Abstract
Background
With the changed therapeutic armamentarium for Crohn's disease (CD) and ulcerative colitis (UC), biomarkers predicting treatment response are urgently needed. We studied whole blood and mucosal expression of genes previously reported to predict outcome to anti-TNF therapy, and investigated if the signature was specific for anti-TNF agents.
Methods
We prospectively included 54 active IBD patients (24CD, 30UC) initiating anti-TNF therapy, as well as 22 CD patients initiating ustekinumab and 51 patients initiating vedolizumab (25CD, 26UC). Whole blood expression of OSM, TREM1, TNF and TNFR2 was measured prior to start of therapy using qPCR, and mucosal gene expression in inflamed biopsies using RNA-sequencing. Response was defined as endoscopic remission (SES-CD ≤ 2 at week 24 for CD and Mayo endoscopic sub-score ≤ 1 at week 10 for UC).
Findings
Baseline whole blood TREM1 was downregulated in future anti-TNF responders, both in UC (FC = 0.53, p = .001) and CD (FC = 0.66, p = .007), as well as in the complete cohort (FC = 0.67, p < .001). Receiver operator characteristic statistics showed an area under the curve (AUC) of 0.78 (p = .001). A similar accuracy could be achieved with mucosal TREM1 (AUC 0.77, p = .003), which outperformed the accuracy of serum TREM1 (AUC 0.58, p = .31). Although differentially expressed in tissue, OSM, TNF and TNFR2 were not differentially expressed in whole blood. The TREM1 predictive signal was anti-TNF specific, as no changes were seen in ustekinumab and vedolizumab treated patients.
Interpretation
We identified low TREM-1 as a specific biomarker for anti-TNF induced endoscopic remission. These results can aid in the selection of therapy in biologic-naïve patients.
Keywords: TREM1, IBD, Anti-TNF, Infliximab, Adalimumab, Personalised medicine, Biomarker, Endoscopic remission
Abbreviations: ADM, adalimumab; AUC, area under the curve; CD, Crohn's disease; CI, confidence interval; IBD, inflammatory bowel disease; IFX, infliximab; IL, interleukin; IQR, interquartile range; LP, lamina propria; OSM, oncostatin M; qPCR, real-time polymerase chain reaction; ROC, receiver operator characteristic; SES-CD, simple endoscopic score for Crohn's disease; TNF, tumour necrosis factor; TNFR2, tumour necrosis factor receptor 2; TREM, triggering receptor expressed on myeloid cells; UC, ulcerative colitis; UST, ustekinumab; VDZ, vedolizumab
Research in context.
Evidence before this study
Biological agents have dramatically changed therapeutic algorithms in inflammatory bowel disease (IBD) and significantly improved disease outcome. However, an individualised approach with predictive biomarkers is urgently awaited, as up to one third of patients never respond to a particular drug and thus a ‘one-size fits all principle’ should no longer be applied. Recently, whole blood TREM1 expression was suggested as a potential biomarker predicting response to anti-TNF therapy in patients with Crohn's disease.
Added value of this study
We validated whole blood TREM1 as the first predictive signal for anti-TNF induced endoscopic remission in a mixed cohort of patients with both Crohn's disease or ulcerative colitis. Additionally, we demonstrated its anti-TNF specificity by studying the similar signature in vedolizumab and ustekinumab treated patients. Finally, we demonstrated that mucosal TREM1 expression is as accurate as whole blood TREM1, whereas serum TREM1 is not a good biomarker for anti-TNF non-responsiveness.
Implications of all the available evidence
Our results can aid in the future selection of therapy in biologic naïve IBD patients and could be translated in the first biomarker-driven randomized trial stratifying patients towards or away from anti-TNF therapy based on TREM1 whole blood expression.
Alt-text: Unlabelled Box
1. Introduction
The introduction of biological therapies in the treatment of inflammatory bowel disease (IBD) has significantly improved disease outcome and altered the natural history of the disease, including less steroid exposure, less hospitalizations, and less major surgeries [1]. Novel insights in IBD pathogenesis led to the development of new compounds with a different mode of action, including anti-adhesion molecules (vedolizumab, VDZ) and interleukin (IL) 12/23 antibodies (ustekinumab, UST) [2]. However, some patients never respond to a particular therapy. For anti-TNF therapy in particular, primary non-response rates vary from 10 to 30%, and the annual risk of secondary loss of response ranges from 13% for infliximab (IFX) to 20% for adalimumab (ADM) [3]. Both from a patient perspective as from a socio-economic perspective, identifying the most suitable therapy for a given patient is key. With many more compounds being tested in phase II and III clinical trials [4], personalised medicine will become even more necessary in future.
During recent years, researchers focused on a better understanding of the working mechanisms of anti-TNF agents [5]. This not only contributed to the development of novel targeted therapies, but also paved the way for biomarker development predicting response to anti-TNF. Gene expression analysis of inflamed biopsies of Crohn's disease (CD) and ulcerative colitis patients (UC) prior to IFX therapy, identified several genes differentially expressed between responders and non-responders [[6], [7], [8]]. Among these, IL13RA2 was the highest ranked common gene for both CD and UC analyses. Co-expression network analysis of the same dataset concluded that TNF-driven pathways are significantly increased at baseline in future non-responders [9]. Recently, expansion of apoptosis-resistant intestinal TNFR2+ IL-23R+ T-cells has been associated with resistance to anti-TNF therapy in CD [10]. Finally, advanced bioinformatic techniques integrated all publically available datasets and identified colonic expression of both oncostatin M (OSM) and Triggering Receptor Expressed on Myeloid cells 1 (TREM1) as key players in and predictors of anti-TNF (non-)responsiveness [[11], [12], [13]]. However, their specificity for anti-TNF agents has not yet been investigated, and therefore it remains to be clarified if these markers are true anti-TNF-specific predictors or just bystanders of inflammation.
So far, no predictive biomarker has found its way into IBD clinical practice yet. Potentially because markers based on gene expression of intestinal biopsies are more complex to translate to clinical practice. In contrast, whole blood biomarkers may be more applicable. Whole blood TREM1 expression looks a promising predictive biomarker for anti-TNF therapy in CD, although conflicting results are currently reported [12,13]. We here studied mucosal biopsies and whole blood expression of IL13RA2, TNF-alpha, TNFR2, OSM, TREM1 and its transcripts in a prospectively collected cohort of CD and UC patients prior to initiation of biological therapy (ADM, IFX, UST, or VDZ) and assessed endoscopic remission as outcome.
