Abstract
Background
We aimed to identify a magnetic resonance imaging (MRI)-based model for assessment of the risk of individual distant metastasis (DM) before initial treatment of nasopharyngeal carcinoma (NPC).
Methods
This retrospective cohort analysis included 176 patients with NPC. Using the PyRadiomics platform, we extracted the imaging features of primary tumors in all patients who did not exhibit DM before treatment. Subsequently, we used minimum redundancy-maximum relevance and least absolute shrinkage and selection operator algorithms to select the strongest features and build a logistic model for DM prediction. The independent statistical significance of multiple clinical variables was tested using multivariate logistic regression analysis.
Findings
In total, 2780 radiomic features were extracted. A DM MRI-based model (DMMM) comprising seven features was constructed for the classification of patients into high- and low-risk groups in a training cohort and validated in an independent cohort. Overall survival was significantly shorter in the high-risk group than in the low-risk group (P < 0·001). A radiomics nomogram based on radiomic features and clinical variables was developed for DM risk assessment in each patient, and it showed a significant predictive ability in the training [area under the curve (AUC), 0·827; 95% confidence interval (CI), 0.754–0.900] and validation (AUC, 0.792; 95% CI, 0.633–0.952) cohorts.
Interpretation
DMMM can serve as a visual prognostic tool for DM prediction in NPC, and it can improve treatment decisions by aiding in the differentiation of patients with high and low risks of DM.
Fund
This research received financial support from the National Natural Science Foundation of China (81571664, 81871323, 81801665, 81771924, 81501616, 81671851, and 81527805); the National Natural Science Foundation of Guangdong Province (2018B030311024); the Science and Technology Planning Project of Guangdong Province (2016A020216020); the Scientific Research General Project of Guangzhou Science Technology and Innovation Commission (201707010328); the China Postdoctoral Science Foundation (2016M600145); and the National Key R&D Program of China (2017YFA0205200, 2017YFC1308700, and 2017YFC1309100).
Keywords: Distant metastasis, Nasopharyngeal carcinoma, Risk assessment, Prognostic tool, Magnetic resonance imaging
Research in context.
Evidence before this study
Distant metastasis (DM) is currently the main cause of treatment failure in patients with nasopharyngeal carcinoma (NPC). Most NPC patients rapidly progress to death due to metastatic progression; therefore, accurate pretreatment risk assessment for DM is urgently needed so that aggressive therapeutic strategies can be used to treat high-risk individuals. Magnetic resonance imaging (MRI)-based radiomics patterns have been used as prognostic biomarkers in various types of cancers. Recently, a few radiomics models were developed for the evaluation of progression-free survival and progression in patients with advanced NPC; these have shown significant prognostic capabilities. However, to the best of our knowledge, a risk assessment model for DM, which is the primary cause for treatment failure in patients with NPC, has not been established.
Added value of this study
In this study, we used high-throughput extraction of data-characterization algorithms to extract specific radiomic features from patients with NPC and created a DM MRI-based model combining radiomic features and clinical variables. This model allowed the classification of patients into high-risk and low-risk groups.
Implications of all the available evidence
A nomogram combining radiomic features and clinical variables serves as a visual tool that aids clinicians in making optimal decisions and identifying patients with a high risk of DM in a timely and cost-effective manner.
Alt-text: Unlabelled Box
1. Introduction
Nasopharyngeal carcinoma (NPC) is an epidemic in South China, Southeast Asia, and North Africa [1,2]. Notably, it is a highly radiosensitive cancer [3], and with advancements in radiotherapy, control of local recurrence has substantially improved. Treatment failure primarily occurs in patients with distant metastasis (DM) [4]. Although advances in chemoradiotherapy have improved DM control, the prognosis remains poor, with a 5-year survival rate of <10% [5]. Most patients rapidly progress to death due to metastatic progression; therefore, accurate pretreatment risk assessment for DM is urgently required so that aggressive therapeutic strategies can be used for the treatment of high-risk individuals.
Currently, clinical treatment decisions for NPC are taken on the basis of the tumor–nodule–metastasis (TNM) stage [6]. However, in a previous study, although patients with the same TNM stage received similar therapies, >20% eventually developed DM and showed poor response to treatment [7]. Treatment failure was attributed to the use of the TNM staging system, which simply reflects the anatomy of tumor invasion and ignores intratumor variations. Previous studies in patients with NPC have expended substantial efforts for predicting DM by using different clinical variables such as the plasma Epstein–Barr virus DNA (EBV DNA) status, C-reactive protein (CRP) level, serum lactate dehydrogenase (LDH) level, and N-classification [1,8,9]. A previous study showed that EBV DNA can be a useful independent biomarker of DM from NPC [10]. However, most predictive models are based on pretreatment examination of blood metabolism, which provides unstable and nonspecific results. Several cellular and genetic parameters reflecting intratumor heterogeneity, such as HOP homeobox (HOPX), microRNA, and gene expression, have also been used to predict metastasis from NPC [3,[11], [12], [13], [14]]. However, further tests are needed before these markers can be used in the clinic, and the high cost of such examinations confines their usage to a small number of patients.
