Abstract
Background
Glioma is the most common and aggressive primary brain tumor with polygenic susceptibility. The cytoplasmic/nuclear shuttling protein, SLC2A4RG (SLC2A4 regulator), has been identified in the 20q13.33 region influencing glioma susceptibility by genome-wide association studies (GWAS) and fine mapping analyses.
Methods
To discover the expression of SLC2A4RG and its relationship with patient prognosis, tissue microarray containing glioma samples and normal brains was constructed followed by immunohistochemical staining. The role of SLC2A4RG on cell proliferation, cell cycle, and apoptosis was evaluated by gain- and loss-of-function assays in vivo, and subcutaneous and intracranial xenografts were performed to assess its functional effects. The mechanism underlying SLC2A4RG was further investigated via luciferase reporter analyses, ChIP, mass spectrometry, Co-IP, immunofluorescence, etc.
Findings
The potential tumor suppressor role of SLC2A4RG was further validated by in vitro and in vivo experiments that SLC2A4RG could attenuate cell proliferation via G2/M phase arrest and induce glioma cell apoptosis by direct transactivation of caspase-3 and caspase-6. Moreover, its function displaying showed to depend on the nuclear transportation of SLC2A4RG, however, bound with 14-3-3θ, it would be sequestered in the cytoplasm followed by reversal effect.
Interpretation
We identify a new pro-oncogenic mechanism whereby 14-3-3θ negatively regulates the nuclear function of the tumor suppressor SLC2A4RG, with significant therapeutic implications for the intervention of human glioma.
Fund
This work was supported by the National Natural Science Foundation of China (81372706, 81572501, and 81372235).
Keywords: Glioma, Prognosis, Apoptosis, SLC2A4RG, 14-3-3θ
Research in context.
Evidence before this study
Glioma is the most common and aggressive primary brain tumor with polygenic susceptibility. The cytoplasmic/nuclear shuttling protein, SLC2A4RG, has been identified as a susceptibility gene in glioma, suggesting it as a potential driver in tumor development and progression.
Added value of this study
Our research revealed a potential tumor suppressor role of SLC2A4RG that it could attenuate cell proliferation via G2/M phase arrest and induce glioma cell apoptosis by direct transactivation of caspase-3 and caspase-6. Furthermore, the interaction with 14-3-3θ will result in sequestration of SLC2A4RG in the cytoplasm and then promote tumor progression.
Implications of all the available evidence
The nuclear SLC2A4RG expression is substantial down-regulated in glioma and associated with poor prognosis of patients. 14-3-3θ negatively regulates the nuclear function of the tumor suppressor SLC2A4RG and maybe a promising therapeutic target of human glioma.
Alt-text: Unlabelled Box
1. Introduction
Glioma represents the most common primary brain tumors occurring in 4.67 to 5.73 per 100,000 persons [1]. Despite aggressive multimodal treatment, gliomas are still associated with a poor prognosis, with the most malignant type, glioblastoma (GBM) having a median overall survival (OS) of 12–15 months [2]. Recently, genomic analyses of glioma have provided new insight into the risk and prognosis. Our understanding of the polygenic susceptibility of glioma has been transformed by genome-wide association studies (GWAS), which have identified single-nucleotide polymorphisms (SNPs) at 13 loci with increased risk, such as 17p13.1 (TP53) for glioma, and 5p15.33 (TERT), 7p11.2 (EGFR), 9p21.3 (CDKN2B-AS1), and 20q13.33 (RTEL1) for GBM [[3], [4], [5]]. Coincidently, our replication study also confirmed 20q13.33 influencing risk [6], and found a SNP rs1058319 located in the 3’-UTR of SLC2A4 regulator (SLC2A4RG) in this region with a 2.06-fold increased risk for glioma (Cox regression P = 6.31E-15, 95%CI = 1.72–2.45) [7]. Further whole exome-wide association study also identified a missense variant rs8957 (E[GAG]233D[GAU]) in SLC2A4RG (Cox regression P = 9.58E-08, HR = 1.43, 95%CI = 1.25–1.63), and its functional contribution to glioma progression has been resolved [8]. As shown in previous results, the susceptibility gene SLC2A4RG probably played a vital role in tumorigenesis and glioma development.
SLC2A4RG was first identified for human transcription factors that bound specifically to Domain I which is located in the region of the SLC2A4 promoter [9], and was demonstrated its activity for modulating SLC2A4 expression [10]. A subsequent study found that SLC2A4RG also interacted with a 7-bp consensus sequence (GCCGGCG), which is an essential cis-regulatory element for Huntington's disease (HD) gene expression in neuronal cells [11]. All these indicate that SLC2A4RG is an important factor required for transcriptional regulation of some genes. Additionally, functional nuclear localization signal (NLS) and nuclear export signal (NES) were found in the structure of SLC2A4RG, and made it be a cytoplasmic/nuclear shuttling protein [11]. Protein localization is a highly dynamic biological process linked to delicate cellular function, but also is correlated with the outcome of cell proliferation and the response to apoptotic stress in cancer [12]. However, how SLC2A4RG functions being regulated in different cellular locations in glioma, as well as the mechanism involved in SLC2A4RG contributing to tumor progression, remains unclear.
Here we confirm that SLC2A4RG expression is down-regulated in glioma and associated with poor prognosis of patients, illustrate that the nucleus-localized SLC2A4RG induces glioma cell apoptosis by direct transactivation of caspase-3 and caspase-6 genes, and further demonstrate that the subcellular localization of SLC2A4RG is regulated by 14-3-3θ in the cytoplasm. These findings highlight the importance of SLC2A4RG in controlling glioma cell fate and its potential to be a therapeutic target for this tumor.
2. Materials and methods
2.1. Cell lines and cell culture
HeLa, Hek293T cells and the human glioma cell lines (U87, U251) were obtained from the American Type Culture Collection (ATCC) and cultured in DMEM with 10% FBS. Transfection was performed as instructed by the manufacturer. All cell lines used in this study were proved to be the absence of mycoplasma contamination.
2.2. Reagents
DMEM and Fetal Bovine Serum (FBS) were obtained from Gibco (Thermo Fisher Scientific, USA). The Annexin V-PE/7-AAD was purchased from BD biosciences (USA). The TRIzol Reagent was acquired from the Invitrogen (Thermo Fisher Scientific, USA). The ReverTra Ace was obtained from the Toyobo (Japan). The dual luciferase reporter assay system was purchased from Promega (USA). The Cell Counting Kit-8 kit was purchased from Dojindo Laboratories (Japan). The Caspase-3/Caspase-6 Activity Assay Kit was obtained from the Cell Signaling Technology (USA). The protease inhibitors cocktail was acquired from the Sigma-Aldrich (USA).
2.3. Patient samples
The first independent cohort of fresh-frozen tissues from 16 LGG and 34 HGG glioma patients and 17 normal controls were obtained from the Department of Neurosurgery in Changzheng Hospital, Second Military Medical University (SMMU) (Shanghai, China) between June 2008 and July 2010. The second independent cohort of tissue microarrays containing 289 gliomas and 16 controls consecutively recruited from the Department of Neurosurgery in Changzheng Hospital, SMMU, between March 1998 and August 2010. The third independent cohort of fresh-frozen tissues from 18 LGG and 24 HGG glioma patients and12 normal controls were obtained from the Department of Neurosurgery in Changzheng Hospital, SMMU, between January 2010 and December 2013. Written informed consent was provided by all patients. The patient sample acquisition was approved by the Specialty Committee on Ethics of Biomedicine Research, Second Military Medical University.
2.4. Acquisition of primary glioma cells
Primary glioma cell G1 was derived from a patient with a glioblastoma, which was confirmed by two independent pathologists. The fresh tumor tissue was immerged in DMEM medium and transferred to the lab within half an hour after the resection of glioblastoma. Then, the mechanical cutting method was used to isolate glioma cells from the tumor tissue, followed by digestion with collagenases (1 mg/ml; Gibico 17,018,029) in 37 °C for 2 h. The 70 mm filter would remove undigested tissues, and the filtrate was collected and centrifuged for 5 min at the speed of 1200 rpm. The obtained cells, also named as G1 cells, would be counted, planted on cell plates, and cultured in DMEM with 10% FBS. Written informed consent was provided by the patient. The primary glioma cells acquisition was approved by the Specialty Committee on Ethics of Biomedicine Research, Second Military Medical University.
2.5. Flow cytometry analysis
Flow cytometry assay was carried out as reported previously [13]. The Cell cycle analysis was performed by propidium iodide staining. The percentage of cellular apoptosis was performed by Annexin V-PE/7-AAD staining.
2.6. RNA extraction and quantitative real-time PCR
The total RNA was extracted from cell lines and clinical specimens using TRIzol Reagent (Invitrogen) following the manufacturer's protocol. First-strand cDNA was synthesized using ReverTra Ace (Toyobo). The quantitative real-time PCR was conducted by ABI PRISM 7900HT Sequence Detection system. All PCR primers used in this study were listed in Supplementary Table S2.