2. Methods
2.1. Patient selection
This prospective study was conducted at the IBD center of the University Hospitals Leuven (Leuven, Belgium). We collected whole blood of 127 IBD patients initiating biologic therapy: 54 CD and UC patients initiating IFX or ADM, 22 CD patients initiating UST and 51 CD and UC patients initiating VDZ (Table 1, Supplementary Table 1). All patients had endoscopy-proven active disease (Mayo endoscopic sub score 2–3 in case of UC; presence of ileal and/or colonic ulcerations in case of CD) and had to be naïve for the drug that was initiated at inclusion.
Table 1.
Disease characteristics of the whole blood, anti-TNF treated cohort.
| Characteristic | Crohn's disease n = 24 |
Ulcerative colitis n = 30 |
|---|---|---|
| Sex, women, n (%) | 12 (50.0) | 18 (60.0) |
| Endoscopic assessment after initiated therapy, n (%) | ||
|
13 (54.2) | 10 (33.3) |
|
11 (45.8) | 20 (66.7) |
| Anti-TNF agent, n (%) | ||
|
10 (41.7) | 12 (40.0) |
|
14 (58.3) | 18 (60.0) |
| Age, years, median (IQR) | 31.9 (26.5–51.5) | 43.5 (29.6–55.7) |
| Disease duration, years, median (IQR) | 7.8 (2.1–22.2) | 5.1 (1.7–17.0) |
| C-reactive protein, mg/L, median (IQR) | 5.7 (0.9–8.5) | 3.9 (1.1–24.6) |
| Faecal calprotectin, μg/g, median (IQR) | 1190 (328–1800) | 1361 (804–1800) |
| Albumin, g/L, median (IQR) | 42.3 (39.2–45.2) | 43.6 (39.4–45.5) |
| Body Mass Index, kg/m2, median (IQR) | 22.1 (20.4–25.1) | 21.6 (19.7–25.9) |
| Disease location Crohn's disease, n (%) | ||
|
6 (25.0) | |
|
6 (25.0) | |
|
12 (50.0) | N.A. |
|
1 (4.2) | |
|
Disease location ulcerative colitis, n (%) |
||
|
3 (10.0) | |
|
19 (63.3) | |
|
8 (26.7) | |
| Disease behaviour Crohn's disease, n (%) | ||
|
15 (62.5) | |
|
6 (25.0) | |
|
3 (12.5) | N.A. |
|
4 (16.7) | |
| Previous IBD related surgery (resection, stricturoplasty) | 10 (41.7) | N.A. |
| Concomitant medication, n (%) | ||
|
8 (33.3) | 8 (26.7) |
|
12 (50.0) | 6 (20.0) |
| Previous biological agents, n (%) | ||
|
8 (33.3) | 10 (33.3) |
|
4 (16.7) | 2 (6.6) |
|
4 (16.7) | 3 (10.0) |
|
4 (16.7) | 7 (23.3) |
|
3 (12.5) | N.A. |
| Smoking, n (%) | ||
|
16 (66.6) | 18 (60.0) |
|
4 (16.7) | 5 (16.7) |
|
4 (16.7) | 7 (23.3) |
All anti-TNF treated patients had to have persistent endoscopic lesions with sufficient drug exposure, defined as a maintenance trough level > 3.0 μg/mL for infliximab or > 5.0 μg/mL for adalimumab before being defined as non-responder. Due the lack of agreement on the targeted threshold for ustekinumab and vedolizumab, if any, we did not include an exposure requirement in the definition of non-response for both drugs.
Whole blood (PAXgene blood RNA tubes, Qiagen, Benelux, Netherlands) samples were collected at baseline, prior to the first infusion/injection, stored overnight at room temperature whereafter they were preserved at −80 °C according to the manufacturer's instructions. Biopsies at the edge of an ulcer in the most inflamed area were taken during endoscopy prior to the start of therapy, stored in RNALater buffer (Ambion, Austin, TX, USA) and preserved at −80 °C. Similarly, serum of all patients initiating anti-TNF therapy was taken prior to first administration, centrifuged and stored at −20 °C.
All included patients had given written consent to participate in the Institutional Review Board approved IBD Biobank (B322201213950/S53684).
2.2. Outcomes
Response was defined based on endoscopic findings as an objective parameter In CD patients, endoscopic remission was assessed after 6 months and defined as a Simple Endoscopic Disease (SES-CD) score ≤ 2 [14,15]. In UC patients, a Mayo endoscopic sub-score of ≤1 was considered as endoscopic remission. Due to national reimbursement criteria, all UC patients were endoscopically evaluated at week 8 (ADM) or week 14 (IFX and VDZ). All endoscopies were performed by the same 3 experienced IBD staff members (GVA, SV, MF).
2.3. Isolation of RNA
Total whole blood RNA was extracted using the PAXgene Blood RNA Kit (Qiagen, Benelux, Netherlands) according to the manufacturer's instructions. Total RNA from inflamed biopsies was extracted using the AllPrep DNA/RNA Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The integrity and quantity of all RNA was assessed with a 2100 Bioanalyzer (Agilent, Waldbronn, Germany) and a Nanodrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Extracted RNA was stored at −80 °C until further processing.
2.4. Quantitative RT-PCR
Gene expression (TREM1, OSM, TNF, TNFR2, IL13RA2) in whole blood was studied trough quantitative real-time polymerase chain reaction (qPCR) analysis. To further unravel the TREM1 predictive signal, the expression of all known TREM1 transcripts, TREM1 transcript variant x1 (TREM1-mb), TREM1 transcript variant x2 (TREM1-x2) and TREM1 transcript variant x3 (TREM1-sv), was studied too. cDNA was synthesized from 0.25 μg of total RNA using the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany) according to the manufacturer's protocol. The primers were synthesized by Sigma-Genosys (Haverhill, UK) (Supplementary Table 2) and 10 μM stock solutions were used to make the reaction mixture (5 μL SybrGreen, 0.2 μM FW & RV primer, 2 μL cDNA sample, 2.8 μL RNAse-free H2O). All samples were amplified in duplicate reactions. Samples were analysed with the Lightcycler 480 (Roche, Basel, Switzerland). The following amplification program was used: 5′ 95 °C, 45 x (10″ 95 °C, 15″ 60 °C, 15″ 72 °C), 5″ 95 °C, 1′ 60 °C, 4 °C. mRNA-levels were normalized to the housekeeping gene β-actin and quantified using the comparative (ΔΔ) Ct method.