Radiological methods, including conventional radiography, computed tomography (CT), magnetic resonance (MR) imaging (MRI), and positron emission tomography/CT (PET/CT), are routinely and widely used in clinical practice because they are noninvasive, repeatable, and inexpensive [15,16]. Radiomics, an emerging field of medical study, involves the transformation of conventional medical images into analyzable quantitative imaging features extracted by data-characterization algorithms. It has been applied in various cancers [head and neck cancer [17], colorectal cancer [18], lung cancer [19,20], and breast cancer [21] and imaging modalities (MRI, CT, and PET/CT) [[22], [23], [24]]. Emerging reports have revealed that radiomics can evaluate the tumor histopathology as well as progression-free survival (PFS), recurrence, metastasis, and other clinical outcomes [15,[25], [26], [27]]. Recently, a few radiomics models were developed for the evaluation of PFS and progression in patients with advanced NPC [28,29]. However, to the best of our knowledge, a model for assessing the risk of DM, which is the primary cause for treatment failure in NPC, has not been established.
Therefore, the aim of this study was to develop and validate an MRI-based model for the prediction of DM before initial treatment in patients with NPC. We also integrated radiomic features with clinical variables to build a visual nomogram with greater accuracy for the identification of individuals with a high risk of DM.
2. Materials and methods
The institutional review board of Guangdong General Hospital approved this retrospective analysis of anonymous data and waived the requirement for informed consent. In total, 176 consecutive patients with previously untreated, biopsy-proven NPC were enrolled between August 2009 and October 2014. The histological tumor subtypes were categorized according to the World Health Organization (WHO) standards [30] as follows: type I (differentiated keratinizing carcinoma), type II (differentiated nonkeratinizing carcinoma), and type III (undifferentiated nonkeratinizing carcinoma). The inclusion and exclusion criteria are described in Supplementary Methods 1. Eligible patients were randomly divided into a training cohort (n = 123) and an independent validation cohort (n = 53) in a ratio of 7:3. Data concerning the following 18 conventional clinical variables were obtained from the medical records: age; sex; T stage; N stage; histological subtype; pretreatment platelet (PLT), white blood cell (WBC), neutrophil (NE), and lymphocyte (LY) counts; and pretreatment levels of plasma EBV DNA, viral capsid antigen immunoglobulin A (VCA-IgA), early antigen immunoglobulin A (EA-IgA), C-reactive protein (CRP), lactate dehydrogenase (LDH), alkaline phosphatase (ALP), albumin (ALB), alanine aminotransferase (ALT), aspartate aminotransferase (AST), and hemoglobin (HGB). We also calculated NE/WBC, NE/LY, and ALB/ALP ratios.
All enrolled patients underwent head and neck MRI, chest radiography, abdominal ultrasound, PET/CT, and/or skeletal scintigraphy. Patients were staged on the basis of the 7th American Joint Committee on Cancer (AJCC) TNM staging manual [31]. If DM was mentioned in any medical reports, the potentially involved sites were subjected to additional examinations, including CT, MRI, PET/CT, and/or biopsy. If the presence of DM was immediately confirmed by these additional examinations, the diagnosis was accepted. If additional examinations were not feasible or yielded negative results, follow-up examinations were performed every 3 months for at least 12 months. Patients were considered to have locoregional disease if the lesion remained unchanged during the follow-up period. If lesion enlargement was observed, DM was considered present.
The primary endpoint of this study was defined as the time from the date of the first MRI study to the date of DM detection or the date of censoring (date on which the patient was last known to be DM-free. Five-year survival was analyzed as a secondary endpoint and defined as the time between the date of the first MRI study and the date of censoring (date on which the patient was last known to be alive or date of death from any cause) or the end of 60 months, whichever was earlier. The minimum follow-up duration for patients without DM was 36 months after the first MRI study, while the maximum follow-up duration was 60 months. Because >90% metastatic NPCs occur in the first 3 years after initial treatment, the 5-year outcomes may reflect the long-term effects of radiation toxicity combined with changes in the tumor biology.
2.1. Treatment
Treatment comprised curative-intent chemoradiotherapy for the nasopharynx, cervical lymph nodes, and adjacent at-risk tissues. Patients with stage I tumors were treated with radiotherapy alone, while those with stage II tumors received concurrent chemoradiotherapy. Advanced stage tumors (stages III, IVA, and IVB) were treated with concurrent chemoradiotherapy and adjuvant chemotherapy [32,33]. The concurrent chemoradiotherapy regimen comprised cisplatin (40 mg/m2 for 1–5 days per week, 6–7 cycles), beginning on the first day of radiotherapy. A cumulative radiation dose of ≥66 Gy was delivered to the primary tumor, while a dose of 60–66 Gy was delivered to the involved cervical regions. Adjacent at-risk tissues received a dose of ≥50 Gy.
2.2. Image acquisition, normalization, and segmentation
In total, 76 patients underwent nasopharyngeal and neck MRI using a 1ꞏ5-T device (Optima, TwinSpeed, GE Healthcare, Milwaukee, WI, USA, n = 42; Achieva, Philips Healthcare, The Netherlands, n = 34), while 100 underwent nasopharyngeal and neck MRI using a 3ꞏ0-T device (DISCOVERY, TwinSpeed, GE Healthcare, n = 32; Ingenia, Philips Healthcare, n = 68). The image acquisition parameters are described in Supplementary Methods 2. We acquired axial T2-weighted (T2-w) Digital Imaging and Communications in Medicine (DICOM) images and contrast-enhanced T1-weighted (CET1-w) DICOM images that had been archived using the Picture Archiving and Communication Systems (PACS), with normalization. Because different MRI devices with various protocols were used in this study, the intensity range of the images was normalized from 0 to 255 using the PyRadiomics platform, which is an open-source platform implemented in Python 3.6.5 (https://www.python.org/).
