Abstract
Background
HIV-1 infection and physiological aging are independently linked to elevated systemic inflammation and changes in enteric microbial communities (dysbiosis). However, knowledge of the direct effect of HIV infection on the aging microbiome and potential links to systemic inflammation is lacking.
Methods
In a cross-sectional study of older people living with HIV (PLWH) (median age 61.5 years, N = 14) and uninfected controls (median 58 years, n = 22) we compared stool microbiota, levels of microbial metabolites (short-chain fatty acid levels, SCFA) and systemic inflammatory biomarkers by HIV serostatus and age.
Findings
HIV and age were independently associated with distinct changes in the stool microbiome. For example, abundances of Enterobacter and Paraprevotella were higher and Eggerthella and Roseburia lower among PLWH compared to uninfected controls. Age-related microbiome changes also differed by HIV serostatus. Some bacteria with inflammatory potential (e.g. Escherichia) increased with age among PLWH, but not controls. Stool SCFA levels were similar between the two groups yet patterns of associations between individual microbial taxa and SCFA levels differed. Abundance of various genera including Escherichia and Bifidobacterium positively associated with inflammatory biomarkers (e.g. soluble Tumor Necrosis Factor Receptors) among PLWH, but not among controls.
Interpretation
The age effect on the gut microbiome and associations between microbiota and microbial metabolites or systemic inflammation differed based on HIV serostatus, raising important implications for the impact of therapeutic interventions, dependent on HIV serostatus or age.
Keywords: Aging, HIV, Microbiome, Inflammation, Short chain fatty acids (SCFA)
Research in context.
Evidence before this study
The development of highly effective antiviral therapies have greatly increased the life expectancy of people living with HIV (PLWH). However, even with long-term effective treatment, a persistent, low-grade inflammation persists in PLWH which has been linked to increased risk for comorbidities such as diabetes and heart disease. The inflammatory profile and comorbid conditions associated with chronic HIV infection are similar to those reported with aging. Thus, many PLWH will experience a double inflammatory “hit” that may accelerate the aging process. Previous studies have highlighted that gut microbial communities (microbiome) are different in both PLWH and in older persons and this dysbiosis is independently associated with inflammation. However, knowledge of the direct effect of HIV infection on the aging microbiome and potential links to inflammation is lacking in older PLWH.
Added value of study
In this present study, we compared the profiles of bacteria in the stool of older PLWH with those of older persons that were not infected with HIV (controls). We also compared levels of short-chain fatty acids (SCFAs), products produced by bacteria that are important for gut health, and various indicators of inflammation between the two groups. HIV and age were independently associated with distinct changes in the stool microbiome, and age-related microbiome changes differed by HIV serostatus. Stool SCFA levels were similar between the two groups yet patterns of associations between microbiota and SCFA levels differed.
Implications of all the available evidence
To the best of our knowledge, this study represents the first description of the combined effects of HIV infection and aging on the stool microbiome. We show that not only are HIV and age independently associated with distinct changes in the stool microbiome in an older population, but HIV infection and age interact together to shape the microbiome. Our observations highlight that responses to microbial-based therapeutic interventions may differ by HIV serostatus and by age.
Alt-text: Unlabelled Box
1. Introduction
With the advent of effective antiretroviral therapy (ART), the life expectancy of people living with HIV-1 infection (PLWH) has dramatically improved, with recent estimates suggesting that 70% of PLWH will be age 50 or older by 2030 [1]. However, even with effective virologic suppression, chronic HIV infection is associated with a low level inflammatory state that has been linked to an increased risk of comorbidities, as well as geriatric syndromes including dementia and frailty [[2], [3], [4]]. Physiological aging is similarly associated with low levels of inflammation (inflammaging), as measured by increased circulating systemic inflammatory markers such as interleukin-6 (IL-6) and C-reactive protein (CRP) [[5], [6], [7], [8]], and is linked to increased comorbidities [9] [10]. Thus, many PLWH will experience a double inflammatory “hit” that may together accelerate the aging process.
One source of inflammation underlying both HIV infection and aging is the human gut [[11], [12], [13]]. Intestinal epithelial barrier damage and associated microbial translocation has been observed with both HIV infection [[14], [15], [16], [17], [18], [19]] and with physiological aging in animal models [[20], [21], [22], [23], [24]] and humans [25,26]. We previously demonstrated an age-related increase in plasma biomarkers of intestinal epithelial barrier damage (intestinal fatty acid binding protein; IFABP) and microbial translocation (lipopolysaccharide; LPS) in study participants without HIV as well as age-related increases in circulating markers of cellular immune activation (sCD14, sCD27) and inflammation (IL-6, CRP), linking gut barrier leakiness to systemic inflammation [26]. Changes in the composition of stool microbial communities (dysbiosis) have also been observed in both PLWH and with aging (reviewed in [[27], [28], [29]]). Although these studies varied by the age of the population, lifestyle (diet, physical activity), and geographic location, some commonalities exist between stool dysbiotic profiles with HIV and with aging. These include changes in diversity [[30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44]], increases in abundances of Proteobacteria [30,31,41,43,[45], [46], [47], [48], [49]] and/or alterations in Prevotella/Bacteroides ratio [39,41,43,48,[50], [51], [52]]. This dysbiosis has been linked to physiologic consequences: stool profiles characterized by increased abundance of taxa containing commensal bacteria with pathogenic potential, termed pathobionts [53,54] (e.g. Enterobacteriaceae) have been associated with markers of systemic immune activation and inflammation in both ART-treated PLWH [45] and with chronological aging [30,55]. Indicators of biological aging such as frailty have also been linked to increased abundance of pathobionts including Enterobacteriaceae [56], Eggerthella [57] and Prevotella [58].
Microbial metabolites, such as the short chain fatty acids (SCFA) butyrate, propionate and acetate, are primarily synthesized following fermentation of dietary fiber [59] and play important roles in gut function including maintaining homeostasis [60]. For example, butyrate serves as a primary source of energy for intestinal epithelial cells and has immunomodulatory properties in the gut immune system [61,62]. Butyrate is produced primarily by bacterial species belonging to the Roseburia, Faecalibacterium and Ruminococcus genera and propionate by members of the Firmicutes (e.g. Blautia, Dialister) and Bacteroidetes (e.g. Bacteroides, Prevotella, Alistipes) phyla, while acetate is produced by a wide variety of enteric bacteria [[63], [64], [65]]. We and others have demonstrated lower abundance of butyrate-producing bacterial taxa (in gut or stool) among both ART-treated and untreated PLWH [32,34,35,48,51,52,66]. Furthermore, a lower abundance of genera containing butyrate-producing species (Faecalibacterium, Roseburia) in the gut mucosa is associated with higher systemic markers of immune activation and microbial translocation [34,51,66]. A lower abundance of butyrate-producing bacteria species has also been seen with both chronological age [30,42,46,47,67] and with measures of frailty [56,57,68,69].
Although these studies suggest similarities in parameters of gut dysfunction in both HIV and in aging, a direct understanding of the collective effect in an individual aging with HIV is an understudied area of research. Therefore, the goals of this pilot study were to investigate the combined effect of age and HIV infection on the stool microbiome, and explore the functional impact of microbiome differences through SCFA production and markers of systemic inflammation.
2. Materials and methods
2.1. Participants
Participants were recruited from two clinical studies. Eight participants with and twenty-two without HIV were recruited from the “Exercise in Healthy Aging” study, a randomized controlled trial examining the effect of an exercise intervention on markers of inflammation and physical function (Clinical Trials #: NCT02404792) [70]. Six additional PLWH were recruited from “Assessing Tenofovir Pharmacology in Older HIV-infected Individuals Receiving Tenofovir-based Antiretroviral Therapy”, an observational study analyzing the pharmacokinetics of tenofovir in older vs younger individuals with HIV (Clinical Trials #: NCT02304263). All participants were between the ages of 50 and 75 years old; PLWH were on ART for ≥2 years, had an HIV-1 RNA <200 copies/mL, and a CD4 count of >200 cells/μL. Key exclusion criteria included active diarrhea, antibiotic use within the last two weeks, active hepatitis C infection, diabetes requiring insulin, BMI <20 or > 40 kg/m2, and chronic steroid usage. All participants provided written, informed consent, and both studies were approved by the Colorado Multiple Institutional Review Board.
2.2. Microbiome analysis
A self-collected stool sample was obtained after consent and prior to any intervention and immediately stored in the participant's home freezer until transfer to a −80C freezer. All samples were processed within 6 months of storage. Stool bacterial profiles were generated by broad-range amplification and sequence analysis of bacterial 16S rRNA genes following established methods [51,[71], [72], [73], [74], [75]]. DNA was extracted using the QIAamp Power Fecal DNA Isolation Kit (QIAGEN, Venlo, Netherlands) and 16S rRNA gene [51,73] amplicons were generated using oligonucleotide primers that target the V3 V4 variable region of the 16S rRNA gene (primers 338F and 805R) and included barcode [76] and Illumina adapter sequences. Illumina paired-end sequencing was performed on the Miseq platform using a 600-cycle (v.3) reagent kit. Paired-end reads were quality-filtered, demultiplexed, merged, and classified using SINA(1.3.0-r23838) [77,78] as previously described [51,[71], [72], [73], [74], [75]]. Operational taxonomic units (OTUs) were produced by clustering sequences with identical taxonomic assignments.
