| SEM. | 2.01 | 0.533 | 0.607 | 0.517 | 0.712 | 1.90± | 5.97± | 2.86± | 4.17± | 1.93± | 15.1± | 8.34± | 5.38± | 5.41± | 4.99± | 5.17± | |
| n | 7 | 7 | 7 | 7 | 7 | 0.598 | 1.50 | 0.807 | 1.03 | 0.667 | 4.22 | 2.66 | 1.60 | 0.730 | 0.686 | 0.929 | |
| Liver weight/body weight index [%] | Mean ± | 5.70± | 5.78± | 6.31± | 7.86± | 5.12± | 6 | 7 | 8 | 11 | 6 | 8 | 10 | 8 | 15 | 11 | 8 |
| SEM. | 0.272 | 0.252 | 0.367 | 0.619 | 0.181 | 5.09± | 8.42± | 7.70± | 5.01± | 4.71± | 9.24± | 7.31± | 4.01± | 4.19± | 11.2± | 7.49± | |
| n | 6 | 7 | 8 | 8 | 7 | 0.0946 | 0.295 | 0.402 | 0.106 | 0.082 | 0.853 | 0.485 | 0.272 | 0.087 | 1.23 | 0.763 | |
| Total area of neoplastic foci per section [%] | Mean ± | – | – | – | – | – | – | 7 | 7 | – | – | 8 | 10 | – | – | 11 | 9 |
| SEM | – | 0.031± | 0± | – | – | 2.03± | 0.560± | – | – | 25.1± | 13.6± | ||||||
| n | – | – | – | – | – | 0.019 | 0 | 1.21 | 0.318 | 7.16 | 7.29 | ||||||
| Number of neoplastic foci per section | Mean ± | – | – | – | – | – | – | 7 | 7 | – | – | 8 | 10 | – | – | 11 | 9 |
| SEM | – | 0.286 | 0 | – | – | 2.00± | 0.80± | – | – | 11.7± | 5.89± | ||||||
| n | – | – | – | – | – | 0.171 | 0 | 0.952 | 0.465 | 2.11 | 2.09 | ||||||
Table 2.
Number of animals and data analysis for the siRNA experiment.
| Parameter | Wt control | LRep1−/− control | LRep1−/− STZ/HFD | |
|---|---|---|---|---|
| Total number of mice | n | 6 | 7 | 7 |
| mRNA expression [Repin1/Gapdh] | Minimum | 1.30 | 0.684 | 0.400 |
| 25% percentile | 1.30 | 0.765 | 0.679 | |
| Median | 1.42 | 3.30 | 0.770 | |
| 75% percentile | 2.47 | 3.68 | 1.02 | |
| Maximum | 3.44 | 7.92 | 1.36 | |
| n | 6 | 7 | 7 | |
| F4/80-positive area [%] | Minimum | 0.550 | 0.490 | 0.160 |
| 25% percentile | 0.573 | 0.540 | 0.170 | |
| Median | 0.645 | 0.620 | 0.220 | |
| 75% percentile | 0.773 | 0.760 | 0.360 | |
| Maximum | 0.930 | 0.770 | 0.460 | |
| n | 6 | 7 | 7 | |
| mRNA expression [F4/80/Gapdh] | Minimum | 1.42 | 1.84 | 0.462 |
| 25% percentile | 1.46 | 1.98 | 0.499 | |
| Median | 1.78 | 2.45 | 0.742 | |
| 75% percentile | 2.33 | 3.61 | 1.02 | |
| Maximum | 2.64 | 3.83 | 1.05 | |
| n | 5 | 7 | 7 | |
| CAE-positive cells [n/HPF] | Minimum | 5.78 | 5.63 | 4.03 |
| 25% percentile | 5.89 | 5.93 | 4.10 | |
| Median | 6.03 | 7.47 | 4.70 | |
| 75% percentile | 8.03 | 8.13 | 5.40 | |
| Maximum | 8.80 | 9.07 | 6.13 | |
| n | 6 | 7 | 7 | |
| Sirius Red-positive area [%] | Minimum | 0.777 | 0.555 | 0.525 |
| 25% percentile | 0.842 | 0.803 | 0.687 | |
| Median | 0.924 | 1.12 | 0.694 | |
| 75% percentile | 1.03 | 1.30 | 0.762 | |
| Maximum | 1.31 | 1.32 | 0.851 | |
| n | 6 | 7 | 7 | |
| mRNA expression [Col1α/Gapdh] | Minimum | 1.51 | 2.06 | 1.86 |
| 25% percentile | 2.02 | 2.49 | 2.40 | |
| Median | 3.56 | 4.21 | 2.55 | |
| 75% percentile | 5.72 | 4.84 | 4.57 | |
| Maximum | 7.12 | 6.48 | 5.05 | |
| n | 5 | 7 | 7 | |
| GLDH [U/l] | Minimum | 28.0 | 48.0 | 21.0 |
| 25% percentile | 35.5 | 64.0 | 22.0 | |
| Median | 64.0 | 99.0 | 40.0 | |
| 75% percentile | 216 | 110 | 70.0 | |
| Maximum | 598 | 144 | 76.0 | |
| n | 6 | 7 | 7 | |