Segmentation for regions of interest (ROIs) was performed using ITK-SNAP software (open source software; https://itk.org/). All images were manually segmented by a radiologist with 11 years of experience in head and neck MR image interpretation (O.Y., reader 1), who repeated the same procedure after 2 weeks. The interobserver reproducibility of each segmentation was evaluated by another radiologist with 10 years of clinical experience (B.G., reader 2). ROI for the entire tumor in each patient was delineated on each slice of both axial T2-w and CET1-w images.
2.3. Feature extraction and selection and model development
Imaging features were extracted using the PyRadiomics platform that extracts standardized radiomic features from medical image data by using a large panel of engineered hard-coded feature algorithms (http://www.radiomics.io/pyradiomics.html) [34]. This platform is used to standardize feature extraction for the achievement of reproducibility and comparability among the results. Furthermore, it is used to explain the meaning and origin of each feature. In the present study, we extracted three standardized feature classes: first-order statistics, shape descriptors, and texture features [including gray level cooccurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), gray level dependence matrix (GLDM), and neighboring gray tone difference matrix (NGTDM)] [[35], [36], [37]]. In total, we extracted 2803 radiomic features from axial T2-w and CET1-w images using the PyRadiomics platform.
We used intra- and interclass correlation coefficient (ICC) to assess the effects of variations in manual segmentation on radiomic feature values and calculated the intra- and interobserver stabilities for each extracted feature. ICC was calculated for the 2803 radiomic features, and a value of >0.75 indicated good agreement.
After ICC calculation, all selected features (ICC > 0.75) were integrated with 22 clinical variables for the construction of a stable and reliable primary dataset. The R package was used for feature selection. We performed initial selection from the primary dataset using univariate analysis with the Mann–Whitney U and χ2 tests. The p-value threshold for the remaining significant variables was set at 0.05 (P < 0.05). Univariate analysis was used during the initial filtering step to filter out many unrelated features; this enabled calculation of the optimal feature set through multivariate analysis. Then, the minimum redundancy-maximum relevance (mRMR) algorithm was used to rank features, and the top 10% features were selected [38]. Importantly, the mRMR algorithm is an entropy-based feature selection method that initially calculates the mutual information (MI) between a set of features and an outcome variable. It ranks input features by maximizing MI with respect to the outcome and subsequently minimizes the average MI for features with higher rankings. The most marginally significant features with the highest area under the curve (AUC) were then selected using the least absolute shrinkage and selection operator (LASSO) algorithm. Use of the LASSO algorithm could result in overfitting and bias; therefore, backward elimination was added to reduce the number of remaining final features. Using the final features, we constructed a classification model called a distant metastasis MRI-based model (DMMM), with coefficients weighted by logistic regression analysis in the training cohort. An optimal cutoff value for classifying the patients into low- and high-risk groups based on the risk of DM was calculated in R with the pROC package [39]. Feature selection and model development were performed in the training cohort, while the independent validation cohort was used to evaluate the performance of the model. An overview of the DMMM development process is presented in Fig. 1.
Fig. 1.
Flow diagram showing the development of a distant metastasis (DM) magnetic resonance (MR) imaging (MRI)-based model (DMMM) for patients with nasopharyngeal carcinoma. The steps include (1) MR image acquisition and segmentation, (2) extraction of features using the PyRadiomics platform, and (3) selection of features and development of the model.
We drafted a nomogram, which was based on coefficients weighted by the logistic regression model, in R with the rms package. Calibration curves were graphically assessed by plotting the observed rates against the nomogram-predicted probabilities via a bootstrap method with 1000-iteration resampling. We also performed multivariate logistic regression analysis to test the independent significance of distinct clinical variables.
2.4. Statistical analysis
Kaplan–Meier survival curves and the log-rank test were used to compare overall survival between the high-risk and low-risk groups. The clinical characteristics of the training and validation cohorts were compared using an independent samples t-test, Fisher's exact test, the χ2 test, or the Mann–Whitney U test, as appropriate. AUC was calculated to assess the predictive ability of the model. A two-tailed P-value of <0.05 was considered statistically significant. All statistical analyses were performed using R software (version 3.2.1) and Statistical Package for Social Sciences (SPSS) software (version 23.0). The R software packages used for our statistical analyses are described in Supplementary Methods 3. Decision curve analysis (DCA) was used to evaluate the clinical usefulness of DMMM by calculation of the net benefit for a range of threshold probabilities [40,41] using the following formula:
where n is the total number of patients in the study and pt is the threshold probability.
3. Results
A total of 176 patients with nonmetastatic, pretreatment NPC was included in our study. The patients' clinical characteristics are summarized in Table 1. The training and validation cohorts were similar in terms of the baseline clinical characteristics (P > 0.05). Among the enrolled patients, 87.5% were treated with chemoradiotherapy and 12.5% were treated without concurrent chemoradiotherapy.
Table 1.