2.7. Luciferase reporter assay
The wild-type and mutated putative SLC2A4RG binding sites within the caspase-3/-6 promoter region (Supplementary Table S2) were cloned into a pGL3-basic luciferase reporter plasmid (Invitrogen). The firefly and sea pansy luciferase activity were determined using the dual luciferase reporter assay system (Promega).
2.8. Immunohistochemistry assay
The immunohistochemistry assay was performed on human specimens, and nude mice xenograft tissue to detect and score SLC2A4RG, caspase-3, caspase-6, and Ki-67 expression by methods described previously [14]. The immunohistochemical staining was scored by two independent pathologists in a blinded manner. In brief, the staining density was graded on a scale of 0–3 (0 for no cells stained, 1 for <25%, 2 for 25–75%, and 3 for >75%), and the staining intensity was also scored 0–3 (0 for no staining, 1 for light-brown staining, 2 for medium-brown staining, and 3 for dark-brown staining). The total immunoreactivity score of staining yielded from the scores for intensity and density. Nuclear SLC2A4RG expression was denoted as low (total score = 0) or high (total score ≥ 1), while Cytoplasmic SLC2A4RG was denoted as low (total score = 0 or 1) or high (total score > 1) to divide the glioma patients into two groups.
2.9. Cell proliferation and colony formation assay
Cells were seeded in 96 well plates (2000 cells/well) in sextuple, and cell proliferation assay was performed by Cell Counting Kit-8 (CCK-8, Dojindo Laboratories) following the manufacturer's instructions. The optical density was measured at 570 nm wavelength using a microplate reader (EL × 800, BIO-TEK, USA). For colony formation assay, 1 × 103 cells were independently seeded onto 60 mm culture plates in triplicate. After about two weeks, cells were fixed with 100% methanol and then stained with crystal violet. The total number of visible colonies in each dish was counted. These experiments were performed at least three times.
2.10. Caspase activity assay
U87 cells were harvested and washed three times with cold PBS. Caspase 3 or caspase-6 activity was measured as described in the protocol of the Caspase-3/Caspase-6 Activity Assay Kit (Cell Signaling Technology).
2.11. Immunofluorescence and confocal microscopy
For immunofluorescence, cells grown on chamber slides were fixed 4% paraformaldehyde at room temperature for 30 min. After washing with PBS, cells were permeabilized with 0.25% Triton X-100 for 10 min. Then the cells were blocked with 1% bovine serum albumin for 20 min before incubation with primary antibodies in PBS at 4 °C for overnight. After washing with PBS, the cells were incubated with fluorescence-labeled secondary antibodies and DAPI for 1 h at room temperature. After washing with PBS, fluorescence-labeled secondary antibodies were applied, and DAPI was counterstained for 1 h at room temperature. The images were captured with a confocal microscope (LSM710, Zeiss).
2.12. Western blot
The tissue and cell samples were homogenized in lysis buffer containing a cocktail of 1% protease and 1% protease inhibitors (Sigma). Protein lysates were separated by 10% SDS-PAGE and transferred onto a PVDF membrane (Invitrogen), which were blocked in 5% milk for 1 h and then treated with the appropriate antibody at 4 °C overnight. The blots were visualized by an enhanced chemiluminescence detection system (Thermo Scientific, USA). ImageJ software (NIH) was used to analyze the Western blot results.
2.13. Protein–protein interaction studies
Co-immunoprecipitation (co-IP) was performed as previously described. Briefly, Whole cell extracts of Flag-SLC2A4RG and Myc-14-3-3θ co-transfected Hek293T cells were prepared in the presence of protease inhibitors cocktail (Sigma-Aldrich). Immunoprecipitation of Flag-SLC2A4RG was conducted by adding 10 μl of anti-Flag M2 affinity sepharose (Sigma) and incubated overnight at 4 °C. Finally, the immune-complex was detected by Western blot. The GST-pulldown assay was conducted as described previously [15].
2.14. Antibodies
The following antibodies were used: SLC2A4RG (AP1422c; Abgent), Histone H3 (9728, Cell Signaling Technology) CDK1 (19532-1-AP; Proteintech), Cyclin E (11554-1-AP; Proteintech), CDKN2A (10883-1-AP; Proteintech), Ki-67 (9449; Cell Signaling Technology), FLAG (M2; Sigma), Myc (9E10; Sigma), HA (MM5-101R; Convance), Actin (AC-74; Sigma), caspase-3 (9662S; Cell Signaling Technology), cleaved-caspase-3 (9664; Cell Signaling Technology), caspase-6 (9762; Cell Signaling Technology), PARP (9542; Cell Signaling Technology), IDH1 R132H (DIA-H09).
2.15. Animal studies
All animal studies were performed using 6- to 7- week-old Balb/c nude mice. For subcutaneous xenograft experiments, U87 cells (1 × 106) infected with SLC2A4RG overexpression or deletion lentiviruses and their corresponding control cells were subcutaneously inoculated into the right flanks nude mice. When tumors became palpable, tumor size was calculated by the following formula: 0.5 × length ×width2. Mice were sacrificed when the length of the largest xenograft reached 3 cm. For intracranial xenograft studies, luciferase-labeled U87 cells (3 × 106) infected with SLC2A4RG overexpression or deletion lentiviruses and their corresponding control cells were inoculated intracranially into the striatum of nude mice via a stereotactic device. Tumor progression was monitored by bioluminescence imaging technique on day 7 and 21 after implantation. Mice were sacrificed when they displayed weight loss, rough coat, and hunching. All animal experiments were conducted with the approval of the Institutional Animal Care and Use Committee of Fudan University. All animal study was performed according to the Ethics Committee guidelines of Fudan University.
2.16. Statistics
All experiments were conducted in triplicate with means and SEM subjected to the Student's t-test or ANOVA in SPSS Statistics 17.0. Analysis of survival was conducted by Kaplan-Meier survival and Cox regression. (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.)
3. Results
3.1. SLC2A4RG is decreased in glioma and serves as a prognostic factor
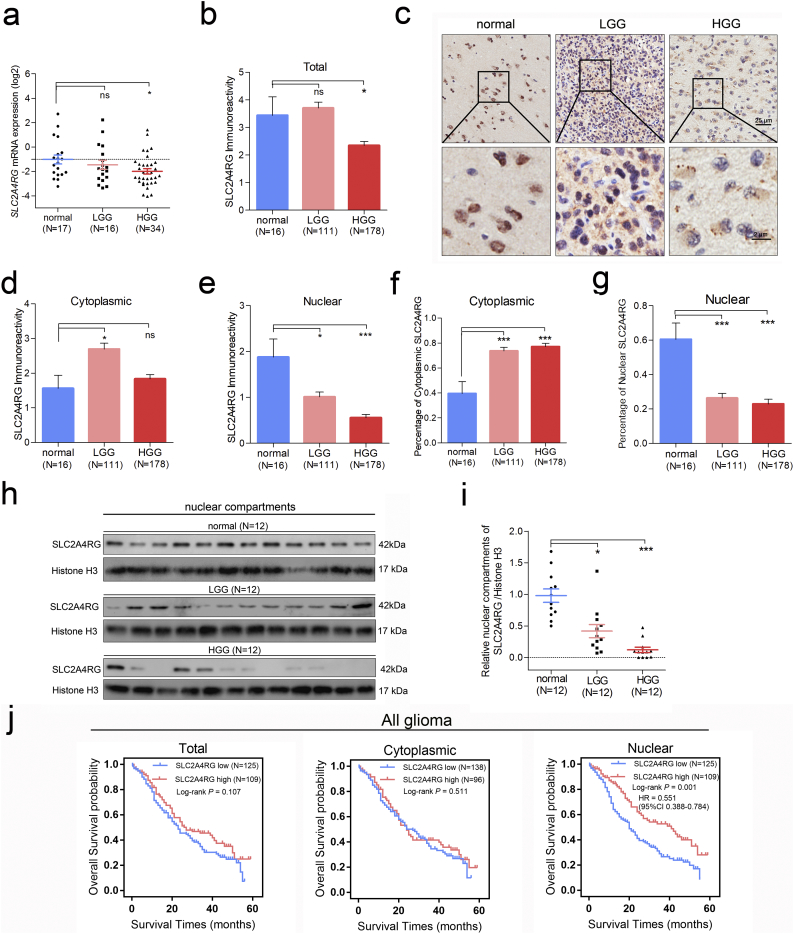
To investigate the expression of SLC2A4RG in glioma individuals, we first measured its mRNA level in a cohort of 16 low grade glioma (LGG) and 34 high grade glioma (HGG) specimens and 17 normal brain tissues via quantitative RT-PCR. Significantly downregulated SLC2A4RG was found in HGG (Student's t-test P = 0.014), and a similar trend could also be seen between normal brain and LGG (Fig. 1a). Furthermore, immunohistochemistry (IHC) was conducted to analyze its protein expression in a tissue microarray containing 289 gliomas and 16 controls. Consistently, the immunoreactivity of SLC2A4RG was also decreased in HGG (P = 0.034), but not in LGG, when compared to normal brains (Fig. 1b-c). Subsequently, the cytoplasmic and the nuclear immunoreactivities were separated, no similar trend of cytoplasmic SLC2A4RG expression were observed among the three groups, but a dramatically downregulated nuclear SLC2A4RG with grade increasing did exist (ANOVA P < 0.0001) (Fig. 1d-e). Similar results were observed under the criteria of IDH1 status and the histology (Supplementary Fig. S1). The percentages of cytoplasmic and nuclear SLC2A4RG were also calculated and showed an upward and a downward trend respectively (Fig. 1f-g). Also, western blot analysis of nuclear fractions from an independent series of 12 normal brain, 12 LGG, and 12 HGG specimens further validated a reduction of nuclear SLC2A4RG in glioma (Fig. 1h-i).