2.3. SCFA extraction and analysis
Stool SCFA was measured using gas chromatography as previously described [79] with the following modification: seven grams of stool taken from disparate sections of each sample were dissolved in purified water (1:5, w/v) and homogenized by vortexing.
2.4. Dietary analysis
Each participant completed a 3-day dietary survey designed to quantify micro and macronutrient intake during the period of stool collection. Raw dietary data was converted to caloric intake by the Nutrition Core at the Colorado Clinical and Translational Sciences Institute [80]. Dietary measurements were averaged over 3 days.
2.5. Soluble biomarker analysis
Plasma levels of soluble CD14 (sCD14), sCD163, IFABP, interleukin-10 (IL-10) and serum levels of tumor necrosis factor alpha (TNFα) and soluble tumor necrosis factor receptor 1 and 2 (sTNFR1 and sTNFR2) were measured by ELISA (R&D Systems; Minneapolis, MN, USA). Plasma CRP and IL-6 were quantitated by ELISA in the Colorado Clinical and Translational Research Center.
2.6. Statistical analysis
Participant characteristics were summarized with frequencies (%) for categorical variables and means (standard deviation) or median (interquartile range [IQR]) for continuous variables. Fisher's exact tests were used to test for differences between PLWH or uninfected controls in categorical variables, and two-sample t-tests with unequal variances were used to test for differences in means among continuous variables. Linear regression was used to evaluate differences in dietary intake or SCFA by HIV serostatus.
Relative abundance (RA) was calculated as the number of sequences for a specific taxa standardized to the total number of sequences. Taxa which were present in <5% of the population were collapsed into a single “other” category. Stacked bar plots visually displayed mean RA by HIV serostatus or predicted RA at ages 50 or 70. Low abundance taxa at <2% were collapsed into a single “other” category for visualization purposes on the stacked bar plots. To obtain predicted RA for ages 50 and 70, the intercept and age estimate regression coefficients from the negative binomial regression models (described below) were used to calculate the expected counts for each taxa, given the specified age. The expected counts were calculated using an assumed offset of 10,000 sequences, and the RA was then calculated by dividing the counts by 10,000. Alpha-diversity measures (Sobs, Shannon diversity [H], and Shannon evenness [H/Hmax]) were compared by HIV serostatus using a linear regression models. Beta-diversity was compared using a permutation-based multiple analysis of variance (PERMANOVA) test and the Bray-Curtis diversity measures calculated at the phylum, family, and genus taxonomic levels. Relationships between HIV serostatus or age with individual taxa were evaluated using generalized linear models assuming a negative binomial distribution and log link function. Interactions between HIV and age were evaluated for each taxa to determine whether the HIV effect differed with age; interaction terms meeting an false discovery rate (FDR) p-value threshold <0.20 were included in final models. A FDR correction was applied to control for multiple comparisons, with significance defined as FDR p-value <.05. Explicet [81] (v2.8.3) was used to store/export data and calculate diversity measures, and statistical comparisons of bacterial communities were performed with R software (v 3.4.4) packages vegan, MASS and gplots [[82], [83], [84]].
3. Results
3.1. Study demographics
The 14 PLWH and 22 uninfected controls included in the study were similar by age, body mass index, race, and alcohol and tobacco use (Table 1). All but 1 study participant identified as male. Male PLWH were more likely to report sex with men compared to the male uninfected controls (p < .001). Among the PLWH, the median CD4 count was 570 (IQR: 364) cells/μl and all had a HIV-1 RNA <50 copies/ml.
Table 1.
Study participant characteristics.
| Characteristic | PLWH (n = 14) | Controls (n = 22) | P value |
|---|---|---|---|
| Male | 14 (100)a | 21 (95)a | 1.0 |
| Age (years) | 61.5 (13.5)b | 58 (8.8)b | 0.81 |
| White Race | 10 (71)a | 17 (77)a | 0.33 |
| Men who have sex with other men | 13 (93)a | 6 (27)a | <0.001 |
| Body mass index (kg/m2) | 27.0 (4)b | 27.5 (6)b | 0.82 |
| Alcohol intake | 1.0 | ||
| >2 drinks/day | 1 (7)a | 2 (9)a | |
| ≤2 drinks/day | 13 (93)a | 20 (91)a | |
| Current tobacco smoker | 3 (21)a | 2 (9)a | 0.11 |
| HIV-1 RNA < 200 copies/mL | 14 (100)a | NA | NA |
| CD4 count, cells/μL | 570 (364)b | NA | NA |
Results presented as frequency (%)a or median (interquartile range)b.
3.2. Effect of HIV serostatus on the stool microbiome
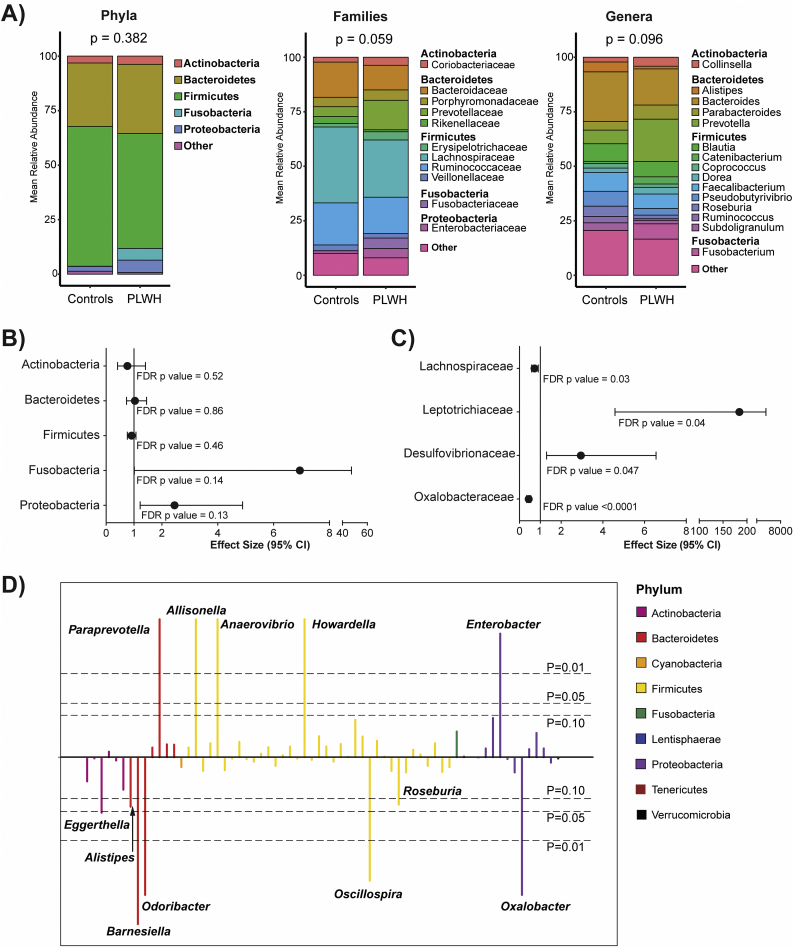
Stool microbiota profiling was successful in all samples (median of 115,568 sequences/sample, median Goods coverage of 99.97%). Measures of microbial alpha diversity (richness, diversity, evenness) were similar between PLWH and controls (Supplemental Fig. 1). PERMANOVA tests of beta diversity to determine overall compositional differences in microbiota between PLWH and controls at the phylum, family and genus levels are shown in Fig. 1A. No significant differences were seen at the phylum level although the profile of PLWH generally reflected higher RA of Fusobacteria and Proteobacteria in conjunction with decreased Firmicutes. Comparisons at the family and genus levels indicated non-significant overall differences in microbiota between PLWH and controls (p = .059 and p = .096 respectively), most prominently reflected by higher RA of Prevotellaceae/Prevotella, Fusobacteriaceae/Fusobacterium and lower RA of Bacteroidaceae/Bacteroides, Faecalibacterium and Roseburia among PLWH.
Fig. 1.
Effect of HIV status on stool microbiota.
(A) Stacked bar charts representing the mean relative abundance at the phylum, family, and genus levels. P-values represent beta-diversity differences between groups at each taxonomic level, compared using a PERMANOVA test of Bray-Curtis diversity measures. Taxa with relative abundance <2% were collapsed into a single “Other” category. (B, C) Forest plots were created displaying effect size and 95% confidence interval (CI) for (B) top 5 most abundant phyla and (C) specific families that were significantly different based on FDR p values <.05 (all data can be found in Supplemental Table 1). Effect size >1 indicates taxa relative abundance is higher with HIV infection. (D) Manhattan plot indicating alterations in genera using negative binomial modeling with genus-level taxa as outcomes and HIV status as the predictor. FDR p-values above the center line represent taxa that are higher in PLWH compared to controls. The colors of each genus bar corresponds with the phylum from which the genus originates (legend).