2.5. RNA sequencing
Next-generation single-end sequencing was performed using the Illumina HiSeq 4000NGS, after library preparation using the TruSeq Stranded mRNA protocol (Illumina, San Diego, USA) according to the manufacturer's instructions. Raw RNA-sequencing data were aligned to the reference genome using Hisat2 version 2.1.0 [16], absolute counts generated using HTSeq [17], whereafter counts were normalized and differential gene expression assessed using the DESeq2 package [18]. RNA-seq data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7604.
2.6. Serum proteins
Serum TREM1 (soluble TREM1, sTREM1, CD 354) was measured using the Human sTREM-1 ELISA kit (HK348, Hycult Biotech, Uden, the Netherlands). Serum TNF was measured using the MesoScale Discovery electrochemiluminescence technology (MSD, Rockville, USA).
2.7. Statistical analysis
All analyses were carried out using IBM SPSS Statistics 24 (IBM SPSS, Costa Mesa, CA, USA) and R version 3.5.0 (R Development Core Team, Vienna, Austria). Continuous variables are expressed as median and interquartile range (IQR). Unpaired data were compared using the Mann-Whitney U test for continuous variables, and with Fisher's exact test for categorical variables. Correlations were assessed using the Spearman r correlation coefficient. Stepwise forward and backward elimination logistic regression modelling was performed to identify independent predictors of the outcome of anti-TNF therapy. Final model selection was based on the most optimal second-order Akaike information criterion. Diagnostic performance was assessed with receiver operating characteristics (ROC) curve analysis. A relevant threshold value was chosen on the ROC curve, based on the performance of the Youden's J statistic and closest top-left method. A two-tailed p-value <.05 was considered significant.
3. Results
3.1. Patient characteristics
Fifty-four actively inflamed patients (24 CD, 30 UC) with a median (IQR) disease duration of 6.8 (1.7–19.6) years were included in this prospective study, prior to their first IFX or ADM administration (Table 1). At time of induction, 16 patients (29.6%) were on corticosteroids and 18 patients (33.3%) received immunomodulatory agents (IFX treated patients only). CD patients were endoscopically evaluated after 27.1 (25.0–29.0) weeks, with an overall endoscopic remission rate of 54.2% (30.0% ADM, 71.4% IFX). In UC patients, an endoscopic remission rate of 33.3% (27.8% ADM, 41.7% IFX) was observed after a median of 8.4 (8.0–10.0) weeks. Similar response rates were observed in patients who were naïve to anti-TNF therapy and in patients who previously failed another anti-TNF agent (43.9% vs. 38.5%, p = .73).
Additionally, whole blood was collected in 51 actively inflamed patients (25 CD, 26 UC) initiating vedolizumab therapy, of whom 9 (17.6%) were entirely anti-TNF naïve. After 6 months (CD) and 14 weeks (UC), vedolizumab induced endoscopic remission in 48.0%, 61.5% of patients respectively. Finally, 22 active CD patients initiated ustekinumab with an endoscopic remission rate of 22.3% after 6 months (Supplementary Table 1).
3.2. Whole blood, comparative analysis between responders and non-responders
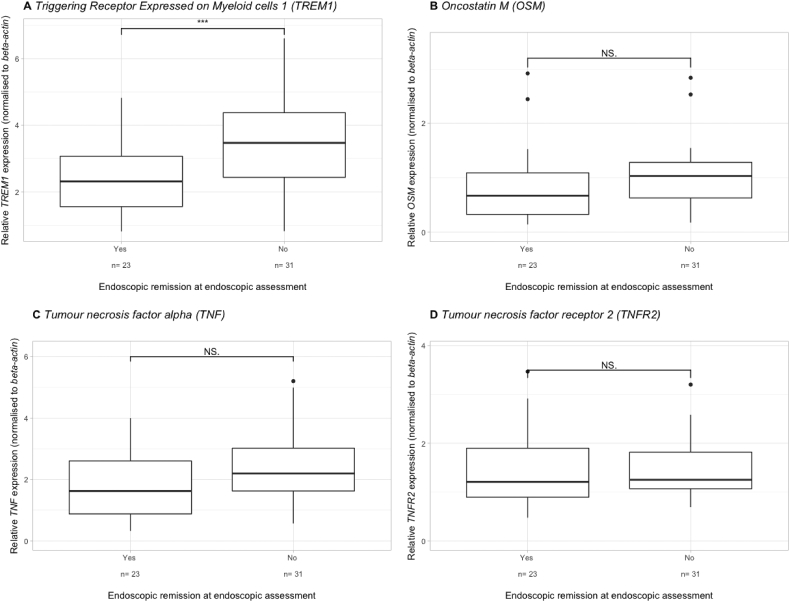
In the anti-TNF cohort, TREM1 was significantly downregulated at baseline in patients achieving endoscopic remission (fold change (FC) = 0.67, p < .001) (Fig. 1A). In contrast, OSM, TNF and TNFR2 expression was not significantly different between future responders and non- responders (FC = 0.61, p = .09; FC = 0.74, p = .13; FC = 0.94, p = .24 respectively) (Fig. 1B-D). Baseline sTREM1 was only numerically lower in future responders (0.28 ng/mL, IQR 0.16–0.56 ng/mL) compared to non-responders (0.40 ng/mL, IQR 0.30–0.83 ng/mL) (FC = 0.70, p = .09). Whole blood IL13RA2 mRNA could not be detected in both the discovery and validation cohort using 2 different pairs of primers (Supplementary Table 2) and using different dilutions of cDNA.
Fig. 1.
Baseline whole blood TREM1 (A), OSM (B), TNF (C) and TNFR2 (D) expression in relation to endoscopic remission later on in both Crohn's disease and ulcerative colitis patients, treated with either adalimumab or infliximab *** p < .001, NS = not significant.
