Abstract
Background
Changes in glycosylation are known to play critical roles during gastric carcinogenesis. Expression of truncated O-glycans, such as the Sialyl-Tn (STn) antigen, is a common feature shared by many cancers and is associated with cancer aggressiveness and poor-prognosis.
Methods
Glycoengineered cell lines were used to evaluate the impact of truncated O-glycans in cancer cell biology using in vitro functional assays, transcriptomic analysis and in vivo models. Tumor patients ‘samples and datasets were used for clinical translational significance evaluation.
Findings
In the present study, we demonstrated that gastric cancer cells expressing truncated O-glycans display major phenotypic alterations associated with higher cell motility and cell invasion. Noteworthy, the glycoengineered cancer cells overexpressing STn resulted in tumor xenografts with less cohesive features which had a critical impact on mice survival. Furthermore, truncation of O-glycans induced activation of EGFR and ErbB2 receptors and a transcriptomic signature switch of gastric cancer cells. The disclosed top activated genes were further validated in gastric tumors, revealing that SRPX2 and RUNX1 are concomitantly overexpressed in gastric carcinomas and its expression is associated with patients' poor-survival, highlighting their prognosis potential in clinical practice.
Interpretation
This study discloses novel molecular links between O-glycans truncation frequently observed in cancer and key cellular regulators with major impact in tumor progression and patients' clinical outcome.
Keywords: Gastric cancer, Sialyl-Tn, RUNX1, SRPX2, Poor-survival
Research in context.
Evidence before this study
Gastric cancer is a silent disease that is often diagnosed at late stages leading to poor patient survival. Gastric tumors displaying high levels of truncated O-glycans have been shown to positively correlate with poor-prognosis of the patients. Some studies have previously reported that Sialyl-Tn expression is present during gastric cancer development and is associated with altered tumor cell behaviour. However, the mechanisms underlying truncated O-glycans driven tumor cell aggressiveness, that ultimately culminates in patients' poor survival, remain unclear.
Added value of this study
We have resourced to glycoengineered gastric cancer cells lines presenting high levels of truncated O-glycans to mechanistically disclose the role of such glycans in modulating cancer cell biology. Altering the O-glycosylation of gastric cancer cells was able per se to induce changes in cell signaling that culminated in the up-regulation of SRPX2 and RUNX1 transcription. These markers were further validated for the first time as co-expressed in gastric cancer tissue and to be correlated with patients' poor survival.
Implications of all the available evidence
Cancer-associated glycosylation modifications have been shown to impact and regulate several biological processes within tumor cells. Future studies should take in consideration the glycosylation status of tumor cells when addressing cancer patients. The disclosure of the induction of SRPX2 and RUNX1 expression concomitant with O-glycans truncation in gastric cancer patients and its association with patients' survival highlights its clinical relevance as novel prognostic factors.
Alt-text: Unlabelled Box
1. Introduction
Gastric cancer is a major threat to public health worldwide, with >700,000 deaths every year [1]. A common characteristic shared by many cancer is glycosylation modifications at cell surface, providing a set of diagnostic biomarkers and therapeutic targets currently being used in clinic, and known to play crucial roles during carcinogenesis [[2], [3], [4]]. Metastasis is often the major cause of cancer-related dead. Specific altered glycosylation patterns have been reported in metastatic tissue due to changes in the expression of the enzymes controlling the synthesis of such glycans. These modifications showed to be crucial for cancer cells to adapt to a new host environment such as the metastatic site [5,6]. Alterations on specific proteins glycosylation disturb cell-cell and cell-extracellular matrix (ECM) adhesion features supporting cancer progression [[7], [8], [9]]. In addition, changes occurring in receptor tyrosine kinases (RTKs) glycosylation have also been reported to play key roles in cancer [[10], [11], [12]]. Particularly, we have previously shown that altered RTKs sialylation resulted in its hyperactivation leading to a pro-invasive phenotype of gastric cancer cells [13,14]. In other models, high levels of sialylation displayed by cancer cells have also contributed to increased invasiveness of tumor cells and concomitant higher metastatic potential [3,[15], [16], [17]]. Truncation of the O-glycosylation pathway, leading to expression of simple O-glycans, is one of the most common features of many cancers [[18], [19], [20], [21], [22]], including gastric cancer [[23], [24], [25]]. These truncated glycan structures include the T (Galβ1-3GalNAcα1-O-Ser/Thr), Tn (GalNAcα1-O-Ser/Thr) and Sialyl-Tn (STn) (NeuAcα2–6GalNAcα1-O-Ser/Thr) antigens (Fig. 1a) [[26], [27], [28]].
Fig. 1.
O-glycans premature truncation affects cellular dynamics and alters gastric cancer cells morphology. Gastric cancer cell lines were genetically manipulated targeting COSMC gene. a) Mutations on COSMC gene lead to an interruption on the elongation of O-glycans and an increased expression of the precursor structure Tn and its sialylated glycoform, STn. b-e) Actin (red) and STn (green) (top) or actin (red) and tubulin (green) (bottom) immunolocalization were assessed in MKN45 and AGS SimpleCell lines (MKN45 SC and AGS SC) and wild-type controls (MKN45 WT and AGS WT). STn labeling was exclusively observed in MKN45 SC and AGS SC. STn induced expression by COSMC knock-out led to marked changes in the gastric cancer cell morphology, independently of the ECM component used to grow the cells, polymer surface (b), collagen IV (c), fibronectin (d) and poly-d-lysine (e). Both glycoengineered MKN45 SC and AGS SC exhibited a more elongated cell shape, displaying more cytoskeletal actin and tubulin projections, which also stained positive for STn antigen. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The truncated STn glycan, a well-known tumor-associated antigen, is highly detected in most gastric carcinomas [[29], [30], [31]] as well as in other tumor tissues [[32], [33], [34]], and its detection is rare or absent in normal tissue [[35], [36], [37]]. The mechanisms underlying STn synthesis include the overexpression of the sialyltransferase ST6GalNAc1 enzyme, responsible for STn biosynthesis (Fig. 1a) [28,30], and lack of expression of the core 1 synthase C1GALT1 private chaperon COSMC, essential for O-glycans elongation. Therefore, mutations or hypermethylation of COSMC gene can also lead to STn overexpression [38,39] (Fig. 1a). In fact, genetically engineered COSMC knock-out gastric cancer cell models, named SimpleCells (SC), display overexpression of truncated O-glycan antigens Tn and STn [40,41]. Cell genetic manipulation forcing STn synthesis, either by overexpression of ST6GalNAc1 enzyme [42] or by dysregulation of C1GalT1 gene [22] led to increased metastasis and decreased survival in mice. On the other hand, silencing of ST6GalNAc1 suppressed invasion in hepatocarcinoma cells [43] and prevented metastatic potential in gastric cancer cells [44]. Although the association between STn and poor survival of cancer patients has been known for decades [[45], [46], [47]], the mechanisms underlying such phenotype remain unknown.
In the present study, we have evaluated the molecular and functional effects of the presence of truncated O-glycans, particularly the STn antigen, in gastric cancer using glycoengineered COSMC knock-out models, disclosing the link between STn cancer-associated phenotype and its consequences for tumor progression.
2. Materials and methods
2.1. Cell culture
The gastric cancer cell lines MKN45 and AGS were obtained from the Japanese Collection of Research Bioresources and ATCC, respectively. MKN45 and AGS SimpleCells (MKN45 SC and AGS SC) were generated by targeting the COSMC gene using zinc finger nuclease precise gene editing as previously described [41]. Briefly, both MKN45 and AGS cells were transfected with 4 μg of compoZr® C1GalT1C1 DNA using an AmaxaTM NucleofectorTM according to cell lines specific manufacture's protocols (Lonza). The cells were grown RPMI in 1640 Glutamax, HEPES medium supplemented with 10% FBS plus 1% penicillin-streptomycin (all from Invitrogen) and maintained at 37 °C in an atmosphere of 5% CO2.
2.2. Antibodies
All the antibodies used in this manuscript, as well as it's protocol details are listed in Table 1.
Table 1.
List of antibodies.
| Antibody | Clone | Source | Application | Dilution |
|---|---|---|---|---|
| Tubulin | DM1A | Sigma | IF/WB | 1:750/1:10000 |
| Actin | I-9 | Santa Cruz | IF/WB | 1:150/1:2000 |
| Sialyl-Tn | 3F1 | [48] | IF/IHC | 1:200/1:5 |
| Phosphorylated FAK (Tyr397) | #3283 | Cell Signaling | WB | 1:1000 |
| FAK | #3285 | Cell Signaling | WB | 1:1000 |
| Phosphorylated EGFR (Tyr1068) | D7A5 | Cell Signaling | WB | 1:1000 |
| EGFR | D38B1 | Cell Signaling | WB | 1:1000 |
| Phosphorylated ErbB2 (Tyr1221/1222) | 6B12 | Cell Signaling | WB | 1:1000 |
| ErbB2 | 29D8 | Cell Signaling | WB | 1:1000 |
| E-cadherin | 4A2C7 | Thermo Fisher | IHC | 1:50 |
| SRPX2 | ABN488 | Millipore | IHC | 1:500 |
| RUNX1 | sc-365,644 | Santa Cruz | IHC | 1:50 |
| CD31 | ab28364 | Abcam | IHC | 1:50 |
Abbreviations: IF – Immunofluorescence; WB – Western Blot; IHC - Immunohistochemistry.
