Abstract
Background
Imaging techniques used to measure hippocampal atrophy are key to understanding the clinical progression of Alzheimer's disease (AD). Various semi-automated hippocampal segmentation techniques are available and require human expert input to learn how to accurately segment new data. Our goal was to compare 1) the performance of our automated hippocampal segmentation technique relative to manual segmentations, and 2) the performance of our automated technique when provided with a training set from two different raters. We also explored the ability of hippocampal volumes obtained using manual and automated hippocampal segmentations to predict conversion from MCI to AD.
Methods
We analyzed 161 1.5 T T1-weighted brain magnetic resonance images (MRI) from the ADCS Donepezil/Vitamin E clinical study. All subjects carried a diagnosis of mild cognitive impairment (MCI). Three different segmentation outputs (one produced by manual tracing and two produced by a semi-automated algorithm trained with training sets developed by two raters) were compared using single measure intraclass correlation statistics (smICC). The radial distance method was used to assess each segmentation technique's ability to detect hippocampal atrophy in 3D. We then compared how well each segmentation method detected baseline hippocampal differences between MCI subjects who remained stable (MCInc) and those who converted to AD (MCIc) during the trial. Our statistical maps were corrected for multiple comparisons using permutation-based statistics with a threshold of p < .01.
Results
Our smICC analyses showed significant agreement between the manual and automated hippocampal segmentations from rater 1 [right smICC = 0.78 (95%CI 0.72–0.84); left smICC = 0.79 (95%CI 0.72–0.85)], the manual segmentations from rater 1 versus the automated segmentations from rater 2 [right smICC = 0.78 (95%CI 0.7–0.84); left smICC = 0.78 (95%CI 0.71–0.84)], and the automated segmentations of rater 1 versus rater 2 [right smICC = 0.97 (95%CI 0.96–0.98); left smICC = 0.97 (95%CI 0.96–0.98)]. All three segmentation methods detected significant CA1 and subicular atrophy in MCIc compared to MCInc at baseline (manual: right pcorrected = 0.0112, left pcorrected = 0.0006; automated rater 1: right pcorrected = 0.0318, left pcorrected = 0.0302; automated rater 2: right pcorrected = 0.0029, left pcorrected = 0.0166).
Conclusions
The hippocampal volumes obtained with a fast semi-automated segmentation method were highly comparable to the ones obtained with the labor-intensive manual segmentation method. The AdaBoost automated hippocampal segmentation technique is highly reliable allowing the efficient analysis of large data sets.
Abbreviations: AD, Alzheimer's disease; MCI, mild cognitive impairment; MCInc, mild cognitive impairment non-converters; MCIc, mild cognitive impairment converters; ADCS, Alzheimer's Disease Cooperative Study; ADAS-Cog, Alzheimer's Disease Assessment Scale, cognitive subscale; CDR, Clinical Dementia Rating; MMSE, Mini-Mental State Examination; NINCDS-ADRDA, National Institute of Neurological and Communicative Diseases and Stroke and the AD and Related Disorders Association
Keywords: Automated segmentation, Manual segmentation, Radial distance mapping, Magnetic resonance imaging, Mild cognitive impairment, Alzheimer's disease
Highlights
-
•
Manual segmentation of subcortical structures is time and labor intensive.
-
•
Reliable automated segmentation techniques can be developed.
-
•
AdaBoost volumes favorably compare to manual volumes.
-
•
Hippocampal volumetry has limited ability to aid clinical prognosis.
1. Introduction
Alzheimer's disease (AD), the most common type of dementia, is a slowly progressing disease affecting a rising number of individuals every year. The structural integrity of AD patients' brains is often compromised decades before they become symptomatic. There are three stages to disease progression: the preclinical/ presymptomatic stage, the prodromal stage of mild cognitive impairment (MCI), and the dementia stage. MCI is the first stage where cognitive decline can be objectively captured by neuropsychology testing using population-derived normative data. While some MCI patients revert to normal cognition, the majority progress to dementia at a rate of 10–15% per year (Petersen, 2007; Petersen et al., 2001). Being able to identify MCI patients who will convert to AD will enable us to effectively halt or slow down the progression of MCI to AD once disease-modifying treatments are available.