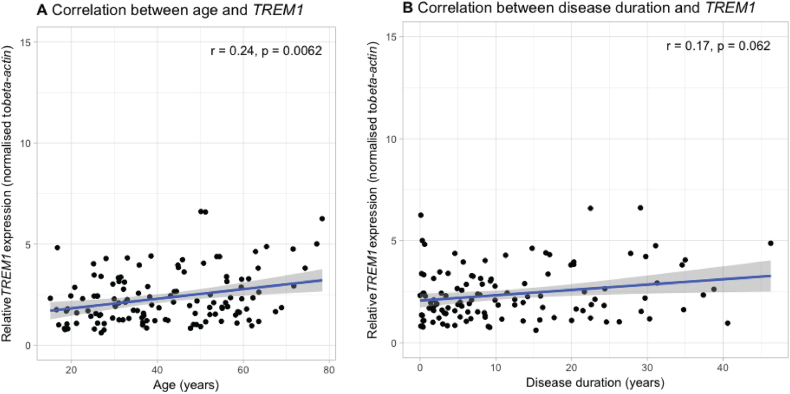
Whole blood TREM1 expression did not significantly correlate with CRP (spearman ρ = −0.08, p = .38), faecal calprotectin (spearman ρ = −0.06, p = .64) or serum TNFα (spearman ρ = − 0.15, p = .63). However, it did significantly increase with increasing age (spearman ρ = 0.24, p = .007) and tended to increase with longer disease duration (spearman ρ = 0.17, p = .06) (Supplementary Fig. 1A-B). No correlation with the total number of previous biological agents previously exposed to could be observed (spearman ρ = 0.08, p = .55).
Supplementary Fig. 1.
Correlation between baseline TREM1 whole blood expression and age (A) or disease duration (B). r = Spearman correlation coefficient.
TREM1 expression did not differ between anti-TNF exposed and anti-TNF naïve patients (FC = 1.24, p = .46). Additionally, both in anti-TNF exposed and in anti-TNF naïve patients, future responders had lower TREM1 levels (FC = 0.48, p = .01; FC = 0.69, p = .01 respectively).
The expression levels of all individual transcripts significantly correlated with each other and with the overall TREM1 expression level (Table 2), suggesting that they all contribute to the overall anti-TNF predictive signature (Fig. 2A-C). Furthermore, total TREM1 mRNA levels in whole blood correlated significantly with sTREM1 protein levels (spearman ρ = 0.36, p = .01), which did not hold true for the individual transcript levels (p = .43, p = .13, p = .15 respectively).
Table 2.
Correlation between the overall TREM1 expression level and the expression of the different transcripts in whole blood.
| Overall TREM1 signal | TREM1-mb | TREM1-sv | |
|---|---|---|---|
| Overall TREM1 signal | |||
| TREM1-mb | ρ = 0.55 (p = 1.6 × 10−5) |
||
| TREM1-sv | ρ = 0.52 (p = 1.0 × 10−4) |
ρ = 0.73 (p = 3.5 × 10−9) |
|
| TREM1-x2 | ρ = 0.72 (p = 2.3 × 10−9) |
ρ = 0.78 (p = 9.9 × 10−12) |
ρ = 0.78 (p = 4.8 × 10−11) |
ρ = Spearman r correlation coefficient.
Fig. 2.
Baseline expression of the different whole blood TREM1 transcripts, including TREM1 mb (A), TREM1 x2 (B) and TREM1 sv (C), in relation to endoscopic remission later on in both CD and UC patients, treated with either adalimumab or infliximab. * p < .05.
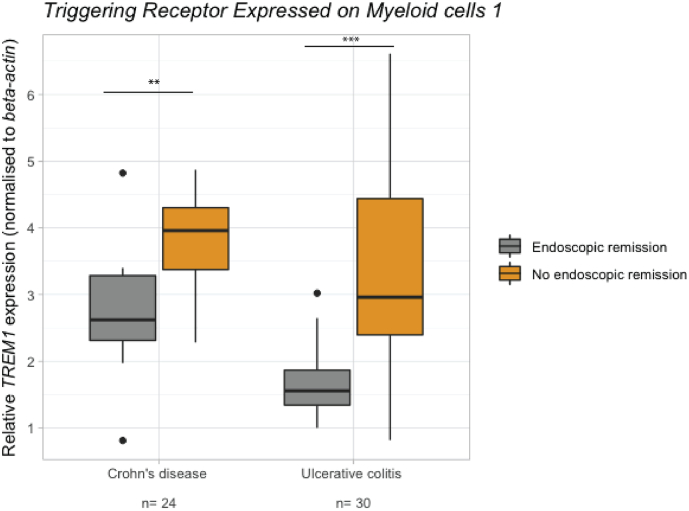
3.3. Similar findings in both Crohn's disease and ulcerative colitis
No significant differences in whole blood TREM1 expression between CD and UC patients could be observed (p = .19). However, the association with endoscopic remission seemed stronger in UC patients than in CD patients (FC = 0.53, p = .001; FC = 0.66, p = .007 respectively) (Fig. 3). Similarly, CD and UC patients did not significantly differ in OSM, TNF or TNFR2 expression (FC = 0.62, p = .06; FC = 1.2, p = .58; FC = 1.25, p = .25 respectively). Although future CD responders did not differ from non-responders in OSM, TNF and TNFR2 expression (FC = 0.97, p = .36; FC = 0.79, p = .41; FC = 1.18, p = .83 respectively), UC responders did for TNFR2 (FC = 0.66, p = .03) but not for OSM or TNF FC = 0.55, p = .34; FC = 0.65, p = .08 respectively).
Fig. 3.
Baseline whole blood TREM1 expression in relation to endoscopic remission later on in both discovery and validation cohort, visualised by diagnosis (B). ** p < .01, *** p < .001.
3.4. Intestinal tissue, comparative analysis between responders and non-responders
To validate previous findings and study the relationship between mucosal and whole blood gene expression levels, we performed RNA-sequencing on 44 inflamed mucosal biopsies of IBD patients prior to the first anti-TNF administration (Supplementary Table 3), including 20 patients of the whole blood cohort. TREM1 was significantly decreased in future non-responders (FC = 0.27, p = .002), as well as OSM (FC = 0.27, p = .007), TNF (FC = 0.57, p = .02), IL13RA2 (FC = 0.20, p = .01) and TNFR2 (FC = 0.72, p = .008) (Fig. 4). Whole blood TREM1 levels correlated significantly with mucosal TREM1 levels (Spearman ρ = 0.79, p = .01, n = 20).
Fig. 4.
Baseline mucosal TREM1 (A), OSM (B), TNF (C), IL13RA2 (D) and TNFR2 (E) expression in relation to endoscopic remission later on in both Crohn's disease and ulcerative colitis patients, treated with either adalimumab or infliximab. * p < .05, ** p < .01.