2.3. Immunofluorescence confocal microscopy
Cells were seeded on ibidi μ-Slide 8-well chambers with different coatings, ibidi-treated surface (polymer-coated), collagen IV, fibronectin and poly-d-Lysine (ibidi) at 70% confluence. The cells were fixed with 4% p-formaldehyde for 20 min and labeled with tubulin and actin antibodies or with sialyl-Tn [48] and actin antibodies overnight at 4 °C (Table 1). Goat anti-rabbit Alexa 549 and goat anti-mouse Alexa 488-conjugated antibodies (Thermo Fisher Scientific) were used for 45 min at room temperature. DAPI (Sigma-Aldrich) was used for nuclei visualization. Cells were visualized using a Zeiss Imager Z.1 microscope (Zeiss).
2.4. EGF treatment
MKN45 WT and SC (5 × 105 cells) were seeded on a 6-well plate in RPMI supplemented with 10% FBS. After 48 h the medium was replaced with non-supplemented RPMI medium and cells were treated with 25 ng/mL of EGF ligand. After EGF incubation for 10 min, cells were collected and protein was extracted to evaluate EGFR phosphorylation status as described in the western blot section. To evaluate gene expression regulation, cells were collected 4 h after EGF incubation. RNA was extracted and real-time quantitative PCR (RT-qPCR) for SRPX2 and RUNX1 genes was performed as described in the transcriptomic analysis section.
2.5. Transcriptomic analysis
Total RNA was extracted from MKN45 WT and SC, AGS WT and SC cell lines using TRI Reagent (Sigma-Aldrich). Ion AmpliSeq Transcriptome Human Gene Expression Kit was used to sequence the mRNAs of over 20,000 primed targets. Ion Chef system was used for templating and the loaded chips were sequenced using the Ion Proton System (Life Technologies). After sequencing, the data was automatically transferred to the dedicated Ion Torrent server and the sequencing reads were generated. Reads quality and trimming was performed using Torrent Server v4.2 before read alignment using TMAP 4.2. The TS plugin Coverage Analysis v4.2 was used to generate read counts. The sequencing was performed in two independent biological replicates and sequence reads were normalized to the total read count. Genes from MKN45 SC and AGS SC were analyzed in comparison with MKN45 WT and AGS WT, respectively. Only the genes presenting >10 reads and at least 2-fold change differences between the two sets after considering its standard deviation, were selected for analysis.
For RT-qPCR gene expression analysis total RNA was extracted from MKN45 WT and SC using TRyzol Reagent (Invitrogen). One μg of RNA was reverse transcribed with random primers using the SuperScript® IV Reverse Transcriptase Kit (Invitrogen). RT-qPCR was performed with 1 μL of cDNA, 10 μM of each primer, 10 μL SYBR® Green Master Mix (1×) (Thermo Fischer Scientific) and ultrapure water to a final volume of 20 μL using the ABI 7500 (Applied Biosystems). The following primers were used, SRPX2 (Fw: ACTGGATTTGCGGCATGTGA; Rv: CCATGTTGAAGTAGGAGCGAGTGA; 146 bp), RUNX1 (Fw: CTGCTCCGTGCTGCCTAC; Rv: AGCCATCACAGTGACCAGAGT; 109 bp). Relative gene expression was normalized to β-actin (Fw: AGAAAATCTGGCACCACACC; Rv: TAGCACAGCCTGGATAGCAA; 173 bp) and DeltaDelta CT was performed to compare different conditions. All conditions were normalized and compared to MKN45 WT non-treated cells. Three independent experiments with three technical replicates per condition were performed. Results are shown as average ± SEM. Two-way ANOVA with Bonferroni correction was used for statistical analysis.
2.6. Western blot
Cells were lysed in RIPA buffer and protein quantified with DC protein assay (BioRad). Total protein extracts were separated by gel electrophoresis and gels transferred onto a nitrocellulose membrane (Amersham). Membranes were probed overnight with phosphorylated FAK and FAK, phosphorylated EGFR and EGFR, and phosphorylated ERBB2 and ERBB2 primary antibodies (Table 1). Membranes were developed with ECL (GE Healthcare Life Sciences). The phospho-protein band quantification was normalized for the respective total protein amount. MKN45 and AGS SC were analyzed in relation with MKN45 and AGS WT, respectively. Actin or tubulin (Table 1) were used as loading control. Three biological independent batches of protein extracts were analyzed for each cell model. Results are shown as average ± SEM. Student t-test was used for statistical analysis.
2.7. Proliferation assay
BrdU Labelling and Detection Kit (Roche) was used according to the manufacturer's instructions. Briefly, cells (1 × 105) were seeded in cover slips. When cells reached a 50% confluence, cells were incubated with BrdU labelling medium for 30 min. The nuclei were stained with DAPI. The percentage of dividing cells was measured by counting the number of positive BrdU cells in five different fields. Pictures were acquired using a fluorescence microscope (Carl Zeiss), and analyzed with Image J software. Three independent biological experiments were performed and cells were seeded in duplicate. Results are shown as average ± SEM. Student T-test was used for statistical analysis.
2.8. Adhesion assay
Adhesion assays were performed using the ibidi culture-inserts family. Cells were plated on ibidi μ-Slide 8-well chambers with different coatings, ibidi-treated surface (polymer-coated), collagen IV, fibronectin and poly-d-Lysine. After 24 h, the slides were washed with phosphate buffered saline (PBS) to remove non-adherent cells. Adherent cells were fixed with acetone/methanol (1:1) and stained with 0.5% crystal violet/20% methanol. At least 4 random pictures acquired at 100× magnification were used to count adherent cells. Cells were seeded in duplicate and two independent biological experiments were performed. Results are shown as average ± SEM. Student T-test was used for statistical analysis.
2.9. In vitro wound-healing assay
Wound-healing assays were performed using the 2-well silicone ibidi inserts from ibidi culture-inserts family. The 2-well ibidi inserts were applied to ibidi μ-Slide 8-well chambers coated either with, ibidi-treated surface (polymer), collagen IV, fibronectin or poly-d-lysine (ibidi). MKN45 WT and SC cells (8 × 104) and AGS WT and SC cells (5 × 104) were seeded in each side of the 2-well and left to adhere for 24 h. The insert was then carefully removed, leaving a defined gap area (wound) between the cells. MN45 WT, MKN45 SC were followed during 58 h and AGS WT, AGS SC cells were followed during 12 h for wound healing. Cells were seeded in duplicate and two independent biological experiments were performed. The percentage of area with no cells was calculated by measuring the free space at each time-point, normalized to the initial area with no cells (right after insert removal) using Image J software. Results are shown as average ± SEM. Two-way ANOVA was used as statistical test.
2.10. In vitro invasion assay
Invasion assays were performed in BD Biocoat Matrigel invasion chamber with 8 μm of diameter pore size, in 24-well plate (BD Biosciences). Inserts were rehydrated for at least 1 h in RPMI medium. Cells (7 × 104) were seeded and RPMI medium supplemented with 1% P/S and 10% FBS was used as chemo-attractive. After 24 h, non-invading cells were carefully removed from the upper part of the insert. The inserts were washed and cells were fixed with cold methanol for 10 min on ice. Total number of invasive nuclei (DAPI labeling) was counted using a Leica DM2000 microscope (Leica). Cells were seeded in duplicate for each cell line and three independent biological replicates were performed. Results are shown as average ± SEM. Student T-test was used for statistical analysis.
2.11. In vivo chorioallantoic membrane (CAM) angiogenic assays
In vivo angiogenic activity of MKN45 WT and SC, AGS WT and SC cells was assessed by the CAM assay. Fertilized chick (Gallus gallus) eggs (8 per group) obtained from commercial sources were incubated horizontally at 37.8 °C in a humidified atmosphere and referred to embryonic day (E). On E3 a square window was opened in the shell after removal of 1.5–2 mL of albumen to allow detachment of the developing CAM. The window was sealed with adhesive tape and the eggs returned to the incubator. At E10, 1 × 106 cells of each cell line were loaded inside a silicon ring under sterile conditions. The eggs were re-sealed and returned to the incubator for an additional 3 days. After removing the ring, the CAM was excised from the embryos, photographed ex ovo under a stereoscope, at 20× magnification (Olympus, SZX16 coupled with a DP71 camera). The number of new vessels (<15 μm diameter) growing radially towards the ring area was counted in a blind fashion manner. Student T-test was used for statistical analysis.
2.12. In vivo survival, tumorigenesis and angiogenesis evaluation in nude mice
N:NIH(s)II:nu/nu nude mice were housed at IPATIMUP Animal's House (Medical Faculty of the University of Porto) under a pathogen-free environment, with controlled light and humidity. Two groups of female N:NIH(s)II:nu/nu nude mice, aged 6–8 weeks, were subcutaneously injected between the scapula with 1 × 106 viable cells of MKN45 WT (5 mice) and MKN45 SC (4 mice). Mice were examined and tumors were measured every two days. The product of 3 major diameters was record as the tumor volume. After 21 days of cell injection, the primary tumors were surgically removed from all animals, except for one animal that died 18 days after cell injection. Surgical procedures were performed under anesthesia. The animals were left after surgery for survival analysis. Their weight was followed 25 days after surgery. The living mice were sacrificed and examined by detailed necropsy and liver, lymph nodes and lung were collected. The organs and the primary tumors were fixed in 10% buffered formalin and then embedded in paraffin for hematoxylin eosin (HE) staining and immunohistochemistry labeling. For morphological evaluation, the tumors were visualized and analyzed under a brightfield microscope and parameters such as cell morphology and tumor cell cohesion were evaluated by two independent pathologists. Necrosis was evaluated as a percentage of the whole tissue. Angiogenesis was measured by vessel number counting after overnight CD31 immunolabeling (Table 1). Kaplan-Meier method by means of Gehan-Breslow-Wilcoxon Test was used for survival curves statistical analysis.
