Abstract
We built and validated a deep learning algorithm predicting the individual diagnosis of Alzheimer's disease (AD) and mild cognitive impairment who will convert to AD (c-MCI) based on a single cross-sectional brain structural MRI scan. Convolutional neural networks (CNNs) were applied on 3D T1-weighted images from ADNI and subjects recruited at our Institute (407 healthy controls [HC], 418 AD, 280 c-MCI, 533 stable MCI [s-MCI]). CNN performance was tested in distinguishing AD, c-MCI and s-MCI. High levels of accuracy were achieved in all the classifications, with the highest rates achieved in the AD vs HC classification tests using both the ADNI dataset only (99%) and the combined ADNI + non-ADNI dataset (98%). CNNs discriminated c-MCI from s-MCI patients with an accuracy up to 75% and no difference between ADNI and non-ADNI images. CNNs provide a powerful tool for the automatic individual patient diagnosis along the AD continuum. Our method performed well without any prior feature engineering and regardless the variability of imaging protocols and scanners, demonstrating that it is exploitable by not-trained operators and likely to be generalizable to unseen patient data. CNNs may accelerate the adoption of structural MRI in routine practice to help assessment and management of patients.
Keywords: Alzheimer's disease, Mild cognitive impairment, Diagnosis, Prediction, Deep learning, Convolutional neural networks
HIGHLIGHTS
-
•
CNNs predict AD and MCI with high accuracy based on a single T1-weighted image
-
•
CNNs discriminate c-MCI from s-MCI patients with an accuracy up to 75%
-
•
CNNs are exploitable by not-trained operators
-
•
CNNs are likely to be generalizable to unseen patient data
1. Introduction
The diagnosis of Alzheimer's disease (AD) can be improved by the use of biomarkers (Albert et al., 2011; Dubois et al., 2014; McKhann et al., 2011). Structural MRI, which provides biomarkers of neuronal loss, is an integral part of the clinical assessment of patients with suspected AD (Albert et al., 2011; Dubois et al., 2014; McKhann et al., 2011). Several studies have shown that atrophy estimates in characteristically vulnerable brain regions, particularly the hippocampus and entorhinal cortex, reflect disease stage and are predictive of progression of mild cognitive impairment (MCI) to AD (Frisoni et al., 2010). The clinical utility of structural MRI in differentiating AD from other diseases, such as vascular or non-AD dementia, has been also established (Frisoni et al., 2010). However, the value of structural MRI will be increased by standardization of acquisition and analysis methods, and by development of robust algorithms for automated assessment. All of these are needed to achieve the ultimate goal of individual patient diagnosis with a single cross-sectional structural MRI scan and for structural MRI to be definitely qualified by regulatory agencies as a biomarker for enrichment of pre-dementia AD trials (Frisoni et al., 2017).
Previous work in computer-aided classification of AD and MCI patients has used several machine learning methods applied to structural MRI (Rathore et al., 2017). The most popular among these methods is Support Vector Machine (SVM) (Rathore et al., 2017). SVM extracts high-dimensional, informative features from MRI to build predictive classification models that facilitate the automation of clinical diagnosis (Rathore et al., 2017). However, feature definition and extraction typically rely on manual/semi-automatic outlining of brain structures, which is laborious and prone to inter- and intra-rater variability, or complex image pre-processing, which is time-consuming and computationally demanding.
An alternative family of machine learning methods, known as deep learning algorithms, are achieving optimal results in many domains such as speech recognition tasks, computer vision and natural language understanding (Lecun et al., 2015) and, more recently, medical analysis (Esteva et al., 2017; Vieira et al., 2017; Xiong et al., 2015). Deep learning algorithms differ from conventional machine learning methods by the fact that they require little or no image pre-processing and can automatically infer an optimal representation of the data from the raw images without requiring prior feature selection, resulting in a more objective and less bias-prone process (LeCun et al., 2015; Vieira et al., 2017). Therefore, deep learning algorithms are better suited for detecting subtle and diffuse anatomical abnormalities (LeCun et al., 2015; Vieira et al., 2017). Recently, deep learning has been successfully applied to the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset to identify AD patients from healthy controls (Table 1) (for a review see (Vieira et al., 2017)). Only one study so far has applied deep learning algorithms, without a priori feature selection (considering gray matter [GM] volumes as input), to the prediction of AD development within 18 months in individuals with MCI using ADNI structural MRI scans (Suk et al., 2017) (Table 1).
Table 1.
Literature review on structural MRI (T1-weighted images) and deep learning in AD and MCI patient classification.
| Studies (chronological order) | Dataset | Sample size | MCI conversion to AD? | Deep learning architecture | Input | Data augmentation | Validation | Transfer learning | Classifications & Accuracy (%) |
|---|---|---|---|---|---|---|---|---|---|
| Natural Image Bases to Represent Neuroimaging Data (Gupta et al., 2013) | ADNI | 200 AD, 232 HC, 411 MCI | NO | Sparse Auto-encoder with Convolutional Neural Network | Normalized 3D T1-weighted images | YES (serial scans from each subject) | Independent sample | NO | AD vs HC: 95.24% MCI vs HC: 92.23% AD vs MCI: 84.07% |
| Predicting Alzheimer's disease: a neuroimaging study with 3D convolutional neural networks (Payan and Montana, 2015) | ADNI | 755 AD, 755 HC, 755 MCI | NO | Sparse Auto-encoder with 3D Convolutional Neural Network | Normalized 3D T1-weighted images | YES (serial scans from each subject) | Independent sample | NO | AD vs HC: 95.4% MCI vs HC: 92.1% AD vs MCI: 86.8% AD vs MCI vs HC: 89.5% |
| Alzheimer's disease diagnostics by a deeply supervised adaptable 3D convolutional network (Hosseini-Asl et al., 2016) | ADNI | 70 from each class (AD, HC, MCI) | NO | 3D convolutional autoencoder | Normalized 3D T1-weighted images | NO | Not specified | YES | AD vs HC: 99.3% MCI vs HC: 94.2% AD vs MCI: 100% AD vs MCI vs HC: 94.8% AD+MCI vs HC: 95.7% |
| DeepAD: Alzheimer's Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI (Sarraf et al., 2016) | ADNI | 91 AD, 211 HC | NO | Convolutional Neural Network | Normalized 3D T1-weighted images | YES (3D to 2D conversion) | Not specified | NO | AD vs HC: 98.84% |
| Deep ensemble learning of sparse regression models for brain disease diagnosis (Suk et al., 2017) | ADNI | 186 AD, 226 HC, 167 converters MCI, 226 stable MCI | YES | Multiple sparse regression models with deep Convolutional Neural Network | GM volumes | NO | 10-fold cross validation | NO | AD vs HC: from 84.69 to 91.02% MCI vs HC: from 66.78 to 73.02% Converters MCI vs stable MCI: from 66.39 to 74.82% |
Abbreviations: AD = Alzheimer's disease; ADNI = Alzheimer's Disease Neuroimaging Initiative; GM = GRAY matter; HC = healthy controls; MCI = Mild Cognitive Impairment.