Magnetic resonance (MR) imaging is an important tool used by medical professionals in the diagnosis of patients with neurodegenerative disorders. It is also used abundantly in clinical research to study disease progression or to examine correlations between atrophy and other variables such as genetic profiles or performance on neuropsychological tests. Hippocampal atrophy is a widely accepted imaging biomarker for AD (Apostolova et al., 2006a, Apostolova et al., 2006b, Apostolova et al., 2010a, Apostolova et al., 2010b; Chetelat et al., 2008; Csernansky et al., 2005; Frisoni et al., 2008; Jack Jr. et al., 2000). Disease history studies and clinical trials are nowadays enrolling hundreds of patients and rely on serial MR imaging to capture brain atrophy rates. Manual hippocampal segmentation is a slow and highly labor-intensive approach. Consequently, it is critical to develop automated brain imaging techniques that can accurately extract hippocampal structures from large datasets while using minimal human operator input.
Several studies have proposed automated hippocampal segmentation techniques. One such study involved the patch-based method, which uses expert traces as priors to segment anatomical structures. In this method, each voxel is labeled individually and its surrounding patch of voxels is compared to patches in the training set in order to match anatomical regions of brain structures (Coupe et al., 2011). Other studies use deformable shape (Yang and Duncan, 2004; Chupin et al., 2007; Shen et al., 2002), appearance-based (Hu et al., 2011; Duchesne et al., 2002), and atlas-based (Hammers et al., 2007; Collins et al., 1994) models for hippocampal segmentation. More recently, segmentation techniques are being developed that incorporate different aspects from these three models (Morra et al., 2008a; Lotjonen et al., 2010; van der Lijn et al., 2008).
Despite that multiple techniques for automated hippocampal segmentation have been developed and embraced by many for analyzing large data sets, there are ongoing concerns in the research community regarding their accuracy given the fact that brain structures, especially the subcortical regions, display significant anatomic complexity and variation. Several groups are therefore working on approaches to overcome these problems. One group has suggested using template sets that are specific to the age of individuals in the cohort that is being studied (Shen et al., 2012). Others have suggested that a common online dataset of segmented hippocampi or other anatomical structures should be developed and used as a validation tool (Jafari-Khouzani et al., 2011; Heckemann et al., 2011). Some researchers state both random and systematic errors in segmentation can be corrected (Wang et al., 2011). Random errors, such as structural abnormalities, can be corrected by combining segmentation data from multiple attempts. Systematic errors caused by the misinterpretation of the manually segmented images that serve as primers can be addressed by creating an algorithm that detects them using model errors (Wang et al., 2011) or by testing the segmentation performance of various primer sets as we have done here.
The challenges brought about by the different factors that may cause incorrect labeling of subcortical structures have led researchers to come up with multiple features that are sensitive to anatomical variation (Morra et al., 2008a; Wang et al., 2011; Tangaro et al., 2014). To label hippocampal tissues correctly, our automated technique takes into account approximately 13,000 features. Among these features are image intensity, gradients, curvatures, tissue classification maps of gray and white matter as well as CSF, means and standard deviations, Haar filters, and combinations of x, y, and z positions (Morra et al., 2008a). The algorithm's performance has been validated in prior reports, and, when labeling new data previously unseen by the algorithm, it has been found to agree with human raters as well as human raters agree with each other (Morra et al., 2008a). It has also been found to favorably compare to the automated hippocampal segmentation method from the Freesurfer packet (Morra et al., 2010).
In this study, we compare manual and automated hippocampal segmentation methods in order to establish the reliability and reproducibility of our machine learning based classification technique. We trained our automated segmentation tool, called AdaBoost, with training sets traced by two different experts and compared the volumetric and 3D shape outputs to each other and to the gold standard – manual hippocampal segmentation of the same dataset. We hypothesized that the three groups of segmentations would produce comparable results. We also explored the ability of hippocampal volumes obtained using manual and automated hippocampal segmentations to predict conversion from MCI to AD.