Characteristics of patients with nasopharyngeal carcinoma (NPC) in the training and validation cohorts. Statistical comparisons between the training and validation cohorts were computed using the Mann–Whitney U test for continuous variables and χ2 test for categorical variables. A P-value of <0.05 indicates a significant difference.
| Patients | Training cohort (n = 123) |
P value | Validation cohort (n = 53) |
P value | |||
|---|---|---|---|---|---|---|---|
| DM | Non-DM | DM | non-DM | ||||
| Age (years) | 0.914 | 0.800 | |||||
| <43 | 93 | 26 | 40 | 7 | 20 | ||
| ≥43 | 83 | 23 | 34 | 5 | 21 | ||
| Sex | 0.435 | 0.806 | |||||
| Male | 135 | 40 | 56 | 8 | 31 | ||
| Female | 41 | 9 | 18 | 4 | 10 | ||
| T stage | 0.306 | 0.160 | |||||
| T1 | 14 | 1 | 7 | 1 | 5 | ||
| T2 | 39 | 14 | 17 | 2 | 6 | ||
| T3 | 79 | 19 | 31 | 4 | 25 | ||
| T4 | 44 | 15 | 19 | 5 | 5 | ||
| N stage | 0.016 | 0.448 | |||||
| N0 | 19 | 2 | 11 | 1 | 5 | ||
| N1 | 56 | 12 | 30 | 4 | 10 | ||
| N2 | 78 | 25 | 27 | 4 | 22 | ||
| N3 | 23 | 10 | 6 | 3 | 4 | ||
| TNM Stage | 0.027 | 0.005 | |||||
| I | 3 | 0 | 1 | 0 | 2 | ||
| II | 21 | 3 | 16 | 2 | 0 | ||
| III | 91 | 25 | 33 | 3 | 30 | ||
| IVA | 38 | 12 | 18 | 3 | 5 | ||
| IVB | 23 | 7 | 8 | 4 | 4 | ||
| Histology | 0.525 | – | |||||
| Differentiated keratinising | 0 | 0 | 0 | 0 | 0 | ||
| Differentiated non-keratinising | 4 | 1 | 3 | 0 | 0 | ||
| Undifferentiated non-keratinising | 172 | 48 | 71 | 12 | 41 | ||
| EBV DNA (copies/mL) | <0.001 | 0.446 | |||||
| <3245 | 88 | 15 | 48 | 4 | 21 | ||
| ≥3245 | 88 | 34 | 26 | 8 | 20 | ||
| VCA-IgA | 0.144 | 0.583 | |||||
| <1:160 | 69 | 26 | 24 | 3 | 16 | ||
| ≥1:160 | 107 | 23 | 40 | 9 | 25 | ||
| EA-IgA | 0.268 | 0.800 | |||||
| <1:20 | 90 | 29 | 35 | 5 | 21 | ||
| ≥1:20 | 86 | 20 | 39 | 7 | 20 | ||
| CRP concentration (mg/L) | 0.882 | 0.687 | |||||
| <2.06 | 87 | 23 | 37 | 5 | 22 | ||
| ≥2.06 | 89 | 26 | 37 | 7 | 19 | ||
| LDH concentration (U/L) | 0.553 | 0.864 | |||||
| <185 | 103 | 27 | 46 | 6 | 24 | ||
| ≥185 | 73 | 22 | 28 | 6 | 17 | ||
| Alkaline phosphatase (U/L) | 0.325 | 0.771 | |||||
| <89 | 113 | 28 | 50 | 9 | 26 | ||
| ≥89 | 63 | 21 | 24 | 3 | 15 | ||
| Albumin (g/L) | 0.827 | 0.775 | |||||
| <44 | 79 | 21 | 29 | 7 | 22 | ||
| ≥44 | 97 | 28 | 45 | 5 | 19 | ||
| Alanine aminotransferase (U/L) | 0.98 | 0.771 | |||||
| <31 | 121 | 33 | 50 | 9 | 29 | ||
| ≥31 | 55 | 16 | 24 | 3 | 12 | ||
| Aspartate aminotransferase (U/L) | 0.525 | 0.930 | |||||
| <25 | 125 | 33 | 55 | 9 | 28 | ||
| ≥25 | 51 | 16 | 19 | 3 | 13 | ||
| WBC, ×109/L | 0.713 | 0.941 | |||||
| <7.13 | 91 | 24 | 40 | 6 | 21 | ||
| ≥7.13 | 85 | 25 | 34 | 6 | 20 | ||
| Neutrophil, ×109/L | 0.906 | 0.965 | |||||
| <4.73 | 101 | 29 | 43 | 6 | 23 | ||
| ≥4.73 | 75 | 20 | 31 | 6 | 18 | ||
| Lymphocytes, ×109/L | 0.656 | 0.639 | |||||
| <1.90 | 96 | 28 | 38 | 8 | 22 | ||
| ≥1.90 | 80 | 21 | 36 | 4 | 19 | ||
| Platelet counts, ×109/L | 0.185 | 0.062 | |||||
| <217 | 88 | 28 | 32 | 3 | 25 | ||
| ≥217 | 88 | 21 | 42 | 9 | 16 | ||
| Hemoglobin concentration (g/L) | 0.966 | 0.869 | |||||
| <140 | 74 | 21 | 32 | 5 | 16 | ||
| ≥140 | 102 | 28 | 42 | 7 | 25 | ||
| Concurrent chemoradiotherapy | 0.838 | 0.375 | |||||
| Yes | 153 | 43 | 64 | 9 | 37 | ||
| No | 23 | 6 | 10 | 3 | 4 | ||
In total, 2803 radiomic features were extracted from axial T2-w and CET1-w images. From these features, we selected 1914 with high stability (ICC > 0.75) and integrated them with 22 clinical variables for analyses using Mann–Whitney U and χ2 tests. A total of 507 features showed a significant association with DM. After the mRMR algorithm was applied, 50 features remained and were subjected to further selection by the LASSO algorithm and backward elimination. The final seven remaining features were used to build DMMM: N stage, “CET1-w_wavelet.HHL_GLSZM_ Zone-Size Non-uniformity Normalized,” “T2-w_wavelet.LLH_GLCM_Correlation,” “T2-w_squareroot_GLCM_Idmn,” “CET1-w_exponential_GLDM_Dependence Variance,” “CET1-w_wavelet.HLL_GLCM_Correlation,” and “T2-w_squareroot_NGTDM_Strength.” We included these features in logistic regression analysis, which yielded DMMM with an AUC of 0.827 [95% confidence interval (CI), 0.754–0.900] in the training cohort and 0.792 (95% CI, 0.633–0.952) in the validation cohort. The receiver operating characteristic curve (ROC) is presented in Fig. 2. We calculated the risk score for each patient using a formula resulting from the seven features weighted by their regression coefficients:
Fig. 2.