The aim of the present study was to build and validate a deep learning algorithm (specifically convolutional neural networks [CNN]) that can predict the individual diagnosis of AD and the development of AD in MCI patients based on a single cross-sectional brain structural MRI scan. A robust diagnostic marker should adapt to various datasets to diminish discrepancies in data distribution and biases toward specific groups (Frisoni et al., 2017). One of the most important caveats of previous works is the single-center origin of imaging data that limits the generalizability of findings. In light of this, one of the main goal and novelty of our study was to overcome this limit by comparing data from different centers, neuroimaging protocols and scanners, in order to reach both reliability and reproducibility of results.
2. METHODS
2.1. Participants
We used the structural brain MRI scans from the ADNI dataset (ADNI.LONI.USC.EDU). The ADNI was launched in 2003 as a public private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, positron emission tomography, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. For up-to-date information, see WWW.ADNI-INFO.ORG. A total of 1409 subjects (294 patients with probable AD, 763 patients with MCI, and 352 healthy controls) were considered in this study (Table 2). Standard 3 T baseline T1-weighted images were included from the ADNI dataset. We included all ADNI1, ADNI2 and ADNI-GO subjects that had baseline 3D T1-weighted scans. After 36 months, 253 MCI patients (33%) converted clinically to AD (c-MCI).
Table 2.
Demographic and clinical features of AD and MCI patients and healthy controls from the ADNI dataset.
| HC | AD | c-MCI | s-MCI | P AD vs HC | P c-MCI vs HC | P s-MCI vs HC | P AD vs c-MCI | P AD vs s-MCI | P c-MCI vs s-MCI | |
|---|---|---|---|---|---|---|---|---|---|---|
| N | 352 | 294 | 253 | 510 | ||||||
| Age [years] | 74.53 ± 6.16 (56.20–89.60) | 75.13 ± 7.75 (55.10–90.90) | 73.80 ± 7.35 (55.00–88.30) | 72.33 ± 7.68 (54.40–91.40) | 1.00 | 1.00 | <0.001 | 0.20 | <0.001 | 0.05 |
| Gender [women/men] | 185/167 | 136/158 | 102/151 | 223/287 | 0.12 | 0.003 | 0.01 | 0.17 | 0.51 | 0.35 |
| Education [years] | 16.30 ± 2.76 (6–20) | 15.14 ± 3.02 (4–20) | 15.76 ± 2.84 (6–20) | 16.02 ± 2.82 (4–20) | <0.001 | 0.13 | 0.93 | 0.07 | <0.001 | 1.00 |
| CDR sum of boxes | 0.03 ± 0.12 (0–1) | 4.46 ± 1.61 (1−10) | 1.95 ± 0.97 (0.5–5.5) | 1.29 ± 0.76 (0.5–4) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| MMSE | 29.07 ± 1.16 (24–30) | 23.12 ± 2.1 (18–27) | 26.91 ± 1.78 (23−30) | 27.99 ± 1.71 (23–30) | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
Values are numbers or means ± standard deviations (range). P values refer to ANOVA models, followed by post-hoc pairwise comparisons (Bonferroni-corrected for multiple comparisons), or Chi-squared test. Abbreviations: AD = Alzheimer's Disease; CDR = Clinical Dementia Rating Scale; HC = healthy controls; MCI = Mild Cognitive Impairment (c = converters; s = stable); MMSE = Mini Mental State Examination; N = Number.
An independent dataset of 3D T1-weighted images were obtained from 229 subjects (hereafter named as “Milan” dataset) including 124 patients with probable AD (McKhann et al., 2011), 50 patients with MCI (Albert et al., 2011), and 55 healthy controls who were recruited consecutively at the Department of Neurology, Scientific Institute and University Vita-Salute San Raffaele, Milan (Table 3). After 36 months, 27 (54%) MCI patients converted clinically to AD. An experienced neurologist blinded to MRI results performed clinical assessments. Healthy controls with no history of neurologic, psychiatric or other major medical illnesses were recruited among friends and spouses of patients and by word of mouth (Table 3).
Table 3.
Demographic, clinical and neuropsychological features of AD and MCI patients and healthy controls from the Milan dataset.