HIV-related effects on the RA of specific taxa between PLWH and controls were explored with negative binomial regression models. No significant differences by HIV serostatus were detected in the RA of any phylum, including the five most abundant phyla: Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria and Proteobacteria (Fig. 1B, Supplemental Table 1). At the family level, Leptotrichaceae (fold change (effect size) 184, p = .04) and Desulfovibrionaeceae (fold change 2.94, p = .047) were significantly higher among PLWH compared to controls, while Lachnospiraceaea (fold change 0.72, p = .03) and Oxalobacteraceae (fold change 0.44, p < .0001) were significantly lower (Fig. 1C, Supplemental Table 1). At the genus level (Fig. 1D, Supplemental Table 1), Eggerthella (Actinobacteria phylum) was significantly lower (0.17 fold, p = .047) in PLWH. Within the phylum Bacteroidetes, lower RA of Barnesiella (0.09 fold, p = .001) and Odoribacter (0.70 fold, p < .0001) were noted while Paraprevotella was higher (1.53 fold, p < .0001) in PLWH. Numerous genera belonging to the phylum Firmicutes were also higher in PLWH including Allisonella (2.85 fold, p < .0001), Anaerovibrio (422 fold, p < .0001) and Howardella (1.74 fold, p < .0001) with lower RA of Oscillospira (0.01 fold, p = .001). Within the phylum Proteobacteria, Enterobacter was significantly higher (73.7 fold, p = .001) while Oxalobacter was significantly lower (0.43 fold, p < .0001) in PLWH compared to controls.
Among genera known to contain SCFA-producing species [[63], [64], [65]], the RA of Roseburia (contains butyrate- and propionate-producing species) and Alistipes (contains propionate-producing species) were lower (Roseburia: 0.44 fold, p = .07; Alistipes: 0.32 fold, p = .06) in PLWH versus controls (Fig. 1D, Supplementary Table 1) but did not reach statistical significance. No significant differences were observed between PLWH and controls in the RA of other genera containing butyrate-producing species (Butyrivibrio, Faecalibacterium, Anaerostipes, Subdoligranulum), propionate-producing bacteria (Bacteroides, Blautia, Akkermansia, Prevotella, Dialister and Phascolarctobacterium) or genera that include species that produce either butyrate or propionate (Coprococcus) (Supplementary Table 1).
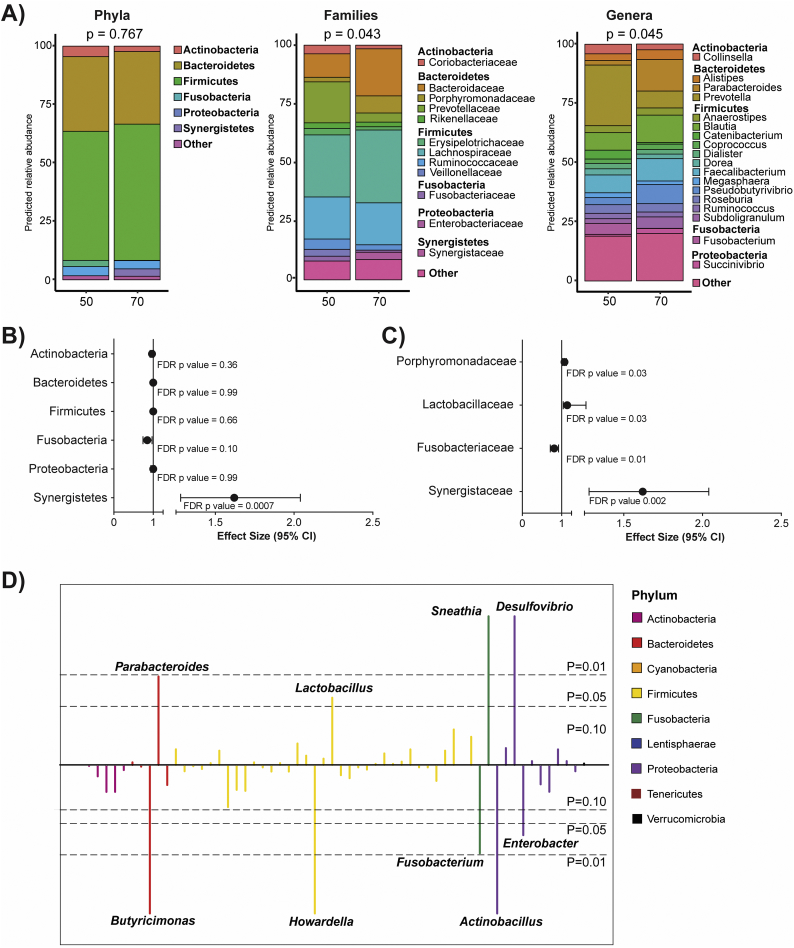
3.3. Effect of age on the stool microbiome
To assess the overall impact of age on the stool microbiome independent of HIV serostatus, the predicted RAs of an individual at age 50 versus age 70 were calculated using data from both groups (Fig. 2A). The predicted profiles suggested that by age 70, microbial communities were different to those at age 50, most notably at family and genus taxonomic levels. Using negative binomial regression models, only the RA of the phylum Synergistetes was found to significantly increase per year of increasing age (1.62 fold; p = .0007) (Fig. 2B, Supplementary Table 2). At the family level (Fig. 2C, Supplemental Table 2), significant increases in the RA of Porphyromonadaceae (1.07 fold; p = .03), Lactobacilliaceae (1.14, p = .03) and Synergistaceae (1.62 fold, p = .002) were observed with increasing age, whereas Fusobacteriaceae decreased (0.81, p = .01) (Fig. 2C, Supplemental Table 2). For each year of increasing age, significant increases in a number of genera were also noted including Parabacteroides (1.09 fold, p = .01), Lactobacillus (1.15 fold, p = .03), Sneathia (1.44, p < .0001) and Desulfovibrio (1.04, p < .0001) (Fig. 2D, Supplementary Table 2). In contrast, Butyricimonas (0.96 fold, p < .0001), Howardella (0.96 fold, p < .0001), Fusobacterium (0.81, p = .01), Actinobacillus (0.10, p < .0001) and Enterobacter (0.75 fold, p = .03) decreased in RA for each year of increasing age (Fig. 2D, Supplementary Table 2). No significant age effects were evident among the butyrate- or propionate-producing genera (all p > .1) (Supplementary Table 2).
Fig. 2.
Effect of age on stool microbiota.
(A) Stacked bar charts representing the predicted relative abundance for an individual at age 50 and 70 at the phylum, family and genus taxonomic levels. P-values represent beta-diversity differences between groups at each taxonomic level, compared using a PERMANOVA test of Bray-Curtis diversity measures. Due to the continuous nature of the age variable, a predicted relative abundance was estimated using coefficients from the negative binomial regression models for each taxa. Taxa with relative abundance <2% were collapsed into a single “Other” category. (B, C) Forest plots were created displaying effect size and 95% confidence interval for (B) top 6 most abundant phyla and (C) families that were significantly different based on FDR p values <.05 (all data can be found in Supplemental Table 1). Effect size >1 indicates when the relative abundance increases per year of increasing age. (D) Manhattan plot indicating alterations in genera using negative binomial modeling with genus-level taxa as outcomes and age as the predictor. FDR p-values above the center line represent taxa that significantly increase with age. The colors of each genus bar corresponds with the phylum from which the genus originates (legend).
3.4. Interacting effect of HIV serostatus and age on stool microbiome composition
To understand whether the age effect on the gut microbiome differed in the presence versus absence of HIV infection, we tested an interaction between age and HIV serostatus. The age effect differed between PLWH and controls across multiple taxa (interaction p-value <.20) (Table 2, Table 3, Supplementary Fig. 2). For example, while Proteobacteria was lower with increasing age among controls, it was not in PLWH (Table 2). Conversely, a number of bacterial families (e.g. Bifidobacteriaceae, Bacteroidaceae, Peptostreptococcaceae) and genera (e.g. Bifidobacterium, Bacteroides) were significantly higher with increasing age in PLWH, but not in controls (Tables 2, Table 3, Supplementary Fig. 2). In some instances, significant changes were observed in both PLWH and controls, but were opposite in direction. For example, Leptotrichiaceae and the genera Butyricimonas, Escherichia and Oxalobacter significantly increased with age in PLWH, but were significantly lower with age in controls (Table 2, Table 3, Supplementary Fig. 2). Significant age and HIV interactions were also observed for some genera known to contain butyrate- or propionate-producing bacteria. Alistipes (containing propionate-producing spp.) increased with age in PLWH, but was not significantly different in controls; Butyrivibrio (butyrate-producers) decreased with age in both PLWH and controls, but to a significantly greater degree in controls (Table 3, Supplementary Fig. 2).
Table 2.