Receiver operating characteristic (ROC) curves for a newly developed distant metastasis (DM) magnetic resonance (MR) imaging (MRI)-based model (DMMM) (a, b), radiomic features (c, d), and clinical variables (e, f) in the training and validation cohorts of patients with nasopharyngeal carcinoma. The ROC curves of DMMM is outperformed than radiomic features and clinical variables alone in both the training (AUC, 0.827 vs. 0.816 vs. 0.652) and validation (AUC, 0.792 vs. 0.713 vs. 0.660) cohorts.
The optimal cutoff value for dividing the patients into high- and low-risk groups was 0.37. Accordingly, 62 (35.2%) and 114 (64.8%) patients were categorized into the high-risk (risk score > 0.37) and low-risk (risk score < 0.37) groups, respectively. The 5-year survival rate was 12% for the high-risk group and 26% for the low-risk group, with a significant difference between groups (P < 0.001; Fig. 3a).
Fig. 3.
Kaplan–Meier curves for 5-year survival in patients with nasopharyngeal carcinoma. For patients with high and low risks of distant metastasis (DM), stratified by our newly developed DM magnetic resonance (MR) imaging (MRI)-based model (DMMM). The 5-year survival rate is 12% for the high-risk group and 26% for the low-risk group, with a significant difference between groups (P < 0.001). b. For high-risk and low-risk patients who received concurrent chemoradiotherapy (stratified using DMMM). The 5-year survival rate of high-risk patients who received concurrent chemoradiotherapy is significantly lower than that of low-risk patients who received the same treatment (P < 0.001). The log-rank test was used to calculate P-values.
Subgroup analysis for 5-year survival was performed for patients with early-stage (stages I and II) and advanced-stage (stages III and IV); there was no significant difference between groups (P = 0.39; data not shown). The 5-year survival rate of high-risk patients who received concurrent chemoradiotherapy was significantly lower than that of low-risk patients who received the same treatment (P < 0.001; Fig. 3b).
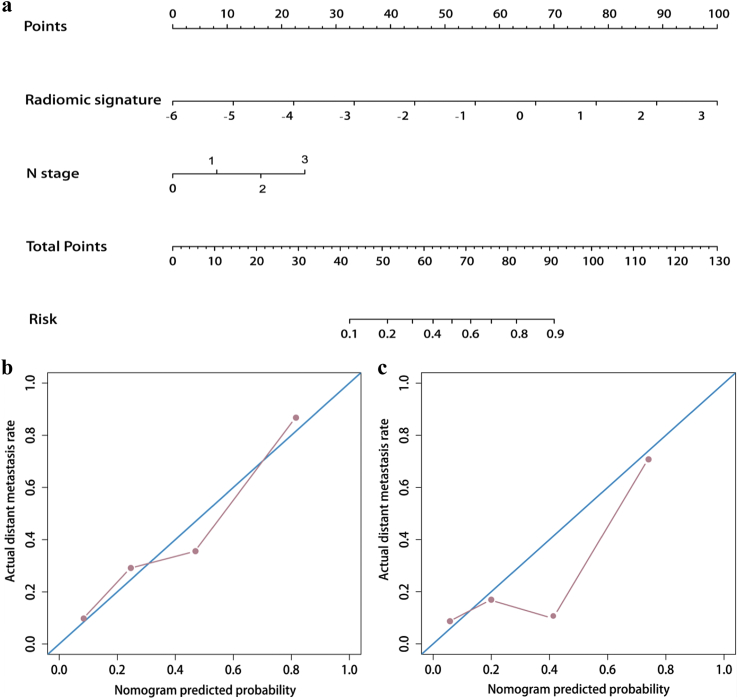
A nomogram was generated on the basis of the seven radiomic features for prediction of the risk of DM in NPC (Fig. 4a). Fig. 4b and c illustrate the calibration curves of the nomogram; there was good calibration in the training and validation cohorts. According to DCA, when the threshold probability for a patient was within the range of 0% to 100%, the nomogram added more net benefit than the “treat all” or “treat none” strategies. DCA for the nomogram is presented in Fig. 5.
Fig. 4.
Nomogram for the prediction of distant metastasis (DM) in patients with nasopharyngeal carcinoma (NPC) (a) Calibration curves of nomograms developed in the training (b) and validation (c) cohorts.
Fig. 5.