2.13. Immunohistochemistry
Expression of STn, E-cadherin, SRPX2, RUNX1 and CD31 (Table 1) were assessed in tumor mice xenograft tissues. Detection of STn, SRPX2 and RUNX1 was measured in 25 cases of human gastric carcinoma. Heat mediated antigen retrieval using sodium citrate buffer (10 mM, pH 6.0) was required for the E-cadherin, RUNX1 and CD31 markers. Endogenous peroxidase blocking was performed in all the samples using 3% hydrogen peroxide. Tissues were incubated overnight with the respective primary antibodies (Table 1), followed by biotin-labeled secondary antibodies (Dako) and with ABC kit (Vector Labs). Sections were stained with DAB (Sigma-Aldrich) containing 0.01% H2O2 and counterstained with Mayers' hematoxylin. Pictures were taken under 400× magnification with a Zeiss Optical Microscope.
2.14. In silico data analysis
The differentially expressed genes between MKN45 SC and WT and between AGS SC and WT cells were analyzed with PANTHER (http://pantherdb.org) [49] for Gene Ontology (GO) functional enrichment in terms of Biological Processes (GO database release 2016.07.29). The number of mapped gene IDs was 752 out of 825 for MKN45 and 375 out of 413 for AGS cell lines. The statistical over-representation was calculated using a binomial test (release 2016.07.15) with Homo sapiens gene set as background reference and the results were considered significant at p < 0.05, after Bonferroni correction. All significantly enriched GO Biological Processes were presented as converted –log p-value and gene frequency and redundant terms clustered manually according to term similarity. Additionally, we performed a similar GO functional enrichment on a list of gene IDs corresponding to proteins previously identified to carry truncated glycans in MKN45 SC and AGS SC [41]. The protein-encoding genes associated to the GO term Adhesion are presented (Supplementary Table 4).
The Oncomine platform (https://www.oncomine.org) [50] was used to evaluate the expression and prognostic value of SRPX2 and RUNX1 in independent datasets of gastric cancer patients. SRPX2 and RUNX1 mRNA levels in normal and tumor samples were assessed in 5 independent cohorts (Wang [51], Cui [52], DErrico [53], Chen [54] and Cho [55]), comprising a total of 453 tissues, including 175 adjacent gastric mucosa and 278 gastric cancer tissues. The prognostic value of SRPX2 and RUNX1 was further evaluated in the dataset with higher number of cases and information for overall survival (Chen [54]). Categorization of patients with high and low expression was based on the log2 median-centered intensity values. The top 25% patient samples with highest expression values (>75% of all patients) were categorized as SRPX2- or RUNX1-high, and all the other patients were grouped as SRPX2- or RUNX1-low. Student t-test was used for the expression statistical analysis. Kaplan-Meier method was used for survival curves statistical analysis.
2.15. Ethics statement
Human tissue samples were obtained from IPATIMUP Diagnostics in accordance with the national regulative law for the handling of biological specimens from tumor banks, and the international Helsinki declaration.
Mice experiments followed the European Directive 2010/63/UE and the corresponding Portuguese law on animal experimentation (Portaria 113/2013), according to the FELASA (Federation of European Laboratory Animal Science Associations) guidelines and recommendations concerning laboratory animal welfare. The protocol was previously approved by the Local Animal Ethics Committee and authorized by the Portuguese Official Veterinary Department (Direccão Geral de Veterinária – DGV).
3. Results
3.1. Expression of truncated O-glycans in gastric cancer cells impacts cell shape
Two gastric carcinoma SimpleCell lines, MKN45 and AGS, were generated by targeting the COSMC gene using zinc-finger nuclease technology [41]. This manipulation resulted in the disruption of O-glycans normal elongation and increased expression of truncated O-glycans, Tn and STn (Fig. 1a).
COSMC knock-out in both MKN45 (MKN45 SC) and AGS (AGS SC) cells led to the overexpression of STn, both in the cytoplasm and plasma membrane of the targeted cells (Fig. 1b–e). Concomitantly, critical changes in MKN45 SC and AGS SC cell morphology could be observed through actin and tubulin labeling when cells were grown in different extracellular matrix (ECM) components, including collagen IV, fibronectin and poly-d-Lysine (Fig. 1b–e). The MKN45 SC and AGS SC presented a mesenchymal-like morphology with more cytoskeletal actin and tubulin protrusions compared to wild-type (WT) cells. Overall, truncation of O-glycans drastically gastric cancer cell morphology.
3.2. Truncation of O-glycans in gastric cancer cells promotes invasive features
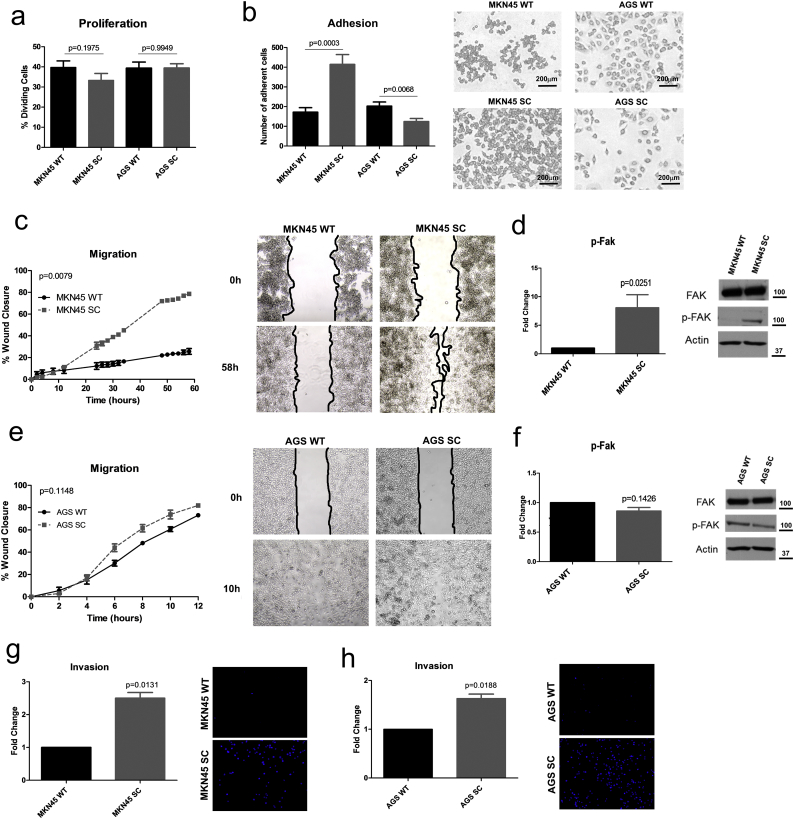
To evaluate the impact of truncated O-glycans on cellular behavior, a set of proliferation, adhesion, migration and invasion assays were performed. MKN45 SC had similar cellular proliferation rates to the WT cell line (Fig. 2a). MKN45 SC displayed higher adhesion capacity, to a polymer coated surface (Fig. 2b) and to collagen type IV and poly-d-Lysine ECM components, whereas no differences where observed when cells were grown on fibronectin (Supplementary Fig. 2a–c). Furthermore, MKN45 SC revealed an increased migration capacity independently of all the tested ECM components (Fig. 2c, Supplementary Fig. 2d-f). The enhanced motility of MKN45 SC, in comparison with MKN45 WT, was accompanied by increased activation of the focal adhesion kinase (Fak) protein (Fig. 2d).
Fig. 2.
Premature truncation of O-glycans promotes an increased aggressiveness phenotypic behavior. a) Proliferation assay evaluated by BrdU labeling showed no differences in cell division rates among the glycoengineered cell lines and the wild-type controls. Results are shown as average ± SEM. Student t-test was used for statistical analysis. b) Adhesion assay revealed that MKN45 SC displayed significantly higher surface adhesion capacity than MKN45 WT, whereas AGS SC showed significant less surface adhesion when compared with the AGS WT. Results are shown as average ± SEM. Student t-test was used for statistical analysis. c) Wound-healing assay demonstrated that MKN45 SC presented significantly higher migration capacity than the MKN45 WT cell line, being able to close the wound in 58 h. d) MKN45 SC revealed an increased Fak activation when compared to MKN45 WT cells. Results are shown as average ± SEM. Student t-test was used for statistical analysis. e) Wound-healing assay showed that AGS SC and AGS WT cell lines did not reached a statistical significant difference in migration capacity, although a trend of AGS SC towards higher migration rates was observed leading to wound closer after 12 h. Results are shown as average ± SEM. Two-way ANOVA was used as statistical test. f) No differences in Fak activation were detected between AGS SC and AGS WT cells. Results are shown as average ± SEM. Student t-test was used for statistical analysis. g) Matrigel invasion assay revealed the enhanced capacity of the MKN45 SimpleCells to degrade and invade the ECM in comparison with the respective wild-type cell line. Results are shown as average ± SEM. Student t-test was used for statistical analysis. h) Matrigel invasion assay revealed that AGS SC have an enhanced capacity to degrade and invade the ECM than the respective wild-type cell line. Results are shown as average ± SEM. Student t-test was used for statistical analysis.