Fig. 1.
The expression and prognostic significance of SLC2A4RG in glioma. (a) SLC2A4RG mRNA expression detected via RT-PCR in normal brains and human glioma specimens with different WHO grades. (b, d, and e) Total, cytoplasmic, and nuclear immunoreactivity scores of SLC2A4RG in the second independent cohort of normal brains, LGGs, and HGG. (c) Representative staining images of SLC2A4RG in normal brains and gliomas. (f and g) The percentages of cytoplasmic SLC2A4RG (f) and nuclear SLC2A4RG (g) in these specimens calculated based on b, d, e. (h) The protein level of nuclear SLC2A4RG analyzed in the third independent cohort containing normal brains, LGG and HGG specimens by western blot. (i) The fold change of SLC2A4RG protein is normalized to Histone 3A. (j) Kaplan-Meier curves were used to analyze the relationships between the total/cytoplasmic/nuclear SLC2A4RG immunoreactivity level and OS in glioma patients from the second cohort. Student's t-test, ns, no significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then explored the relationship between the SLC2A4RG expression and clinical prognosis of glioma patients. Kaplan-Meier analysis was performed and found that patients with the low nuclear, but not the total and cytoplasmic, SLC2A4RG expression had a markedly shorter overall survival (OS) than those with high expression of nuclear SLC2A4RG (Fig. 1j and Supplementary Fig. S2).
3.2. SLC2A4RG regulates glioma proliferation and apoptosis in vitro
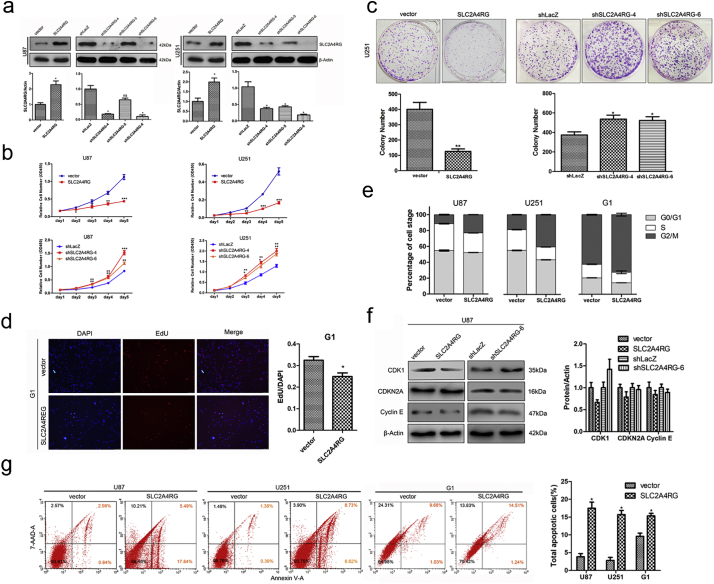
To further address the biological importance of SLC2A4RG in glioma, we stably increased or depleted its expression in U87 and U251 cells with optimized lentivirus vectors (Fig. 2a). As determined by CCK-8 assay, both of SLC2A4RG-overexpressed U87 and U251 cells displayed a slower growth rate than that of controls, while targeting SLC2A4RG by shRNA-4 and shRNA-6 could markedly promote cell proliferation (Fig. 2b). Similar results in U251 cells were observed in colony formation assay (Fig. 2c). Meanwhile, primary tumor cells derived from a piece of fresh glioma tissue was also applied to verify the biological function of SLG2A4RG. As determined by the BrdU assay, cell proliferation was depressed by exogenous SLG2A4RG in primary human glioma G1 cells (Fig. 2d, supplementary Fig. S3). Then we examined whether cell cycle, as well as several important molecules involved in this process, was changed by SLC2A4RG in glioma. A significant elevation in the percentage of G2/M phase was identified in U87,U251, and G1 cells with SLC2A4RG overexpression (U87, 11.31% vs. 22.95%, Student's t-test P = 4.51E-05; U251, 18.80% vs. 40.56%, Student's t-test P = 3.647E-12; G1, 62.33% vs. 72.37%, Student's t-test P = 0.006), which also showed a reduction of CDK1 expression (Fig. 2e-f) and increased number of cells with sub-G1 phase (Supplementary Fig. S4). Conversely, both SLC2A4RG-depleted cells displayed an increased expression of CDK1when compared to the control respectively (Fig. 2f). These results indicated that SLC2A4RG might regulate glioma cell proliferation through cell cycle via CDK1. Moreover, overexpression of SLC2A4RG was also observed to substantially induce a higher proportion of apoptotic U87 (3.85% vs. 21.85%, Student's t-test P = 2.05E-04), U251 (1.80% vs. 15.69%, Student's t-test P = 3.28E-04) and G1 (9.60% VS.15.34%, Student's t-test P = 0.008) cells as judged by Annexin V/PE staining (Fig. 2g).
Fig. 2.
SLC2A4RG controls glioma cell survival in vitro. (a) Western blot was used to examine the expression efficiency of SLC2A4RG in U87 and U251 cells with SLC2A4RG overexpression or knockdown. β-Actin serves as the loading control. (b) The growth curves of U87 and U251 cells with overexpression or depletion of SLC2A4RG described from day 1 to day 5. (c) The Growth of U251 cells with SLC2A4RG overexpression or depletion observed by colony formation assay. (d) The Edu/DAPI rate was calculated in primary human glioma G1 cells transfected with vector or SLC2A4RG. (e) Flow cytometry was detecting the cell cycle changes of U87, U251, and G1 cells with overexpressed or depleted SLC2A4RG. (f) The effects of SLC2A4RG overexpression or depletion on the expression of cell cycle-related proteins examined by Western blot. β-Actin serves as the loading control. (g) Flow cytometry with Annexin V and 7-AAD staining determining the changes of apoptosis in U87 and U251 cells under the condition of SLC2A4RG overexpression. Results analyzed by t-test presented as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.3. SLC2A4RG inhibits glioma growth in vivo
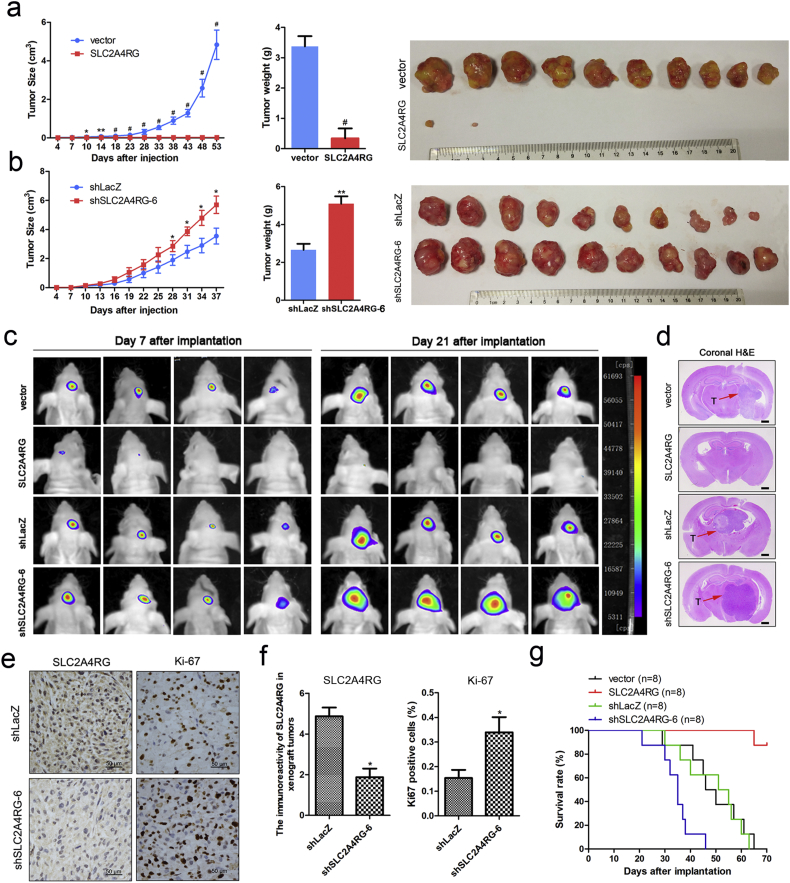
Our data demonstrated that SLC2A4RG negatively regulated glioma cell survival in vitro and suggested that it might play a major role in the modulation of glioma progression. To confirm the suppressor function of SLC2A4RG in glioma, we performed in vivo experiments with subcutaneous and intracranial xeno-transplanted tumor models. As shown in Fig. 3a, it was surprising that there was extremely little growth of tumors in nude mice injected subcutaneously with SLC2A4RG-transduced U87 cells, and the average weight of tumors developed from SLC2A4RG -overexpressing cells (0.342 ± 0.146 g) was also significantly lighter than those from the control cells (3.372 ± 0.339 g, Student's t-test P = 0.003). In contrast, tumors from SLC2A4RG-depleted U87 cells displayed faster growth rates and heavier tumor weight compared with the shLacZ group (shSLC2A4RG 4.780 ± 0.466 g vs. shLacZ 2.803 ± 0.410 g, Student's t-test P = 0.005; Fig. 3b). Subsequently, the role of SLC2A4RG in glioma could be better presented in intracranial xenograft mice, whose tumor progression was monitored by bioluminescence imaging technique on day 7 and 21 after implantation. Xenografts carrying SLC2A4RG-overexpressed U87 cells also showed a noticeable regression of tumor growth in comparison to the control group. On the contrary, mice bearing SLC2A4RG-depleted U87 cells showed a drastic increase in signal compared with the shLacZ group (Fig. 3c). Representative images for paralleled HE-stained tumor cytostructure were given in Fig. 3d, and immunostaining analysis revealed an elevation of Ki-67 expression (2.125 ± 0.427 vs. 5.125 ± 0.515, Student's t-test P = 0.004) in tumors derived from SLC2A4RG-depleted U87 cells (Fig. 3e-f). In addition, survival rates of different groups of intracranial xenograft mice were described by the survival curve that mice injected with SLC2A4RG-overexpressed cells exhibited the longest OS time (Fig. 3g).