Statistical significance was set at P < 0.05. Normality was assessed by the Shapiro-Wilk test. Outliers were identified using Grubb’s test and were excluded if P < 0.05. All statistical analyses were performed using SigmaPlot 12.0 (Systat Software Inc., Erkrath, Germany) or GraphPad Prism 6.05 (GraphPad Software, La Jolla, California, USA).
Results
General aspects
Mice treated with STZ/HFD developed progressive liver disease over a period of 20 wks. Blood glucose levels constantly increased in STZ/HFD-treated mice over 20 wks, with no marked differences between the Wt and LRep1−/− mice (Fig. 1b). Compared to the healthy control mice, STZ/HFD-treated Wt and LRep1−/− mice exhibited a lower body weight throughout the study time course (Fig. 1c).
Systemic and hepatic lipid content
In both genotypes, STZ/HFD treatment resulted in significantly increased plasma triglyceride levels when compared to control, particularly at 8 and 12 wks (Fig. 2a). Of interest, LRep1−/− STZ/HFD mice showed significantly lower plasma triglyceride concentrations than Wt STZ/HFD mice at 8 wks of age. Similarly, quantitative analysis of hepatic fat deposition by Oil Red O staining revealed a significant increase in the fat-positive area in STZ/HFD mice at all time points compared to healthy controls, reaching area percentages of approximately 25–40% (Fig. 2b). Whereas fat accumulation reached a maximum at 8 wks in Wt STZ/HFD mice, this value held nearly constant over the whole observation period in LRep1−/− STZ/HFD mice and was significantly lower than in Wt mice at 8 wks (Fig. 2b and c).
Fig. 2.
(a) Analysis of plasma triglycerides and (b) quantitative analysis of lipids by Oil Red O staining in livers of 6-, 8-, 12-, and 20-wk-old control and STZ/HFD-treated wild-type (Wt) and LRep1−/− mice. (c) Representative images of Oil Red O-stained liver sections (200× magnification). Values are presented as the mean ± SEM. Group differences were tested by two-way ANOVA with the Holm-Sidak post hoc test. n.d., not detectable.
To characterize intrahepatic lipid droplets in more detail, we performed electron microscopy analysis of livers from STZ/HFD-treated Wt and LRep1−/− mice at the ages of 8 and 12 wks (Fig. 3a–f). At both time points, there were no differences in diameter or the calculated size of hepatocellular lipid droplets between the genotypes (Fig. 3a–c). In general, small lipid droplets with a diameter of 1 to 2 µm were most prominent (Fig. 3a). Further analysis revealed a markedly reduced number and, ultimately, a significantly decreased total area of hepatic lipid droplets in Repin1-deficient mice compared to Wt mice at 12 wks (Fig. 3d–f). Representative electron microscopic images (Fig. 3f) show this distinct difference in hepatocellular fat content between the two genotypes.
Next, we examined the expression of the hepatic lipid transporter CD36 (Fig. 3g). Cd36 mRNA expression was significantly upregulated from the 8th wk onwards in Wt mice under STZ/HFD conditions. However, Cd36 mRNA expression was only moderately increased in LRep1−/− STZ/HFD mice, with significantly lower values than those in Wt STZ/HFD mice, particularly at 20 wks (Fig. 3g).