| HC | AD | c-MCI | s-MCI | P AD vs HC | P c-MCI vs HC | P s-MCI vs HC | P AD vs c-MCI | P AD vs s-MCI | P c-MCI vs s-MCI | |
|---|---|---|---|---|---|---|---|---|---|---|
| N | 55 | 124 | 27 | 23 | ||||||
| Age [years] | 67.1 ± 6.8 (56.1–81.8) | 68.3 ± 8.1 (48.5–85.6) | 71.6 ± 7.5 (55.3–85.7) | 68.2 ± 6.4 (52.1–80.7) | 1.00 | 0.08 | 1.00 | 0.25 | 1.00 | 0.71 |
| Gender [women/men] | 29/26 | 69/55 | 13/14 | 10/13 | 0.42 | 0.44 | 0.31 | 0.31 | 0.20 | 0.48 |
| Education [years] | 12.2 ± 4.7 (5–24) | 9.5 ± 4.5 (1–18) | 10.4 ± 4.4 (4–18) | 11.3 ± 3.7 (5–18) | 0.001 | 0.56 | 1.00 | 1.00 | 0.38 | 1.00 |
| Disease duration [years] | – | 3.5 ± 2.0 (0.0–10.2) | 3.0 ± 1.5 (1.0–6.1) | 3.0 ± 1.7 (0.6–6.0) | – | – | – | 0.65 | 0.64 | 1.00 |
| CDR | – | 1.2 ± 0.6 (0.5–3) | 0.5 ± 0.2 (0.5–1) | 0.5 ± 0.1 (0.5–1) | – | – | – | <0.001 | <0.001 | 1.00 |
| CDR sum of boxes | – | 5.1 ± 2.2 (2−12) | 2.2 ± 1.0 (1–4.5) | 2.4 ± 1.1 (1–4.5) | – | – | – | <0.001 | <0.001 | 1.00 |
| MMSE | 29.1 ± 1.0 (26–30) | 19.8 ± 4.5 (9–27) | 26.8 ± 1.7 (24–30) | 27.4 ± 2.0 (23–30) | <0.001 | 0.17 | 0.64 | <0.001 | <0.001 | 1.00 |
| Verbal memory | ||||||||||
| RAVLT, immediate recall | 43.4 ± 9.0 (25–60) | 15.0 ± 7.1 (0–40) | 19.6 ± 6.0 (8–32) | 23.7 ± 7.8 (10–34) | <0.001 | <0.001 | <0.001 | 0.11 | 0.001 | 0.62 |
| RAVLT, delayed recall | 8.9 ± 3.3 (4–15) | 0.4 ± 0.9 (0–3) | 1.2 ± 1.9 (0–6) | 1.9 ± 2.1 (0–7) | <0.001 | <0.001 | <0.001 | 0.53 | 0.04 | 1.00 |
| Digit span, forward | 5.9 ± 1.1 (4–9) | 4.6 ± 1.1 (0–7) | 4.9 ± 0.8 (3–6) | 5.4 ± 1.2 (3–8) | <0.001 | 0.01 | 0.47 | 0.99 | 0.02 | 1.00 |
| Memory prose | 9.7 ± 7.0 (3–17) | 2.2 ± 2.5 (0–14) | 5.5 ± 3.1 (0−11) | 7.3 ± 3.6 (2–15) | <0.001 | 0.14 | 1.00 | <0.001 | <0.001 | 0.30 |
| Visuospatial memory | ||||||||||
| Spatial span, forward | 5.1 ± 1.0 (4–7) | 3.0 ± 1.2 (0–6) | 4.2 ± 0.7 (3–6) | 4.5 ± 0.5 (4–5) | <0.001 | 0.02 | 0.46 | <0.001 | <0.001 | 1.00 |
| Rey's figure, recall | 17.7 ± 5.9 (9–33) | 2.8 ± 3.8 (0–26) | 5.5 ± 3.0 (0−13) | 10.0 ± 5.5 (2−21) | <0.001 | <0.001 | <0.001 | 0.03 | <0.001 | 0.002 |
| Visuospatial abilities | ||||||||||
| Rey's figure, copy | 33.2 ± 2.4 (27–36) | 15.3 ± 10.0 (0–35) | 24.3 ± 9.1 (0–36) | 27.7 ± 5.3 (15–36.0) | <0.001 | 0.001 | 0.15 | <0.001 | <0.001 | 0.99 |
| Clock Drawing Test | 8.9 ± 0.9 (8–10) | 3.5 ± 3.9 (0−10) | 7.1 ± 3.2 (0–10) | 8.0 ± 3.1 (0–10) | <0.001 | 1.00 | 1.00 | 0.002 | <0.001 | 1.00 |
| Attention and executive functions | ||||||||||
| Attentive matrices | 48.8 ± 7.6 (32–57) | 31.0 ± 12.7 (2–56) | 43.3 ± 8.2 (30–56) | 46.5 ± 7.9 (33–58) | <0.001 | 0.43 | 1.00 | <0.001 | <0.001 | 1.00 |
| Raven coloured progressive matrices | 29.9 ± 3.8 (22–35) | 16.8 ± 8.2 (2–35) | 23.3 ± 5.5 (10−31) | 26.1 ± 5.9 (9–33) | <0.001 | 0.01 | 0.37 | <0.001 | <0.001 | 1.00 |
| Semantic fluency | 42.4 ± 8.9 (27–60) | 19.0 ± 9.0 (3–55) | 29.5 ± 7.0 (16–42) | 32.7 ± 10.3 (12–55) | <0.001 | <0.001 | 0.001 | <0.001 | <0.001 | 1.00 |
| Phonemic fluency | 36.7 ± 10.4 (18–55) | 16.6 ± 10.3 (0–43) | 24.0 ± 10.3 (11–48) | 30.6 ± 13.8 (10–66) | <0.001 | <0.001 | 0.32 | 0.01 | <0.001 | 0.22 |
| Language | ||||||||||
| Token test | 33.2 ± 2.1 (29–36) | 25.5 ± 5.8 (5–36) | 30.5 ± 3.1 (24–35) | 31.6 ± 2.3 (25–34) | <0.001 | 0.26 | 1.00 | <0.001 | <0.001 | 1.00 |
Values are numbers or means ± standard deviations (range). Disease duration was defined as years from onset to date of MRI scan. P values refer to ANOVA models, followed by post-hoc pairwise comparisons (Bonferroni-corrected for multiple comparisons), or Chi-squared test. Abbreviations: AD = Alzheimer's Disease; CDR = Clinical Dementia Rating Scale; HC = healthy controls; MCI = Mild Cognitive Impairment (c = converters; s = stable); MMSE = Mini Mental State Examination; N = Number; RAVLT = Rey Auditory Verbal Learning Test.
In both datasets (ADNI and Milan), the conversion from MCI to dementia was established clinically. This was a judgment made by skilled clinicians on the determination of whether or not there was significant interference in the ability to function at work or in usual daily activities based on the information obtained from the patient and from a knowledgeable caregiver.