Fig. 3.
SLC2A4RG regulates glioma growth in subcutaneous and intracranial xenografts. (a and b) Growth curves, weights, and sizes of subcutaneous tumors derived from overexpressed (a) and depleted (b) SLC2A4RG U87 cells comparing to that of respective control groups. (c) Representative pseudocolor bioluminescence images of intracranial mice bearing SLC2A4RG overexpressed or depleted cells as well as control cells on the days as indicated. (d) Representative H&E staining for a pathological form of the intracranial tumor from each group; T, tumor. (e and f) IHC analysis of SLC2A4RG and Ki-67 expression in intracranial tumors developed from SLC2A4RG silenced or control U87 cells. (g) Kaplan-Meier curve determining the survival rate of mice with intracranial xenografts derived from overexpressed, depleted SLC2A4RG, and control U87 cells. Results analyzed by t-test presented as mean ± SEM. *, P < 0.05; ***, P < 0.001; #, P < 0.0001.
3.4. Ectopic of nuclear SLC2A4RG induces glioma cell apoptosis
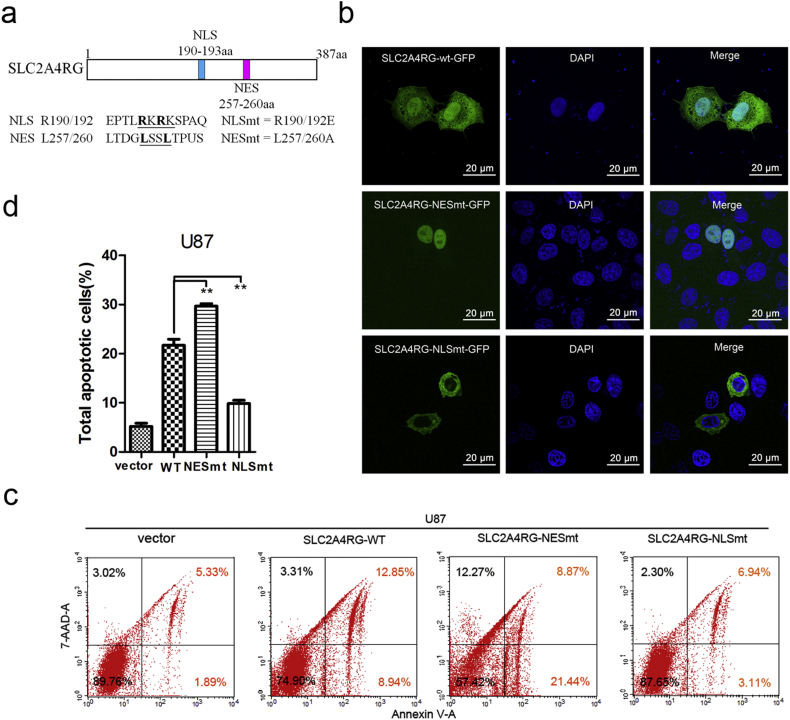
SLC2A4RG protein was reported to be equipped with a NES and a NLS [11]. As mentioned above, the survival analysis defined nuclear SLC2A4RG, but not cytoplasmic SLC2A4RG, serving as a prognostic factor for glioma patients. Within this context, we postulated that the subcellular location of SLC2A4RG might be essential for its function. We began by mutating both Arg-190 and Arg-192 to glutamic acid residues and both Leu-257 and Leu-260 to alanine residues to generate mutant NLS SLC2A4RG (SLC2A4RG-NLSmt = R190/192E) and mutant NES SLC2A4RG (SLC2A4RG-NESmt = L257/260A), respectively (Fig. 4a). C-terminal fusions of SLC2A4RG-wt, SLC2A4RG-NLSmt, and SLC2A4RG-NESmt to GFP were continued to be constructed to investigate the subcellular localization of SLC2A4RG. After HeLa cell transfection, SLC2A4RG-wt-GFP was localized in both the nucleus and cytoplasm and the majority was nuclear localization. Obviously, SLC2A4RG-NLSmt-GFP was accumulated exclusively in the cytoplasm, while SLC2A4RG-NESmt-GFP only localized in the nucleus (Fig. 4b). Collectively, these data indicated the functional NLS and NES of SLC2A4RG. Then the rate of apoptosis was further detected by flow cytometry analysis. When compared to the SLC2A4RG-wt vector, SLC2A4RG-NESmt could significantly increase the percentage of apoptotic U87 cells (13.37% vs. 18.66%, Student's t-test P = 0.006), whereas SLC2A4RG-NLSmt substantially reduced the apoptotic rate (13.37% vs. 8.17%, Student's t-test P = 0.016, Fig. 4c and d). Collectively, these data indicate that SLC2AR4G could induce glioma cell apoptosis, and its function mainly depended on the nuclear localization.
Fig. 4.
Nuclear SLC2A4RG mainly induces glioma cell apoptosis. (a) A schematic drawing of SLC2A4RG protein with the positions of NLS and NES. (b) Confocal microscopy showing localizations of SLC2A4RG in Hela cells transfected with SLC2A4RG-wt-GFP, SLC2A4RG-NESmt-GFP, or SLC2A4RG-NLSmt-GFP. (c and d) Flow cytometry with Annexin V and 7-AAD staining determining the changes in U87 cells apoptosis induced by different localizations of SLC2A4RG. Results analyzed by t-test presented as mean ± SEM. ns, no significant; **, P < 0.01; ***, P < 0.001.