2. Methods
2.1. Subjects
The Alzheimer's Disease Cooperative Study (ADCS) MCI Donepezil/Vitamin E trial, conducted between March 1999 and January 2004, enrolled 769 amnestic MCI subjects at 69 different sites in the United States and Canada (Petersen et al., 2005). Qualifying study participants were between the ages of 55 and 90 years, presented with impaired memory performance on the Alzheimer's Disease Assessment Scale, cognitive subscale (ADAS-Cog), and had a Clinical Dementia Rating (CDR) score of 0.5. Additional eligibility criteria were Mini-Mental State Examination (MMSE) score between 24 and 30 and Logical Memory delayed-recall score within 1.5–2.0 standard deviations below education-adjusted norms (Petersen et al., 2005). Participants were also assessed using the ADCS Mild Cognitive Impairment Activities of Daily Living Scale, the Global Deterioration Scale, and a neuropsychological battery which included the New York University paragraph-recall test, the Symbol Digit Modalities Test, the category-fluency test, a number-cancellation test, the Boston Naming Test, the digits-backward test, the clock-drawing test, and the Maze test (Petersen et al., 2005). Demographic details of the entire sample have been previously described (Petersen et al., 2005). The study excluded subjects who met the National Institute of Neurological and Communicative Diseases and Stroke and the AD and Related Disorders Association (NINCDS-ADRDA) criteria for AD (McKhann et al., 1984). The same criteria were used by the ADCS investigators' team in the course of the trial to define conversion to possible or probable AD.
Of the 69 sites, 24 opted in the magnetic resonance images (MRI) sub-study and obtained T1-weighted brain MRI images from consenting clinical trial subjects. Subjects with contraindications to MRI (i.e. pacemakers, claustrophobia, etc.) were excluded. The full MRI sample included 194 subjects. Of these 194 subjects, 161 had both a baseline scan of sufficient quality to allow for accurate and reliable hippocampal tracing and sufficient data to be identified as MCI converters (MCIc) or MCI subjects who remained stable (MCInc). All 161 subjects were included in our manual segmentation analyses. 20 randomly selected subjects were used as the training set for our automated technique (see section Automated segmentation below). These subjects were excluded from the automated segmentation analyses.
Of the 161 subjects two subjects discontinued prior to their first follow-up. The manual segmentation sample size consisted of 62 MCIc and 99 MCInc. The automated segmentation dataset included 52 MCIc and 89 MCInc excluding the 10 MCIc and 10 MCInc subjects who were used in the training set.
2.2. MRI acquisition, processing, and analyses
Of the 24 sites, 14 used General Electric, 9 used Siemens and one used Philips scanners. T1-weighted scans with minimum full-time echo and repetition time, 124 partitions, 25 degree flip and 1.6 mm slice thickness were obtained. Details about the individual protocols have been previously published (Jack Jr. et al., 2008). T2-weighted scans were inspected for abnormalities such as strokes and major white matter hyperintensities at the Mayo Clinic in Rochester, MN. The data was checked for compliance with the imaging protocol and for quality as explained elsewhere (Jack Jr. et al., 2008). Only scans that passed quality control assessment at Mayo Clinic, Rochester were shared with the University of California Los Angeles (UCLA) research team.
Using a 9-parameter linear transformation, each image was separately registered to the ICBM53 standardized brain template (Collins et al., 1994). All scans were intensity normalized (Shattuck et al., 2001).
2.2.1. Manual segmentation
One expert rater [AEW, intrarater reliability = 0.98, interrater reliability relative to the senior author (LGA) = 0.897 tested on a set of 12 hippocampi that were not part of this project] manually traced the hippocampi of the 161 subjects with scans of sufficient quality to allow manual tracing on gapless coronal slices following a detailed hippocampal tracing protocol (Bartzokis et al., 1998). The contours included the hippocampus proper, the subiculum, and the dentate gyrus. AEW remained blinded to the subjects' conversion status and demographic information. Hippocampal volumes were extracted.
2.2.2. Automated segmentation
The automated method that we used in this study is described in detail by Morra et al. (Morra et al., 2008b, Morra et al., 2008a, Morra et al., 2009, Morra et al., 2009a, Morra et al., 2009b, Morra et al., 2009c) 20 subjects (10 MCIc and 10 MCInc) were randomly selected as the training dataset. The manual hippocampal traces of these 20 subjects from rater 1 (AEW) and rater 2 (JS, interrater reliability = 0.96, intrarater reliability relative to LGA = 0.896; tested on a set of 12 hippocampi that were not part of this project] were used to develop the semi-automated hippocampal segmentation approach. The tracers remained blind to demographic and conversion information.