This cross-sectional study was approved by the Local Ethical Committee on human studies and written informed consent from all subjects was obtained prior to their enrolment.
2.2. MRI acquisition protocol
Details about the ADNI MRI data acquisition protocol can be seen in ADNI's official webpage (ADNI.LONI.USC.EDU). Patients and healthy controls from the Milan dataset underwent a 3.0 T MR scan using a Philips Medical Systems Intera machine. The following sequences were acquired: (i) T2-weighted spin echo (SE) (repetition time [TR] = 3000 ms, echo time [TE] = 85 ms, flip angle = 90°, echo train length = 15, thickness = 3 mm, 46 contiguous axial slices, field of view [FOV] = 230 × 208 mm2, matrix size = 256 × 242); (ii) fluid-attenuated inversion recovery (FLAIR) (TR = 11,000 ms, TE = 120 ms, inversion time = 2800 ms, flip angle = 90°, echo train length = 21, thickness = 3 mm, 46 contiguous axial slices, FOV = 230 × 183 mm2, matrix size =256 × 192); and (iii) 3D T1-weighted fast field echo (TR = 25 ms, TE = 4.6 ms, flip angle = 30°, thickness = 1 mm, 220 contiguous axial slices, and in-plane resolution 0.89 × 0.89 mm2, FOV = 230 × 230 mm2, matrix size = 256 × 256).
2.3. MRI analysis
An experienced observer, blinded to patients' identity, performed MRI analysis. MRI analysis and CNN procedures were performed on a Dell Powerdge T630 Linux, including high-performance GPU NVIDIA Tesla K40, with 2880 CUDA cores and High Frequency Intel Xeon E5–2623 v3 with 78 GB memory overall.
3D T1-weighted images from both datasets were normalized to the MNI space using Statistical Parametric Mapping (SPM12; HTTP://WWW.FIL.ION.UCL.AC.UK/SPM/) and the Diffeomorphic Anatomical Registration Exponentiated Lie Algebra (DARTEL) registration method (Ashburner, 2007). Briefly, (i) T1-weighted images were segmented to produce GM, white matter (WM) and cerebrospinal fluid (CSF) tissue probability maps in the Montreal Neurological Institute (MNI) space; (ii) the segmentation parameters obtained from the step (i) were imported in DARTEL; (iii) the rigidly aligned version of the images previously segmented (i) was generated; (iv) the DARTEL template was created and the obtained flow fields were applied to the modulated 3D T1-weighted images of single subjects (generated by the segmentation step) to warp them to the common DARTEL space and then modulated using the Jacobian determinants. Since the DARTEL process warps to a common space that is smaller than the MNI space, we performed an additional transformation as follows: (v) the modulated 3D T1-weighted images from DARTEL were normalized to the MNI template using an affine transformation estimated from the DARTEL GM template and the a priori GM probability map without resampling (HTTP://BRAINMAP.WISC.EDU/NORMALIZEDARTELTOMNI).
2.4. Convolutional neural networks
Mimicking how the human brain processes information, the building blocks of deep learning networks, known as ‘artificial neurons’, are organized in layers in which each ‘neuron’ is fully connected to all ‘neurons’ in the next layer through weighted connections (Lecun et al., 2015). Briefly, deep learning networks (i) ‘learn’ from a series of inputs that are the data inputted into the model, (ii) propagate learned information through the network from the input to the output layer, (iii) calculate the error signal (i.e., difference between the network output and target value), and (iv) propagate the error signal back. After that, deep learning networks adjust their weights and repeat all the steps from (i) to (iv) until the error becomes as small as possible. Finally, trained networks are used to blind-predict the class of new (unseen) observations.
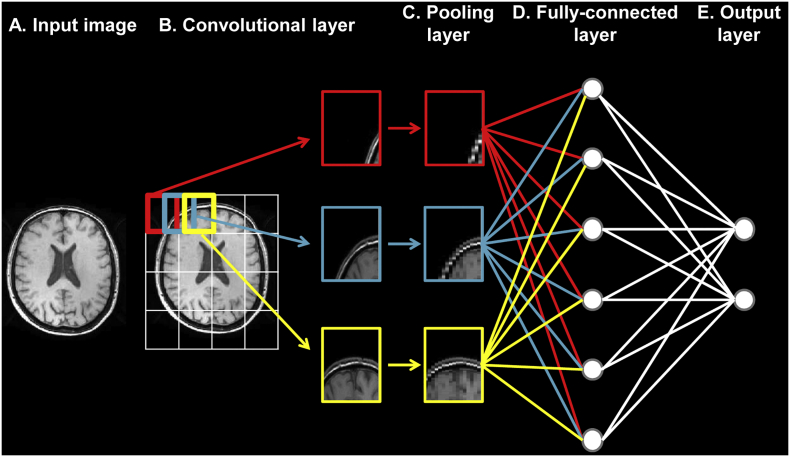
There are several architectures currently used for deep learning (Vieira et al., 2017). CNNs are a special type of feedforward neural networks that were initially designed to process images regardless various distortions, and as such are biologically-inspired by the visual cortex (Lecun et al., 1998). As illustrated in Fig. 1, standard CNNs typically alternate convolutional and max-pooling layers followed by a small number of fully-connected layers, in addition to the input and output layers. In the convolutional layer, which is the first neuronal layer receiving an input signal, neurons identify the main features that characterize the images, storing the information into a ‘feature map’ containing the relationship between the neurons and their features. Immediately after each convolutional layer, it is convention to apply a nonlinear layer (or activation layer). This layer, which just changes all the negative activations to 0, increases the nonlinear properties of the model and the overall network without affecting the receptive fields of the convolutional layer. The most common activation function is the Rectified Linear Unit, due to its faster training speed. A pooling (or subsampling) layer follows, which performs a downsampling operation along the spatial dimension. The last layers in the network are the fully-connected layers, where the neurons are connected to all neurons from the previous layer. CNN properties reduce the number of parameters that must be learned, thus improving training performance upon general deep learning algorithms (Lecun et al., 2015).