3.5. Nuclear SLC2A4RG regulates apoptosis via transactivation of caspase-3 and caspase-6
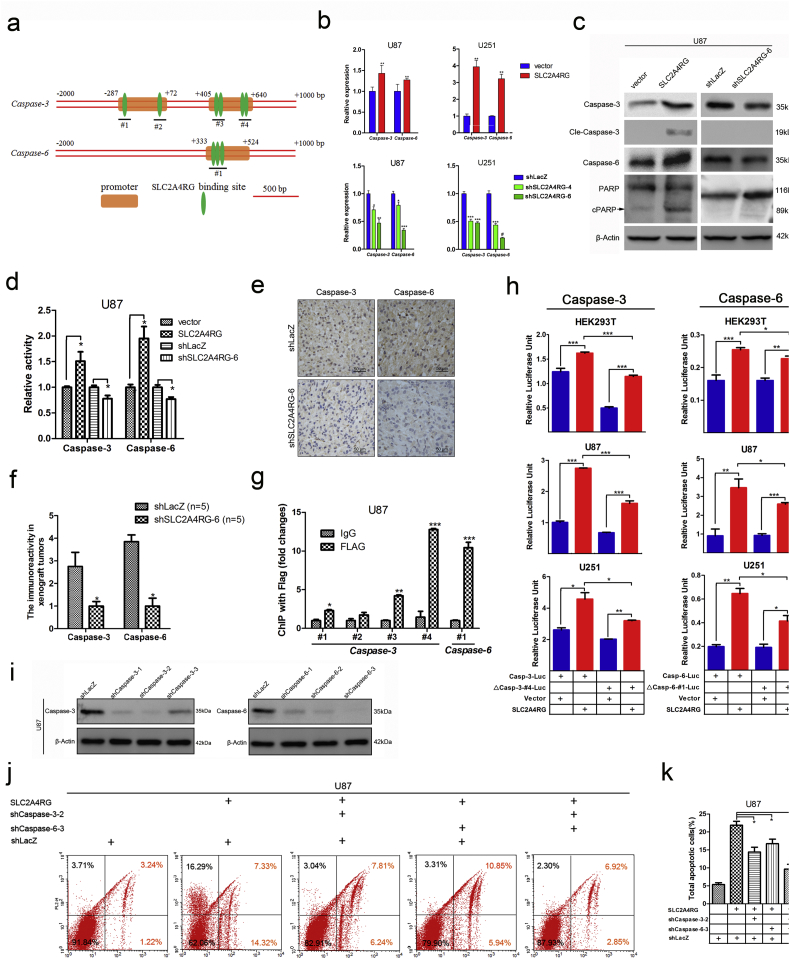
Potent effects of SLC2A4RG on cell apoptosis promoted us to investigate the mechanism underlying the downstream of SLC2A4RG. Previously, SLC2A4RG has been found to bind to the 7-bp consensus sequence (GCCGGCG) in a gene promoter region as a transcription factor [11]. We then performed a high-throughput screen of candidate genes with the GCCGGCG sequence located in the genomic region of −2000 - +1000 bp using the Ensembl database. The generated candidate genes were further selected by the correlation analysis (Pearson r > 0.4) with SLC2A4RG in TCGA glioblastoma database and narrowed down to 186 potential targets (Supplementary Fig. S5). The DAVID pathway analysis showed that these overlapping genes were significantly enriched in cellular processes such as apoptosis and cell death (Supplementary Table S1). This result combined with our finding that SLC2A4RG participated in glioma cell apoptosis, impelled us to focus on candidate genes involved in the pathway. Then the two important apoptotic effector genes caspase-3 and caspase-6 were ferreted out. Several potential SLC2A4RG DNA binding sites in the promoter regions of caspase-3 or caspase-6 were predicted in the genomatix website (http://www.genomatix.de, Fig. 5a). Among these sites, site #4 of caspase-3 and site #1 of caspase-6 contained the full sequence of GCCGGCG. Accordingly, we examined the mRNA and protein expressions of caspase-3 and caspase-6, as well ascaspase-7, in SLC2A4RG-overexpressed and -depleted glioma cells to explore the relationship between SLC2A4RG and caspase-3 /caspase-6. As expected, both the mRNA and protein expressions of caspase-3 or caspase-6 were positively correlated with SLC2A4RG changes between SLC2A4RG-overexpressed and -depleted groups. In contrast, both the mRNA and protein expressions of caspase-7 didn't have a significant correlation with SLC2A4RG expression in these groups. (Fig. 5b, c and supplementary Fig. S6). The enzymatic activities of caspase-3 and caspase-6 were also substantially elevated by overexpression of SLC2A4RG but could be diminished in SLC2A4RG-depleted glioma cells (Fig. 5d). Besides, overexpressed SLC2A4R induced an increase of cleaved PARP, which was regarded as a classical substrate for caspase-3 and revealed an enhanced enzymatic activity of caspase-3 (Fig. 5c). The immunohistochemistry examination of caspase-3 and caspase-6 in the xenograft specimens consistently confirmed reduced expressions in the SLC2A4RG-depleted groups (Fig. 5e and f). All these findings pointed to that SLC2A4RG might regulate caspase-3 and caspase-6 in glioma, and the ChIP-PCR data further validated the mechanism underlying SLC2A4RG directly binding to promoters of these two caspase genes. As shown in Fig. 5g, in comparison to the IgG group, anti-FLAG antibody was markedly enriched by the detected site #4 of caspase-3 and site #1 of caspase-6 in the FLAG-SLC2A4RG infected U87 cells. Then a firefly luciferase reporter whose expression was turned on by the caspase-3 promoter (casp-3-Luc) or caspase-6 promoter (casp-6-Luc) was constructed and transiently transfected into HEK293T, U87, or U251 cells. Co-transfection of casp-3-Luc or casp-6-Luc with the vector expressing SLC2A4RG in all groups significantly increased the fluorescence strength as depicted inFig. 5 h. However, deletion of the site #4 from the caspase-3 promoter (△casp-3-#4-Luc) and site #1 from the caspase-6 promoter (△casp-6-#1-Luc) made apparent deficiency of luciferase expression when co-transfected with SLC2A4RG-expressing vector (Fig. 5h). Interestingly, the luciferase fluorescence strength was still higher in SLC2A4GR group than the control. This results suggested that the SLC2A4RG transactivated caspase-3 and caspase-6 mainly through binding to site #4 in the caspase-3 promoter and site #1 in the caspase-6 promoter. Meanwhile, SLC2A4RG also weakly bound to some other functional sites. Subsequently, shcaspase-3-2 and shcaspase-6-3 (Fig. 5i) targeting caspase-3 and caspase-6 respectively were chosen for the following study to investigate whether SL2A4RG-mediated cell apoptosis was triggered by directly regulating caspase-3 or caspase-6. In SLC2A4RG-infected U87 cells, single knockout of caspase-3 or caspase-6 resulted in a significant decrease of cell apoptotic rate. Moreover, co-depletion of caspase-3 and caspase-6 strongly reversed the effect of SL2A4RG-mediated cell apoptosis (Fig. 5j and k).
Fig. 5.
SLC2A4RG induces cellular apoptosis via direct transactivation of caspase-3 and caspase-6. (a) A schematic drawing of potential SLC2A4RG DNA binding sites in the promoter region for caspase-3 or caspase-6. (b) The mRNA expressions of caspase-3 and caspase-6 in U87 and U251 cells with overexpressed or depleted SLC2A4RG detected by RT-PCR. (c) Western blot confirming the protein levels of caspase-3, caspase-6, and PARP in SLC2A4RG-overexpressed or -silenced U87 cells. β-Actin serves as the loading control. (d) Detection of the relative enzymatic activity of caspase-3 and caspase-6 in U87 cells with overexpression or knockdown of SLC2A4RG. (e) IHC analysis of caspase-3 and caspase-6 in intracranial tumors developed from SLC2A4RG silenced or control U87 cells. (f) The expression scores of caspase-3 or caspase-6 in the two groups. (g) Exploration and validation of SLC2A4RG binding sites depicted in (a) with ChIP-PCR. (h) Dual-luciferase reporter assay determining the function of #4 site or #1 site on the expression of caspase-3 or caspase-6 when regulated by SLC2A4RG in HEK293T, U87, and U251 cells. (i) Western blot is analyzing the efficiency of shRNAs targeting caspase-3 or caspase-6 in U87 cells. β-Actin serves as the loading control. (j and k) Flow cytometry with Annexin V and 7-AAD staining determining the changes of SLC2A4RG-induced apoptosis in U87 cells after inhibiting caspase-3 or/and caspase-6. Results analyzed by t-test presented as mean ± SEM. ns, no significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
3.6. Subcellular localization of SLC2A4RG is modulated by 14-3-3θ
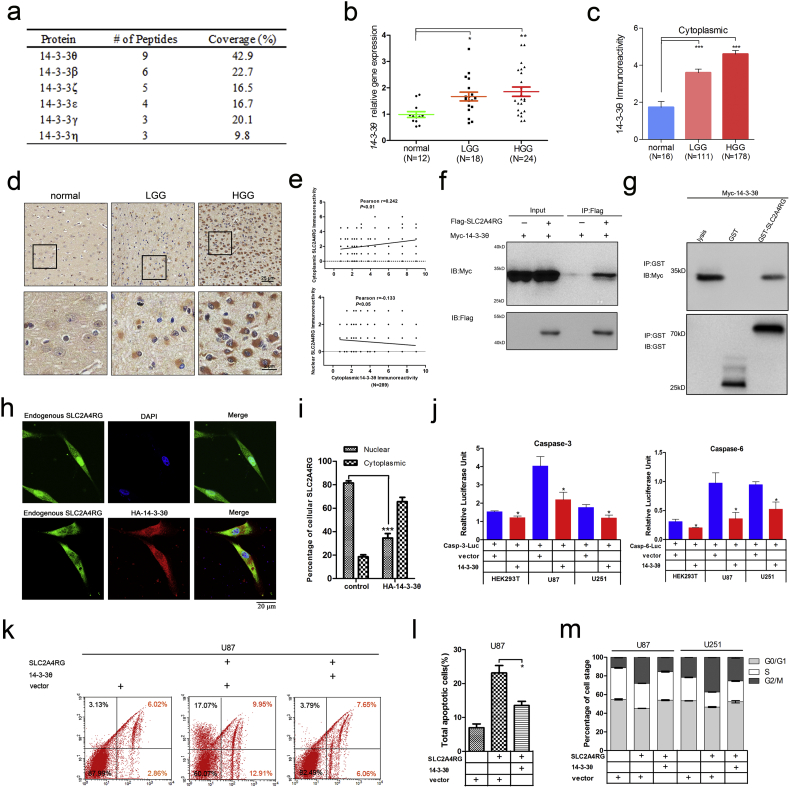
As demonstrated, the nuclear localization and the transcriptional function of SLC2A4RG were vital for SLC2A4RG-induced cell apoptosis in glioma. However, a decreased percentage of nuclear SLC2A4RG was found in our glioma specimens, especially in HGG. Increasing evidence suggests that malignant gliomas are characterized by an intrinsic resistance to apoptosis to evade elimination [16]. It is deduced that there may be some mediators restrain the transportation of SLC2A4RG from the cytoplasm into the nucleus. Co-IP and mass spectrometry hereby were performed to identify the potential mediators of SLC2A4RG. Among the proteins immunoprecipitated by FLAG-SLC2A4RG, six out of seven 14-3-3 isoforms were found to interact with SLC2A4RG probably (Fig. 6a). 14-3-3 proteins are well-known universal regulators binding a vast number of client proteins, and the interaction can have effects on the stability, activity, and localization of their partners [17]. Among the six isoforms, 14-3-3θ has the maximum number of peptides with the largest coverage rate. Moreover, previous studies have found that overexpression of 14-3-3θ significantly suppressed apoptosis induced by proteasome inhibitors in human glioma cells [18]. Collectively, 14-3-3θ was inferred to have the potential of interacting with SLC2A4RG.