AGS SC maintained similar proliferation rates (Fig. 2a), compared to WT AGS. However, AGS SC showed decreased adhesion capacity when compared with WT cells (Fig. 2B and Supplementary Fig. 3a–c). AGS SC showed no major impact on the migration capability (Fig. 2e) except for a decreased motility noted on fibronectin component (Supplementary Fig. 3d–e). In contrast with MKN45 SC, AGS SC cells did not show Fak activation when compared with its parental cell line (Fig. 2f).
A PANHER gene ontology (GO) analysis was performed using previously identified proteins carrying truncated glycans in MKN45 SC and AGS SC [41]. We have analyzed the truncated O-glycosylated adhesion-related proteins present in our samples (Supplementary table 4), combining the information with the RNAseq values of the respective genes. Although some proteins were found to be commonly O-glycosylated in both models, several proteins were found to be specific of each cell line model. Interestingly, proteins belonging to the cadherin family were specifically O-glycosylated in MKN45 SC. Particular laminin and integrin subunits were found to be the O-glycosylation carriers in AGS SC. The correspondent RNAseq values corroborated the difference in expression levels of the genes belonging to the adhesion GO term in MKN45 SC and AGS SC. Regardless of the differences found in the adhesion and migration capacity between MKN45 SC and AGS SC, both models revealed an increased ability to degrade and invade the ECM in comparison to the respective parental cell line (Fig. 2g–h). Overall, these results demonstrated that premature truncation of O-glycans per se was capable of promoting a pro-invasive phenotypic behavior on gastric cancer cells.
3.3. Glycoengineering of gastric cancer cells triggers a switch in the cell transcriptomic signature
In order to identify the genetic signature underlying the increased invasive phenotype observed in the glycoengineered cell models, a transcriptomic analysis on >20,000 genes was performed in both MKN45 and AGS cell models. It was observed that COSMC knock-out resulted in a significant alteration in the transcription profile of both gastric cancer cell models when compared to their respective parental cells. From the genes that showed significant transcription alterations, 185 (63.4%) genes were upregulated in MKN45 SC and 99 (33.9%) genes in AGS SC, with 8 (2.7%) of those genes being commonly upregulated in both models. The analysis of the downregulated genes revealed 606 (66.4%) in MKN45 SC and 280 (30.7%) in AGS SC, with 26 (2.9%) genes downregulated in both SimpleCell models (Fig. 3a and Supplementary Table 1). The differentially expressed genes were analyzed with PANTHER, for functional enrichment analysis, in order to determine the GO biological processes significantly affected in the SimpleCells. Our analysis showed that the genes significantly altered were over-represented in GO categories clustered as development and motility processes (Fig. 3b). Significantly upregulated and downregulated genes in both MKN45 SC and AGS SC were represented in a heatmap, with SRPX2 (Gene ID: 27286) and RUNX1 (Gene ID: 861) being the two most upregulated genes in the two COSMC knock-out gastric carcinoma cell models. The expression patterns of the downregulated genes were dissimilar between the two cell models (Fig. 3c). This transcriptomic analysis showed that targeting the O-glycosylation pathway led to significant changes in the transcriptomic profile between the isogenic cell lines in both MKN45 and AGS cell models.
Fig. 3.
Transcriptomic expression profile of MKN45 SC and AGS SC discloses an enrichment in genes associated with development and motility. a) Venn diagram of upregulated (upper panel) and downregulated (lower panel) genes between MKN45 SC and MKN45 WT (in blue) and between AGS SC and AGS WT (in grey). The results are represented as gene count (and percentage) with the intersecting values indicating the common genes between MKN45 and AGS cell lines. Premature truncation of O-glycans lead to transcriptional changes in about 60% and 30% of both up- and down-regulated genes in MKN45 SC and in AGS SC model, respectively. Only about 3% of the genes were either commonly up- or down-regulated in both cell models. b) Functional enrichment analysis of GO Biological Processes for the set of differentially expressed genes between MKN45 SC and MKN45 WT (in blue) and between AGS SC and AGS WT (in grey). The results are represented as –log p-value (left axis) and gene frequency (right axis) per functional category with categories clustered based on term similarity. The genes differentially expressed were annotated to significantly enriched GO terms related to development, motility and organism processes. The statistical over-representation was calculated using a binomial test (release 2016.07.15) with Homo sapiens gene set as background reference and the results were considered significant at p < 0.05, after Bonferroni correction. c) Heatmap of the common differentially expressed genes between MKN45 and AGS cell lines. The results are represented as fold change ranked as percentile, indicated by the graded colour scale. SRPX2 and RUNX1 genes were the most upregulated genes in the two cell line models. Two biological replicates per cell line were used for transcriptomic analysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Induced O-glycosylation truncation activates receptor tyrosine kinases (RTK) in gastric cancer cells
We have previously showed that RTKs could be activated due to changes in cell glycosylation [13,14]. To evaluated if in our model of gastric cancer cells expressing truncated O-glycans some particular RTKs were activated, we have analyzed the phosphorylation status of both EGFR and ErbB2 RTKs. We found that glycoengineering of gastric cancer cells led to activation of both receptors (Fig. 4a). Furthermore, we evaluated if the activation of these receptors could be responsible for inducing up-regulation of SRPX2 and RUNX1 genes. By treating MKN45 WT cells with the EGFR ligand (EGF) to force receptor activation we demonstrated that both RUNX1 and SRPX2 were up-regulated to the same levels of expression found in the SC model (Fig. 4b, c). The same treatment did not induce further transcription activation of SRPX2 and RUNX1 genes in the MKN45 SC model, where both EGFR and ErbB2 were found to be constitutively activated. These results shed light on the mechanism by which O-glycosylation truncation in our cells could lead to a switch in the cell transcriptomic profile of SRPX2 and RUNX1 genes.
Fig. 4.
Glycoengineering of gastric cancer cells activates receptor tyrosine kinases that culminate in up-regulation of SRPX2 and RUNX1 genes. a) Induced O-glycosylation truncation on MKN45 cells led to both EGFR and ErbB2 RTKs phosphorylation. Results are shown as average ± SEM. Student t-test was used for the expression statistical analysis. b) Treating MKN45 WT cells with EGF ligand induced EGFR activation after 10 min. c) Activation of EGFR through EGF treatment for 4 h in MKN45 WT cells caused up-regulation of RUNX1 (not reaching statistical significance) and a significant up-regulation of SRPX2 gene. No effect was noted on the expression level of these genes when MKN45 SC were treated with EGF ligand in the same conditions as MKN45 WT cells. Results are shown as average ± SEM. Two-way ANOVA with Bonferroni correction was used for statistical analysis.
3.5. Truncated O-glycans impact tumor features and survival in a mouse model
MKN45 SC and WT cells were injected in nude mice to evaluate the impact of truncated O-glycans in an in vivo environment. After 16 days of cell heterotopic injection, MKN45 SC-injected mice started to lose weight, being significantly lower at day 21 when compared with MKN45 WT-injected mice (Fig. 5a and c). Both MKN45 SC and WT cells induced tumor formation in vivo and no significant differences in tumor size were observed over time (Fig. 5b, d and e). The animals were kept after tumor removal for prognostic value evaluation and all the tumors were assessed for several histopathologic features. MKN45 SC tumors presented larger and more elongated cells characterized by a higher pleomorphism, and a large number of atypical mitosis compared to MKN45 WT tumors (Fig. 5f). Additionally, a less cohesive cellular pattern was observed in MKN45 SC tumors when comparing to the WT xenografts. This decreased cell-cell adhesion pattern was accompanied by a mislocalization of E-cadherin from the membrane to the cytoplasm on MKN45 SC tumors (Fig. 5f), whereas MKN45 WT tumors showed a typical membrane localization of E-cadherin. IHC analysis showed that MKN45 SC tumors presented increased STn expression both at cytoplasmic and membrane cell compartments when compared to MKN45 WT tumor cells (Fig. 5f and Supplementary Table 2). Furthermore, MKN45 SC xenografts displayed less necrotic areas (Fig. 5g) together with less angiogenesis (Fig. 5h), in comparison to MKN45 tumors. A standard assay to measure angiogenesis in vivo was used to assess the capacity of both SimpleCell models to induce de novo vessel formation. CAM assay confirmed that both MKN45 SC and AGS SC had decreased capability to promote angiogenesis in vivo than their respective WT parental cells (Fig. 5j). Remarkably, concomitant with all the differences found between MKN45 SC and MKN45 WT tumors, mice injected with MKN45 SC revealed a significant poor-survival rate when compared to the mice injected with MKN45 WT cells (Fig. 5i). Nevertheless, it is important to consider that the aggressiveness of MKN45 SC tumor model resulted in a very short survival period precluding the time required for metastasis development. Altogether, in vivo data support that premature truncation of O-glycans with concomitant overexpression of STn O-glycoform, resulted in tumors with more oncogenic features as well as in mice poor-survival.
Fig. 5.