Fig. 1.
Architecture of a typical convolutional neural network. a) Input layer: the data is given to the network. b) Convolutional layer: neurons identify the main features that characterize the images, storing the information into a ‘feature map’ (e.g., red, blue and yellow blocks). c) Pooling layer: the size of each feature map is reduced with a downsampling operation along the spatial dimension (e.g., red, blue and yellow blocks). d) Fully-connected layer: the neurons are connected to all neurons from the previous layer. e) Output layer: the step that returns the probability of the input data to belong to each class.
Here, we introduce in detail the CNNs implemented in our study. First, given the volumetric nature of MR images, a network architecture that uses 3D convolutions was developed. The inputs were normalized 3D T1-weighted images and the outputs to be predicted were subject groups. The architecture of the network contains: 12 repeated blocks of convolutional layers (2 blocks with 50 kernels of size 5 × 5 × 5 with alternating strides 1 and 2 and 10 blocks with 100 to 1600 kernels of size 3 × 3 × 3 with alternating strides 1 and 2); a Rectified Linear Unit (activation layer); a fully-connected layer; and one output (logistic regression) layer. The network used in our study differs from the standard CNNs as max-pooling layers were replaced by standard convolutional layers with stride of 2 (‘all convolutional network’(Springenberg et al., 2015)). The "all convolutional network" is a basic architecture reaching good performance without the need for complicated activation functions, any response normalization or max-pooling (Springenberg et al., 2015). All software was written in Python using Theano, a scientific computing library with support for machine learning and GPU computing.
2.5. Experiments
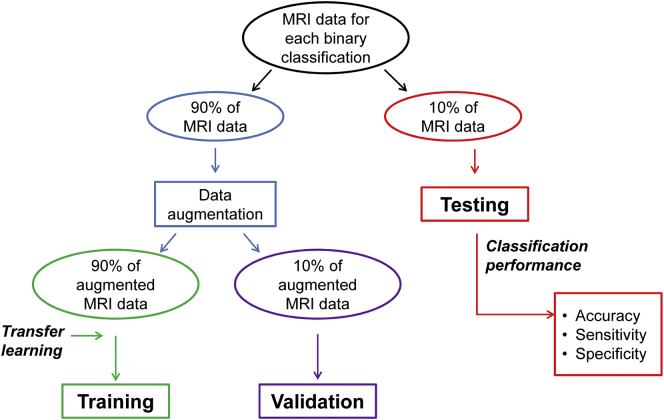
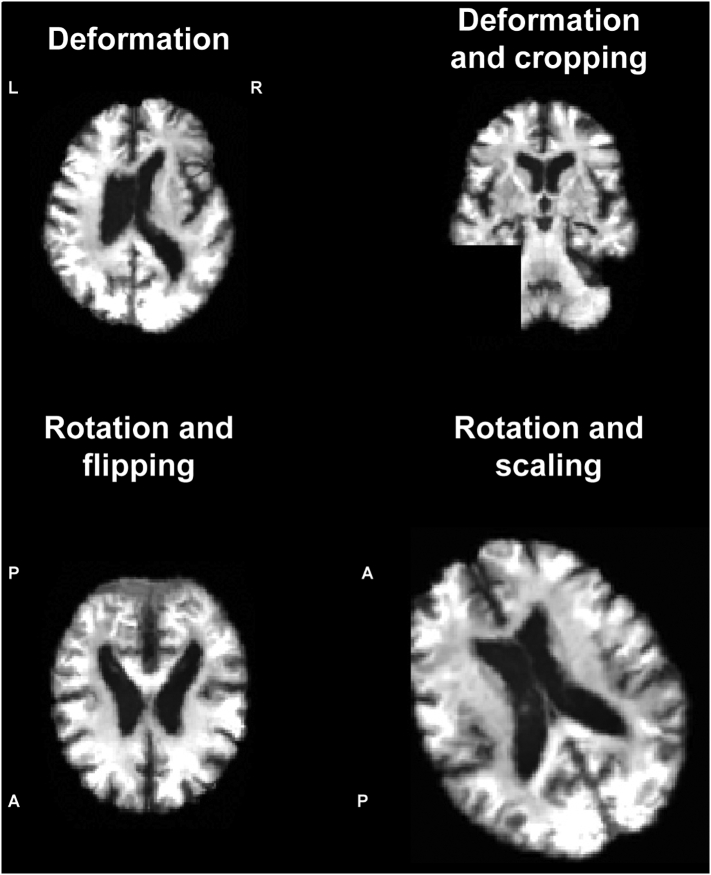
Performance of the 3D CNN was validated and tested on patients and controls, with six binary classifications: AD vs HC, c-MCI vs HC, stable MCI (s-MCI) vs HC, AD vs c-MCI, AD vs s-MCI, c-MCI vs s-MCI. For each classification, the CNN was evaluated firstly on ADNI dataset and then on ADNI + Milan dataset (12 classifications in total). Each classification included three steps (Fig. 2): (i) training, (ii) validation, and (iii) testing. First, MRI data of each classification dataset was randomly split into a large training and validation set (90% of images) and a testing set (10% of images). Data augmentation was then applied on images selected for training and validation (not testing) in order to generate additional artificial images and consequently prevent overfitting, which can occur when a fully connected layer occupies most of the parameters. Providing a CNN with more training and validation examples can reduce overfitting. Data augmentation strategy consisted of deformation, flipping, scaling, cropping and rotation of images (see examples in Fig. 3). We augmented the dataset of each subject group in any of the 12 classifications up to 1000. Each augmented dataset was randomly split into two subsets (90% for training and 10% for validation). For each classification, (i) CNN was trained on the augmented dataset and (ii) validated using a 10-fold cross validation. To improve the performance of our classifier, a so-called transfer learning was applied, i.e., weights of the CNN used to classify ADNI AD vs HC were transferred to the other CNNs and used as (pre-trained) initial weights (Hosseini-Asl et al., 2016). “Transferring” the learned features reduces training time and increases the network efficiency.