Decision curve analysis for our newly developed distant metastasis (DM) magnetic resonance (MR) imaging (MRI)-based model (DMMM) for the prediction of DM in patients with nasopharyngeal carcinoma. The y-axis measures the net benefit. The red line represents DMMM with integrated radiomic features and clinical variables. The net benefit is calculated by adding the benefits (true-positive results) and subtracting the risks (false-positive results), with the latter weighted by a factor related to the harm of an undetected cancer relative to the harm of unnecessary treatment. Our DMMM shows the highest net benefit when compared with simple strategies [e.g., follow-up of all patients (yellow line) or no patients (horizontal black line)] across the entire range of threshold probabilities at which a patient could choose to undergo follow-up imaging studies. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We additionally built a clinical model based on the clinical variables. After multivariate analysis, only N stage and the plasma EBV DNA level remained as independent predictors of DM. The clinical model yielded an AUC of 0.652 (95% CI, 0.553–0.752) in the training cohort and 0.660 (95% CI, 0.503–0.818) in the validation cohort. We also built a radiomic signature based on the imaging features alone. After multivariate analysis, five imaging features were selected to build the radiomic signature, which showed an AUC of 0.816 (95% CI, 0.738–0.895) in the training cohort and 0.713 (95% CI, 0.554–0.872) in the validation cohort. Because of an AUC bias of 0.103 between the training and validation cohorts, we performed cross-validation using out-of-bag bootstrapping (described in Supplementary Methods 4). The results revealed that the bias was not an outlier, thus showing the potential overfitting of the tumor phenotype on the small-scale dataset. The predictive value of DMMM was increased by incorporating both clinical variables and imaging features.
4. Discussion
In the present study, we used high-throughput extraction of data-characterization algorithms to extract specific radiomic features and construct DMMM with combined radiomic features and clinical variables for the prediction of DM before initial treatment in patients with NPC. The patients could be classified into high-risk and low-risk groups by using our DMMM. Patients in the high-risk group exhibited a significantly lower 5-year overall survival rate (12%) than did patients in the low-risk group (26%). We also demonstrated that the radiomic features constituted independent risk factors for DM.
The DM-related radiomic features with the maximum significance in the present study were different from those in a previous study [28], with the exception of the “CET1-w_wavelet.HLL_ GLCM_Correlation” feature, which is extracted from CET1-w images and considered to reflect intratumor heterogeneity from the perspective of grayscale extension. However, this feature was negatively associated with the risk of DM in our study. In contrast, the remaining five radiomic features were specific for predicting the risk of DM. Notably, we found a strong positive correlation between the “CET1-w_wavelet.HHL_GLSZM_ Zone-Size Non-uniformity Normalized” feature, which is related to variability in gray values in GLSZM, and the risk of DM. Typically, greater variability in gray values is associated with greater intratumor heterogeneity and a higher risk of DM. For instance, Li et al. [26] extracted quantitative features from breast MR images and showed that radiomic features included the tumor size and enhancement texture, which indicated tumor heterogeneity. Furthermore, they showed that a smaller enhancement texture was associated with greater tumor heterogeneity, which appears to indicate a higher risk of recurrence. Kwan et al. [42] and Vallieres et al. [43] also suggested that radiomic features could reflect the characteristics of intratumor heterogeneity and were associated with DM. Other DM-related radiomic features reflect intratumor heterogeneity in terms of local heterogeneity, grayscale extension, variance, and gray value differences in different matrices. Thus, we demonstrated that our DMMM could reflect intratumor heterogeneity, with an AUC of 0.827 in the training cohort and 0.792 in the validation cohort.
We determined that N stage and the EBV DNA level were outstanding clinical predictors of DM in patients with NPC. Moreover, N stage was the only clinical feature that could predict DM in combination with the six identified radiomic features. Although the pretreatment EBV DNA level is an important risk factor and a predictor of the tumor response in patients with metastatic NPC [1,5], our results suggested a much stronger positive correlation between N stage and DM risk in patients with NPC. We also found that DMMM performed better than the clinical model in both the training (AUC, 0.827 vs. 0.652) and validation (AUC, 0.792 vs. 0.660) cohorts. Therefore, DMMM developed in the present study is not only a simple combination of radiomic features but also a synergy between intratumor heterogeneity and clinical variables.
Our data showed that patients in the low-risk group could achieve greater benefit from concurrent chemoradiotherapy than could patients in the high-risk group. This result is consistent with that of a previous study where a DM gene signature (DMGN) was built for locoregionally advanced NPC [3]. The findings of that study suggested that patients considered to be at low risk as per the DMGN tool experienced benefits from concurrent chemoradiotherapy, whereas patients considered to be at high risk did not experience any benefit. Chen et al. [32] demonstrated that concurrent chemoradiotherapy is not the most effective choice for patients with high-risk DM; instead, aggressive therapeutic strategies such as high-dose radiation, adjuvant therapy, and molecular target therapy, should be considered for these patients. Accordingly, we constructed a nomogram based on DMMM for prediction of the metastatic risk and management of therapeutic strategies for each patient with NPC. The parameters of the nomogram can be easily acquired. For instance, N stage is a conventional predictive factor and a component of the TNM system in patients with malignancies. Moreover, the radiomic features could be obtained via common algorithms, which could transform nasopharyngeal and neck MR imaging features into analytical quantitative features. In summary, our study showed that the nomogram can serve as either a scoring system or a visualization tool for DM prediction in patients with NPC, thus aiding physicians in rapid evaluation of the metastatic risk via a simple calculation method in the clinic.
This study has several limitations. First, it is known that the AUC metric is generally used to assess the predictive accuracy of clinical models, although it does not contain information about follow-up or clinical consequences. Second, our study included a small number of enrolled patients; therefore, despite our efforts to prevent overfitting and increase the generalizability (univariate analysis, 10-fold cross-validation in LASSO), the prediction model can be further optimized with larger datasets. Accordingly, multicenter prospective trials with larger patient samples are needed in both NPC-endemic and nonendemic areas in order to improve the clinical efficacy of our model [41]. Third, various MRI devices were used for the extraction of radiomic features. We noted that the use of different field strengths was beneficial in terms of applicability to other sites and centers; however, many radiomic features may have been filtered out despite superior performance at a particular field strength.