Fig. 6.
SLC2A4RG-induced cell apoptosis is regulated by 14-3-3θ in human glioma. (a) The table is showing the results of 14–3-3 proteins found in Co-IP & mass spectrometry experiments. (b) 14-3-3θ mRNA expression detected via RT-PCR in normal controls and gliomas of different WHO grades. (c) Immunoreactivity scores of cytoplasmic 14-3-3θ in the second independent cohort of tissue microarray. (d) Representative staining images of cytoplasmic 14-3-3θ in normal brains and gliomas. (e) The correlation between cytoplasmic 14-3-3θ and cytoplasmic/nuclear SLC2A4RG levels. (f) Western blot analysis of cell lysates and Co-IP samples from HEK293T cells transfected with vectors as depicted. (g) Physical interaction of SLC2A4RG and 14-3-3θ in vitro validated by GST pull-down assay. (h) Representative images of immunofluorescence stained with SLC2A4RG, HA-14-3-3θ, and DAPI in U87 cells transfected with indicated vectors. (i) Percentages of U87 cells transfected with HA-14-3-3θ or control calculated by cytoplasmic or nuclear SLC2A4RG fluorescence. (j) Relative fluorescence intensity analyzed by luciferase reporter assay in HEK293T, U87, and U251 cells transfected with different combinations of vectors to explore the effect of 14-3-3θ on caspase-3 and caspase-6 expressions. (k and l) Detection of the apoptotic U87 cells transfected with vectors as depicted and labeled with Annexin V and 7-AAD by flow cytometry. (m) Flow cytometry detecting changes in cell cycle distribution of SLC2A4RG-infected U87 and U251 cells in the absence or presence of 14-3-3θ. Results analyzed by t-test presented as mean ± SD. *, P < 0.05; **, P < 0.01.
A cohort of 12 normal brains, 18 LGGs, and 24 HGGs was then used for detecting the mRNA expression level of 14-3-3θ. Significant upregulation of 14-3-3θ was found in glioma specimens and showed the positive association with pathological grade (Fig. 6b). Moreover, IHC was conducted to analyze its protein expression in the tissue microarray. The 14-3-3θ expresses exclusively in the cytoplasm, and the immunoreactivity of 14-3-3θ was also significantly increased in LGG (Student's t-test P = 3.200E-04) and HGG (Student's t-test P = 1.345E-05), when compared to normal brains (Fig. 6c and d). In addition, Kaplan-Meier analysis found that patients with high cytoplasmic 14-3-3θ expression had a markedly shorter OS than those with low expression of cytoplasmic 14-3-3θ (39 vs. 54 months, P = 0.010, Supplementary Fig. S7). Intriguingly, we found that cytoplasmic 14-3-3θ levels were positively correlated with cytoplasmic SLC2A4RG (Pearson r = 0.242, P < 0.01, N = 289), but negatively correlated with nuclear SLC2A4RG (Pearson r = −0.133, P < 0.05, N = 289, Fig. 6e). Co-immunoprecipitation (co-IP) assay in HEK293T cells co-transfected with FLAG-SLC2A4RG and Myc-14-3-3θ was further performed to examine whether 14-3-3θ interacts with SLC2A4RG. As shown in Fig. 6f, Myc-14-3-3θ was distinctly immunoprecipitated by FLAG-SLC2A4RG, which was also confirmed by a GST pull-down assay that GST-SLC2A4RG, but not GST alone, specifically pulled down Myc-14-3-3θ (Fig. 6g). Next, immunofluorescence was introduced to detect the subcellular distribution of SLC2A4RG and 14-3-3θ. Endogenous SLC2A4RG was localized in both cytoplasm and nuclei of U87 cells, 75.47% of which showed stronger nuclear fluorescence (Fig. 6h and i). Intriguingly, exogenous HA-14-3-3θ changed the present cellular pattern and recruited more SLC2A4RG to co-localize to the cytoplasm followed by a significant decrease in the percentage of cells with strong nuclear SLC2A4RG fluorescence (75.47% vs. 49.53%, Student's t-test P = 0.016, Fig. 6h and i). In accordance with these findings in glioma, 98.33% of regular Hela cells displayed stronger nuclear SLC2A4RG fluorescence, which was only detected in 9.30% of Hela cells after transfected with HA-14-3-3θ (Student's t-test P = 6.18E-09; Supplementary Fig. S8).
Then we analyzed the expression levels of SLC2A4RG and 14-3-3θ in normal controls, IDH1 mutation (grade I, II, III), IDH1 wild type (grade I, II, III), and GBM samples. Compared to the normal controls, we found no significant difference of the total SLC2A4RG expression in IDH1 mutation and IDH1 wild type samples, whereas the cytoplasmic and nuclear SLC2A4RG showed a substantially upward and a considerable downward trend, respectively. Interestingly, we observed the cytoplasmic 14-3-3θ has a significant increase in these two groups compared with the normal controls (Supplementary Fig. 1A). This result further validated the phenomenon that we observed in the immunofluorescence experiment. We found that the increase of cytoplasmic 14-3-3θ could restrain the cytoplasmic SLC2A4RG into the nucleus, leading an increase of cytoplasmic SLC2A4RG and a decrease of nuclear SLC2A4RG (Fig. 6H and Supplementary Fig. S8). Similar effects were seen under the criteria of the glioma grade and histology (Fig. 1B, D, E, and Supplemental Fig. S1B). Accordingly, these data supported the effect of SLC2A4RG-14-3-3θ interaction on subcellular localization of SLC2A4RG.
To address the function of 14-3-3θ in regulating SLC2A4RG-induced phenotype in glioma cells, we first detected the expression changes of caspase-3 and caspase-6 in the presence of 14-3-3θ by luciferase assay. As expected, casp-3-Luc or casp-6-Luc in conjunction with 14-3-3θ considerably reduced the luciferase expression in Hek293T, U87, and U251 cells compared to the control group, respectively (Fig. 6j). The high percentage of the apoptotic rate of U87 cells caused by SLC2A4RG was also substantially inhibited after infection of exogenous 14-3-3θ (23.12% vs. 13.52%, Student's t-test P = 0.018, Fig. 6k and l). Furthermore, 14-3-3θ mainly reversed SLC2A4RG-induced G2/M arrest in both U87 and U251 cells (Fig. 6m).