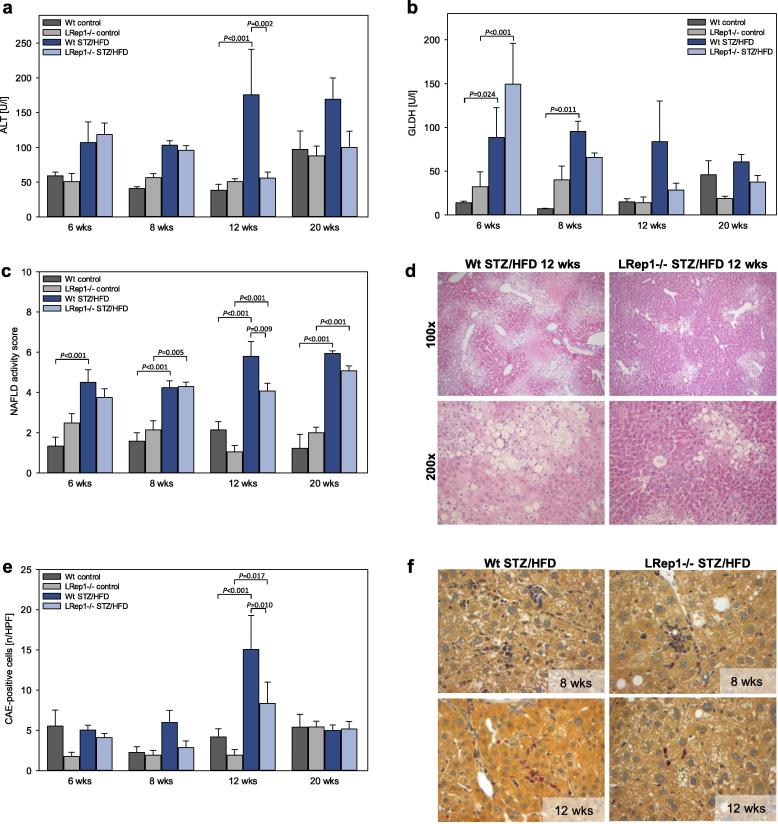
Liver injury and NAFLD progression
STZ/HFD-induced liver injury, as indicated by moderately increased ALT and GLDH activity in plasma of STZ/HFD-treated mice, was diminished at 12 wks in LRep1−/− mice compared to Wt mice (Fig. 4a and b). Additionally, the increase in NAS with age confirmed NAFLD progression in STZ/HFD mice. At 12 wks, LRep1−/− STZ/HFD mice had a significantly lower NAS, suggesting a decrease in disease severity at this time point (Fig. 4c). Liver histology (Fig. 4d) showed increased liver injury in Wt STZ/HFD mice compared to LRep1−/− mice at 12 wks, characterized by a higher number of inflammatory cells, ballooned and steatotic hepatocytes and increased necrosis (Fig. 4d).
Fig. 4.
Analysis of (a) alanine aminotransferase (ALT) and (b) glutamate dehydrogenase (GLDH) in the plasma of control and STZ/HFD-treated wild-type (Wt) and LRep1−/− mice at 6, 8, 12, and 20 wks of age. (c) NAFLD activity score (NAS) of livers in control and STZ/HFD-treated Wt and LRep1−/− mice at 6, 8, 12, and 20 wks of age. The NAS (score 0–8) was calculated as the sum of three different scores (steatosis (score 0–3), lobular inflammation (score 0–3), and hepatocellular ballooning (score 0–2)). (d) Representative H&E-stained liver sections from 12-wk-old STZ/HFD-treated mice showing less liver injury in LRep1−/− mice than in Wt mice (100×/200× magnification). (e) Quantitative analysis of CAE-positive cells (presented as cells/high power field (HPF)) in liver sections of control and STZ/HFD-treated Wt and LRep1−/− mice at 6, 8, 12, and 20 wks of age. (f) Representative images of CAE-stained liver tissue of 8- and 12-wk-old STZ/HFD-treated Wt and LRep1−/− mice with granulocytes (red; 400× magnification). Values are presented as the mean ± SEM. Differences between groups were tested by two-way ANOVA with the Holm-Sidak post hoc test.