3.5. Prediction of response to anti-TNF therapy
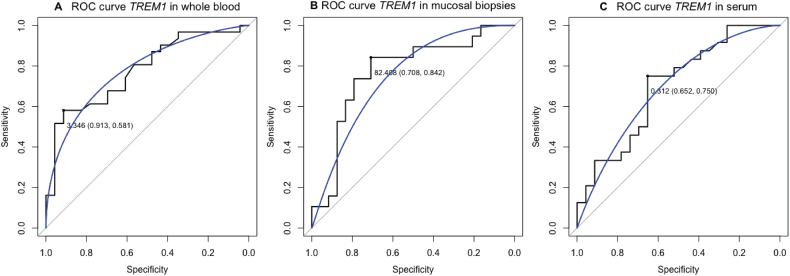
Logistic regression analysis identified total whole blood TREM1 mRNA expression as the only significant predictor of anti-TNF induced endoscopic remission (p = .02). ROC analysis based on baseline TREM1 mRNA levels in the anti-TNF cohort, gave an area under the curve (AUC) of 77.7% (95% CI 65.2–90.1%, p = .001). Similar, mucosal TREM1 mRNA levels seemed to have good predictive accuracy with an AUC of 76.8% (95% CI 61.6–91.9%, p = .003). In contrast, sTREM1 could not accurately predict anti-TNF induced endoscopic remission (AUC 58.3%, p = .31) (Supplementary Fig. 2).
Supplementary Fig. 2.
Receiver operating characteristic (ROC) statistics predicting endoscopic remission based on whole blood TREM1 expression (A), mucosal TREM1 expression (B) or serum TREM1 (C). Every highlighted point on the ROC curve represents the optimal cut-off with associated specificity and sensitivity.
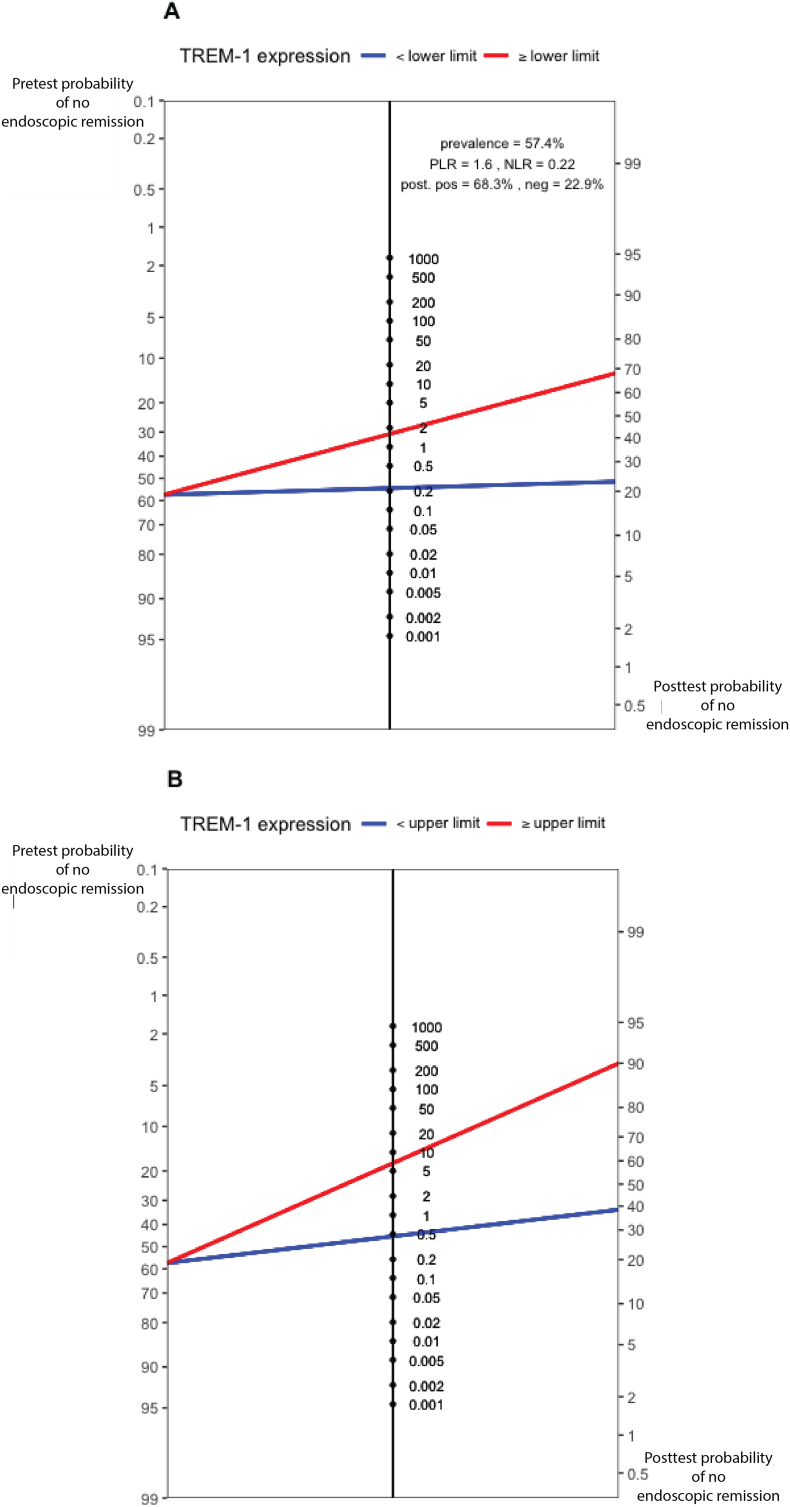
In the anti-TNF cohort with an overall pre-test probability for response and non-response of 42.6%, 57.4% respectively, predictive cut-offs were determined. Based on either 90.0% sensitivity or 90.0% specificity (the latter also representing the Youden statistic), a post-test probability of 77.1% for achieving endoscopic remission (for values below the lower limit) (Fig. 5A) and a post-test probability of 90.0% for non-response (for values above the upper limit) (Fig. 5B) could be achieved. Only 1 out of 5 patients (20.4%) had an intermediate TREM1 level in between both thresholds.
Fig. 5.
Fagan nomogram demonstrating the post-test probability of non-response in anti-TNF exposed patients, based on a lower (A) and upper (B) defined threshold of baseline TREM1 expression with a sensitivity and specificity of 90.0% respectively. Pre-test probability representing the non-response rate in the included cohort.