In conclusion, we developed a novel DMMM combining radiomic features and clinical variables for the prediction of DM before initial treatment in patients with NPC and the differentiation of patients with high and low risks of DM. Our nomogram can serve as a visual prognostic tool that can aid clinicians in identifying patients with a high risk of DM and accordingly optimizing their therapeutic strategies.
Data sharing statement
The data are not available for public access because of patient privacy concerns but are available from the corresponding author on reasonable request approved by the institutional review boards of Guangdong General Hospital.
Funding sources
This research received financial support from the National Natural Science Foundation of China (81571664, 81871323, 81801665, 81771924, 81501616, 81671851, and 81527805); the National Natural Science Foundation of Guangdong Province (2018B030311024); the Science and Technology Planning Project of Guangdong Province (2016A020216020); the Scientific Research General Project of Guangzhou Science Technology and Innovation Commission (201707010328); the China Postdoctoral Science Foundation (2016M600145); and the National Key R&D Program of China (2017YFA0205200, 2017YFC1308700, and 2017YFC1309100).
Declaration of interests
The authors declare no conflicts of interest.
Author contributions
Literature search: Lu Zhang, Hailin Li, and Bin Zhang.
Study design: Lu Zhang, Shuixing Zhang, Hailin Li, Di Dong, Bin Zhang, and Jie Tian.
Data collection: Fusheng Ouyang, Baoliang Guo, Zhouyang Lian, Shufang Pei, Yuhao Dong, Xiaokai Mo, and Wenhui Huang.
Data analysis: Hailin Li, Lu Zhang, Xiaokai Mo, Di Dong, and Jie Tian.
Manuscript editing: Lu Zhang, Hailin Li, and Di Dong.
Manuscript review: Shuixing Zhang, Jing Liu, Di Dong, and Changhong Liang.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.013.
Appendix A. Supplementary data
Supplementary material
References
- 1.An X., Wang F.H., Ding P.R. Plasma Epstein-Barr virus DNA level strongly predicts survival in metastatic/recurrent nasopharyngeal carcinoma treated with palliative chemotherapy. Cancer. 2011;117(16):3750–3757. doi: 10.1002/cncr.25932. [DOI] [PubMed] [Google Scholar]
- 2.Peng H., Tang L.L., Chen B.B. Optimizing the induction chemotherapy regimen for patients with locoregionally advanced nasopharyngeal carcinoma: a big-data intelligence platform-based analysis. Oral Oncol. 2018;79:40–46. doi: 10.1016/j.oraloncology.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Tang X.R., Li Y.Q., Liang S.B. Development and validation of a gene expression-based signature to predict distant metastasis in locoregionally advanced nasopharyngeal carcinoma: a retrospective, multicentre, cohort study. Lancet Oncol. 2018;19(3):382–393. doi: 10.1016/S1470-2045(18)30080-9. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard P., Lee A., Marguet S. Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC-NPC meta-analysis. Lancet Oncol. 2015;16(6):645–655. doi: 10.1016/S1470-2045(15)70126-9. [DOI] [PubMed] [Google Scholar]
- 5.Wang W.Y., Twu C.W., Chen H.H. Plasma EBV DNA clearance rate as a novel prognostic marker for metastatic/recurrent nasopharyngeal carcinoma. Clin Cancer Res. 2010;16(3):1016–1024. doi: 10.1158/1078-0432.CCR-09-2796. [DOI] [PubMed] [Google Scholar]
- 6.Chan A.T., Gregoire V., Lefebvre J.L. Nasopharyngeal cancer: EHNS-ESMO-ESTRO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl. 7):vii83–5. doi: 10.1093/annonc/mds266. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L., Huang Y., Hong S. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 2016;388(10054):1883–1892. doi: 10.1016/S0140-6736(16)31388-5. [DOI] [PubMed] [Google Scholar]
- 8.Xia W.X., Zhang H.B., Shi J.L. A prognostic model predicts the risk of distant metastasis and death for patients with nasopharyngeal carcinoma based on pre-treatment serum C-reactive protein and N-classification. Eur J Cancer. 2013;49(9):2152–2160. doi: 10.1016/j.ejca.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 9.Yang L., Xia L., Wang Y. Development and external validation of nomograms to predict the risk of skeletal metastasis at the time of diagnosis and skeletal metastasis-free survival in nasopharyngeal carcinoma. BMC Cancer. 2017;17(1):628. doi: 10.1186/s12885-017-3630-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang J., Fogg M., Wirth L.J. Epstein-Barr virus-specific adoptive immunotherapy for recurrent, metastatic nasopharyngeal carcinoma. Cancer. 2017;123(14):2642–2650. doi: 10.1002/cncr.30541. [DOI] [PubMed] [Google Scholar]
- 11.Ren X., Yang X., Cheng B. HOPX hypermethylation promotes metastasis via activating SNAIL transcription in nasopharyngeal carcinoma. Nat Commun. 2017;8 doi: 10.1038/ncomms14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N., Chen N.Y., Cui R.X. Prognostic value of a microRNA signature in nasopharyngeal carcinoma: a microRNA expression analysis. Lancet Oncol. 2012;13(6):633–641. doi: 10.1016/S1470-2045(12)70102-X. [DOI] [PubMed] [Google Scholar]
- 13.Wang H.Y., Sun B.Y., Zhu Z.H. Eight-signature classifier for prediction of nasopharyngeal [corrected] carcinoma survival. J Clin Oncol. 2011;29(34):4516–4525. doi: 10.1200/JCO.2010.33.7741. [DOI] [PubMed] [Google Scholar]
- 14.Bruce J.P., Hui A.B., Shi W. Identification of a microRNA signature associated with risk of distant metastasis in nasopharyngeal carcinoma. Oncotarget. 2015;6(6):4537–4550. doi: 10.18632/oncotarget.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lambin P., Leijenaar R.T.H., Deist T.M. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017 Dec;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. Epub 2017 Oct 4. [DOI] [PubMed] [Google Scholar]