Accordingly, the number of infiltrating granulocytes was significantly lower at 12 wks in LRep1−/− STZ/HFD mice than in Wt STZ/HFD mice, with the highest number of CAE-positive cells at this time point (Fig. 4e). Representative CAE-stained liver sections (Fig. 4f) showed differences in the number of infiltrating leukocytes between genotypes at 8 and 12 wks.
Tumour prevalence and mortality
For each mouse, both the presence of macroscopically visible tumours or nodules and survival were recorded. Upon STZ/HFD treatment, tumours of different types, such as highly differentiated HCC and undifferentiated dysplastic nodules with or without massive fat accumulation (Fig. 5e), were first evident at 8 wks, with no difference between genotypes (Fig. 5a). Interestingly, at 12 wks, 62.5% of the Wt STZ/HFD mice but only 33.3% of the LRep1−/− STZ/HFD mice had tumours. This lower tumour prevalence in Repin1-deficient mice persisted at 20 wks. Whereas tumours were detected in almost every Wt STZ/HFD mouse (91.7%), they were observed in only 77.8% of the LRep1−/− STZ/HFD mice (Fig. 5a). Microscopic analysis of neoplastic foci revealed significantly fewer neoplastic foci per histologic liver section in STZ/HFD-treated LRep1−/− mice than in STZ/HFD-treated Wt mice at 20 wks (Fig. 5b). At 8 and 12 wks, the same trend was observed. Accordingly, the total area of neoplastic foci per histologic liver section tended to be higher in Wt STZ/HFD mice than in LRep1−/− STZ/HFD mice at all time points (Fig. 5c).
In general, the STZ/HFD model was associated with a high mortality rate (Fig. 5d). However, compared to Wt STZ/HFD mice (Fig. 5d, dark blue), LRep1−/− STZ/HFD mice (Fig. 5d, light blue) appeared to show improved survival between 8 and 12 wks, but there was no significant effect on overall outcome.
Furthermore, NAFLD progression was accompanied by a significantly increased liver weight/body weight index in both STZ/HFD groups compared to the healthy groups (Fig. 5f). In contrast to Wt STZ/HFD mice, which showed a marked rise in liver weight/body weight index over time up to ∼12%, the liver weight/body weight index of LRep1−/− STZ/HFD mice did not change over time; this value was constant (∼7–8%) throughout the 20-wk observation period and was significantly lower than that in Wt STZ/HFD mice at 12 and 20 wks (Fig. 5f). In situ images of the liver (Fig. 5g) clearly displayed the progression of liver disease in STZ/HFD mice. The liver volume and occurrence of nodular structures or tumours appeared to be less prominent in LRep1−/− STZ/HFD mice than in Wt STZ/HFD mice (Fig. 5g).
siRNA-induced Repin1 deficiency
To confirm the observations in LRep1−/− mice, we evaluated a therapeutic approach in the early phase of NAFLD using Repin1 siRNA (Fig. 6). Repetitive Repin1 siRNA applications over a period of 14 days resulted in significantly lower hepatic Repin1 mRNA expression in 6-wk-old Wt STZ/HFD mice (Fig. 6b). Generally, 6 wk-old siRNA treated STZ/HFD animals showed lipid accumulation, NAS, and liver weight/body weight index comparable to LRep1−/− STZ/HFD mice at 6 wks (data not shown). Interestingly, siRNA-induced Repin1 deficiency had beneficial effects on the development of chronic inflammation. The quantification of Kupffer cells (Fig. 6c–e) and infiltrating granulocytes (Fig. 6f) revealed significantly lower numbers/values in siRep1-treated mice than in siLuci- or buffer-treated mice. Moreover, Repin1 siRNA-treated STZ/HFD mice exhibited a smaller hepatic Sirius Red-positive area (Fig. 6g) and reduced collagen 1α mRNA expression (Fig. 6h) in comparison to buffer- and siLuci-treated STZ/HFD mice. Finally, these molecular events led to reduced liver damage, as indicated by significantly reduced GLDH activity in siRep1-treated mice (Fig. 6i).