Premature truncation of O-glycans is associated in vivo with poor-survival. Two groups of mice were injected subcutaneously between the scapula with 1 × 106 viable cells of MKN45 SC or MKN45 WT gastric cancer cell models. Five animals were injected with MKN45 WT and four animals were injected with MKN45 SC cells. a) During the experiment, MKN45 SC-injected mice lost a higher percentage of body weight than MKN45 WT-injected mice with a significant body weight loss 21 days after cells injection (red box). Mice body weight was recovered after tumor removal (red box) by surgery. Percentage of body weight loss was calculated taking into consideration the initial body weight of each mice. Results are shown as average ± SEM. Student t-test was used for body weight loss statistical analysis 21 days after cells injection. b) Analysis of tumorigeneses showed that the tumor formation between the two groups was not significantly different in size during the experience time course. Results are shown as average ± SEM. Two-way ANOVA was used as statistical test. c) Mice images were taken at day 21 (day of tumor removal by surgery). d) Mice tumor xenografts images after being removed by surgery, 21 days after cell injection. e) Measurement of tumor size after mice surgery confirmed no differences in tumor volume. Results are shown as average ± SEM. Student t-test was used for statistical analysis. f) Xenografts from MKN45 SC-injected mice clearly presented poorly cohesive cells (HE), showed overexpression of STn and a mislocalized E-cadherin when compared with xenografts from MKN45 WT-injected mice, together with higher expression of SRPX2 and RUNX1 proteins. Pictures were taken under 400× magnification. g) Tumor xenografts from mice injected with MKN45 SC cells showed significant less necrotic areas when compared with MKN45 WT tumors. Results are shown as average ± SEM. Student t-test was used for statistical analysis. h) Xenografts from MKN45 SC-injected tumors revealed fewer vessels, based on CD31 immunolabeling, when compared with MKN45 WT tumors. Results are shown as average ± SEM. Student t-test was used for statistical analysis. i) Survival curve analysis revealed that mice injected with MKN45 SC had a significant poor-survival rate when compared with mice injected with MKN45 WT. To be noticed that one of the MKN45 SC-injected mice died 18 days after cell injection. Kaplan-Meier method by means of Gehan-Breslow-Wilcoxon Test was used for survival curves statistical analysis. j) In vivo neoangiogenesis of MKN45 WT, MKN45 SC, AGS WT and AGS SC cells, was assessed by the CAM assay. Both MKN45 SC and AGS SC displayed significantly less vessel formation than MKN45 WT and AGS WT, respectively. Results are shown as average ± SEM. Student t-test was used for statistical analysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Glycoengineered cells expressing truncated O-glycan revealed new potential biomarkers for gastric cancer patient survival
In order to translate our previous findings into a clinical setting, we tested whether the top upregulated genes in our cell models would also be upregulated in human gastric tumors and whether the identified genes hold potential to be used as novel prognostic biomarkers in gastric cancer. For this analysis, we selected the SRPX2 and RUNX1 genes, since these markers showed the highest fold increase in both MKN45 SC and AGS SC (Fig. 3c).
Firstly, we evaluated the expression of SRPX2 and RUNX1 in different datasets of human gastric tissues, from Oncomine database, including both adjacent mucosa and tumor samples. Through a microarray expression data analysis, it was possible to determine that both SRPX2 and RUNX1 mRNA levels were significantly upregulated in gastric cancer tissues when compared with adjacent gastric mucosa (Fig. 6a and Supplementary Fig. 3a and b). These findings were further validated at the protein level in a cohort of gastric cancer tissue samples. SRPX2 showed higher expression in the cytoplasm of both intestinal and diffuse subtypes of gastric tumor cells in comparison with adjacent mucosa. Similarly, high RUNX1 expression was observed in the nucleus of intestinal and diffuse gastric carcinoma cells, whereas adjacent mucosa showed low RUNX1 expression. Finally, STn expression on the evaluated tumors varied from negative or low to high expression, being absent in adjacent mucosa (Fig. 6e and Supplementary Table 3). Co-expression of SRPX2, RUNX1 and STn was observed in 84% of the analyzed human gastric carcinoma tissue samples (Fig. 6e and Supplementary Table 3). The analysis of ST6GALNAC1, C1GALT1 and C1GALT1C1 from Oncomine database did not show major differences in the expression levels between adjacent mucosa and gastric carcinoma samples (Supplementary Fig. 3d). However, an inverse correlation was observed between SRPX2 and C1GALT1C1, and between RUNX1 and C1GALT1C1 expression levels in gastric carcinoma samples (Fig. 6d). Furthermore, a correlation analysis based on microarray expression data on tumor and adjacent mucosa of the same patient samples from the Oncomine database corroborated the tissue co-expression results, showing a significant correlation between SRPX2 and RUNX1 expression levels within gastric cancer (Fig. 6b and Supplementary Fig. 3c) but not in adjacent gastric mucosa (Fig. 6c). Similarly, the in vivo tumor MKN45 SC xenografts displaying high STn expression levels also showed high SRPX2 and RUNX1 expression (Fig. 4f and Supplementary Table 2).
Fig. 6.
O-glycans premature truncation revealed gastric cancer patients' survival biomarkers. a) Analysis of microarray expression data for SRPX2 and RUNX1 levels from the Oncomine database [54]. Expression values are represented as Log2 median-centered intensity. SRPX2 and RUNX1 expression levels were significantly higher in gastric cancer when compared with adjacent gastric mucosa. Student t-test was used for the expression statistical analysis. b) Microarray expression data for SRPX2 and RUNX1 levels from the Wang dataset [51] extracted from the Oncomine database. Normalized expression values are shown for adjacent mucosa and tumor of the same patients. c) Correlation analysis based on Pearson statistical test demonstrated the highly significant correlation between SRPX2 and RUNX1 expression levels in gastric cancer. d) Correlation analysis based on Pearson statistical test demonstrated an inverse correlation between SRPX2 and C1GALT1C1 and between RUNX1 and C1GALT1C1 expression levels in gastric cancer. e) Immunohistochemistry of STn, SRPX2 and RUNX1 markers on adjacent mucosa, intestinal- and diffuse-type gastric carcinoma. Pictures were taken under 400× magnification. f) Overall survival analysis of gastric cancer patients based upon SRPX2 or RUNX1 expression levels. Analyses were performed on all cases of which gastric cancer patients' survival and gene expression data were available on Oncomine, namely 81 and 75 cases for SRPX2 and RUNX1, respectively [54]. Cohort was divided into high- (black line) and low- (green line) expressing subgroups. A log-rank test was used to evaluate differences between groups. Increased expression of both SRPX2 and RUNX1 genes significantly correlated with poor-prognosis in gastric cancer. Kaplan-Meier method was used for survival curves statistical analysis. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Finally, the prognostic value of SRPX2 and RUNX1 was evaluated in a dataset with survival data available for both genes. This analysis revealed that patients with high expression of SRPX2 or RUNX1 presented a worse prognosis when compared to patients with low SRPX2 or RUNX1 levels (Fig. 6f). These results indicate that SRPX2 and RUNX1 genes, associated with O-glycan truncation, could serve as novel prognostic biomarkers in gastric cancer patients.
4. Discussion
Glycosylation is involved in several regulatory mechanisms controlling cellular physiological and pathological processes. Tumor cells commonly express truncated O-glycans, as a consequence of the impairment of the normal extension of O-glycans, usually present in healthy tissues [19,27,28,30,56]. Sialylation is a major glycosylation modification which has critical impact in cell recognition, adhesion, signaling and is remarkably elevated in metastatic tumor cells [13,14,16,57]. The overexpression of the truncated STn has been described to occur in many epithelial cancers, including gastric tumors [[58], [59], [60]], and to be associated with more aggressive tumors and patients' poor-prognosis [[45], [46], [47]]. However, the molecular mechanisms underlying these high malignancy features remain unclear.
In order to evaluate the biological impact of truncation of O-glycans in gastric cancer cells behavior we have performed a systematic analysis of genetically engineered gastric cancer cell models expressing homogeneous truncated O-glycans. Our findings revealed that aberrant expression of the simple O-glycan antigen STn induced major phenotypic alterations in gastric cancer cells, characterized by the acquisition of a mesenchymal-like morphology, together with a less cohesive cell monolayer characteristic of an epithelial to mesenchymal transition [61]. The enhanced expression of STn observed in the cellular membrane extensions of the SimpleCells highlights its relevance in cell-cell and cell-ECM interactions, suggesting that STn might be responsible for mediating interactions between tumor cells and the surrounding environment during cancer progression. In agreement, STn overexpressing cells displayed reduced cell-cell adhesion contacts in the in vivo tumor xenograft model. This phenotype was accompanied with a mislocalization of E-cadherin from the membrane to the cytoplasm. Overall, these observations are in accordance with previous studies showing that expression of STn reduces cell-cell adhesion and impairs the formation of cohesive solid tumors [62].