Age Effect Among People Living with HIV (PLWH) and Controls, at Phylum and Family Taxonomic Ranks.
| Age effect among PLWH |
Age effect among Controls |
Age and HIV Interaction |
|||||
|---|---|---|---|---|---|---|---|
| Effect Size | 95% CI | P value | Effect Size | 95% CI | P value | P value⁎ | |
| Phylum | |||||||
| Cyanobacteria | 0.74 | 0.60, 0.90 | 0.022 | 0.98 | 0.84, 1.12 | 0.82 | 0.087 |
| Proteobacteria | 1.02 | 0.95, 1.09 | 0.68 | 0.90 | 0.84, 0.96 | 0.016 | 0.032 |
| Family | |||||||
| Phylum Actinobacteria | |||||||
| Bifidobacteriaceae | 1.16 | 1.02, 1.30 | 0.043 | 0.99 | 0.88, 1.10 | 0.91 | 0.10 |
| Phylum Bacteroidetes | |||||||
| Bacteroidaceae | 1.15 | 1.08, 1.22 | 0.0001 | 1.00 | 0.94, 1.06 | 0.99 | 0.005 |
| Phylum Firmicutes | |||||||
| Peptostreptococcaceae | 1.28 | 1.16, 1.41 | <0.0001 | 1.00 | 0.91, 1.09 | 0.99 | 0.001 |
| Phylum Fusobacteria | |||||||
| Leptotrichiaceae | 1.36 | 1.33, 1.38 | <0.0001 | 0.35 | 0.14, 0.84 | 0.043 | 0.008 |
| Phylum Proteobacteria | |||||||
| Alcaligenaceae | 1.04 | 0.94, 1.13 | 0.53 | 0.93 | 0.85, 1.01 | 0.14 | 0.13 |
| Enterobacteriaceae | 0.97 | 0.84, 1.12 | 0.80 | 0.82 | 0.72, 0.93 | 0.012 | 0.13 |
| Oxalobacteraceae | 1.12 | 1.08, 1.15 | <0.0001 | 0.95 | 0.93, 0.96 | <0.0001 | <0.0001 |
Taxa with an HIV-age interaction of FDR p value of ≤0.20 are included, with findings suggesting that the age effect differs by HIV serostatus. The effect size can be interpreted as the per year increase (if >1) or decrease (if <1) in relative abundance of each taxa. CI = confidence interval.
Table 3.
Age Effects Among People Living with HIV (PLWH) and Controls, by Genus Taxonomic Rank.
| Age effect among PLWH |
Age effect among Controls |
Age and HIV |
|||||
|---|---|---|---|---|---|---|---|
| Effect Size | 95% CI | P value | Effect Size | 95% CI | P value | Interaction⁎ | |
| Phylum Actinobacteria | |||||||
| Bifidobacterium | 1.16 | 1.02, 1.31 | 0.044 | 0.99 | 0.88, 1.23 | 0.92 | 0.10 |
| Eggerthella | 1.04 | 0.90, 1.18 | 0.67 | 0.89 | 0.78, 1.00 | 0.10 | 0.14 |
| Phylum Bacteroidetes | |||||||
| Alistipes | 1.15 | 1.05, 1.25 | 0.0058 | 1.00 | 0.92, 1.08 | 0.98 | 0.046 |
| Bacteroides | 1.15 | 1.08, 1.22 | 0.0001 | 1.00 | 0.94, 1.06 | 0.99 | 0.004 |
| Barnesiella | 1.14 | 1.02, 1.27 | 0.038 | 0.99 | 0.89, 1.09 | 0.92 | 0.096 |
| Butyricimonas | 1.02 | 1.00, 1.02 | 0.0025 | 0.95 | 0.93, 0.95 | <0.0001 | <0.0001 |
| Paraprevotella | 0.99 | 0.98, 1.00 | 0.30 | 0.88 | 0.86, 0.89 | <0.0001 | <0.0001 |
| Phylum Firmicutes | |||||||
| Anaerostipes | 1.05 | 0.97, 1.13 | 0.30 | 0.96 | 0.89, 1.03 | 0.37 | 0.15 |
| Butyrivibrio | 0.95 | 0.94, 0.96 | <0.0001 | 0.75 | 0.74, 0.76 | <0.0001 | <0.0001 |
| Dorea | 0.95 | 0.95, 0.95 | <0.0001 | 1.01 | 1.01, 1.01 | <0.0001 | <0.0001 |
| Flavonifractor | 1.23 | 1.08, 1.40 | 0.0041 | 0.99 | 0.87, 1.11 | 0.92 | 0.035 |
| Holdemania | 1.11 | 1.01, 1.21 | 0.044 | 0.98 | 0.90, 1.06 | 0.69 | 0.074 |
| Howardella | 0.94 | 0.92, 0.94 | <0.0001 | 0.99 | 0.97, 0.99 | 0.059 | <0.0001 |
| Pseudobutyrivibrio | 0.96 | 0.87, 1.05 | 0.43 | 1.09 | 0.99, 1.18 | 0.096 | 0.087 |
| Phylum Proteobacteria | |||||||
| Escherichia | 1.37 | 1.19, 1.56 | <0.0001 | 0.82 | 0.73, 0.93 | 0.005 | <0.0001 |
| Oxalobacter | 1.12 | 1.08, 1.15 | <0.0001 | 0.95 | 0.93, 0.96 | <0.0001 | <0.0001 |
Taxa with an HIV-age interaction of FDR p value of <0.20 are listed, with findings suggesting that the age effect differs by HIV serostatus. The effect size can be interpreted as the per year increase (if >1) or decrease (if <1) in relative abundance of each taxa. CI = confidence interval.
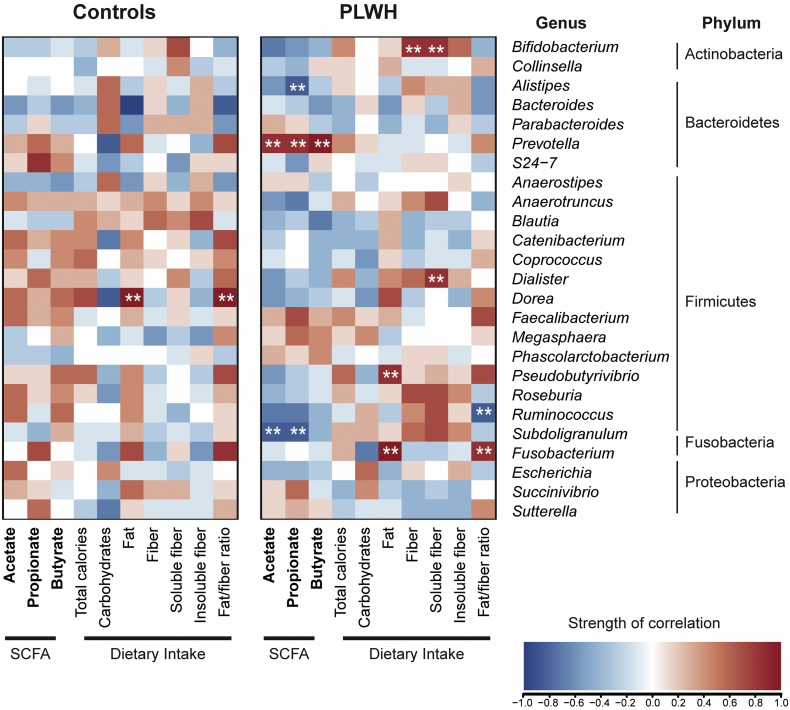
3.5. Stool SCFA levels, dietary intake and associations with stool microbiota
Levels of stool SCFAs and dietary intake were compared between PLWH and controls using linear regression with HIV serostatus as the predictor (Table 4). Stool propionate levels were lower among PLWH (mean 10.2 mmol/g) compared to controls (14.0 mmol/g), although this did not reach statistical significance (p = .075). Acetate (PLWH: 37.7 mmol/g, Controls: 42.0 mmol/g; p = .35) and butyrate levels (PLWH: 7.7 mmol/g, Controls: 7.3 mmol/g; p = .87) were not significantly different by HIV serostatus. Dietary intake was generally similar between the two groups with the exception of soluble fiber intake which was slightly higher in PLWH compared to controls (9.5 g vs 7.1 g; p = .073).
Table 4.
Stool Short Chain Fatty Acid (SCFA) and dietary intake differences between People Living with HIV (PLWH) and controls.
| PLWH (n = 14) | Controls (n = 22) | P value | |
|---|---|---|---|
| SCFA Total (mmol/g) | 55.6 (22.3) | 63.4 (21.9) | 0.31 |
| Acetate (mmol/g) | 37.7 (14.1) | 42.0 (11.7) | 0.35 |
| Propionate (mmol/g) | 10.2 (4.9) | 14.0 (7.6) | 0.075 |
| Butyrate (mmol/g) | 7.7 (6.1) | 7.3 (5.0) | 0.87 |
| PLWH (n = 13) | Controls (n = 22) | P value | |
|---|---|---|---|
| Average daily energy intake (kcal) | 2229 (629) | 2034 (647) | 0.39 |
| % Carbohydrates | 43.3 (6.6) | 44.80 (10.9) | 0.67 |
| % Fat | 39.6 (6.0) | 36.5 (8.5) | 0.25 |
| Fiber (g) | 23.0 (8.8) | 20.5 (7.5) | 0.36 |
| Soluble fiber (g) | 9.5 (5.2) | 7.1 (2.3) | 0.073 |
| Insoluble fiber (g) | 13.2 (4.5) | 12.8 (6.3) | 0.86 |
| Fat/fiber ratio | 4.9 (2.0) | 4.7 (2.1) | 0.82 |
Levels of SCFAs or dietary intake measurements were compared between PLWH and controls using linear regressions with HIV status as the predictor. Data are presented as mean (standard deviation). Dietary intake is missing for one PLWH.