3.6. An anti-TNF specific marker in IBD therapy
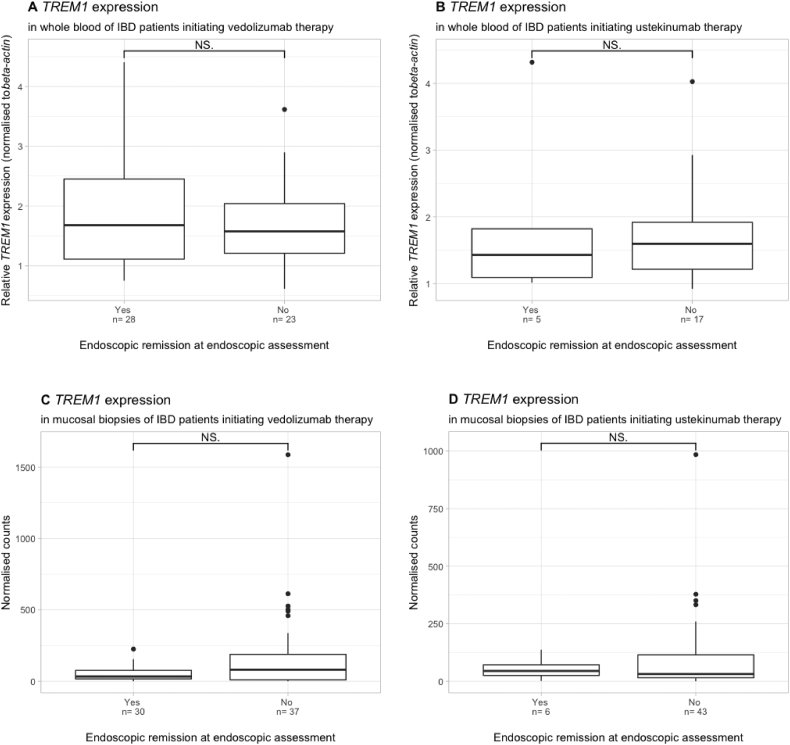
Baseline whole blood TREM1 expression was not associated with endoscopic remission in patients treated with either vedolizumab (n = 51, p = .53) or ustekinumab (n = 22, p = .82) (Fig. 6) (Supplementary Table 3). Similarly, no association between baseline mucosal TREM1 expression and endoscopic remission could be observed in vedolizumab (n = 67, p = .24) or ustekinumab (n = 51, p = 1.0) treated patients (Fig. 6). Finally, no difference in TREM1 could be observed in patients with (82.4%) or without (17.6%) prior anti-TNF exposure (p = .78) at the mucosal level.
Fig. 6.
Baseline whole blood TREM1 expression in relation to endoscopic remission in both Crohn's disease and ulcerative colitis patients, treated with either vedolizumab (A) or ustekinumab (B). Baseline mucosal TREM1 expression in relation to endoscopic remission later on in both Crohn's disease and ulcerative colitis patients, treated with either vedolizumab (C) or ustekinumab (D). NS = not significant.
4. Discussion
This is the first prospective study examining the predictive value of whole blood mRNA transcripts from genes previously identified in tissue as key in the prediction of anti-TNF therapy in patients with IBD. We validated whole blood TREM1 expression as an accurate anti-TNF specific predictor for endoscopic remission in patients with CD and UC. In contrast to the observed moderate endoscopic remission rates with anti-TNF agents in current clinical practice, remission rates may be improved by prioritizing anti-TNF therapy to those patients with low TREM1 expression. Based on our results, pre-test probabilities for primary (non-)response to anti-TNF therapy could be optimized using TREM1 expression, resulting in post-test probabilities of 77.1% for endoscopic remission in the patients with low TREM-1 expression (34.5% increase compared to pre-test probability) and 90.0% for non-response in the patients with high TREM-1 expression (32.6% increase compared to pre-test probability) respectively.
TREM1 is a receptor expressed on innate immune cells, known to amplify inflammatory signals that are initially triggered by Toll-like receptors and thus contributing to the pathophysiology of many acute and chronic inflammatory conditions [19]. Elevated serum levels of TREM1 have been documented in IBD patients, but sTREM1 does not correlate with the degree of endoscopic disease activity [20]. Similarly, TREM1 mRNA and sTREM1 protein levels did not correlate with CRP or faecal calprotectin in our cohort, suggesting that the TREM1 signal we observed is not purely reflecting a higher inflammatory state.
Increased TREM1 levels have been linked earlier to anti-TNF induced clinical response in a retrospective Israelian cohort of 28 patients with CD [12]. We here observed the opposite signal, namely a significant increase in TREM1 in whole blood, both at the protein as at the mRNA level, in non-responders. Because our response criteria were based on more stringent endoscopic criteria and in view of the poor association between clinical symptoms and endoscopic disease activity [21,22], these opposite results may not come as a surprise. As different transcripts could also contribute to the discrepancies between both studies, we not only focussed on the overall TREM1 signal but also measured all known protein coding transcripts individually. Interestingly, all transcripts were significantly upregulated in future non-responders, and thus confirmed our belief of a true biological and clinically relevant signal. The limited number of patients in the original cohort by Gaujoux et al. and the different ethnicity could also contribute to this conflicting observation. However, the current validation of our previous findings (with increased TREM1 expression in future non-responders) [13] in the current extended cohort, together with the absence of the same signal in an UST and VDZ treated cohort, raises the potential clinical applicability of measuring whole blood TREM1 as an anti-TNF specific biomarker in IBD.
Tissue biomarkers may be perceived as a better reflection of what is really going on in patients from a pathophysiological point of view. But, when it comes down to the translation to daily practice, a simple blood sample is less invasive than colonoscopy and easier to implement on a broader scale. In this study we showed that the accuracy of mucosal TREM1 expression is similar to the accuracy of whole blood TREM1 levels. In homeostatic conditions, the vast majority of resident intestinal macrophages completely lack TREM1 expression. In contrast, in patients with active IBD, TREM1 expression is mainly upregulated on intestinal macrophages with only limited TREM1-expressing intestinal neutrophils [23]. Immunophenotyping revealed a higher number of recruited TREM1+ CD14+ HLA-DRint macrophages, and not resident CD14+ HLA-DRhi lamina propria macrophages (LP), among CD45+ LP cells in the inflamed mucosa of patients with IBD (compared to uninflamed regions) [24], explaining why the TREM1 mucosal signal could be picked up in whole blood as a (surrogate) biomarker.