Fig. 2.
Flowchart of the main steps of the experiments performed. MRI data of each classification dataset (AD vs HC, c-MCI vs HC, s-MCI vs HC, AD vs c-MCI, AD vs s-MCI, c-MCI vs s-MCI) were randomly split into a large training and validation set (90% of images) and a testing set (10% of images). Data augmentation was applied on images selected for training and validation. See text for further details.
Fig. 3.
Examples of images after data augmentation, i.e., deformation, cropping, rotation, flipping, and scaling. Axial and coronal images are shown. A = anterior; L = left; P = posterior; R = right.
CNN was finally used to classify raw images of the testing set (iii). CNN's performance was evaluated by several performance measures, i.e. sensitivity, specificity and accuracy. Sensitivity measures the proportion of true positives correctly identified, whereas specificity refers to the proportion of true negatives correctly identified. The accuracy of a classifier represents the overall proportion of correct classifications.
3. RESULTS
Table 4 reports binary classification performances of the CNNs in the testing datasets. The results demonstrated that high levels of accuracy were achieved in all the comparisons. Highest accuracy, sensitivity and specificity (higher than 98%) were obtained in the AD vs HC classification tests using both the ADNI dataset and the combined ADNI + Milan dataset (Table 4). CNNs were also able to discriminate between c-MCI patients and HC with an optimal performance (accuracy, sensitivity and specificity values higher than 86%; Table 4). In distinguishing c-MCI from s-MCI subjects, CNNs reached an accuracy up to about 75%, with no differences between ADNI and non-ADNI images (Table 4).
Table 4.
Binary classification results on testing datasets.
| Accuracy | Sensitivity | Specificity | ||
|---|---|---|---|---|
| AD vs HC | ADNI dataset | 99.2% | 98.9% | 99.5% |
| ADNI + Milan dataset | 98.2% | 98.1% | 98.3% | |
| c-MCI vs HC | ADNI dataset | 87.1% | 87.8% | 86.5% |
| ADNI + Milan dataset | 87.7% | 87.3% | 88.1% | |
| s-MCI vs HC | ADNI dataset | 76.1% | 75.1% | 77.1% |
| ADNI + Milan dataset | 76.4% | 75.1% | 77.8% | |
| AD vs c-MCI | ADNI dataset | 75.4% | 74.5% | 76.4% |
| ADNI + Milan dataset | 75.8% | 74.8% | 77.1% | |
| AD vs s-MCI | ADNI dataset | 85.9% | 83.6% | 88.3% |
| ADNI + Milan dataset | 86.3% | 84.0% | 88.7% | |
| c-MCI vs s-MCI | ADNI dataset | 75.1% | 74.8% | 75.3% |
| ADNI + Milan dataset | 74.9% | 75.8% | 74.1% |
Abbreviations: AD = Alzheimer's disease; ADNI = Alzheimer's Disease Neuroimaging Initiative; HC = healthy controls; MCI = Mild Cognitive Impairment (c = converters; s = stable).
4. DISCUSSION
Effective and accurate AD diagnosis is critical for early treatment. Therefore many researchers have devoted their efforts to develop a computer-aided system, which can diagnose AD in the early stages and on an individual basis (Rathore et al., 2017; Vieira et al., 2017). In this study, we built and validated a deep learning algorithm that predicts the individual diagnosis of AD and MCI who will convert to AD based on a single cross-sectional brain structural MRI scan. Results showed that our CNN was highly-performing in differentiating AD and MCI patients from healthy controls and good-performing in predicting conversion to AD within 36 months. Importantly, our algorithm performed well without any prior feature engineering and regardless the variability of imaging protocols and scanners, demonstrating that it is exploitable by not-trained operators and likely to be generalizable to unseen patient data.
The strengths of our approach relative to previous deep learning studies in AD (Vieira et al., 2017) (Table 1) are several. First, heterogeneous MRI data proved to be a challenge for all evaluated models, with performance deteriorating more when images were obtained using different MR protocols and areas of the images known to be important for identity inference are inhomogeneous, deformed or lacking (Han et al., 2006; Takao et al., 2014). Structured programs aimed at standardizing and harmonizing MRI acquisition and analysis for AD diagnosis and management are ongoing in research settings (Frisoni et al., 2015; Reijs et al., 2015; Weiner et al., 2017). However, data obtained in these selected frameworks might not be representative of real-world populations. This is one of the main reasons why current diagnostic criteria for AD are extremely cautious on recommending the use of MRI in a clinical setting (Albert et al., 2011; Dubois et al., 2014; McKhann et al., 2011). In our experiments, CNN was trained, validated and tested using two datasets obtained by different MR protocols and scanners in order to capture the full spectrum of heterogeneity among data and provide a less dataset-specific approach. In fact, our approach overcomes the caveats of previous works, which have obtained data from single-center datasets leading to a limited reproducibility of findings.
We also observed that the studied model is not affected by image quality to different degrees as provided by data augmentation. Second, transfer learning from the AD vs controls ADNI comparison was applied for computational efficiency (Hosseini-Asl et al., 2016). Models trained with AD and control subjects can be particularly effective when attempting to distinguish c-MCI and s-MCI patients, as the differences among MCI groups are expected to be smaller than those between AD and controls (Bozzali et al., 2006). Therefore, a pre-trained model is the ideal tool to be used in routine clinical practice because it is a less time-consuming task and can provide high performance in distinguishing only slightly different images. Our approach is finally unique as we used a simplified CNN architecture called “all convolutional network”, which is optimized to achieve state-of-the-art performances with the minimum necessary CNN components (Springenberg et al., 2015). The great advantage of such a network model relative to standard CNNs is that it greatly reduces the number of network parameters and thus serves as a form of regularization (Springenberg et al., 2015).