Discussion
Besides cardiovascular diseases, hyperlipidaemia and type 2 diabetes, NAFLD is the most important comorbidity of metabolic syndrome, even though NAFLD can occur in patients with a normal basic metabolic index [17]. Although diet and intensive lifestyle interventions can lead to significant improvements, no safe and effective drug treatment for NAFLD is currently available [18], [19], [20].
Due to its complexity, the pathophysiological mechanisms of NAFLD are not fully understood, which hampers drug discovery. However, most of the pharmacologic agents currently being tested in clinical trials target steatosis, lipotoxicity, oxidative stress, fibrogenesis, energy homeostasis and inflammatory signals [18], [19], [21], [22]. Although promising improvements in steatosis and inflammation have been observed, long-term efficacy and safety have yet to be proven. Several trials have shown that the treatment of metabolic risk factors may alleviate NAFLD progression [23], [24], [25]. For example, pioglitazone and vitamin E proved to be effective at reducing hepatic steatosis and lobular inflammation to some extent but were not effective in reducing the overall NAS [20], [23], [24].
In this study, we demonstrate in a mouse model of progressive NAFLD that liver-specific Repin1 deletion attenuates the progression of fatty liver disease, as indicated by significantly less intrahepatic lipid accumulation and decreased chronic inflammation and liver injury. Consequently, and most strikingly, Repin1-deficient mice had a lower tumour prevalence, reductions in the number and total area of neoplastic foci, and a lower liver weight/body weight index. The beneficial effects of Repin1 siRNA treatment confirmed the potential of Repin1 as a target gene in NAFLD therapy.
Based on initial studies in congenic and subcongenic rat strains as well as in human subjects, Repin1 was established as a candidate gene for obesity and metabolic syndrome [5], [7], [26]. Comprehensive studies in mice with Repin1 deletion in either adipose tissue, liver or the whole body have demonstrated that Repin1 is a crucial modulator of lipid and glucose metabolism [6], [10], [11]. Phenotypic characterization of Repin1-knockout mice revealed significant improvements in the metabolic profile. Therein, it was demonstrated that Repin1 regulates the expression of genes involved in adipogenesis, lipid droplet formation and fusion, and glucose and fatty acid transport in adipocytes [7]. Similarly, positive consequences of Repin1 deficiency were also observed in the livers of STZ/HFD mice. Moreover, regardless of the treatment, LRep1−/− mice displayed a slightly lower body weight than Wt mice, which was also observed in previous studies, suggesting greater physical activity and energy expenditure in Repin1-deficient mice [6], [10], [11].
The pathogenesis of NAFLD includes metabolic stress to the liver associated with cell stress and the activation of inflammatory and fibrotic pathways, particularly in response to hepatic fat accumulation [27], [28], finally resulting in NASH. Based on this, we suggest that decreased lipid load in Repin1-deficient mice at an early stage possibly has beneficial effects at later time points. It is known that increased hepatic lipid accumulation, particularly of saturated fatty acids, induces endoplasmic reticulum (ER) stress and lipotoxicity [29], [30], [31], [32]; thus, lower concentrations of intrahepatic lipids and plasma triglycerides may contribute to the reduced tissue inflammation and liver injury in LRep1−/− mice. In fact, neutrophil recruitment and migration to the liver were reduced under conditions of Repin1 deficiency, resulting in an overall improved NAS. Several mouse studies and clinical trials have shown that lipid-lowering agents and substances that improve insulin resistance also decrease hepatic steatosis [33]. Hepatic uptake of free fatty acids, among others, involves the fatty acid transporter CD36, which is a target gene of Repin1 [10]. Previous in vivo and in vitro siRNA studies showed reduced expression of CD36 in mice or adipocytes with reduced Repin1 expression, resulting in significantly impaired fatty acid uptake [6], [7], [10], [11]. Impaired and delayed uptake of fatty acids, resulting in lower hepatic triglycerides, was also present in CD36-deficient mice [34], [35]. Similarly, STZ/HFD-treated LRep1−/− mice showed reduced hepatic Cd36 expression. Thus, impaired CD36-mediated fatty acid transport or uptake and storage in the liver might contribute to the decreased lipid load in STZ/HFD-treated LRep1−/− mice. Further in vivo and in vitro studies by our group support the involvement of Repin1 in hepatocellular fat accumulation by demonstrating attenuated lipid accumulation in Repin1-deficient primary hepatocytes under steatotic conditions and decreased transient steatosis after liver resection in LRep1−/− mice [36].