We trained AdaBoost - our automated machine-learning hippocampal segmentation algorithm (Morra et al., 2008a, Morra et al., 2009b) separately with rater 1 and rater 2 hand-traced hippocampal training sets. From each training set, AdaBoost developed a set of classification rules to distinguish hippocampal from non-hippocampal tissue. These classification rules were comprised of mathematical combinations of as many of the ~13,000 different features as needed to achieve the most accurate segmentation. Examples of such features include image intensity, position, curvatures, gradients and tissue classification maps. The performance of the segmentation models was tested in a testing dataset and subjected to careful visual inspection by both raters and the senior author (LGA) prior to segmentation of the full dataset. Hippocampal volumes were extracted.
2.2.3. Radial distance mapping
The hippocampal segmentations produced with each of the three methods (manual rater 1, automated rater 1 and automated rater 2) were transformed into 3D parametric surface mesh models to normalize the spatial frequency of the digitized surface points. The medial core - a curve threading down the center of each hippocampus, was computed and the radial distance from this medial core to every surface point of each hippocampus was determined and mapped onto each point of the hippocampal surface producing a distance map. Radial distance – a measure of hippocampal thickness, was compared between MCIc and MCInc for each segmentation dataset (manual rater 1, automated rater 1 and automated rater 2) (Apostolova et al., 2006a).
2.3. Statistical methods
We compared the demographic and conversion status characteristics between the manual and automated cohorts using Student's t-test for continuous and Chi-Squared test for categorical variables. We used single measure intra-class correlation coefficient (smICC) to compare the segmentation agreement between the three methods. Next, we analyzed in 3D the ability of each method to detect hippocampal atrophy between MCIc and MCInc. Our 3D significance maps were adjusted for multiple comparisons using permutation-based statistics with a threshold of p < .01 (Thompson et al., 2004).
Sensitivity, specificity and area under the curve (AUC) for conversion to dementia using each of the segmentation methods were estimated using logistic regression models. Sensitivity and specificity were derived using a threshold set to equal the conversion rate in the sample.
3. Results
There were no statistical differences in age, gender, education, apolipoprotein E (ApoE ε4) genotype distribution and MMSE between the manual (N = 161) and automated (N = 141) cohorts (Table 1). We found no significant differences in age, gender, education and ApoE ε4 genotype frequency between MCIc vs MCInc in the manual and automated cohorts (Table 2). As expected, MMSE was significantly lower in MCIc vs MCInc, respectively, in both samples (mean ± SD: manual – 26.7 ± 1.9 vs. 28.0 ± 1.7, p < .001 and automated - 26.8 ± 1.8 vs. 28.0 ± 1.7, p < .001).
Table 1.
Demographic comparisons between samples.
| Variable | Manual dataset N = 161 |
Automated dataset N = 141 |
P-values |
|---|---|---|---|
| Age, years, mean (SD) | 72.9 (6.8) | 72.6 (6.9) | p = .73 |
| Gender, M:F | 88:73 | 79:62 | p = .45 |
| Education, years, mean (SD) | 15.1 (3.0) | 15.06 (3.0) | p = .98 |
| ApoE ε4, noncarriers:carriers | 82:79 | 74:67 | p = .44 |
| MCIc:MCInc | 62:99 | 52:89 | p = .43 |
| MMSE, mean (SD) | 27.5 (1.9) | 27.5 (1.8) | p = .96 |
Table 2.
Demographic comparisons between diagnostic groups within each sample.
| Variable | Manual dataset |
Automated dataset |
||||
|---|---|---|---|---|---|---|
| MCInc N = 99 |
MCIc N = 62 |
p-value | MCInc N = 89 |
MCIc N = 52 |
p-value | |
| Age, years, mean (SD) | 72.3 (6.7) | 73.8 (6.8) | 0.17 | 72.3 (6.9) | 73.2 (6.9) | 0.47 |
| Education, years, mean (SD) | 15.2 (2.8) | 14.9 (3.3) | 0.57 | 15.1 (2.7) | 15.1 (3.5) | 0.97 |
| Gender, M:F | 56:43 | 32:30 | 0.63 | 51:38 | 28:24 | 0.73 |
| ApoE ε4, noncarriers:carriers | 47:52 | 35:27 | 0.33 | 44:45 | 30:22 | 0.39 |
| MMSE, mean (SD) | 28.0 (1.7) | 26.7 (1.9) | p < .001 | 28.0 (1.7) | 26.8 (1.8) | p < .001 |
3.1. Manual versus automated segmentation
Single measure intra-class correlation coefficient (smICC) showed significant agreement between the manual versus automated segmentations of rater 1 [right smICC = 0.78 (95% CI 0.72–0.84); left smICC = 0.79 (95% CI 0.72–0.85)]. Comparing manual rater 1 to automated rater 2 segmentations also yielded favorable results [right smICC = 0.78 (95% CI 0.7–0.84); left smICC = 0.78 (95% CI 0.71–0.84)].