4. Discussion
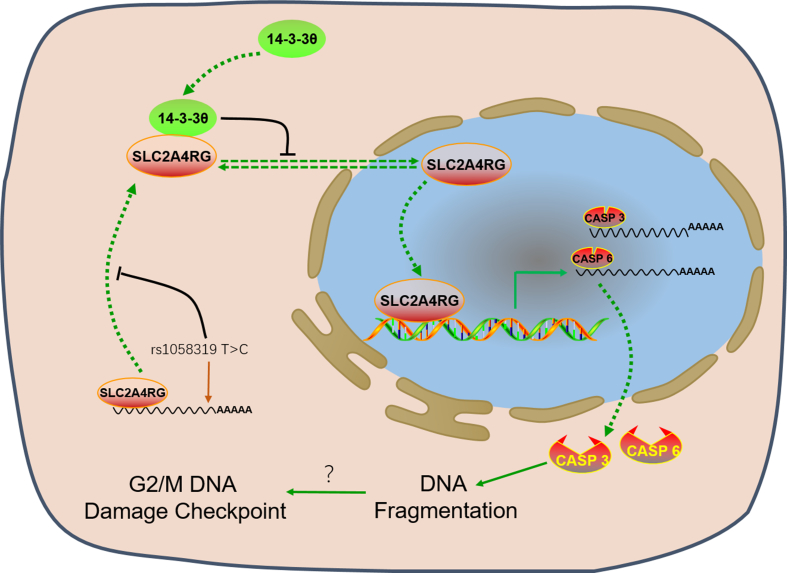
In our previous and the current studies, we have addressed the roles of SLC2A4RG gene susceptibility in gliomagenesis, gene expression in predicting prognosis, and tumor suppressor function in glioma progression [[7], [8], [9], [10], [11]]. Our findings underscore nuclear SLC2A4RG in regulating glioma cell survival. We found that nuclear SLC2A4RG could transactivate directly caspase-3 and caspase-6 genes to induce cell apoptosis in glioma. However, this effect was attenuated by decreased of nuclear SLC2A4RG expression, and the cytoplasmic 14-3-3θ could negatively mediate nuclear localization of SLC2A4RG (Fig. 7). These data suggest that the SLC2A4RG and 14-3-3θ system may be a driver of glioma development and a potential target for cancer treatment.
Fig. 7.
A proposed model for the role of SLC2A4RG in glioma cell.
As shown in the fine mapping analysis of GWAS, the rs1058319 T > C variant in the 3’UTR of SLC2A4RG was associated with increased glioma risk [7]. Since 3’-UTR plays a pivotal role in modulating stability and translation of mRNA [19,20], our experiment confirmed that the risk C allele decreased the expression of SLC2A4RG compared with wild-type T (Supplementary Fig. S9). The risk allele of rs1058319 may change the structural fold of SLC2A4RG mRNA that affects its stability or leads to the impaired polyadenylation process, which may explain why SLC2A4RG is down-regulated in glioma and needs further investigation [[21], [22], [23], [24]]. Moreover, the possible etiology of glioma associated with SLC2A4RG is also presented by another result that the G > T transversion at rs8957 (E[GAG]233D[GAU]) might impair the nuclear export signal or protein folding of SLC2A4RG, resulting in excess accumulation of unphysiological protein in the nucleus [8].
Shuttling between the nucleus and the cytoplasm is inextricably linked to protein functions, while the transportation is regulated by various stimuli such as phosphorylation/dephosphorylation and stress response [[25], [26], [27], [28]]. The structural analysis reveals phosphoserine sites in the SLC2A4RG (NP_064446), which are the putative 14-3-3θ binding sites mediating the protein-protein interaction. 14-3-3 proteins participate in a wide range of biological processes through diverse regulatory mechanisms by binding to phosphoserine-containing motifs, such as signal transduction, cell cycle control, DNA repair, and apoptosis [[29], [30], [31], [32]], and a variety of their target proteins can be observed changes in subcellular localization, conformation, and enzymatic activity [33]. Furthermore, 14-3-3θ has been found in the majority of astrocytoma samples, and its expression was positively increased with pathologic grade [34,35]. Consistently, we also found increased cytoplasmic 14-3-3θ and reduced nuclear SLC2A4RG expressions in the glioma cells. Taken together, our experiment indicated that 14-3-3θ would bind SLC2A4RG, and restrain SLC2A4RG in the cytoplasm in a phosphorylation-dependent manner in the process of glioma development.
Currently, nuclear/cytoplasmic shuttling of proteins has been regarded as a key mechanism for apoptotic regulation [12,36]. Translocation of such proteins, especially a major of tumor suppressors, including p53, FOXO, and PTEN, plays an important role in tumor apoptosis [37], as well as cancer progression and treatment resistance [38,39]. Our data validated that SLC2A4RG could act as nuclear/cytoplasmic shuttling and tumor suppressor protein regulating apoptosis of glioma cells. Besides, the activation of diverse of intermediate molecules involved in two central pathways: the extrinsic (death receptor) pathway and the intrinsic (mitochondrial) pathway, can stimulate the apoptotic cascade [40,41]. After the signaling triggered, both the pathways converge on the same execution factors, mainly containing caspase-3, which will lead to cell death [42]. However, SLC2A4RG was showed to induce glioma cell apoptosis through the direct activation of two effectors caspase-3 and caspase-6 in our study. Given that the SLC2A4RG DNA binding sites were identified in the promoter regions of caspase-3 and caspase-6, SLC2A4RG could directly transactivate caspase-3 and caspase-6. Additionally, reduced glioma cell proliferation with G2/M phase cell cycle arrest was also found to be induced by SLC2A4RG. In the condition of cell apoptosis, caspase-3 and caspase-6 activation are thought to occur early and then triggers DNA fragmentation [[43], [44], [45]]. Whereas, cell mitosis will be prevented by G2/M DNA damage repair checkpoint when damaged DNA exists [46]. Presumably, the DNA fragmentation induced by caspase-3 and caspase-6 activation may contribute to the G2/M phase arrest. However, it should be noted that there are several limitations to this study. One is that the critical role of how SLC2A4RG-mediated regulation of caspase-3 and caspase-6 is still needed to be further studied. For example, the author should detect whether other cellular factors jointly interact with SLC2A4RG to mediate the expression of downstream targets. For another, this study showed that 14-3-3θ negatively regulates the nuclear function of the tumor suppressor SLC2A4RG and maybe a promising therapeutic target of human glioma. This mechanism needs to be further validated with 14-3-3θ inhibitor in vitro and in vivo.
Collectively, our results demonstrate that nuclear SLC2A4RG induces cell apoptosis in a new manner via direct transactivation of caspase-3 caspase-6 in glioma, but the interaction with 14-3-3θ will result in sequestration of SLC2A4RG in the cytoplasm and then promote tumor progression. Further understanding of the novel mechanism involved by SLC2A4RG in gliomagenesis and development may contribute to a useful therapeutic intervention.
Acknowledgments
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81372706, 81572501, and 81372235). We thank all participates recruited for this study. We also thank Dr. Chenji Wang (School of Life Sciences, Fudan University) for his help on the revision of the manuscript; Bo Zhu, and Hao Cai for animal studies; and Qinglong Guo for GST-pull down assays.
Role of the funding source
The study sponsor had no role in study design, data collection, analysis, interpretation of data, writing of the report, or the decision to submit the paper for publication.
Declaration of interests
The authors have no conflict of interests to declare.
Author contributions
DRL, HYC, and JXC conceived the project and designed research. DPY, HXW, and YQW performed experiments. DPY, YYC, and YYJ wrote the manuscript. HXW and QLH provided clinical data. DPY, ZPZ, and JWM performed the animal analysis. All the authors have read and approved the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.030.
Contributor Information
Juxiang Chen, Email: juxiangchen@smmu.edu.cn.
Hongyan Chen, Email: chenhy@fudan.edu.cn.