Next, we explored associations between stool SCFA levels and dietary intake measures with the RA of the top 25 bacterial genera (based on all study participants) and noted differences in patterns of significantly strong associations, based on HIV status (Fig. 3). For instance, the RA of Prevotella was strongly positively correlated with stool levels of all 3 SCFAs while Subdoligranulum and Alistipes were inversely correlated with acetate and propionate levels in PLWH, but not controls. Furthermore, in PLWH, but not controls, Bifidobacterium and Dialister RA were positively correlated with dietary soluble fiber intake. Fusobacterium RA was positively correlated with fat/fiber consumption whereas Ruminococcus was inversely associated with this dietary measure. Few correlations between bacterial genera and stool SCFA or dietary intake were observed in controls. The RA of Dorea was strongly correlated with fat intake in controls, but not in PLWH.
Fig. 3.
Associations between stool SCFA levels and dietary intake with stool microbiota. Associations between the top 25 most abundant genera based on all study participants with levels of stool SCFA levels (bolded text) and dietary intake in controls (N = 22) and in PLWH (N = 13; dietary intake was unavailable from one study participant). Genera are grouped by phylum. Pearson correlations were determined with red shading representing positive association and blue shading indicating a negative association. Correlations that were both significant (p < .05) and strong correlations based on r > 0.6 or < −0.6 are indicated with white asterisks (**). Units for SCFA measurements are mmol/g. Macronutrients are calculated relative to total energy intake: Carbohydrates (%), Fat (%), Soluble and Insoluble fiber (g). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
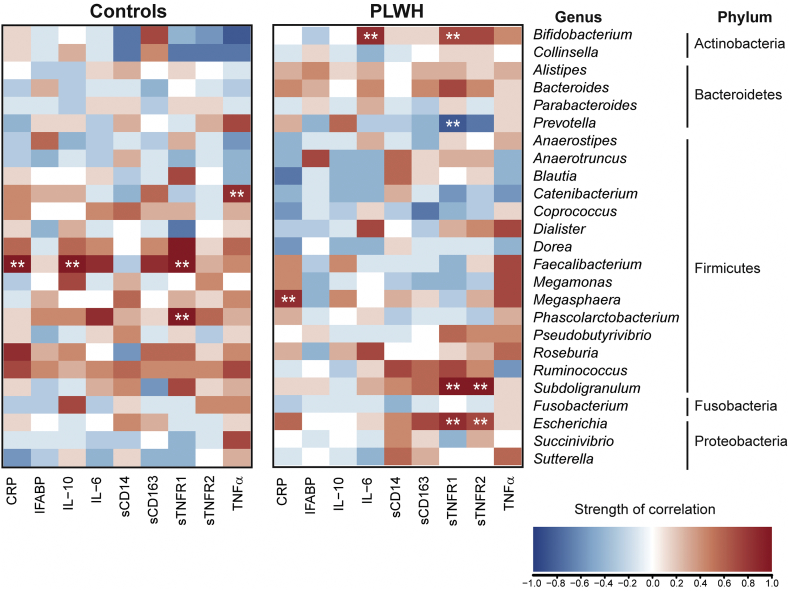
3.6. Impact of HIV infection on systemic biomarkers and associations with stool microbiota
PLWH had higher levels of sCD163, sCD14, and sTNFR2 compared to uninfected controls (Table 5). Levels of IL-10, IFABP, TNFα, sTNFR1, CRP, and IL-6 were not statistically different (Table 5). Again, major differences were noted in the patterns of associations of microbiota and inflammatory biomarkers between PLWH and controls. For example, among PLWH, Escherichia and Subdoligranulum were positively correlated with sTNFR1 and sTNFR2, Bifidobacterium with TNFR1 and IL-6, and Prevotella inversely correlated with sTNFR1 levels (Fig. 4). Among controls, but not PLWH, Faecalibacterium positively correlated with levels of CRP, sTNFR1 and IL-10.
Table 5.
HIV effect on systemic biomarker levels.
| Biomarker | HIV Effect | 95% CI | P value |
|---|---|---|---|
| IL-6 (pg/ml) | 0.64 | −0.49, 1.77 | 0.28 |
| TNFα (pg/ml) | 0.21 | −0.08, 0.5 | 0.17 |
| sTNFR1 (pg/ml) | 199.7 | −11.3, 410.7 | 0.079 |
| sTNFR2 (pg/ml) | 744.2 | 283.1, 1205.2 | 0.0034 |
| CRP (mg/L) | 1.50 | −0.44, 3.43 | 0.14 |
| sCD163 (ng/ml) | 134.0 | 27.6, 240.3 | 0.019 |
| sCD14 (ng/ml) | 469.7 | 226.1, 713.3 | 0.0006 |
| IFABP (pg/ml) | 555.9 | −1596.0, 2707.7 | 0.62 |
| IL-10 (pg/ml) | 0.17 | −1.98, 2.32 | 0.88 |
Levels of biomarkers were compared between PLWH and controls using linear regressions with HIV status as the predictor. Effect size and confidence (CI) intervals are shown with effect size >1 indicating higher levels in PLWH.
Fig. 4.
Associations between systemic inflammatory and immune activation biomarkers with stool microbiota.
Associations between the top 25 most abundant genera based on all study participants with levels of systemic biomarkers indicative of inflammation and immune activation in controls (N = 22) and in PLWH (N = 14). Genera are grouped by phylum. Pearson correlations were determined with red shading representing positive association and blue shading indicating a negative association. Correlations that were both significant (p < .05) and strong correlations based on r > 0.6 or <−0.6 are indicated with white asterisks (**). CRP = C reactive protein; IFABP = intestinal fatty acid binding protein; IL-10 = interleukin 10; IL-6 = interleukin 6; sCD14 = soluble CD14, sCD163 = soluble CD163, sTNFR = soluble tumor necrosis factor receptor [1] and [2]; TNFα = tumor necrosis factor alpha. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
An emerging body of evidence suggests that gut dysbiosis occurs in both the normal aging process and among PLWH, and that these alterations in gut microbial communities may contribute to the low grade chronic inflammatory state that underlies age-associated co-morbidities [11,27,85]. However, evaluation of the effect of HIV infection on the aging microbiome, and the potential links to systemic inflammation is lacking. To the best of our knowledge, this study represents the first description of the combined effects of aging and HIV infection on the stool microbiome and demonstrates that, not only are HIV and age independently associated with distinct changes in the stool microbiome in an older population, but HIV infection and age interact to shape the microbiome. Furthermore, within this older population, associations between microbes and stool metabolites, dietary intake characteristics, and inflammatory biomarkers differed based on HIV serostatus.
Interaction analyses highlighted that numerous bacterial taxa increased with age in PLWH, but were either decreased or not significantly changed among controls. Many of these bacteria have been linked to gut inflammatory diseases. For example, Escherichia belongs to the family Enterobacteriaceae, facultative anaerobes that are commonly increased in abundance in inflammatory bowel disease (IBD) and colorectal cancer [[86], [87], [88]]. Similarly, Peptostreptococcaceae was reported to be overrepresented in persons with colorectal cancer [86,89]. Intriguingly, Bacteroides spp. also increased with age in PLWH, but not in controls. This genus typically plays an important role in immune regulation including the induction of regulatory T cells [90]; however certain strains of B. fragilis have also been linked to gut diseases including colorectal cancer [86]. Notably, a number of the bacterial species that increased with age among PLWH, but not in controls, such as Oxalobacter, Alistipes and Bifidobacterium, potentially have health-promoting properties. Oxalobacter spp. are important in degrading oxalate, excessive amounts of which are associated with pathological conditions including kidney stone formation [91,92]. Alistipes contains propionate-producing organisms and Bifidobacterium, a frequent component of probiotic mixtures, has anti-inflammatory properties [93]. Some of these findings may be related to dietary differences. For example, PLWH in our study tended to have higher fiber intake; Bifidobacterium utilize dietary fiber as a growth substrate [94], and, indeed, Bifidobacterium RA was associated with greater fiber intake among PLWH. Increased abundance may also be a direct result of probiotic use; however probiotic use was not routinely collected in this study.