3.2. Rater 1 versus Rater 2 automated segmentation
Automated rater 1 versus rater 2 segmentations showed an extremely high level of agreement with each other [right smICC = 0.97 (95% CI 0.96–0.98); left smICC = 0.97 (95% CI 0.96–0.98)]. This indicates that AdaBoost performs in a very stable and reliable fashion independent of the inadvertent rater-introduced noise in segmentations of the training dataset.
3.3. MCIc versus MCInc hippocampal atrophy comparisons
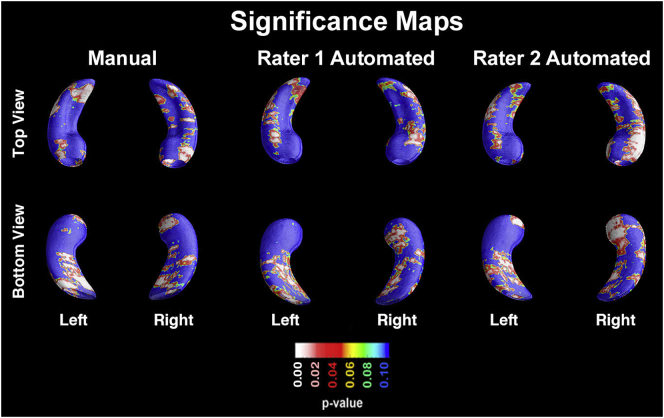
Significance maps showing diagnostic differences between MCIc and MCInc were created using all three segmentation outputs (Fig. 1). Statistical comparison of hippocampal radial distance between MCIc and MCInc demonstrated the expected bilateral atrophy of the CA1 and subiculum regardless of the segmentation method (manual rater 1: right pcorrected = 0.0112, left pcorrected = 0.0006; automated rater 1: right pcorrected = 0.0318, left pcorrected = 0.0302; automated rater 2: right pcorrected = 0.0029, left pcorrected = 0.0166).
Fig. 1.
Significance maps showing the atrophy pattern of MCIc compared to MCInc in the manual (left column), automated rater 1 (middle column) and automated rater 2 (right column) datasets.
3.4. Prediction of conversion from MCI to dementia
Hippocampal volumes by themselves were not able to accurately predict future conversion to dementia regardless of the segmentation method (Table 3). The sensitivity of hippocampal volume to discriminate MCIc from MCInc ranged from 51.9% to 61.5%, specificity from 56.2% to 62.9% and the area under the curve from 0.602 to 0.677.
Table 3.
Prediction of conversion to dementia.
| Method | Side | Sensitivity, % | Specificity, % | AUC |
|---|---|---|---|---|
| Manual | Left hippocampus | 51.9 | 56.2 | 0.602 |
| Right hippocampus | 57.7 | 57.3 | 0.639 | |
| Automated rater 1 | Left hippocampus | 55.8 | 61.8 | 0.630 |
| Right hippocampus | 61.5 | 61.8 | 0.668 | |
| Automated rater 2 | Left hippocampus | 55.8 | 62.9 | 0.619 |
| Right hippocampus | 61.5 | 62.9 | 0.677 |
4. Discussion
Our analyses show that the AdaBoost automated hippocampal segmentation technique accurately contours hippocampi and can be used as a reliable substitute for manual delineation of the hippocampal structure, which currently remains the gold standard for hippocampal segmentation. We were able to demonstrate that the volumes obtained from the manual and the two automated segmentation approaches were in significant agreement with each other. This suggests that AdaBoost performs comparably to manual segmentation, as previously established (Morra et al., 2008a, Morra et al., 2009b). We also demonstrate a highly stable and robust AdaBoost performance when provided with training sets traced by different raters, and performance stability in spite of tracer-introduced segmentation noise. Both the manual and automated segmentations proved sensitive to structural hippocampal differences between MCI individuals who converted to AD, and those who remained stable, as previously demonstrated in this trial population (Apostolova et al., 2010b). We detected atrophy in the CA1 and subiculum in MCIc compared to MCInc, a finding that was previously reported by our group in several cohorts (Apostolova et al., 2006b, Apostolova et al., 2010b, Apostolova et al., 2012; Morra et al., 2009c; Chow et al., 2015). Our predictive analyses however highlighted that hippocampal atrophy measured by hippocampal volume by itself is a poor predictor of future conversion to dementia. This is not an unexpected finding as hippocampal neurodegeneration is common among the elderly with MCI with and without AD pathology (Wisse et al., 2018).