Low-grade chronic inflammation resulting from chronic intracellular oxidative stress and the presence of local inflammatory cells, particularly Kupffer cells and neutrophils, has been suggested to play a crucial role in the progression to NAFLD-related HCC [37], [38], [39]. Of particular interest, STZ/HFD-treated LRep1−/− mice had relatively lighter livers than Wt mice, which were increased in size proportionally to body weight. This finding is most likely due to the lower tumour load, the presence of smaller tumours, or the overall lower tumour prevalence. Several mechanisms could explain these observations in LRep1−/− mice, e.g., an overall improved metabolic profile, decreased hepatic lipid accumulation and hence alleviation of chronic inflammation or thus far unrecognized Repin1 target genes that regulate inflammation, tumourigenesis or energy metabolism in cancer.
To test a clinically relevant therapeutic approach, we induced hepatic Repin1 deficiency in the early phase of NAFLD using a liver-specific siRNA-lipid formulation. Impressively, a two-week treatment of STZ/HFD mice with Repin1 siRNA had positive effects on inflammatory and fibrotic parameters in the early phase of NAFLD. These results confirm the observations in Repin1-knockout mice and suggest direct Repin1-dependent regulation of genes involved in inflammation and fibrogenesis.
Conclusions
Taken together, the current findings indicate that Repin1 is an emerging player in the progression of STZ/HFD-induced NAFLD. Repin1 deficiency improved key morphological features of progressive NAFLD/NASH, such as steatosis, inflammation and tumour development. As treatment of NAFLD with a single targeted agent will be ineffective and Repin1 deficiency has multiple beneficial metabolic effects, the Repin1 pathway might be an attractive therapeutic target for the treatment of NAFLD. Further studies should clarify whether Repin1 might also be an important genetic factor in determining the phenotypic manifestation and overall risk for NAFLD.
Conflict of interest
The authors have declared no conflict of interest.
Acknowledgements
We thank Maren Nerowski, Eva Lorbeer-Rehfeldt, Dorothea Frenz, Berit Blendow, Ute Schulz, Laura Grüner and Dr Armin Springer for their excellent technical assistance. This research was supported by a grant from the Deutsche Forschungsgemeinschaft, Germany (AB 453/2-1, NK SFB1052/2-B4).
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Younossi Z.M., Koenig A.B., Abdelatif D., Fazel Y., Henry L., Wymer M. Global epidemiology of nonalcoholic fatty liver disease—meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 2.Wree A., Broderick L., Canbay A., Hoffman H.M., Feldstein A.E. From NAFLD to NASH to cirrhosis—new insights into disease mechanisms. Nat Rev Gastroenterol Hepatol. 2013;10:627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 3.Glen J, Floros L, Day C, Pryke R, Group GD Non-alcoholic fatty liver disease (NAFLD): summary of NICE guidance. BMJ. 2016;354:i4428. doi: 10.1136/bmj.i4428. [DOI] [PubMed] [Google Scholar]
- 4.Bahr J., Klöting N., Wilke B., Klöting I., Follak N. High-fat diet protects BB/OK rats from developing type 1 diabetes. Diabetes Metab Res Rev. 2011;27:552–556. doi: 10.1002/dmrr.1207. [DOI] [PubMed] [Google Scholar]
- 5.Klöting N., Wilke B., Klöting I. Triplet repeat in the Repin1 3′-untranslated region on rat chromosome 4 correlates with facets of the metabolic syndrome. Diabetes Metab Res Rev. 2007;23:406–410. doi: 10.1002/dmrr.713. [DOI] [PubMed] [Google Scholar]