Numerous studies have demonstrated that PLWH manifest stool dysbiosis, despite effective viral suppression with ART (reviewed in [85]). Indeed, recent studies have shown that ART drugs themselves may contribute to dysbiosis, although potential mechanisms by which this occurs remain to be fully elucidated (reviewed in [95]). Therefore, in addition to determining if the impact of age differed among PLWH and controls, the design of our study also permitted investigation into the interrelationships between HIV, the gut microbiome, and functional outcomes (SCFA production and systemic inflammation) in this older cohort of PLWH. Similar to a number of previous studies of stool microbiota of ART-treated PLWH, we observed higher RA of bacteria belonging to the Enterobacteriaceae (Enterobacter) and Prevotellaceae (Paraprevotella) families and lower relative abundance of Barnesiella, Alistipes, and Roseburia in our participants with HIV [45,48,49,52,96]. The latter two genera are of particular relevance given that they contain species with the capacity to produce butyrate or propionate [[63], [64], [65]]. Indeed, PLWH tended to have lower levels of stool propionate than controls potentially due to lower abundances of the propionate-producing species Alistipes putredinis or Roseburia inulinivorans [[63], [64], [65]]. However, in PLWH, the RA of the genus Alistipes was inversely correlated with levels of stool propionate; thus additional studies with species identification are required to fully delineate the role decreased Alistipes may play in HIV-related pathogenesis. While Roseburia contains a number of species with butyrate-producing ability, levels of butyrate were similar between the two groups. However, it is important to note that butyrate is produced following fermentation of soluble dietary fibers [94], and the higher soluble fiber intake among PLWH may account for the lack of difference in butyrate levels. Although not significantly different between PLWH and controls, Subdoligranulum, known to contain the butyrate-producing bacteria S. variabile [63,64], inversely correlated with levels of acetate and propionate in PLWH. This observation may reflect the process of bacterial cross-feeding whereby the metabolic end products produced by one bacteria are utilized by another as has been reported for other known butyrate-producing bacteria and the utilization of acetate for butyrate formation [97].
Few studies similarly investigating stool microbiomes in ART-treated PLWH have detailed alterations in the remaining genera that we observed to be lower (Odoribacter, Oscillospira, Oxalobacter, Eggerthella) or higher (Allisonella, Anaerovibrio, Howardella) in PLWH versus controls. Lozupone and colleagues noted lower abundance of Odoribacter in ART-treated PLWH relative to uninfected controls [96]. Eggerthella was higher among PLWH receiving protease inhibitors in combination with nucleoside/tide reverse transcriptase inhibitors (NRTI) relative to uninfected controls, but was not different among those on other NRTI combination therapies, suggesting associations with both ART regimen and HIV [38]. It is also important to note that Allisonella, Anaerovibrio and Howardella were enriched in MSM versus non-MSM from prior studies [44]; given that our controls were not matched for sexual behavior, the overall higher abundance of these specific genera may reflect sexual behavior rather than HIV infection itself.
As expected, we observed that a number of biomarkers of systemic immune activation and inflammation were higher among PLWH. Some biomarkers did not differ significantly by HIV serostatus (e.g. IL-6, CRP): as age is a major driver of both IL-6 and CRP, any HIV-related differences may have been attenuated by the age effects. Furthermore, we identified expected and unexpected associations between the stool microbiome and systemic inflammation in PLWH, with similar associations not observed in controls. In keeping with its role as a potentially inflammatory bacterium, Escherichia RA was associated with levels of sTNFR1 and sTNFR2 in PLWH whereas Prevotella, a genus also known to contain species linked to inflammatory diseases (reviewed in [98]), inversely correlated with sTNFR1 in PLWH. Moreover, Bifidobacterium RA positively associated with sTNFR1 and IL-6 levels, an additional unexpected finding given its previously described anti-inflammatory capabilities and the prominent use of Bifidobacterium strains in probiotics. In older persons without HIV infection, few associations between microbiota taxa and inflammatory biomarkers were noted with the exception of Faecalibacterium which positively correlated with IL-10, CRP and sTNFR1. The association with IL-10 is congruent with the reported ability of strains of F. prausnitzii to induce IL-10 production from human peripheral blood immune cells and myeloid dendritic cells in vitro [99,100], yet positive correlations with CRP and sTNFR1, indicators of systemic inflammation, contrast with the generally accepted anti-inflammatory role of F. prausnitzii [101,102]. Taken together, these observations suggest some intriguing links between microbiota and systemic inflammation in older persons both with and without HIV infection; however, it is essential to emphasize that these associations do not indicate causality. Further studies will be required to determine if the association holds in a larger study population and if they are direct associations driven by individual species or indirectly driven by alterations in networks of bacterial communities. It will also be important to evaluate if, like enterotoxigenic strains of E. coli and B. fragilis, certain species traditionally considered to reflect a healthy microbiome, have undergone adaptive evolution due to environmental pressures (i.e., bacterial community changes or interactions with host immune system) resulting in positive selection of virulence genes [[103], [104], [105]].
A number of study limitations should be noted: the sample size was small, which limited our ability to adjust for numerous confounders, including sexual behaviors [44,106,107] and prebiotic/probiotic use, that may influence the microbiome. Few women or minorities were enrolled therefore generalizability is limited. Lastly, bacterial classification was limited to genus-level identification due to short read sequencing. Overall, our findings are preliminary and need to be confirmed in larger, more diverse participant populations.
In summary, this study has demonstrated that not only are alterations in the gut microbiome present in older PLWH, but HIV influenced age-associated effects on the gut microbiome. Furthermore, associations between the stool microbiome and microbial metabolite levels, dietary factors and inflammatory biomarkers differed by HIV serostatus. A ‘one-bug-fits-all’ interpretation may therefore not be applicable, because microbiome changes in one population may not have the same implications as in another population. This also raises important implications for therapeutic interventions: targeted replacement of certain microbial taxa may have different effects depending on the host. Further research is needed to confirm our findings in a larger sample size with a wider age range, and to test whether the response to therapeutic interventions such as prebiotics, probiotics or fecal microbial transplantation, differ by HIV serostatus and by age.
Funding sources
This work was supported by the National Institute of Aging of the National Institutes of Health [K23AG050260] to KME and T32 AG279-15 to JL, the NCATS Colorado CTSA Grant Number UL1TR002535 to KME and SS, the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [T32 AI007447-25] to SS, the University of Colorado GI and Liver Innate Immune Program (JL, KME, CW, DNF, CER, DI), and the Gilead Sciences Research Scholars Program in HIV (to KME). The funding sources had no a role in data collection, analysis, or interpretation; trial design; or patient recruitment. No payments were made in the writing of this manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Declaration of interests
KME has received consult payments from Gilead Sciences and ViiV (paid to the University of Colorado), and grant funding from Merck.
Author contributions
KME, JL, SS, DF, CW, and MK contributed to the study design. JL, KME, and SS contributed to data collection. RJ and MK led the statistical analysis, with input from JL, CW, and KME. JL, DF, CR, DI, YT, XZ, and BH analyzed samples for the microbiome and/or SCFA outcomes; JH analyzed samples for the dietary outcomes. JL prepared the initial draft with substantial input from SD, CW, and KME. All authors provided input on the manuscript and have reviewed and approved the final manuscript.
Acknowledgements
We gratefully acknowledge all the study participants and study staff.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.033.
Contributor Information
Cara C. Wilson, Email: cara.wilson@ucdenver.edu.
Kristine M. Erlandson, Email: kristine.erlandson@ucdenver.edu.