As in previous supervised and unsupervised machine learning studies (Rathore et al., 2017; Vieira et al., 2017) (Table 1), accuracy in identifying c-MCI from s-MCI patients was not as high as when classifying AD or MCI patients from healthy controls. Using deep neural networks, combined with sparse regression models, a recent structural MRI study obtained a similar accuracy in identifying c-MCI patients (Suk et al., 2017). Importantly, multiple biomarker modalities may help enhance the diagnostic accuracy in MCI population. The most widely accepted diagnostic criteria for AD assume that the greatest accuracy can be achieved with a combination of amyloidosis and neurodegeneration biomarkers (Albert et al., 2011; Dubois et al., 2014; McKhann et al., 2011). It is worth noting that the accuracy achieved by our algorithm is also comparable to that of previous studies applying deep learning algorithms on multimodal datasets (e.g., clinical, cognitive, CSF, MRI, and PET (Vieira et al., 2017)), thus suggesting that there may be a huge margin of improvement using our simplified deep learning architecture in a multimodal biomarker framework. In light of this, in particular for the crucial comparison between c-MCI and s-MCI, future studies should consider to add other MRI sequences (such as functional MRI and/or diffusion tensor imaging), PET and CSF biomarkers together with neuropsychological scores and genetic information in order to improve the power of classification.
There are some limitations that need to be considered. First, we cannot exclude the presence of future c-MCI among s-MCI patients. Indeed, a longer clinical follow up may improve clinical diagnosis and thus our algorithm performance. Second, as previously mentioned, our model should be tested in combination with clinical, cognitive, genetic, PET and CSF biomarkers to improve the prediction of full-blown dementia development in MCI patients. Third, AD is a clinically heterogeneous disease and this should not be ignored. Effective diagnostic tools should be developed that can deal with atypical AD presentations, like posterior cortical atrophy and logopenic variant of primary progressive aphasia. Finally, neurodegeneration due to AD occurs years, even decades, before the clinical onset (Jack Jr. and Holtzman, 2013). Future studies are warranted to test the accuracy of the procedure in identifying subjects in the preclinical phase of the disease and, potentially, as a screening tool in the general population to identifying people at high risk of developing dementia.
In conclusion, CNNs show promises for building a model for the automated, individual and early detection of AD and thus accelerating the adoption of structural MRI in routine practice to help assessment and management of patients.
Acknowledgments
Acknowledgment
The authors thank the patients and their families for the time and effort they dedicated to the research.
Funding
This study was supported by the Italian Ministry of Health (grant number: GR-2011-02351217).
Disclosure statement
S. Basaia, L. Wagner, R. Santangelo and G. Magnani report no disclosures.
F. Agosta is Section Editor of NeuroImage: Clinical; has received speaker honoraria from Biogen Idec and Novartis; and receives or has received research supports from the Italian Ministry of Health, AriSLA (Fondazione Italiana di Ricerca per la SLA), and the European Research Council.
E. Canu has received research supports from the Italian Ministry of Health.
M. Filippi is Editor-in-Chief of the Journal of Neurology; received compensation for consulting services and/or speaking activities from Biogen Idec, Merck-Serono, Novartis, Teva Pharmaceutical Industries; and receives research support from Biogen Idec, Merck-Serono, Novartis, Teva Pharmaceutical Industries, Roche, Italian Ministry of Health, Fondazione Italiana Sclerosi Multipla, and ARiSLA (Fondazione Italiana di Ricerca per la SLA).
Acknowledgements
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award numberW81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (WWW.FNIH.ORG). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
REFERENCES
- Albert M.S., DeKosky S.T., Dickson D., Dubois B., Feldman H.H., Fox N.C., Gamst A., Holtzman D.M., Jagust W.J., Petersen R.C., Snyder P.J., Carrillo M.C., Thies B., Phelps C.H. The diagnosis of mild cognitive impairment due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J. A fast diffeomorphic image registration algorithm. NEUROIMAGE. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Bozzali M., Filippi M., Magnani G., Cercignani M., Franceschi M., Schiatti E., Castiglioni S., Mossini R., Falautano M., Scotti G., Comi G., Falini A. The contribution of voxel-based morphometry in staging patients with mild cognitive impairment. NEUROLOGY. 2006;67:453–460. doi: 10.1212/01.wnl.0000228243.56665.c2. [DOI] [PubMed] [Google Scholar]
- Dubois B., Feldman H.H., Jacova C., Hampel H., Molinuevo J.L., Blennow K., DeKosky S.T., Gauthier S., Selkoe D., Bateman R., Cappa S., Crutch S., Engelborghs S., Frisoni G.B., Fox N.C., Galasko D., Habert M.O., Jicha G.A., Nordberg A., Pasquier F., Rabinovici G., Robert P., Rowe C., Salloway S., Sarazin M., Epelbaum S., de Souza L.C., Vellas B., Visser P.J., Schneider L., Stern Y., Scheltens P., Cummings J.L. Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol. 2014;13:614–629. doi: 10.1016/S1474-4422(14)70090-0. [DOI] [PubMed] [Google Scholar]
- Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M., Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. NATURE. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B., Fox N.C., Jack C.R., Jr., Scheltens P., Thompson P.M. The clinical use of structural MRI in Alzheimer disease. NAT. REV. NEUROL. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B., Jack C.R., Jr., Bocchetta M., Bauer C., Frederiksen K.S., Liu Y., Preboske G., Swihart T., Blair M., Cavedo E., Grothe M.J., Lanfredi M., Martinez O., Nishikawa M., Portegies M., Stoub T., Ward C., Apostolova L.G., Ganzola R., Wolf D., Barkhof F., Bartzokis G., DeCarli C., Csernansky J.G., DeToledo-Morrell L., Geerlings M.I., Kaye J., Killiany R.J., Lehericy S., Matsuda H., O'Brien J., Silbert L.C., Scheltens P., Soininen H., Teipel S., Waldemar G., Fellgiebel A., Barnes J., Firbank M., Gerritsen L., Henneman W., Malykhin N., Pruessner J.C., Wang L., Watson C., Wolf H., DeLeon M., Pantel J., Ferrari C., Bosco P., Pasqualetti P., Duchesne S., Duvernoy H., Boccardi M. The EADC-ADNI harmonized protocol for manual hippocampal segmentation on magnetic resonance: evidence of validity. Alzheimers Dement. 2015;11:111–125. doi: 10.1016/j.jalz.2014.05.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisoni G.B., Boccardi M., Barkhof F., Blennow K., Cappa S., Chiotis K., Demonet J.F., Garibotto V., Giannakopoulos P., Gietl A., Hansson O., Herholz K., Jack C.R., Jr., Nobili F., Nordberg A., Snyder H.M., Ten Kate M., Varrone A., Albanese E., Becker S., Bossuyt P., Carrillo M.C., Cerami C., Dubois B., Gallo V., Giacobini E., Gold G., Hurst S., Lonneborg A., Lovblad K.O., Mattsson N., Molinuevo J.L., Monsch A.U., Mosimann U., Padovani A., Picco A., Porteri C., Ratib O., Saint-Aubert L., Scerri C., Scheltens P., Schott J.M., Sonni I., Teipel S., Vineis P., Visser P.J., Yasui Y., Winblad B. Strategic roadmap for an early diagnosis of Alzheimer's disease based on biomarkers. Lancet Neurol. 2017;16:661–676. doi: 10.1016/S1474-4422(17)30159-X. [DOI] [PubMed] [Google Scholar]
- Gupta A., Ayhan M., Maida A. Proceedings of the 30th International Conference on Machine Learning (ICML-13) 2013. Natural Image Bases to Represent Neuroimaging Data; pp. 987–994. [Google Scholar]
- Han X., Jovicich J., Salat D., van der Kouwe A., Quinn B., Czanner S., Busa E., Pacheco J., Albert M., Killiany R., Maguire P., Rosas D., Makris N., Dale A., Dickerson B., Fischl B. Reliability of MRI-derived measurements of human cerebral cortical thickness: the effects of field strength, scanner upgrade and manufacturer. NEUROIMAGE. 2006;32:180–194. doi: 10.1016/j.neuroimage.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Hosseini-Asl, E., Gimel'farb, G., El-Baz, A., 2016. Alzheimer's Disease Diagnostics by a Deeply Supervised Adaptable 3D Convolutional Network. arXiv:1607.00556v1 [cs.LG]. [DOI] [PubMed]
- Jack C.R., Jr., Holtzman D.M. Biomarker modeling of Alzheimer's disease. NEURON. 2013;80:1347–1358. doi: 10.1016/j.neuron.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y., Bottou L., Bengio Y., Haffner P. Gradient-Based learning applied to document recognition. PROC. IEEE. 1998;86:2278–2324. [Google Scholar]
- LeCun Y., Bengio Y., Hinton G. Deep learning. NATURE. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- McKhann G.M., Knopman D.S., Chertkow H., Hyman B.T., Jack C.R., Jr., Kawas C.H., Klunk W.E., Koroshetz W.J., Manly J.J., Mayeux R., Mohs R.C., Morris J.C., Rossor M.N., Scheltens P., Carrillo M.C., Thies B., Weintraub S., Phelps C.H. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payan, A., Montana, G., 2015. Predicting Alzheimer's disease: a neuroimaging study with 3D convolutional neural networks. arXiv:1502.02506 [cs.CV].
- Rathore S., Habes M., Iftikhar M.A., Shacklett A., Davatzikos C. A review on neuroimaging-based classification studies and associated feature extraction methods for Alzheimer's disease and its prodromal stages. NEUROIMAGE. 2017;155:530–548. doi: 10.1016/j.neuroimage.2017.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijs B.L., Teunissen C.E., Goncharenko N., Betsou F., Blennow K., Baldeiras I., Brosseron F., Cavedo E., Fladby T., Froelich L., Gabryelewicz T., Gurvit H., Kapaki E., Koson P., Kulic L., Lehmann S., Lewczuk P., Lleo A., Maetzler W., de Mendonca A., Miller A.M., Molinuevo J.L., Mollenhauer B., Parnetti L., Rot U., Schneider A., Simonsen A.H., Tagliavini F., Tsolaki M., Verbeek M.M., Verhey F.R., Zboch M., Winblad B., Scheltens P., Zetterberg H., Visser P.J. The Central Biobank and Virtual Biobank of BIOMARKAPD: A resource for studies on neurodegenerative diseases. FRONT. NEUROL. 2015;6:216. doi: 10.3389/fneur.2015.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf S., Tofighi G., for the Alzheimer's Disease Neuroimaging Initiative DeepAD: Alzheimer's Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI. 2016. HTTPS://WWW.BIORXIV.ORG/CONTENT/EARLY/2016/08/21/070441
- Springenberg, J.T., Dosovitskiy, A., Brox, T., M., R., 2015 Striving for Simplicity: The All Convolutional Net. ARXIV:1412.6806 [Cs.LG] ICLR-2015 Workshop.
- Suk H.I., Lee S.W., Shen D. Deep ensemble learning of sparse regression models for brain disease diagnosis. Medical Image Anal. 2017;37:101–113. doi: 10.1016/j.media.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao H., Hayashi N., Ohtomo K. Effects of study design in multi-scanner voxel-based morphometry studies. NEUROIMAGE. 2014;84:133–140. doi: 10.1016/j.neuroimage.2013.08.046. [DOI] [PubMed] [Google Scholar]
- Vieira S., Pinaya W.H., Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: Methods and applications. NEUROSCI. BIOBEHAV. REV. 2017;74:58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Weiner M.W., Veitch D.P., Aisen P.S., Beckett L.A., Cairns N.J., Green R.C., Harvey D., Jack C.R., Jr., Jagust W., Morris J.C., Petersen R.C., Saykin A.J., Shaw L.M., Toga A.W., Trojanowski J.Q. Recent publications from the Alzheimer's Disease Neuroimaging Initiative: Reviewing progress toward improved AD clinical trials. Alzheimers Dement. 2017;13:e1–e85. doi: 10.1016/j.jalz.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K., Hua Y., Gueroussov S., Najafabadi H.S., Hughes T.R., Morris Q., Barash Y., Krainer A.R., Jojic N., Scherer S.W., Blencowe B.J., Frey B.J. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. SCIENCE. 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]