- 6.Kunath A., Hesselbarth N., Gericke M., Kern M., Dommel S., Kovacs P. Repin1 deficiency improves insulin sensitivity and glucose metabolism in db/db mice by reducing adipose tissue mass and inflammation. Biochem Biophys Res Commun. 2016;478:398–402. doi: 10.1016/j.bbrc.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 7.Ruschke K., Illes M., Kern M., Klöting I., Fasshauer M., Schön M.R. Repin1 maybe involved in the regulation of cell size and glucose transport in adipocytes. Biochem Biophys Res Commun. 2010;400:246–251. doi: 10.1016/j.bbrc.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 8.Heiker J.T., Klöting N. Replication initiator 1 in adipose tissue function and human obesity. Vitam Horm. 2013;91:97–105. doi: 10.1016/B978-0-12-407766-9.00005-5. [DOI] [PubMed] [Google Scholar]
- 9.Krüger J., Berger C., Weidle K., Schleinitz D., Tönjes A., Stumvoll M. Metabolic effects of genetic variation in the human REPIN1 gene. Int J Obes. 2018 doi: 10.1038/s41366-018-0123-0. [DOI] [PubMed] [Google Scholar]
- 10.Kern M., Kosacka J., Hesselbarth N., Brückner J., Heiker J.T., Flehmig G. Liver-restricted Repin1 deficiency improves whole-body insulin sensitivity, alters lipid metabolism, and causes secondary changes in adipose tissue in mice. Diabetes. 2014;63:3295–3309. doi: 10.2337/db13-0933. [DOI] [PubMed] [Google Scholar]
- 11.Hesselbarth N., Kunath A., Kern M., Gericke M., Mejhert N., Rydén M. Repin1 deficiency in adipose tissue improves whole-body insulin sensitivity, and lipid metabolism. Int J Obes. 2017;41:1815–1823. doi: 10.1038/ijo.2017.172. [DOI] [PubMed] [Google Scholar]
- 12.Fujii M., Shibazaki Y., Wakamatsu K., Honda Y., Kawauchi Y., Suzuki K. A murine model for non-alcoholic steatohepatitis showing evidence of association between diabetes and hepatocellular carcinoma. Med Mol Morphol. 2013;46:141–152. doi: 10.1007/s00795-013-0016-1. [DOI] [PubMed] [Google Scholar]
- 13.Liebig M., Hassanzada A., Kämmerling M., Genz B., Vollmar B., Abshagen K. Microcirculatory disturbances and cellular changes during progression of hepatic steatosis to liver tumors. Exp Biol Med. 2018;243:1–12. doi: 10.1177/1535370217738730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew Chem - Int Ed. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belliveau N.M., Huft J., Lin P.J., Chen S., Leung A.K., Leaver T.J. Microfluidic synthesis of highly potent limit-size lipid nanoparticles for in vivo delivery of siRNA. Mol Ther - Nucleic Acids. 2012;1:e37. doi: 10.1038/mtna.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ofosu A., Ramai D., Reddy M. Non-alcoholic fatty liver disease: controlling an emerging epidemic, challenges, and future directions. Ann Gastroenterol. 2018;31:288–295. doi: 10.20524/aog.2018.0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J., Zhang X., Li H. Progress and challenges in the prevention and control of nonalcoholic fatty liver disease. Med Res Rev. 2018:1–21. doi: 10.1002/med.21515. [DOI] [PubMed] [Google Scholar]
- 19.Ilan Y. Future of treatment for nonalcoholic steatohepatitis: can the use of safe, evidence - based, clinically proven supplements provide the answer to the unmet need? Dig Dis Sci. 2018;63:1726–1736. doi: 10.1007/s10620-018-5080-x. [DOI] [PubMed] [Google Scholar]
- 20.Konerman M.A., Jones J.C., Harrison S.A. Pharmacotherapy for NASH: current and emerging. J Hepatol. 2018;68:362–375. doi: 10.1016/j.jhep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 21.Friedman S., Sanyal A., Goodman Z., Lefebvre E., Gottwald M., Fischer L. Efficacy and safety study of cenicriviroc for the treatment of non-alcoholic steatohepatitis in adult subjects with liver fibrosis: CENTAUR Phase 2b study design. Contemp Clin Trials. 2016;47:356–365. doi: 10.1016/j.cct.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Safadi R., Konikoff F.M., Mahamid M., Zelber-Sagi S., Halpern M., Gilat T. Pancreas, biliary tract, and liver the fatty acid – bile acid conjugate Aramchol reduces liver fat content in patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2014;12:2085–2091. doi: 10.1016/j.cgh.