Appendix A. Supplementary data
Supplementary material
References
- 1.Smit M., Brinkman K., Geerlings S., Smit C., Thyagarajan K., Sighem A. Future challenges for clinical care of an ageing population infected with HIV: a modelling study. Lancet Infect Dis. 2015;15(7):810–818. doi: 10.1016/S1473-3099(15)00056-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erlandson K.M., Allshouse A.A., Jankowski C.M., Lee E.J., Rufner K.M., Palmer B.E. Association of functional impairment with inflammation and immune activation in HIV type 1-infected adults receiving effective antiretroviral therapy. J Infect Dis. 2013;208(2):249–259. doi: 10.1093/infdis/jit147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erlandson K.M., Ng D.K., Jacobson L.P., Margolick J.B., Dobs A.S., Palella F.J., Jr. Inflammation, Immune Activation, Immunosenescence, and Hormonal Biomarkers in the Frailty-Related Phenotype of men with or at risk for HIV Infection. J Infect Dis. 2017;215(2):228–237. doi: 10.1093/infdis/jiw523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolick J.B., Bream J.H., Martinez-Maza O., Lopez J., Li X., Phair J.P. Frailty and circulating markers of inflammation in HIV+ and HIV- men in the multicenter AIDS cohort study. J Acquir Immune Defic Syndr. 2017;74(4):407–417. doi: 10.1097/QAI.0000000000001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franceschi C., Bonafe M., Valensin S., Olivieri F., De Luca M., Ottaviani E. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 6.Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl. 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 7.Puzianowska-Kuznicka M., Owczarz M., Wieczorowska-Tobis K., Nadrowski P., Chudek J., Slusarczyk P. Interleukin-6 and C-reactive protein, successful aging, and mortality: the PolSenior study. Immunity Ageing I & A. 2016;13:21. doi: 10.1186/s12979-016-0076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei J., Xu H., Davies J.L., Hemmings G.P. Increase of plasma IL-6 concentration with age in healthy subjects. Life Sci. 1992;51(25):1953–1956. doi: 10.1016/0024-3205(92)90112-3. [DOI] [PubMed] [Google Scholar]
- 9.Cesari M., Penninx B.W., Newman A.B., Kritchevsky S.B., Nicklas B.J., Sutton-Tyrrell K. Inflammatory markers and cardiovascular disease (the Health, Aging and Body Composition [Health ABC] Study) Am J Cardiol. 2003;92(5):522–528. doi: 10.1016/s0002-9149(03)00718-5. [DOI] [PubMed] [Google Scholar]
- 10.Il'yasova D., Colbert L.H., Harris T.B., Newman A.B., Bauer D.C., Satterfield S. Circulating levels of inflammatory markers and cancer risk in the health aging and body composition cohort. Cancer Epidemiol Biomarkers Prev. 2005;14(10):2413–2418. doi: 10.1158/1055-9965.EPI-05-0316. [DOI] [PubMed] [Google Scholar]
- 11.Deeks S.G., Tracy R., Douek D.C. Systemic effects of inflammation on health during chronic HIV infection. Immunity. 2013;39(4):633–645. doi: 10.1016/j.immuni.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guigoz Y., Dore J., Schiffrin E.J. The inflammatory status of old age can be nurtured from the intestinal environment. Curr Opin Clin Nutr Metab Care. 2008;11(1):13–20. doi: 10.1097/MCO.0b013e3282f2bfdf. [DOI] [PubMed] [Google Scholar]
- 13.Man A.L., Gicheva N., Nicoletti C. The impact of ageing on the intestinal epithelial barrier and immune system. Cell Immunol. 2014;289(1–2):112–118. doi: 10.1016/j.cellimm.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12(12):1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 15.Cao W., Mehraj V., Vyboh K., Li T., Routy J.P. Antiretroviral therapy in primary HIV-1 infection: influences on immune activation and Gut mucosal barrier dysfunction. AIDS Rev. 2015;17(3):135–146. [PubMed] [Google Scholar]
- 16.Hunt P.W., Sinclair E., Rodriguez B., Shive C., Clagett B., Funderburg N. Gut epithelial barrier dysfunction and innate immune activation predict mortality in treated HIV infection. J Infect Dis. 2014;210(8):1228–1238. doi: 10.1093/infdis/jiu238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchetti G., Tincati C., Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26(1):2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Somsouk M., Estes J.D., Deleage C., Dunham R.M., Albright R., Inadomi J.M. Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS. 2015;29(1):43–51. doi: 10.1097/QAD.0000000000000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tugizov S. Human immunodeficiency virus-associated disruption of mucosal barriers and its role in HIV transmission and pathogenesis of HIV/AIDS disease. Tissue Barriers. 2016;4(3) doi: 10.1080/21688370.2016.1159276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollander D., Tarnawski H. Aging-associated increase in intestinal absorption of macromolecules. Gerontology. 1985;31(3):133–137. doi: 10.1159/000212694. [DOI] [PubMed] [Google Scholar]
- 21.Kim K.A., Jeong J.J., Yoo S.Y., Kim D.H. Gut microbiota lipopolysaccharide accelerates inflamm-aging in mice. BMC Microbiol. 2016;16:9. doi: 10.1186/s12866-016-0625-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rera M., Clark R.I., Walker D.W. Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc Natl Acad Sci U S A. 2012;109(52):21528–21533. doi: 10.1073/pnas.1215849110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thevaranjan N., Puchta A., Schulz C., Naidoo A., Szamosi J.C., Verschoor C.P. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(4):455–466. doi: 10.1016/j.chom.2017.03.002. [e4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran L., Greenwood-Van Meerveld B. Age-associated remodeling of the intestinal epithelial barrier. J Gerontol A Biol Sci Med Sci. 2013;68(9):1045–1056. doi: 10.1093/gerona/glt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Man A.L., Bertelli E., Rentini S., Regoli M., Briars G., Marini M. Age-associated modifications of intestinal permeability and innate immunity in human small intestine. Clin Sci (Lond) 2015;129(7):515–527. doi: 10.1042/CS20150046. [DOI] [PubMed] [Google Scholar]
- 26.Steele A.K., Lee E.J., Vestal B., Hecht D., Dong Z., Rapaport E. Contribution of intestinal barrier damage, microbial translocation and HIV-1 infection status to an inflammaging signature. PloS One. 2014;9(5) doi: 10.1371/journal.pone.0097171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buford T.W. (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome. 2017;5(1):80. doi: 10.1186/s40168-017-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dillon S.M., Frank D.N., Wilson C.C. The gut microbiome and HIV-1 pathogenesis: a two-way street. AIDS. 2016;30(18):2737–2751. doi: 10.1097/QAD.0000000000001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Williams B., Frank D., Dillon S.M., Wilson C.C., Landay A.L. Inside out: HIV, the gut microbiome, and the mucosal immune system. J Immunol. 2017;198(2):605–614. doi: 10.4049/jimmunol.1601355. [DOI] [PubMed] [Google Scholar]
- 30.Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PloS One. 2010;5(5) doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y., Zhang F., Zhang R., Shen Y., Liu L., Wang J. Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count. Emerg Microbes Infections. 2018;7(1):113. doi: 10.1038/s41426-018-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McHardy I.H., Li X., Tong M., Ruegger P., Jacobs J., Borneman J. HIV infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monaco C.L., Gootenberg D.B., Zhao G., Handley S.A., Ghebremichael M.S., Lim E.S. Altered virome and bacterial microbiome in human immunodeficiency virus-associated acquired immunodeficiency syndrome. Cell Host Microbe. 2016;19(3):311–322. doi: 10.1016/j.chom.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mutlu E.A., Keshavarzian A., Losurdo J., Swanson G., Siewe B., Forsyth C. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10(2) doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowak P., Troseid M., Avershina E., Barqasho B., Neogi U., Holm K. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29(18):2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 36.Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.Z. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto-Cardoso S., Lozupone C., Briceno O., Alva-Hernandez S., Tellez N., Adriana A. Fecal bacterial communities in treated HIV infected individuals on two antiretroviral regimens. Sci Rep. 2017;7:43741. doi: 10.1038/srep43741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villanueva-Millan M.J., Perez-Matute P., Recio-Fernandez E., Lezana Rosales J.M., Oteo J.A. Differential effects of antiretrovirals on microbial translocation and gut microbiota composition of HIV-infected patients. J Int AIDS Soc. 2017;20(1):21526. doi: 10.7448/IAS.20.1.21526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woodmansey E.J., McMurdo M.E., Macfarlane G.T., Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol. 2004;70(10):6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., Ou Z., Tang X., Zhou Y., Xu H., Wang X. Alterations in the gut microbiota of patients with acquired immune deficiency syndrome. J Cell Mol Med. 2018;22(4):2263–2271. doi: 10.1111/jcmm.13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zwielehner J., Liszt K., Handschur M., Lassl C., Lapin A., Haslberger A.G. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp Gerontol. 2009;44(6–7):440–446. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 43.Lozupone C.A., Li M., Campbell T.B., Flores S.C., Linderman D., Gebert M.J. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14(3):329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noguera-Julian M., Rocafort M., Guillén Y., Rivera J., Casadellà M., Nowak P. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinh D.M., Volpe G.E., Duffalo C., Bhalchandra S., Tai A.K., Kane A.V. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211(1):19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mueller S., Saunier K., Hanisch C., Norin E., Alm L., Midtvedt T. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl Environ Microbiol. 2006;72(2):1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rampelli S., Candela M., Turroni S., Biagi E., Collino S., Franceschi C. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging. 2013;5(12):902–912. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Y., Ma Y., Lin P., Tang Y.W., Yang L., Shen Y. Fecal bacterial microbiome diversity in chronic HIV-infected patients in China. Emerg Microbes Inf. 2016;5 doi: 10.1038/emi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volpe G.E., Ward H., Mwamburi M., Dinh D., Bhalchandra S., Wanke C. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs. 2014;75(2):347–357. doi: 10.15288/jsad.2014.75.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Claesson M.J., Cusack S., O'Sullivan O., Greene-Diniz R., de Weerd H., Flannery E. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl. 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dillon S.M., Lee E.J., Kotter C.V., Austin G.L., Dong Z., Hecht D.K. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7(4):983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vazquez-Castellanos J.F., Serrano-Villar S., Latorre A., Artacho A., Ferrus M.L., Madrid N. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015 Jul;8(4):760–772. doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- 53.Chow J., Tang H., Mazmanian S.K. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23(4):473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zechner E.L. Inflammatory disease caused by intestinal pathobionts. Curr Opin Microbiol. 2017;35:64–69. doi: 10.1016/j.mib.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Cattaneo A., Cattane N., Galluzzi S., Provasi S., Lopizzo N., Festari C. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging. 2017;49:60–68. doi: 10.1016/j.neurobiolaging.2016.08.019. [DOI] [PubMed] [Google Scholar]