There are both strengths and limitations of the study that should be recognized. Strengths include the selection of a large, well-defined, prospectively collected, and well-characterized longitudinal MCI sample, and the use of two sensitive, state-of-the-art hippocampal analytic techniques (i.e. AdaBoost and radial distance mapping). One weakness of the study relates to uneven sample sizes, with a relatively small number of MCIc compared to MCInc included in the analysis. Uneven sample sizes were the result of observed natural disease history and conversion rates in the ADCS dataset, however a more balanced number of subjects in each outcome group may have produced more significant findings in terms of structural differences in the hippocampal formations. A limitation that is common among clinical trials is that participants are frequently healthier than the general population. Clinical trials have preset strict inclusion/exclusion criteria, which when imposed at enrollment can lead to limited disease heterogeneity in the sample, as may be the case for the current study. While epidemiological MRI cohorts may include a more representative sample of the population, and may therefore be better positioned to answer questions about disease course, there remains tremendous value in studying clinical trial samples such as the one studied here. In order to test the generalization of our findings however, it would be informative to repeat these analyses in other datasets. Furthermore, AdaBoost has been used for the delineation of other subcortical structures such as the basal ganglia (Apostolova et al., 2010c), highlighting its potential for use in the delineation of various subcortical structures in AD and other neurodegenerative diseases.
This work was completed before the Harmonized Protocol for Hippocampal Segmentation (HarP) was developed (Bocchetta et al., 2015). The protocol we used (Bartzokis et al., 1998) is thus not the result of a strategically implemented Delphi procedure considering and carefully weighing in various segmentation options employed by the most common protocols as is the case with the HarP. A more detailed protocol such as the HarP will further reduce the possibility of subjective choice in defining regions which boundaries are not defined unequivocally.
In summary, the AdaBoost automated hippocampal segmentation technique provides reproducible results, which are comparable to manually segmented hippocampi. Techniques such as this could be easily employed to study normal disease progression and to detect therapeutic effects on the neurodegenerative aspects of AD in clinical trials. Creating reliable automated techniques to delineate subcortical structures could significantly increase the momentum at which research breakthroughs are made.
Disclosures
Sona Hurtz has nothing to disclose.
Nicole Chow has nothing to disclose.
Amity E. Watson has nothing to disclose.
Johanne H. Somme has nothing to disclose.
Naira Goukasian has nothing to disclose.
Kristy S. Hwang has nothing to disclose.
John Morra has nothing to disclose.
David Elashoff has nothing to disclose.
Ronald C. Petersen received research funding from Roche, Inc., Merck, Inc., Genentech, Inc., Biogen, Inc., Eli Lilly and Company: consultant.
Paul S. Aisen has nothing to disclose.
Paul M. Thompson has nothing to disclose.
Liana G. Apostolova received research funding from GE Healthcare. Dr. Apostolova served on the speaker's Bureau of Eli Lilly and Piramal, Inc. Dr. Apostolova serves on an Advisory Board Panel for Eli Lilly.
Acknowledgements
The analyses reported in this manuscript were funded by the National Institutes of Health (NIA R01 AG040770, NIA K02 AG048240, NIA P50 AG16570 and NIA P30 AG010133) and the Easton Consortium for Alzheimer's Drug Discovery and Biomarker Development.