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 23.Bell L.N., Wang J., Muralidharan S., Chalasani S., Fullenkamp A.M., Wilson L.A. Relationship between adipose tissue insulin resistance and liver histology in nonalcoholic steatohepatitis: a pioglitazone versus vitamin E versus placebo for the treatment of nondiabetic patients with nonalcoholic steatohepatitis trial follow-up study. Hepatology. 2012;56:1311–1318. doi: 10.1002/hep.25805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanyal A.J., Chalasani N., Kowdley K.V., Mccullough A., Diehl A.M., Bass N.M. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. N Engl J Med. 2010;362:1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin H., Yang S., Chuckaree C., Kuhajda F., Ronnet G., Am D. Metformin reverses fatty liver disease in obese, leptin-deficient mice. Nat Med. 2000;6:998–1003. doi: 10.1038/79697. [DOI] [PubMed] [Google Scholar]
- 26.Kovacs P., Klöting I. Quantitative trait loci on chromosomes 1 and 4 affect lipid phenotypes in the rat. Arch Biochem Biophys. 1998;354:139–143. doi: 10.1006/abbi.1998.0686. [DOI] [PubMed] [Google Scholar]
- 27.Issa D., Patel V., Sanyal A.J. Future therapy for non-alcoholic fatty liver disease. Liver Int. 2018;38:56–63. doi: 10.1111/liv.13676. [DOI] [PubMed] [Google Scholar]
- 28.Chiang J.Y.L. Targeting bile acids and lipotoxicity for NASH treatment. Hepatol Commun. 2017;1:1002–1004. doi: 10.1002/hep4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiappini F., Desterke C., Bertrand-Michel J., Guettier C., Le Naour F. Hepatic and serum lipid signatures specific to nonalcoholic steatohepatitis in murine models. Sci Rep. 2016;6:31587. doi: 10.1038/srep31587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiappini F., Coilly A., Kadar H., Gual P., Tran A., Desterke C. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci Rep. 2017;7:46658. doi: 10.1038/srep46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendez-Sanchez N., Cruz-Ramon V.C., Ramirez-Perez O.L., Hwang J.P., Barranco-Fragoso B., Cordova-Gallardo J. New aspects of lipotoxicity in nonalcoholic steatohepatitis. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19072034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mantzaris M.D., Tsianos E.V., Galaris D. Interruption of triacylglycerol synthesis in the endoplasmic reticulum is the initiating event for saturated fatty acid-induced lipotoxicity in liver cells. FEBS J. 2010;278:519–530. doi: 10.1111/j.1742-4658.2010.07972.x. [DOI] [PubMed] [Google Scholar]
- 33.Konikoff F., Gilat T. Effects of fatty acid bile acid conjugates (FABACs) on biliary lithogenesis: potential consequences for non-surgical treatment of gallstones. Curr Drug Targets Immune Endocr Metab Disord. 2005;5:171–175. doi: 10.2174/1568008054064904. [DOI] [PubMed] [Google Scholar]
- 34.Febbraio M., Abumrad N.A., Hajjar D.P., Sharma K., Cheng W., Pearce S.F. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 35.Hajri T., Han X.X., Bonen A., Abumrad N.A. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–1389. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abshagen K., Degenhardt B., Liebig M., Wendt A., Genz B., Schaeper U. Liver-specific Repin1 deficiency impairs transient hepatic steatosis in liver regeneration. Sci Rep. 2018;8:16858. doi: 10.1038/s41598-018-35325-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajendran P., Chen Y.-F., Chen Y.-F., Chung L.-C., Tamilselvi S., Shen C.-Y. The multifaceted link between inflammation and human diseases. J Cell Physiol. 2018;233:6458–6471. doi: 10.1002/jcp.26479. [DOI] [PubMed] [Google Scholar]
- 38.Wilson C.L., Jurk D., Fullard N., Banks P., Page A., Luli S. NFκ B1 is a suppressor of neutrophil-driven hepatocellular carcinoma. Nat Commun. 2015;6:1–13. doi: 10.1038/ncomms7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balsano C., Porcu C., Sideri S., Tavolaro S. Fat and hepatocellular carcinoma. Hepatoma Res. 2018;4:38. [Google Scholar]