In mice, inhibition of TREM1 attenuated the severity of colitis clinically, endoscopically and histologically by restoring impaired autophagy and endoplasmic reticulum stress [25]. As anti-TNF induced macrophages (Mϕind), have increased levels of autophagy and an intact autophagy pathway seems crucial for an optimal response to anti-TNF therapy [26], we hypothesize that the lower TREM1 levels observed in future anti-TNF induced responders are indeed associated with a better functioning autophagy pathway and thus a higher chance to achieve endoscopic remission after anti-TNF exposure.
The lower serum TREM1 levels in responders to anti-TNF are not reflecting a higher membrane TREM1 expression. In contrast, future responders have significantly lower membrane bound TREM1, suggesting that their downstream proinflammatory TNF burden is lower, as has been reported earlier [9]. Although there exist a specific splicing variant coding sTREM1 (TREM1-sv) [27,28], sTREM1 can also originate after cleavage of the membrane bound protein, as membrane TREM1 contains a matrix metalloproteinase 9 (MMP-9) cleavage site [29,30]. As MMP-9 is known to be significantly upregulated in serum of active IBD patients [31], an increased cleavage of TREM1-mb is plausible, resulting in higher sTREM1 level than expected based on the TREM1-sv transcript alone and explaining why only the overall TREM1 expression level, but not the individual transcript levels, are correlating with sTREM1.
Although this is the largest prospective study investigating potential whole blood biomarkers for anti-TNF therapy in IBD, we do realize the need for validation in bigger, independent cohorts, also allowing clear cut-offs prior to translation into daily clinical practice. Finally, this study focused only on genes previously suggested as potential mucosal biomarkers for anti-TNF responsiveness. Using an unbiased, genome-wide approach through RNA-sequencing of whole blood may therefore be even better to detect novel, outstanding predictive biomarkers in blood.
In conclusion, we validated baseline whole blood and mucosal TREM1 expression in IBD patients as an anti-TNF specific predictive biomarker for endoscopic remission. Larger, randomized studies will need to validate these findings and define a whole blood TREM1 threshold bringing personalised medicine in IBD therapy one step closer.
The following are the supplementary data related to this article.
Supplementary material
Acknowledgments
Acknowledgements
The authors would like to thank Eline Vandeput, Sophie Organe, Nooshin Ardeshir Davani and Tamara Coopmans for an excellent job in maintaining the Biobank database; and Vanessa Brys, Jens Van Bouwel, Wim Meert, Alvaro Cortes Calabuig, Céline Helsmoortel and Wouter Bossuyt (Genomics Core Facility, University Hospitals Leuven, Belgium) for the technical assistance with the RNA-sequencing library preparation and data generation.
This work is patented by KU Leuven Research & Development, GB1819065.2.
Funding
B Verstockt is a doctoral fellow and G Van Assche, S Vermeire and M Ferrante are Senior Clinical Investigators of the Research Foundation Flanders (FWO), Belgium. B Verstockt has also received research grants by the Belgium Week of Gastroenterology, the Belgian IBD Research and Development (BIRD) and the IBD Patient's Association Flanders (CCV VZW) and support from the Vlaamse Vereniging voor Gastro-enterologie (VVGE). C Breynaert is supported by the Clinical Research Fund KOF (University Hospitals Leuven, Leuven, Belgium).
Part of this research has been funded by the European Crohn's and Colitis Organization Research Grant 2017, awarded to B Verstockt. This work was also partially supported by an Advanced European Research Council (ERC) Grant (ERC-2015-AdG), awarded to S Vermeire. The funders did not have any role in the study design, data collection, data analysis, interpretation or writing of the report.
Declaration of interest
B Verstockt received financial support for research from Pfizer; lecture fees from Abbvie, Ferring Pharmaceuticals, Janssen, R-biopharm and Takeda; consultancy fees from Janssen.
G Van Assche received financial support for research from Abbott and Ferring Pharmaceuticals; lecture fees from Janssen, MSD and Abbott; consultancy fees from PDL BioPharma, UCB Pharma, Sanofi-Aventis, Abbott, Abbvie, Ferring, Novartis, Biogen Idec, Janssen Biologics, NovoNordisk, Zealand Pharma A/S, Millenium/Takeda, Shire, Novartis and Bristol Mayer Squibb.
S Vermeire received financial support for research from MSD, Abbvie, Janssen and UCB Pharma; lecture fees from Abbott, Abbvie, Merck Sharpe & Dohme, Ferring Pharmaceuticals and UCB Pharma; consultancy fees from Pfizer, Ferring Pharmaceuticals, Shire Pharmaceuticals Group, Merck Sharpe & Dohme, and AstraZeneca Pharmaceuticals.
M Ferrante received financial support for research from Takeda and Janssen; lecture fees from Ferring, Boehringer- Ingelheim, Chiesi, Merck Sharpe & Dohme, Tillotts, Janssen Biologics, Abbvie, Takeda, Mitsubishi Tanabe, Zeria; consultancy fees from Abbvie, Boehringer-Ingelheim, Ferring, Merck Sharpe & Dohme, and Janssen Biologics.
SaV, JD, VB, WJW, HB and CB declare no conflicts of interest.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclose.
Ethical approval
All patients included in the analysis had given written consent to participate in the Institutional Review Board approved IBD Biobank (B322201213950/S53684), collecting serum and clinical characteristics among other items.
Author contributions
BV: study design, data acquisition and interpretation, statistical analysis and drafting of the manuscript. SaV: technical assistance qPCR and critical revision of the manuscript. JD: technical assistance primer design qPCR and critical revision of the manuscript. VB: sample recruitment and critical revision of the manuscript. WJW: technical assistance qPCR. HB: technical assistance ELISA. CB: critical revision of the manuscript. GVA: patient recruitment and critical revision of the manuscript. SV and MF: study design, data interpretation, supervision and critical revision of the manuscript. All authors agreed with the final version of the manuscript prior to submission.