Importantly, truncation of O-glycans showed to impact gastric cancer cell-matrix adhesion and motility. It is important to highlight that COSMC KO affects both STn and Tn levels, as we previously reported [41], resulting in a different ratio of STn/Tn expression in both MKN45 SC and AGS SC. This ratio was found to be higher in MKN45 SC cells together with increased Fak activation. These results help to explain why MKN45 SC acquired an enhanced capacity to adhere and migrate on different components of the ECM, in contrast with the AGS SC cell line. Although STn expression usually accompanies with increase cell adhesion capacities, opposite results showing decrease cell-ECM adhesion in breast cancer cells have been reported [63]. The origin of these discrepancies might be related to the specific cell surface O-glycosylated proteins that are affected after glycoengineering of the cells. Supporting this hypothesis, the list of adhesion-related proteins previously identified as carrying truncated O-glycans in MKN45 SC and AGS SC models was found to be distinct [41]. Glycoproteins belonging to the cadherin family of proteins were O-glycosylated in MKN45 SC whereas in AGS SC, O-glycosylation was present in some particular laminin and integrin subunits [41]. Further biochemical and functional studies would help to clarify this matter. Independently of the genetic background of the gastric cell model, both MKN45 and AGS SimpleCells acquired a significant higher capacity to degrade and invade the ECM. This observation is in agreement with other COSMC modulation cell models overexpressing simple O-glycans that also acquired in vitro and in vivo oncogenic features [39,64]. On the same direction, silencing ST6GalNAc1, the enzyme responsible for STn production, reduced tumor growth, cell migration and invasion on hepatocellular cells [43], as well as in gastric carcinoma cells [44].
Our in vivo assays revealed that tumor xenografts from MKN45 SC displayed less necrotic areas accompanied with reduced tumor neoangiogenic features. These findings suggest that STn on tumor cells, besides increasing aggressiveness, supports enhanced biological capacity to survive in a less angiogenic environment. This was corroborated by the in vivo CAM model, where both MKN45 SC and AGS SC cells showed decreased capacity to induce de novo vessel formation. Although the molecular mechanism behind such findings is still unknown, SimpleCells appeared to be able to survive on the pre-existing blood vessels, avoiding the need to form new vessels. Expression of truncated O-glycans revealed to promote survival by decreasing apoptosis in pancreatic cancer cells after COSMC knock-down [64]. It is possible that the massive O-glycosylation alterations occurring in a variety of proteins after COSMC KO, changes the transcription of several genes and affecting new vessel formation.
Supporting the clinical implications discussed above, truncation of O-glycosylation in gastric cancer cells had a remarkable impact in the transcriptomic signature of the glycoengineered cell lines. The enrichment analysis of the differentially expressed genes for GO biological processes showed a significant association with development and motility processes. These genetic signatures support the oncogenic characteristics found in both MKN45 SC and AGS SC cancer cells and demonstrate that truncation of O-glycans affects multiple systems simultaneously that sustain tumor progression. Similarly, genes involved in cellular movement and proliferation, together with genes linked to differentiation and apoptosis, were also found to be significantly changed in pancreatic and in human immortalized keratinocyte HaCaT SimpleCell models [39]. The effect of truncated O-glycans on these different cancer cell models showed that aberrant glycosylation was able to influence several important pathways in tissue homeostasis and oncogenesis, including adhesive and signaling molecules [39]. Our transcriptomic analysis showed that SRPX2 and RUNX1 were the genes with highest fold change in both MKN45 and AGS SimpleCells. Both SRPX2 and RUNX1 have been previously implicated in cancer progression, but their association with O-glycan expression has, to our knowledge, never been addressed. We observed that changing the O-glycosylation by COSMC KO resulted in activation of EGFR and ErbB2 RTKs. Interestingly, we demonstrayed in our model that EGFR activation regulated both SRPX2 and RUNX1 genes transcription. A recent report showed that RUNX1 was able to positively regulate ErbB2 signaling [65]. This raises the hypothesis of a potential loop of signaling between this cell receptor and the RUNX1 gene. Furthermore, both SRPX2 and RUNX1 proteins were highly present in the gastric cancer tissues when compared with adjacent non-neoplastic gastric mucosa. Interestingly, co-expression of SRPX2, RUNX1 and STn was observed within gastric carcinoma samples. Also, an inverse correlation between C1GALT1C1 and both SRPX2 and RUNX1 was found in the analyzed gastric cancer tissues. Our findings, describe one possible mechanism by which changing the glycosylation affects several cellular biological processes. Alterations in O-glycosylation occurring in tumors can influence cell signaling leading to the transcription activation of specific genes that confer increased oncogenic features favoring the process of cancer progression.
In accordance, SRPX2 has been previously described as upregulated in gastric cancer [54,66,67]. SRPX2 was found to promote cell motility and adhesion through Fak phosphorylation [66], and capable of inducing cell invasion and aggressiveness in several types of cancer [66,[68], [69], [70]]. RUNX1, a hematopoietic regulator, is known to be expressed in the isthmus of human stomach, where stem cells reside [71]. Interestingly, RUNX1 gene was found to be a master regulator of carbohydrate metabolism and knocking down this gene led to decreased glucose metabolic activity [72]. Other authors have reported RUNX1 expression to be decreased in gastric cancer [73]. In order to scale up the correlation between the expression of both SRPX2 and RUNX1 in gastric cancer, we analyzed the transcriptomic data based on independent cohorts from the Oncomine database. Both SRPX2 and RUNX1 expression levels were significantly upregulated in gastric cancer in multiple gastric carcinoma datasets. Interestingly, we show for the first time a significant correlation between SRPX2 and RUNX1 expression levels in gastric cancer patients through a microarray expression data analysis from Oncomine database. Furthermore, our analysis showed that SRPX2 and RUNX1 expression was significantly associated with poor-survival of gastric cancer patients, highlighting their importance in tumor biology and in cancer progression. These results were further supported by the decreased mouse survival observed in the MKN45 SC-injected mice that also displayed high SRPX2 and RUNX1 expression levels. Taking into consideration our findings, we envision that inhibitors targeting tumor specific glycosylation pathways and/or glycan epitopes could harbor potential for therapeutic strategies. Further studies addressing this hypothesis are warranted.
Altogether, our results demonstrate a novel molecular link between premature truncation of O-glycans and concomitant STn overexpression occurring in gastric tumors and its relation with patients' poor-survival (Fig. 7). Our work sheds light on the mechanisms underlying the biological advantage of STn expressing cancer cells through regulation of key biological processes, highlighting the impact of changing cell glycosylation per se in a cancer context. These findings also revealed new genes with potential clinical application as prognostic biomarkers in gastric cancer patients.
Fig. 7.
Truncation of O-glycans alters cancer cell biology promoting a poor-survival phenotype. During cancer progression micro-environmental changes, epigenetic and/or genetic mutations promote alterations in glycosyltransferases activity that culminate in several modifications in the cancer cell glycosylation profile. These glycophenotypic changes, namely premature truncation of O-glycans and concomitant STn overexpression occurring in gastric tumors, affected tumor cell morphology and ECM interactions leading to increased invasion. In our model, O-glycans truncation of gastric cancer cells promoted a switch in the cancer cell transcriptomic signature as well as activation of receptor tyrosine kinase (RTKs). We showed that EGFR and HER2 RTKs were activated after COSMC KO of gastric cancer cells, leading to up-regulation of SRPX2 and RUNX1 genes. These genes were further validated as highly expressed in gastric cancer patients, as co-expressed with STn and correlated with patients' poor-survival.
Acknowledgments
Acknowledgements
We thank Catharina Steentoft and Henrik Clausen for the cell line models and helpful scientific discussions. We are grateful for Nuno Mendes's technical support. The authors acknowledge the support of José Luis Costa and Mafalda Rocha from the i3S Genomics Platform (GenCore), and Maria G Lazaro from the Bioimaging I3S Scientific Platform, member of the PPBI (PPBI-POCI-01-0145-FEDER-022122).
Funding sources
This work was funded by FEDER funds through the Operational Programme for Competitiveness Factors-COMPETE (POCI-01-0145-FEDER-016585; POCI-01-0145-FEDER-007274; POCI-01-0145-FEDER-028489) and National Funds through the Foundation for Science and Technology (FCT), under the projects: PTDC/BBB-EBI/0567/2014 (to CAR), PTDC/MED-ONC/28489/2017 (to AM) and UID/BIM/04293/2013; and the project NORTE-01-0145-FEDER-000029, supported by Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). The authors acknowledge the support by Gastric Glyco Explorer Initial Training Network (European Union Seventh Framework Programme GastricGlycoExplorer project, grant number 316929). DF acknowledges the FCT PhD Programmes and Programa Operacional Potencial Humano (POPH), specifically the BiotechHealth Programe (Doctoral Programme on Cellular and Molecular Biotechnology Applied to Health Sciences), with the reference PD/0016/2012 funded by FCT. Grants were received from FCT, POPH and FSE (Fundo Social Europeu): SFRH/BD/110636/2015 to DF and SFRH/BPD/115730/2016 to FP.
Declaration of interest
All the authors have nothing to disclose.
Author contributions
DF designed and performed the experimental work, analyzed the data and wrote the manuscript. DC developed the cell line models. DC, FP, RM and AM performed experimental work. JG and MTP performed in vivo assays. SM contributed for the transcriptomic analysis and illustrated the model schematic figure. FP, JM and SM conducted bioinformatics analysis. AP and FG performed histopathology and analyzed IHC results. AM and CAR conceived the hypothesis, interpreted the data and wrote the manuscript. All authors read, reviewed and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.017.
Contributor Information
Ana Magalhães, Email: amagalhaes@ipatimup.pt.
Celso A. Reis, Email: celsor@ipatimup.pt.