- 56.van Tongeren S.P., Slaets J.P., Harmsen H.J., Welling G.W. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71(10):6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson M.A., Jeffery I.B., Beaumont M., Bell J.T., Clark A.G., Ley R.E. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8(1):8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ticinesi A., Milani C., Lauretani F., Nouvenne A., Mancabelli L., Lugli G.A. Gut microbiota composition is associated with polypharmacy in elderly hospitalized patients. Sci Rep. 2017;7(1):11102. doi: 10.1038/s41598-017-10734-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res. 2013;54(9):2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Levy M., Thaiss C.A., Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev. 2016;30(14):1589–1597. doi: 10.1101/gad.284091.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ploger S., Stumpff F., Penner G.B., Schulzke J.D., Gabel G., Martens H. Microbial butyrate and its role for barrier function in the gastrointestinal tract. Ann N Y Acad Sci. 2012;1258:52–59. doi: 10.1111/j.1749-6632.2012.06553.x. [DOI] [PubMed] [Google Scholar]
- 62.Vinolo M.A., Rodrigues H.G., Nachbar R.T., Curi R. Regulation of inflammation by short chain fatty acids. Nutrients. 2011;3(10):858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Louis P., Flint H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294(1):1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 64.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19(1):29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 65.Louis P., Hold G.L., Flint H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 66.Dillon S.M., Kibbie J., Lee E.J., Guo K., Santiago M.L., Austin G.L. Low abundance of colonic butyrate-producing bacteria in HIV infection is associated with microbial translocation and immune activation. AIDS. 2017;31(4):511–521. doi: 10.1097/QAD.0000000000001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hippe B., Zwielehner J., Liszt K., Lassl C., Unger F., Haslberger A.G. Quantification of butyryl CoA:acetate CoA-transferase genes reveals different butyrate production capacity in individuals according to diet and age. FEMS Microbiol Lett. 2011;316(2):130–135. doi: 10.1111/j.1574-6968.2010.02197.x. [DOI] [PubMed] [Google Scholar]
- 68.Claesson M.J., Jeffery I.B., Conde S., Power S.E., O'Connor E.M., Cusack S. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 69.Haran J.P., Bucci V., Dutta P., Ward D., McCormick B. The nursing home elder microbiome stability and associations with age, frailty, nutrition and physical location. J Med Microbiol. 2018;67(1):40–51. doi: 10.1099/jmm.0.000640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Erlandson K.M., MaWhinney S., Wilson M., Gross L., McCandless S.A., Campbell T.B. Physical function improvements with moderate or high-intensity exercise among older adults with or without HIV infection. AIDS. 2018;32(16):2317–2326. doi: 10.1097/QAD.0000000000001984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brumbaugh D.E., Arruda J., Robbins K., Ir D., Santorico S.A., Robertson C.E. Mode of delivery determines neonatal pharyngeal bacterial composition and early intestinal colonization. J Pediatr Gastroenterol Nutr. 2016;63(3):320–328. doi: 10.1097/MPG.0000000000001124. [DOI] [PubMed] [Google Scholar]
- 72.Kuhn K.A., Schulz H.M., Regner E.H., Severs E.L., Hendrickson J.D., Mehta G. Bacteroidales recruit IL-6-producing intraepithelial lymphocytes in the colon to promote barrier integrity. Mucosal Immunol. 2018;11(2):357–368. doi: 10.1038/mi.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(Pt 1):257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 74.Nycz B.T., Dominguez S.R., Friedman D., Hilden J.M., Ir D., Robertson C.E. Evaluation of bloodstream infections, Clostridium difficile infections, and gut microbiota in pediatric oncology patients. PloS One. 2018;13(1) doi: 10.1371/journal.pone.0191232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandrea I., Xu C., Stock J.L., Frank D.N., Ma D., Policicchio B.B. Antibiotic and antiinflammatory therapy transiently reduces inflammation and hypercoagulation in acutely SIV-infected pigtailed Macaques. PLoS Pathog. 2016;12(1) doi: 10.1371/journal.ppat.1005384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frank D.N. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformat. 2009;10:362. doi: 10.1186/1471-2105-10-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pruesse E., Peplies J., Glockner F.O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28(14):1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bishehsari F., Engen P.A., Preite N.Z., Tuncil Y.E., Naqib A., Shaikh M. Dietary fiber treatment corrects the composition of gut microbiota, promotes SCFA production, and suppresses colon carcinogenesis. Genes (Basel) 2018;9(2) doi: 10.3390/genes9020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kriska A., El Ghormli L., Copeland K.C., Higgins J., Ievers-Landis C.E., Levitt Katz L.E. Impact of lifestyle behavior change on glycemic control in youth with type 2 diabetes. Pediatr Diabetes. 2018;19(1):36–44. doi: 10.1111/pedi.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Robertson C.E., Harris J.K., Wagner B.D., Granger D., Browne K., Tatem B. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29(23):3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oksanen J KR, Legendre P, O'Hara B, Simpson GL, Solymos P, Stevens MHH, Wagner H. Vegan: Community Ecology Package. R package version 1.15–1. http://cranr-projectorg/, http://veganr-forger-projectorg [Internet]. 2008.
- 83.Venables W.R.B. 4. Springer; New York: 2002. Modern Applied Statistics with S. [Google Scholar]
- 84.Warnes G BB, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Macehler M, Magnusson A, Moeler S, Schwartz M and Venables B. gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1. https://CRANR-projectorg/package=gplots [Internet]. 2016.
- 85.Zevin A.S., McKinnon L., Burgener A., Klatt N.R. Microbial translocation and microbiome dysbiosis in HIV-associated immune activation. Curr Opin HIV AIDS. 2016;11(2):182–190. doi: 10.1097/COH.0000000000000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gagniere J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E. Gut microbiota imbalance and colorectal cancer. World J Gastroenterol: WJG. 2016;22(2):501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zeng M.Y., Inohara N., Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10(1):18–26. doi: 10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang W., Wu N., Wang X., Chi Y., Zhang Y., Qiu X. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Round J.L., Mazmanian S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci U S A. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu M., Koh H., Kurtz Z.D., Battaglia T., PeBenito A., Li H. Oxalobacter formigenes-associated host features and microbial community structures examined using the American Gut Project. Microbiome. 2017;5(1):108. doi: 10.1186/s40168-017-0316-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stewart C.S., Duncan S.H., Cave D.R. Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol Lett. 2004;230(1):1–7. doi: 10.1016/S0378-1097(03)00864-4. [DOI] [PubMed] [Google Scholar]
- 93.Groeger D., O'Mahony L., Murphy E.F., Bourke J.F., Dinan T.G., Kiely B. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes. 2013;4(4):325–339. doi: 10.4161/gmic.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Makki K., Deehan E.C., Walter J., Backhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–715. doi: 10.1016/j.chom.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 95.Pinto-Cardoso S., Klatt N.R., Reyes-Teran G. Impact of antiretroviral drugs on the microbiome: unknown answers to important questions. Curr Opin HIV AIDS. 2018;13(1):53–60. doi: 10.1097/COH.0000000000000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lozupone C.A., Rhodes M.E., Neff C.P., Fontenot A.P., Campbell T.B., Palmer B.E. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5(4):562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 97.Rios-Covian D., Ruas-Madiedo P., Margolles A., Gueimonde M., de Los Reyes-Gavilan C.G., Salazar N. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Larsen J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology. 2017;151(4):363–374. doi: 10.1111/imm.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rossi O., van Berkel L.A., Chain F., Tanweer Khan M., Taverne N., Sokol H. Faecalibacterium prausnitzii A2-165 has a high capacity to induce IL-10 in human and murine dendritic cells and modulates T cell responses. Sci Rep. 2016;6:18507. doi: 10.1038/srep18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L.G., Gratadoux J.J. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferreira-Halder C.V., Faria A.V.S., Andrade S.S. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. 2017;31(6):643–648. doi: 10.1016/j.bpg.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 102.Miquel S., Martin R., Rossi O., Bermudez-Humaran L.G., Chatel J.M., Sokol H. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16(3):255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 103.Lefebure T., Stanhope M.J. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8(5):R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Petersen L., Bollback J.P., Dimmic M., Hubisz M., Nielsen R. Genes under positive selection in Escherichia coli. Genome Res. 2007;17(9):1336–1343. doi: 10.1101/gr.6254707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoshizaki S., Umemura T., Tanaka K., Watanabe K., Hayashi M., Muto Y. Genome-wide evidence of positive selection in Bacteroides fragilis. Comput Biol Chem. 2014;52:43–50. doi: 10.1016/j.compbiolchem.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 106.Kelley C.F., Kraft C.S., de Man T.J., Duphare C., Lee H.W., Yang J. The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 2017;10(4):996–1007. doi: 10.1038/mi.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Neff C.P., Krueger O., Xiong K., Arif S., Nusbacher N., Schneider J.M. Fecal Microbiota Composition Drives Immune Activation in HIV-infected individuals. EBioMedicine. 2018;30:192–202. doi: 10.1016/j.ebiom.2018.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material