Daru Lu, Email: drlu@fudan.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Ostrom Q., Bauchet L., Davis F., Deltour I., Fisher J., Langer C. The epidemiology of glioma in adults: a "state of the science" review. Neuro Oncol. 2014;16(7):896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H.X., Xu T., Jiang Y., Xu H.C., Yan Y., Fu D. The challenges and the promise of molecular targeted therapy in malignant gliomas. Neoplasia. 2015;17(3):239–255. doi: 10.1016/j.neo.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melin B.S., Barnholtz-Sloan J.S., Wrensch M.R., Johansen C., Il'yasova D., Kinnersley B. Genome-wide association study of glioma subtypes identifies specific differences in genetic susceptibility to glioblastoma and non-glioblastoma tumors. Nat Genet. 2017;49(5):789–+. doi: 10.1038/ng.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrensch M., Jenkins R., Chang J., Yeh R., Xiao Y., Decker P. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–908. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shete S., Hosking F., Robertson L., Dobbins S., Sanson M., Malmer B. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009;41(8):899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H., Chen Y., Zhao Y., Fan W., Zhou K., Liu Y. Association of sequence variants on chromosomes 20, 11, and 5 (20q13.33, 11q23.3, and 5p15.33) with glioma susceptibility in a Chinese population. Am J Epidemiol. 2011;173(8):915–922. doi: 10.1093/aje/kwq457. [DOI] [PubMed] [Google Scholar]
- 7.Song X., Zhou K.K., Zhao Y.J., Huai C., Zhao Y., Yu H.J. Fine mapping analysis of a region of 20q13.33 identified five independent susceptibility loci for glioma in a Chinese Han population. Carcinogenesis. 2012;33(5):1065–1071. doi: 10.1093/carcin/bgs117. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y., Yun D., Zou X., Jiang T., Li G., Hu L. Whole exome-wide association study identifies a missense variant in SLC2A4RG associated with glioblastoma risk. Am J Cancer Res. 2017;7(9):1937–1947. [PMC free article] [PubMed] [Google Scholar]
- 9.Oshel K.M., Knight J.B., Cao K.T., Thai M.V., Olson A.L. Identification of a 30-base pair regulatory element and novel DNA binding protein that regulates the human GLUT4 promoter in transgenic mice. J Biol Chem. 2000;275(31):23666–23673. doi: 10.1074/jbc.M001452200. [DOI] [PubMed] [Google Scholar]
- 10.Knight J.B., Eyster C.A., Griesel B.A., Olson A.L. Regulation of the human GLUT4 gene promoter: interaction between a transcriptional activator and myocyte enhancer factor 2A. Proc Natl Acad Sci U S A. 2003;100(25):14725–14730. doi: 10.1073/pnas.2432756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka K., Shouguchi-Miyata J., Miyamoto N., Ikeda J.E. Novel nuclear shuttle proteins, HDBP1 and HDBP2, bind to neuronal cell-specific cis-regulatory element in the promoter for the human Huntington's disease gene. J Biol Chem. 2004;279(8):7275–7286. doi: 10.1074/jbc.M310726200. [DOI] [PubMed] [Google Scholar]
- 12.Wang S.C., Hung M.C. Cytoplasmic/nuclear shuttling and tumor progression. Ann N Y Acad Sci. 2005;1059:11–15. doi: 10.1196/annals.1339.002. [DOI] [PubMed] [Google Scholar]
- 13.Li X., Wan X., Chen H., Yang S., Liu Y., Mo W. Identification of miR-133b and RB1CC1 as independent predictors for biochemical recurrence and potential therapeutic targets for prostate cancer. Clin Cancer Res. 2014;20(9):2312–2325. doi: 10.1158/1078-0432.CCR-13-1588. [DOI] [PubMed] [Google Scholar]
- 14.Ohuchida K., Mizumoto K., Ishikawa N., Fujii K., Konomi H., Nagai E. The role of S100A6 in pancreatic cancer development and its clinical implication as a diagnostic marker and therapeutic target. Clin Cancer Res. 2005;11(21):7785–7793. doi: 10.1158/1078-0432.CCR-05-0714. [DOI] [PubMed] [Google Scholar]
- 15.Muller A., Ritzkowsky A., Steger G. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J Virol. 2002;76(21):11042–11053. doi: 10.1128/JVI.76.21.11042-11053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ziegler D.S., Kung A.L., Kieran M.W. Anti-apoptosis mechanisms in malignant gliomas. J Clin Oncol. 2008;26(3):493–500. doi: 10.1200/JCO.2007.13.9717. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan A., Ottmann C., Fournier A.E. 14-3-3 adaptor protein-protein interactions as therapeutic targets for CNS diseases. Pharmacol Res. 2017;125:114–121. doi: 10.1016/j.phrs.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Yan Y., Xu Y., Gao Y., Zong Z., Zhang Q., Li C. Implication of 14-3-3ε and 14-3-3θ/τ in proteasome inhibition-induced apoptosis of glioma cells. Cancer Sci. 2013;104(1):55–61. doi: 10.1111/cas.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuersten S., Goodwin E.B. The power of the 3 ' UTR: translational control and development. Nat Rev Genet. 2003;4(8):626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 20.Mazumder B., Seshadri V., Fox P.L. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem Sci. 2003;28(2):91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 21.Shen L., Basilion J., Stanton V. Single-nucleotide polymorphisms can cause different structural folds of mRNA. Proc Natl Acad Sci U S A. 1999;96(14):7871–7876. doi: 10.1073/pnas.96.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan X., Steitz J. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17(12):3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ford L.P., Wilusz J. 3'-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′-->5′ exonuclease in vitro. Nucleic Acids Res. 1999;27(4):1159–1167. doi: 10.1093/nar/27.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey S.N., Sulem P., Jonasdottir A., Masson G., Gudmundsson J., Gudbjartsson D.F. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–1103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin K.C., Moroishi T., Meng Z.P., Jeong H.S., Plouffe S.W., Sekido Y. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol. 2017;19(8):996–+. doi: 10.1038/ncb3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng W.N., Li J., Wang S.S., Cao S.S., Jiang J.W., Chen C. Phosphorylation controls the nuclear-cytoplasmic shuttling of influenza A virus nucleoprotein. J Virol. 2015;89(11):5822–5834. doi: 10.1128/JVI.00015-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munsie L., Desmond C., Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci. 2012;125(Pt 17):3977–3988. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- 28.Eijkelenboom A., Burgering B.M.T. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14(2):83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 29.Yan Y., Xu Y., Gao Y.Y., Zong Z.H., Zhang Q., Li C. Implication of 14-3-3epsilon and 14-3-3theta/tau in proteasome inhibition-induced apoptosis of glioma cells. Cancer Sci. 2013;104(1):55–61. doi: 10.1111/cas.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison D.K. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009;19(1):16–23. doi: 10.1016/j.tcb.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Obsilova V., Silhan J., Boura E., Teisinger J., Obsil T. 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res. 2008;57(Suppl. 3):S11–S21. doi: 10.33549/physiolres.931598. [DOI] [PubMed] [Google Scholar]
- 32.Tzivion G., Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277(5):3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- 33.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381(Pt 2):329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao L., Cao W., Zhang W., Lin H., Yang X., Zhen H. Identification of 14-3-3 protein isoforms in human astrocytoma by immunohistochemistry. Neurosci Lett. 2008;432(2):94–99. doi: 10.1016/j.neulet.2007.11.071. [DOI] [PubMed] [Google Scholar]
- 35.Yang X., Cao W., Lin H., Zhang W., Lin W., Cao L. Isoform-specific expression of 14-3-3 proteins in human astrocytoma. J Neurol Sci. 2009;276(1–2):54–59. doi: 10.1016/j.jns.2008.08.040. [DOI] [PubMed] [Google Scholar]
- 36.Chu C., Plowey E., Wang Y., Patel V., Jordan-Sciutto K. Location, location, location: altered transcription factor trafficking in neurodegeneration. J Neuropathol Exp Neurol. 2007;66(10):873–883. doi: 10.1097/nen.0b013e318156a3d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salmena L., Pandolfi P. Changing venues for tumour suppression: balancing destruction and localization by monoubiquitylation. Nat Rev Cancer. 2007;7(6):409–413. doi: 10.1038/nrc2145. [DOI] [PubMed] [Google Scholar]
- 38.Fulda S. Seminars in Cancer Biology. Vol. 31. 2015. Targeting apoptosis for anticancer therapy; pp. 84–88. [DOI] [PubMed] [Google Scholar]
- 39.Fulda S. Tumor resistance to apoptosis. Int J Cancer. 2009;124(3):511–515. doi: 10.1002/ijc.24064. [DOI] [PubMed] [Google Scholar]
- 40.Taylor R., Cullen S., Martin S. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9(3):231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 41.Ichim G., Tait S.W.G. A fate worse than death: apoptosis as an oncogenic process. Nat Rev Cancer. 2016;16(8):539–548. doi: 10.1038/nrc.2016.58. [DOI] [PubMed] [Google Scholar]
- 42.Fulda S., Klaus-Michael D. Targeting apoptosis pathways in cancer therapy. Curr Cancer Drug Tar. 2004;4(7):569–576. doi: 10.2174/1568009043332763. [DOI] [PubMed] [Google Scholar]
- 43.Karamitopoulou E., Cioccari L., Jakob S., Vallan C., Schaffner T., Zimmermann A. Active caspase 3 and DNA fragmentation as markers for apoptotic cell death in primary and metastatic liver tumours. Pathology. 2007;39(6):558–564. doi: 10.1080/00313020701684375. [DOI] [PubMed] [Google Scholar]
- 44.Enari M., Sakahira H., Yokoyama H., Okawa K., Iwamatsu A., Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391(6662):43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 45.Sakahira H., Enari M., Nagata S. Cleavage of CAD inhibitor in CAD activation and DNA degradation during apoptosis (vol 391, pg 96, 1998) Nature. 2015;526(7575):728. doi: 10.1038/nature15532. [DOI] [PubMed] [Google Scholar]
- 46.Rhind N., Russell P. Mitotic DNA damage and replication checkpoints in yeast. Curr Opin Cell Biol. 1998;10(6):749–758. doi: 10.1016/s0955-0674(98)80118-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material