References
- Apostolova L.G., Dinov I.D., Dutton R.A. 3D comparison of hippocampal atrophy in amnestic mild cognitive impairment and Alzheimer's disease. Brain. 2006;129:2867–2873. doi: 10.1093/brain/awl274. [DOI] [PubMed] [Google Scholar]
- Apostolova L.G., Dutton R.A., Dinov I.D. Conversion of mild cognitive impairment to Alzheimer disease predicted by hippocampal atrophy maps. Arch. Neurol. 2006;63:693–699. doi: 10.1001/archneur.63.5.693. [DOI] [PubMed] [Google Scholar]
- Apostolova L.G., Mosconi L., Thompson P.M. Subregional hippocampal atrophy predicts Alzheimer's dementia in the cognitively normal. Neurobiol. Aging. 2010;31:1077–1088. doi: 10.1016/j.neurobiolaging.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Thompson P.M., Green A.E. 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum. Brain Mapp. 2010;31:786–797. doi: 10.1002/hbm.20905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Beyer M., Green A.E. Hippocampal, caudate, and ventricular changes in Parkinson's disease with and without dementia. Mov. Disord. 2010;25:687–695. doi: 10.1002/mds.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova L.G., Green A.E., Babakchanian S. Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment (MCI), and Alzheimer Disease. Alzheimer Dis. Assoc. Disord. 2012;26:17–27. doi: 10.1097/WAD.0b013e3182163b62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G., Altshuler L.L., Greider T., Curran J., Keen B., Dixon W.J. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Res. 1998;82:11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- Bocchetta M., Boccardi M., Ganzola R. Harmonized benchmark labels of the hippocampus on magnetic resonance: the EADC-ADNI project. Alzheimers Dement. 2015;11:151–160. doi: 10.1016/j.jalz.2013.12.019. (e155) [DOI] [PubMed] [Google Scholar]
- Chetelat G., Fouquet M., Kalpouzos G. Three-dimensional surface mapping of hippocampal atrophy progression from MCI to AD and over normal aging as assessed using voxel-based morphometry. Neuropsychologia. 2008;46:1721–1731. doi: 10.1016/j.neuropsychologia.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Chow N., Hwang K.S., Hurtz S. Comparing 3T and 1.5T MRI for mapping hippocampal atrophy in the Alzheimer's Disease Neuroimaging Initiative. AJNR Am. J. Neuroradiol. 2015;36:653–660. doi: 10.3174/ajnr.A4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupin M., Mukuna-Bantumbakulu A.R., Hasboun D. Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: Method and validation on controls and patients with Alzheimer's disease. NeuroImage. 2007;34:996–1019. doi: 10.1016/j.neuroimage.2006.10.035. [DOI] [PubMed] [Google Scholar]
- Collins D.L., Neelin P., Peters T.M., Evans A.C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 1994;18:192–205. [PubMed] [Google Scholar]
- Coupe P., Manjon J.V., Fonov V., Pruessner J., Robles M., Collins D.L. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Csernansky J.G., Wang L., Swank J. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. NeuroImage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- van der Lijn F., den Heijer T., Breteler M.M., Niessen W.J. Hippocampus segmentation in MR images using atlas registration, voxel classification, and graph cuts. NeuroImage. 2008;43:708–720. doi: 10.1016/j.neuroimage.2008.07.058. [DOI] [PubMed] [Google Scholar]
- Duchesne S., Pruessner J., Collins D.L. Appearance-based segmentation of medial temporal lobe structures. NeuroImage. 2002;17:515–531. [PubMed] [Google Scholar]
- Frisoni G.B., Ganzola R., Canu E. Mapping local hippocampal changes in Alzheimer's disease and normal ageing with MRI at 3 Tesla. Brain. 2008;131:3266–3276. doi: 10.1093/brain/awn280. [DOI] [PubMed] [Google Scholar]
- Hammers A., Heckemann R., Koepp M.J. Automatic detection and quantification of hippocampal atrophy on MRI in temporal lobe epilepsy: a proof-of-principle study. NeuroImage. 2007;36:38–47. doi: 10.1016/j.neuroimage.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Heckemann R.A., Keihaninejad S., Aljabar P. Automatic morphometry in Alzheimer's disease and mild cognitive impairment. NeuroImage. 2011;56:2024–2037. doi: 10.1016/j.neuroimage.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Coupe P., Pruessner J.C., Collins D.L. Appearance-based modeling for segmentation of hippocampus and amygdala using multi-contrast MR imaging. NeuroImage. 2011;58:549–559. doi: 10.1016/j.neuroimage.2011.06.054. [DOI] [PubMed] [Google Scholar]