Appendix A. Supplementary data
Supplementary material
References
- 1.Stewart B.W., Wild C.P. 2014. World Cancer Report 2014. [Google Scholar]
- 2.Pinho S.S., Reis C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15(9):540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 3.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99(16):10231–10233. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taniguchi N., Hancock W., Lubman D.M., Rudd P.M. The second golden age of glycomics: from functional glycomics to clinical applications. J Proteome Res. 2009;8(2):425–426. doi: 10.1021/pr801057j. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal P., Fontanals-Cirera B., Sokolova E., Jacob S., Vaiana C.A., Argibay D. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell. 2017;31(6):804–19 e7. doi: 10.1016/j.ccell.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magalhaes A., Duarte H.O., Reis C.A. Aberrant glycosylation in cancer: a novel molecular mechanism controlling metastasis. Cancer Cell. 2017;31(6):733–735. doi: 10.1016/j.ccell.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Bassaganas S., Carvalho S., Dias A.M., Perez-Garay M., Ortiz M.R., Figueras J. Pancreatic cancer cell glycosylation regulates cell adhesion and invasion through the modulation of alpha2beta1 integrin and E-cadherin function. PLoS One. 2014;9(5):e98595. doi: 10.1371/journal.pone.0098595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H., Nagy T., Pierce M. Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in Apc(min/+) mice through altered Wnt receptor signaling. J Biol Chem. 2014;289(45):31534–31549. doi: 10.1074/jbc.M114.602680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart G.W., Copeland R.J. Glycomics hits the big time. Cell. 2010;143(5):672–676. doi: 10.1016/j.cell.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau K.S., Partridge E.A., Grigorian A., Silvescu C.I., Reinhold V.N., Demetriou M. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 11.Contessa J.N., Bhojani M.S., Freeze H.H., Rehemtulla A., Lawrence T.S. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008;68(10):3803–3809. doi: 10.1158/0008-5472.CAN-07-6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duarte H.O., Balmana M., Mereiter S., Osorio H., Gomes J., Reis C.A. Gastric cancer cell glycosylation as a modulator of the ErbB2 oncogenic receptor. Int J Mol Sci. 2017;18(11) doi: 10.3390/ijms18112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mereiter S., Magalhaes A., Adamczyk B., Jin C., Almeida A., Drici L. Glycomic analysis of gastric carcinoma cells discloses glycans as modulators of RON receptor tyrosine kinase activation in cancer. Biochim Biophys Acta. 2016;1860(8):1795–1808. doi: 10.1016/j.bbagen.2015.12.016. [DOI] [PubMed] [Google Scholar]
- 14.Gomes C., Osorio H., Pinto M.T., Campos D., Oliveira M.J., Reis C.A. Expression of ST3GAL4 leads to SLe(x) expression and induces c-Met activation and an invasive phenotype in gastric carcinoma cells. PLoS One. 2013;8(6):e66737. doi: 10.1371/journal.pone.0066737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schultz M.J., Swindall A.F., Bellis S.L. Regulation of the metastatic cell phenotype by sialylated glycans. Cancer Metastasis Rev. 2012;31(3–4):501–518. doi: 10.1007/s10555-012-9359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinho S., Marcos N.T., Ferreira B., Carvalho A.S., Oliveira M.J., Santos-Silva F. Biological significance of cancer-associated sialyl-Tn antigen: modulation of malignant phenotype in gastric carcinoma cells. Cancer Lett. 2007;249(2):157–170. doi: 10.1016/j.canlet.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Amado M., Carneiro F., Seixas M., Clausen H., Sobrinho-Simoes M. Dimeric sialyl-Le(x) expression in gastric carcinoma correlates with venous invasion and poor outcome. Gastroenterology. 1998;114(3):462–470. doi: 10.1016/s0016-5085(98)70529-3. [DOI] [PubMed] [Google Scholar]
- 18.Reis C.A., Osorio H., Silva L., Gomes C., David L. Alterations in glycosylation as biomarkers for cancer detection. J Clin Pathol. 2010;63(4):322–329. doi: 10.1136/jcp.2009.071035. [DOI] [PubMed] [Google Scholar]
- 19.Kudelka M.R., Ju T., Heimburg-Molinaro J., Cummings R.D. Simple sugars to complex disease--mucin-type O-glycans in cancer. Adv Cancer Res. 2015;126:53–135. doi: 10.1016/bs.acr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campos D., Freitas D., Gomes J., Reis C.A. Glycoengineered cell models for the characterization of cancer O-glycoproteome: an innovative strategy for biomarker discovery. Expert Rev Proteomics. 2015;12(4):337–342. doi: 10.1586/14789450.2015.1059758. [DOI] [PubMed] [Google Scholar]
- 21.Ju T., Wang Y., Aryal R.P., Lehoux S.D., Ding X., Kudelka M.R. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteomics Clin Appl. 2013;7(9–10):618–631. doi: 10.1002/prca.201300024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chugh S., Barkeer S., Rachagani S., Nimmakayala R.K., Perumal N., Pothuraju R. Disruption of C1galt1 gene promotes development and metastasis of pancreatic adenocarcinomas in mice. Gastroenterology. 2018;155(5):1608–1624. doi: 10.1053/j.gastro.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinho S.S., Carvalho S., Marcos-Pinto R., Magalhaes A., Oliveira C., Gu J. Gastric cancer: adding glycosylation to the equation. Trends Mol Med. 2013;19(11):664–676. doi: 10.1016/j.molmed.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Mereiter S., Balmana M., Gomes J., Magalhaes A., Reis C.A. Glycomic approaches for the discovery of targets in gastrointestinal cancer. Front Oncol. 2016;6:55. doi: 10.3389/fonc.2016.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldus S.E., Zirbes T.K., Monig S.P., Engel S., Monaca E., Rafiqpoor K. Histopathological subtypes and prognosis of gastric cancer are correlated with the expression of mucin-associated sialylated antigens: Sialosyl-Lewis(a), Sialosyl-Lewis(x) and sialosyl-Tn. Tumour Biol. 1998;19(6):445–453. doi: 10.1159/000030036. [DOI] [PubMed] [Google Scholar]
- 26.Springer G.F.T. Tn, general carcinoma autoantigens. Science. 1984;224(4654):1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- 27.Brockhausen I., Yang J., Lehotay M., Ogata S., Itzkowitz S. Pathways of mucin O-glycosylation in normal and malignant rat colonic epithelial cells reveal a mechanism for cancer-associated Sialyl-Tn antigen expression. Biol Chem. 2001;382(2):219–232. doi: 10.1515/BC.2001.029. [DOI] [PubMed] [Google Scholar]
- 28.Marcos N.T., Pinho S., Grandela C., Cruz A., Samyn-Petit B., Harduin-Lepers A. Role of the human ST6GalNAc-I and ST6GalNAc-II in the synthesis of the cancer-associated sialyl-Tn antigen. Cancer Res. 2004;64(19):7050–7057. doi: 10.1158/0008-5472.CAN-04-1921. [DOI] [PubMed] [Google Scholar]
- 29.Baldus S.E., Hanisch F.G. Biochemistry and pathological importance of mucin-associated antigens in gastrointestinal neoplasia. Adv Cancer Res. 2000;79:201–248. doi: 10.1016/s0065-230x(00)79007-5. [DOI] [PubMed] [Google Scholar]
- 30.Marcos N.T., Bennett E.P., Gomes J., Magalhaes A., Gomes C., David L. ST6GalNAc-I controls expression of sialyl-Tn antigen in gastrointestinal tissues. Front Biosci (Elite Ed) 2011;3:1443–1455. doi: 10.2741/e345. [DOI] [PubMed] [Google Scholar]
- 31.Werther J.L., Rivera-MacMurray S., Bruckner H., Tatematsu M., Itzkowitz S.H. Mucin-associated sialosyl-Tn antigen expression in gastric cancer correlates with an adverse outcome. Br J Cancer. 1994;69(3):613–616. doi: 10.1038/bjc.1994.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itzkowitz S., Kjeldsen T., Friera A., Hakomori S., Yang U.S., Kim Y.S. Expression of Tn, sialosyl Tn, and T antigens in human pancreas. Gastroenterology. 1991;100(6):1691–1700. doi: 10.1016/0016-5085(91)90671-7. [DOI] [PubMed] [Google Scholar]
- 33.Itzkowitz S.H., Yuan M., Montgomery C.K., Kjeldsen T., Takahashi H.K., Bigbee W.L. Expression of Tn, sialosyl-Tn, and T antigens in human colon cancer. Cancer Res. 1989;49(1):197–204. [PubMed] [Google Scholar]
- 34.Inoue M., Ton S.M., Ogawa H., Tanizawa O. Expression of Tn and sialyl-Tn antigens in tumor tissues of the ovary. Am J Clin Pathol. 1991;96(6):711–716. doi: 10.1093/ajcp/96.6.711. [DOI] [PubMed] [Google Scholar]
- 35.Yonezawa S., Tachikawa T., Shin S., Sato E. Sialosyl-Tn antigen. Its distribution in normal human tissues and expression in adenocarcinomas. Am J Clin Pathol. 1992;98(2):167–174. doi: 10.1093/ajcp/98.2.167. [DOI] [PubMed] [Google Scholar]