- 16.Aerts H.J. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2(12):1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 17.Bogowicz M., Riesterer O., Ikenberg K. Computed tomography radiomics predicts HPV status and local tumor control after definitive radiochemotherapy in head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2017;99(4):921–928. doi: 10.1016/j.ijrobp.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y.Q., Liang C.H., He L. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 19.Coroller T.P., Grossmann P., Hou Y. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rios Velazquez E., Parmar C., Liu Y. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77(14):3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Y., Feng Q., Yang W. Preoperative Prediction of Sentinel Lymph Node Metastasis in Breast Cancer Based on Radiomics of T2-Weighted Fat-Suppression and Diffusion-Weighted MRI. Eur Radiol. 2018;28(2):582–591. doi: 10.1007/s00330-017-5005-7. [DOI] [PubMed] [Google Scholar]
- 22.Kirienko M., Cozzi L., Antunovic L. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. 2018;45(2):207–217. doi: 10.1007/s00259-017-3837-7. [DOI] [PubMed] [Google Scholar]
- 23.Song J, Shi J, Dong D, et al. A new approach to predict progression-free survival in stage IV EGFR-mutant NSCLC patients with EGFR-TKI therapy. Clin Cancer Res 2018;24(15): 3583-92. [DOI] [PubMed]
- 24.Wu S., Zheng J., Li Y. Development and validation of an MRI-based radiomics signature for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine. 2018;34:76–84. doi: 10.1016/j.ebiom.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coroller T.P., Agrawal V., Narayan V. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119(3):480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Zhu Y., Burnside E.S. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 gene assays. Radiology. 2016;281(2):382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H., Lim Y. Radiomics signature on magnetic resonance imaging: association with disease-free survival in patients with invasive breast cancer. Clin Cancer Res. 2018;24(19):4705–4714. doi: 10.1158/1078-0432.CCR-17-3783. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B., Tian J., Dong D. Radiomics features of multiparametric MRI as novel prognostic factors in advanced nasopharyngeal carcinoma. Clin Cancer Res. 2017;23(15):4259–4269. doi: 10.1158/1078-0432.CCR-16-2910. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B., Ouyang F., Gu D. Advanced nasopharyngeal carcinoma: pre-treatment prediction of progression based on multi-parametric MRI radiomics. Oncotarget. 2017;8(42):72457–72465. doi: 10.18632/oncotarget.19799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou S.H., Zell J.A., Ziogas A. Epidemiology of nasopharyngeal carcinoma in the United States: improved survival of Chinese patients within the keratinizing squamous cell carcinoma histology. Ann Oncol. 2007;18(1):29–35. doi: 10.1093/annonc/mdl320. [DOI] [PubMed] [Google Scholar]
- 31.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 32.Chen L., Hu C.S., Chen X.Z. Concurrent chemoradiotherapy plus adjuvant chemotherapy versus concurrent chemoradiotherapy alone in patients with locoregionally advanced nasopharyngeal carcinoma: a phase 3 multicentre randomised controlled trial. Lancet Oncol. 2012;13(2):163–171. doi: 10.1016/S1470-2045(11)70320-5. [DOI] [PubMed] [Google Scholar]
- 33.Chua M.L., Chan A.T. Gemcitabine: a game changer in nasopharyngeal carcinoma. Lancet. 2016;388(10054):1853–1854. doi: 10.1016/S0140-6736(16)31394-0. [DOI] [PubMed] [Google Scholar]
- 34.van Griethuysen J.J.M., Fedorov A., Parmar C. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21) doi: 10.1158/0008-5472.CAN-17-0339. e104-e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC-3(6):610–621. [Google Scholar]
- 36.Galloway M.M. Texture analysis using gray level run lengths. Comput Graph Image Process. 1975;4(2):172–179. [Google Scholar]
- 37.Chu A., Sehgal C.M., Greenleaf J.F. Use of gray value distribution of run lengths for texture analysis. Pattern Recogn Lett. 1990;11(6):415–419. [Google Scholar]
- 38.De Jay N., Papillon-Cavanagh S., Olsen C. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29(18):2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- 39.Camp R.L., Dolled-Filhart M., Rimm D.L. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10(21):7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 40.Vickers A.J., Elkin E.B. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making. 2006;26(6):565–574. doi: 10.1177/0272989X06295361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vickers A.J., Cronin A.M., Elkin E.B. Extensions to decision curve analysis, a novel method for evaluating diagnostic tests, prediction models and molecular markers. BMC Med Inform Decis Mak. 2008;8:53. doi: 10.1186/1472-6947-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwan J.Y.Y., Su J., Huang S.H. Radiomic biomarkers to refine risk models for distant metastasis in HPV-related oropharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2018;102(4):1107–1116. doi: 10.1016/j.ijrobp.2018.01.057. [DOI] [PubMed] [Google Scholar]
- 43.Vallieres M., Freeman C.R., Skamene S.R. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material