References
- 1.Mandel M.D., Miheller P., Mullner K., Golovics P.A., Lakatos P.L. Have biologics changed the natural history of Crohn's disease? Dig Dis. 2014;32(4):351–359. doi: 10.1159/000358135. [DOI] [PubMed] [Google Scholar]
- 2.Verstockt B., Ferrante M., Vermeire S., Van Assche G. New treatment options for inflammatory bowel diseases. J Gastroenterol. 2018;53(5):585–590. doi: 10.1007/s00535-018-1449-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flamant M., Roblin X. Inflammatory bowel disease: towards a personalized medicine. Ther Adv Gastroenterol. 2018;11 doi: 10.1177/1756283X17745029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argollo M., Fiorino G., Hindryckx P., Peyrin-Biroulet L., Danese S. Novel therapeutic targets for inflammatory bowel disease. J Autoimmun. 2017;85:103–116. doi: 10.1016/j.jaut.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Levin A.D., Wildenberg M.E., van den Brink G.R. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. 2016;10(8):989–997. doi: 10.1093/ecco-jcc/jjw053. [DOI] [PubMed] [Google Scholar]
- 6.Arijs I., Li K., Toedter G. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58(12):1612–1619. doi: 10.1136/gut.2009.178665. [DOI] [PubMed] [Google Scholar]
- 7.Arijs I., Quintens R., Van Lommel L. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn's disease. Inflamm Bowel Dis. 2010;16(12):2090–2098. doi: 10.1002/ibd.21301. [DOI] [PubMed] [Google Scholar]
- 8.Toedter G., Li K., Marano C. Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol. 2011;106(7):1272–1280. doi: 10.1038/ajg.2011.83. [DOI] [PubMed] [Google Scholar]
- 9.Verstockt B., Verstockt S., Creyns B. Mucosal IL13RA2 expression predicts non-response to anti-TNF therapy in Crohn's disease. Aliment Pharmacol Ther. 2018 doi: 10.1111/apt.15126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt H., Billmeier U., Dieterich W. Expansion of IL-23 receptor bearing TNFR2+ T cells is associated with molecular resistance to anti-TNF therapy in Crohn's disease. Gut. 2018 May 30 doi: 10.1136/gutjnl-2017-315671. gutjnl-2017-315671, [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West N.R., Hegazy A.N., Owens B.M.J. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaujoux R., Starosvetsky E., Maimon N. Cell-centred meta-analysis reveals baseline predictors of anti-TNFalpha non-response in biopsy and blood of patients with IBD. Gut. 2018 Apr 4 doi: 10.1136/gutjnl-2017-315494. gutjnl-2017-315494, [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verstockt B., Verstockt S., Blevi H. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn's disease patients? Gut. 2018 doi: 10.1136/gutjnl-2018-316845. gutjnl-2018-316845, [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Sturm A., Maaser C., Calabrese E. ECCO-ESGAR guideline for diagnostic assessment in inflammatory bowel disease. J Crohns Colitis. 2018 Aug 23 [Epub ahead of print] [Google Scholar]
- 15.Maaser C., Sturm A., Vavricka S.R. ECCO-ESGAR guideline for diagnostic assessment in inflammatory bowel disease. J Crohns Colitis. 2018 Aug 23 [Epub ahead of print] [Google Scholar]
- 16.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrasco K., Boufenzer A., Jolly L. TREM-1 multimerization is essential for its activation on monocytes and neutrophils. Cell Mol Immunol. 2018 Mar 22 doi: 10.1038/s41423-018-0003-5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saurer L., Rihs S., Birrer M. Elevated levels of serum-soluble triggering receptor expressed on myeloid cells-1 in patients with IBD do not correlate with intestinal TREM-1 mRNA expression and endoscopic disease activity. J Crohns Colitis. 2012;6(9):913–923. doi: 10.1016/j.crohns.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Peyrin-Biroulet L., Reinisch W., Colombel J.F. Clinical disease activity, C-reactive protein normalisation and mucosal healing in Crohn's disease in the SONIC trial. Gut. 2014;63(1):88–95. doi: 10.1136/gutjnl-2013-304984. [DOI] [PubMed] [Google Scholar]
- 22.Colombel J.F., Keir M.E., Scherl A. Discrepancies between patient-reported outcomes, and endoscopic and histological appearance in UC. Gut. 2017;66(12):2063–2068. doi: 10.1136/gutjnl-2016-312307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schenk M., Bouchon A., Seibold F., Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J Clin Invest. 2007;117(10):3097–3106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brynjolfsson S.F., Magnusson M.K., Kong P.L. An Antibody against triggering Receptor Expressed on Myeloid Cells 1 (TREM-1) Dampens Proinflammatory Cytokine Secretion by Lamina Propria Cells from patients with IBD. Inflamm Bowel Dis. 2016;22(8):1803–1811. doi: 10.1097/MIB.0000000000000822. [DOI] [PubMed] [Google Scholar]
- 25.Kokten T., Gibot S., Lepage P. TREM-1 inhibition restores impaired autophagy activity and reduces colitis in mice. J Crohns Colitis. 2018;12(2):230–244. doi: 10.1093/ecco-jcc/jjx129. [DOI] [PubMed] [Google Scholar]
- 26.Levin A.D., Koelink P.J., Bloemendaal F.M. Autophagy contributes to the induction of anti-TNF induced macrophages. J Crohns Colitis. 2016;10(3):323–329. doi: 10.1093/ecco-jcc/jjv174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gingras M.C., Lapillonne H., Margolin J.F. TREM-1, MDL-1, and DAP12 expression is associated with a mature stage of myeloid development. Mol Immunol. 2002;38(11):817–824. doi: 10.1016/s0161-5890(02)00004-4. [DOI] [PubMed] [Google Scholar]
- 28.Baruah S., Keck K., Vrenios M. Identification of a novel splice variant isoform of TREM-1 in human neutrophil granules. J Immunol. 2015;195(12):5725–5731. doi: 10.4049/jimmunol.1402713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiss G., Lai C., Fife M.E. Reversal of TREM-1 ectodomain shedding and improved bacterial clearance by intranasal metalloproteinase inhibitors. Mucosal Immunol. 2017;10(4):1021–1030. doi: 10.1038/mi.2016.104. [DOI] [PubMed] [Google Scholar]
- 30.Kumar S., Ratnikov B.I., Kazanov M.D., Smith J.W., Cieplak P. CleavPredict: a platform for reasoning about matrix metalloproteinases proteolytic events. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0127877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Bruyn M., Arijs I., De Hertogh G. Serum neutrophil gelatinase B-associated lipocalin and matrix metalloproteinase-9 complex as a surrogate marker for mucosal healing in patients with Crohn's Disease. J Crohns Colitis. 2015;9(12):1079–1087. doi: 10.1093/ecco-jcc/jjv148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material