- 36.Julien S., Picco G., Sewell R., Vercoutter-Edouart A.S., Tarp M., Miles D. Sialyl-Tn vaccine induces antibody-mediated tumour protection in a relevant murine model. Br J Cancer. 2009;100(11):1746–1754. doi: 10.1038/sj.bjc.6605083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kjeldsen T., Clausen H., Hirohashi S., Ogawa T., Iijima H., Hakomori S. Preparation and characterization of monoclonal antibodies directed to the tumor-associated O-linked sialosyl-2----6 alpha-N-acetylgalactosaminyl (sialosyl-Tn) epitope. Cancer Res. 1988;48(8):2214–2220. [PubMed] [Google Scholar]
- 38.Ju T., Lanneau G.S., Gautam T., Wang Y., Xia B., Stowell S.R. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008;68(6):1636–1646. doi: 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- 39.Radhakrishnan P., Dabelsteen S., Madsen F.B., Francavilla C., Kopp K.L., Steentoft C. Immature truncated O-glycophenotype of cancer directly induces oncogenic features. Proc Natl Acad Sci U S A. 2014;111(39):E4066–E4075. doi: 10.1073/pnas.1406619111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steentoft C., Vakhrushev S.Y., Vester-Christensen M.B., Schjoldager K.T., Kong Y., Bennett E.P. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8(11):977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 41.Campos D., Freitas D., Gomes J., Magalhaes A., Steentoft C., Gomes C. Probing the O-glycoproteome of gastric cancer cell lines for biomarker discovery. Mol Cell Proteomics. 2015;14(6):1616–1629. doi: 10.1074/mcp.M114.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozaki H., Matsuzaki H., Ando H., Kaji H., Nakanishi H., Ikehara Y. Enhancement of metastatic ability by ectopic expression of ST6GalNAcI on a gastric cancer cell line in a mouse model. Clin Exp Metastasis. 2012;29(3):229–238. doi: 10.1007/s10585-011-9445-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu X., Wu Q., Wang L., Zhao Y., Zhang Q., Meng Q. Silencing of ST6GalNAc I suppresses the proliferation, migration and invasion of hepatocarcinoma cells through PI3K/AKT/NF-kappaB pathway. Tumour Biol. 2016;37(9):12213–12221. doi: 10.1007/s13277-016-5086-y. [DOI] [PubMed] [Google Scholar]
- 44.Tamura F., Sato Y., Hirakawa M., Yoshida M., Ono M., Osuga T. RNAi-mediated gene silencing of ST6GalNAc I suppresses the metastatic potential in gastric cancer cells. Gastric Cancer. 2016;19(1):85–97. doi: 10.1007/s10120-014-0454-z. [DOI] [PubMed] [Google Scholar]
- 45.David L., Carneiro F., Sobrinho-Simoes M. Sialosyl Tn antigen expression is associated with the prognosis of patients with advanced gastric cancer. Cancer. 1996;78(1):177–178. doi: 10.1002/(SICI)1097-0142(19960701)78:1<177::AID-CNCR24>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 46.Nakagoe T., Sawai T., Tsuji T., Jibiki M., Nanashima A., Yamaguchi H. Pre-operative serum levels of sialyl Tn antigen predict liver metastasis and poor prognosis in patients with gastric cancer. Eur J Surg Oncol. 2001;27(8):731–739. doi: 10.1053/ejso.2001.1199. [DOI] [PubMed] [Google Scholar]
- 47.Victorzon M., Nordling S., Nilsson O., Roberts P.J., Haglund C. Sialyl Tn antigen is an independent predictor of outcome in patients with gastric cancer. Int J Cancer. 1996;65(3):295–300. doi: 10.1002/(SICI)1097-0215(19960126)65:3<295::AID-IJC3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen A.L., Reis C.A., Tarp M.A., Mandel U., Ramachandran K., Sankaranarayanan V. Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology. 2006;16(2):96–107. doi: 10.1093/glycob/cwj044. [DOI] [PubMed] [Google Scholar]
- 49.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Q., Wen Y.G., Li D.P., Xia J., Zhou C.Z., Yan D.W. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 52.Cui J., Chen Y., Chou W.C., Sun L., Chen L., Suo J. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39(4):1197–1207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D'Errico M., de Rinaldis E., Blasi M.F., Viti V., Falchetti M., Calcagnile A. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45(3):461–469. doi: 10.1016/j.ejca.2008.10.032. [DOI] [PubMed] [Google Scholar]
- 54.Chen X., Leung S.Y., Yuen S.T., Chu K.M., Ji J., Li R. Variation in gene expression patterns in human gastric cancers. Mol Biol Cell. 2003;14(8):3208–3215. doi: 10.1091/mbc.E02-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho J.Y., Lim J.Y., Cheong J.H., Park Y.Y., Yoon S.L., Kim S.M. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17(7):1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7(6):599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schultz M.J., Holdbrooks A.T., Chakraborty A., Grizzle W.E., Landen C.N., Buchsbaum D.J. The tumor-associated glycosyltransferase ST6Gal-I regulates stem cell transcription factors and confers a cancer stem cell phenotype. Cancer Res. 2016;76(13):3978–3988. doi: 10.1158/0008-5472.CAN-15-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohuchi N., Thor A., Nose M., Fujita J., Kyogoku M., Schlom J. Tumor-associated glycoprotein (TAG-72) detected in adenocarcinomas and benign lesions of the stomach. Int J Cancer. 1986;38(5):643–650. doi: 10.1002/ijc.2910380505. [DOI] [PubMed] [Google Scholar]
- 59.Munkley J. The role of Sialyl-Tn in cancer. Int J Mol Sci. 2016;17(3):275. doi: 10.3390/ijms17030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David L., Nesland J.M., Clausen H., Carneiro F., Sobrinho-Simoes M. Simple mucin-type carbohydrate antigens (Tn, sialosyl-Tn and T) in gastric mucosa, carcinomas and metastases. APMIS Suppl. 1992;27:162–172. [PubMed] [Google Scholar]
- 61.Heerboth S., Housman G., Leary M., Longacre M., Byler S., Lapinska K. EMT and tumor metastasis. Clin Transl Med. 2015;4:6. doi: 10.1186/s40169-015-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Munkley J.E., Sugars D.J. Cell adhesion: the role of ST6GalNAc1 in prostate cancer progression. Cancer Cell Microenviron. 2016;3 [Google Scholar]
- 63.Julien S., Lagadec C., Krzewinski-Recchi M.A., Courtand G., Le Bourhis X., Delannoy P. Stable expression of sialyl-Tn antigen in T47-D cells induces a decrease of cell adhesion and an increase of cell migration. Breast Cancer Res Treat. 2005;90(1):77–84. doi: 10.1007/s10549-004-3137-3. [DOI] [PubMed] [Google Scholar]
- 64.Hofmann B.T., Schluter L., Lange P., Mercanoglu B., Ewald F., Folster A. COSMC knockdown mediated aberrant O-glycosylation promotes oncogenic properties in pancreatic cancer. Mol Cancer. 2015;14:109. doi: 10.1186/s12943-015-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitsuda Y., Morita K., Kashiwazaki G., Taniguchi J., Bando T., Obara M. RUNX1 positively regulates the ErbB2/HER2 signaling pathway through modulating SOS1 expression in gastric cancer cells. Sci Rep. 2018;8(1):6423. doi: 10.1038/s41598-018-24969-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tanaka K., Arao T., Maegawa M., Matsumoto K., Kaneda H., Kudo K. SRPX2 is overexpressed in gastric cancer and promotes cellular migration and adhesion. Int J Cancer. 2009;124(5):1072–1080. doi: 10.1002/ijc.24065. [DOI] [PubMed] [Google Scholar]
- 67.Tanaka K., Arao T., Tamura D., Aomatsu K., Furuta K., Matsumoto K. SRPX2 is a novel chondroitin sulfate proteoglycan that is overexpressed in gastrointestinal cancer. PLoS One. 2012;7(1):e27922. doi: 10.1371/journal.pone.0027922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang H., Zhao J., Zhang L., Zhao J., Zhuang Y., Liang P. SRPX2 enhances the epithelial-mesenchymal transition and temozolomide resistance in glioblastoma cells. Cell Mol Neurobiol. 2016;36(7):1067–1076. doi: 10.1007/s10571-015-0300-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamada T., Oshima T., Yoshihara K., Sato T., Nozaki A., Shiozawa M. Impact of overexpression of Sushi repeat-containing protein X-linked 2 gene on outcomes of gastric cancer. J Surg Oncol. 2014;109(8):836–840. doi: 10.1002/jso.23602. [DOI] [PubMed] [Google Scholar]
- 70.Liu K.L., Wu J., Zhou Y., Fan J.H. Increased Sushi repeat-containing protein X-linked 2 is associated with progression of colorectal cancer. Med Oncol. 2015;32(4):99. doi: 10.1007/s12032-015-0548-4. [DOI] [PubMed] [Google Scholar]
- 71.Matsuo J., Kimura S., Yamamura A., Koh C.P., Hossain M.Z., Heng D.L. Identification of stem cells in the epithelium of the stomach corpus and antrum of mice. Gastroenterology. 2017;152(1):218–31 e14. doi: 10.1053/j.gastro.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 72.Peng X., Chen Z., Farshidfar F., Xu X., Lorenzi P.L., Wang Y. Molecular characterization and clinical relevance of metabolic expression subtypes in human cancers. Cell Rep. 2018;23(1):255–69 e4. doi: 10.1016/j.celrep.2018.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li N., Zhang Q.Y., Zou J.L., Li Z.W., Tian T.T., Dong B. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget. 2016;7(4):4817–4828. doi: 10.18632/oncotarget.6736. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material