- Jack C.R., Jr., Petersen R.C., Xu Y. Rates of hippocampal atrophy correlate with change in clinical status in aging and AD. Neurology. 2000;55:484–489. doi: 10.1212/wnl.55.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Petersen R.C., Grundman M. Longitudinal MRI findings from the vitamin E and donepezil treatment study for MCI. Neurobiol. Aging. 2008;29:1285–1295. doi: 10.1016/j.neurobiolaging.2007.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari-Khouzani K., Elisevich K.V., Patel S., Soltanian-Zadeh H. Dataset of magnetic resonance images of nonepileptic subjects and temporal lobe epilepsy patients for validation of hippocampal segmentation techniques. Neuroinformatics. 2011;9:335–346. doi: 10.1007/s12021-010-9096-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotjonen J.M., Wolz R., Koikkalainen J.R. Fast and robust multi-atlas segmentation of brain magnetic resonance images. NeuroImage. 2010;49:2352–2365. doi: 10.1016/j.neuroimage.2009.10.026. [DOI] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E.M. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G. Validation of a fully automated 3D hippocampal segmentation method using subjects with Alzheimer's disease mild cognitive impairment, and elderly controls. NeuroImage. 2008;43:59–68. doi: 10.1016/j.neuroimage.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G. 2008. Mapping Hippocampal Degeneration in 400 Subjects With a Novel Automated Segmentation Approach. [Google Scholar]
- Morra J.H., Tu Z., Toga A.W., Thompson P.M. Machine learning for brain image segmentation. In: Gonzalez F., Romero E., editors. Biomedical Image Analysis and Machine Learning Technologies. 2009. [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G. Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum. Brain Mapp. 2009;30:2766–2788. doi: 10.1002/hbm.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G. Automated mapping of hippocampal atrophy in 1-year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. NeuroImage. 2009;45:S3–15. doi: 10.1016/j.neuroimage.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Toga A.W., Thompson P.M. Comparison of AdaBoost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans. Med. Imaging. 2009;29:30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra J.H., Tu Z., Apostolova L.G., Green A.E., Toga A.W., Thompson P.M. Comparison of AdaBoost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans. Med. Imaging. 2010;29:30–43. doi: 10.1109/TMI.2009.2021941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R.C. Mild cognitive impairment. Continuum. 2007;13:13–36. doi: 10.1212/CON.0000000000000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen R.C., Doody R., Kurz A. Current concepts in mild cognitive impairment. Arch. Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- Petersen R.C., Thomas R.G., Grundman M. Vitamin E and donepezil for the treatment of mild cognitive impairment. N. Engl. J. Med. 2005;352:2379–2388. doi: 10.1056/NEJMoa050151. [DOI] [PubMed] [Google Scholar]
- Shattuck D.W., Sandor-Leahy S.R., Schaper K.A., Rottenberg D.A., Leahy R.M. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13:856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- Shen D., Moffat S., Resnick S.M., Davatzikos C. Measuring size and shape of the hippocampus in MR images using a deformable shape model. NeuroImage. 2002;15:422–434. doi: 10.1006/nimg.2001.0987. [DOI] [PubMed] [Google Scholar]
- Shen Q., Zhao W., Loewenstein D.A. Comparing new templates and atlas-based segmentations in the volumetric analysis of brain magnetic resonance images for diagnosing Alzheimer's disease. Alzheimer's Dement. 2012;8:399–406. doi: 10.1016/j.jalz.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Tangaro S., Amoroso N., Boccardi M. Automated voxel-by-voxel tissue classification for hippocampal segmentation: methods and validation. Phys. Med. 2014;30:878–887. doi: 10.1016/j.ejmp.2014.06.044. [DOI] [PubMed] [Google Scholar]
- Thompson P.M., Hayashi K.M., De Zubicaray G.I. Mapping hippocampal and ventricular change in Alzheimer disease. NeuroImage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- Wang H., Das S.R., Suh J.W. A learning-based wrapper method to correct systematic errors in automatic image segmentation: consistently improved performance in hippocampus, cortex and brain segmentation. NeuroImage. 2011;55:968–985. doi: 10.1016/j.neuroimage.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisse L.E.M., Das S.R., Davatzikos C. Defining SNAP by cross-sectional and longitudinal definitions of neurodegeneration. NeuroImage Clin. 2018;18:407–412. doi: 10.1016/j.nicl.2018.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Duncan J.S. 3D image segmentation of deformable objects with joint shape-intensity prior models using level sets. Med. Image Anal. 2004;8:285–294. doi: 10.1016/j.media.2004.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]