Abstract
Background
Tumor cells benefit from tumor-associated macrophages (TAMs) promoting tumor growth and modulating functions of other cells in tumor microenvironment (TME). However, how tumor cells regulate the property of TAMs during tumor invasion remains to be defined.
Methods
Mouse tumor models and cancer patients' samples were analyzed to determine LAMP2a expression in TAMs. In vitro mouse primary macrophages were used to assess LAMP2a-modulated macrophage activation, and to verify LAMP2a's target proteins. The effect of LAMP2a-knockdown on tumor progression and TME maintaining was determined by using mouse tumor models.
Findings
Lysosome associated membrane protein type 2A (LAMP2a) is upregulated in TAMs by tumor cells and important for tumor progression. LAMP2a expression in TAMs, but not in tumor cells, is associated with poor prognosis in breast cancer. LAMP2a inactivation induced by either shRNA or CRISPR/Cas9 prevents TAMs activation and tumor growth. LAMP2a degrades PRDX1 (peroxiredoxin 1) and CRTC1 (CREB-regulated transcription coactivator 1) to promote macrophage pro-tumorigenic activation.
Interpretation
Our study suggests that tumor cells utilize LAMP2a-PRDX1/CRTC1 axis to modulate TAMs activation and promote tumor growth, reveals the role of LAMP2a in macrophage study and TAM-targeting tumor immunotherapy.
Fund
National Natural Science Foundation of China (No. 81602492); National Key Research and Development Program of China (No. 2016YFA0201402).
Keywords: LAMP2a, Tumor-associated macrophage, Chaperone-mediated autophagy, Macrophage activation, Tumor microenvironment
Research in context.
Evidence before this study
Tumor-associated macrophage (TAM) serves as a predominant immune cell type in tumor microenvironment (TME) and induces a wide variety of immunosuppressive function in most cancer types. Also, TAMs amounts are positively correlated with poor prognosis in many solid tumor types. LAMP2a mostly locates in lysosome membrane and contributes essentially to chaperone-mediated autophagy (CMA). In CMA process, LAMP2a binds to its specific substrate proteins and transports them into lysosome to “selectively” remove target proteins, leaving neighboring proteins unperturbed. Current studies about LAMP2a in cancers mostly focus its role in tumor cells.
Added value of this study
In this study we demonstrated that LAMP2a modulates macrophage activation and function. LAMP2a-inhibition alters immunosuppressive macrophages activation and suppresses tumor growth. We also found LAMP2a degrades PRDX1 (peroxiredoxin 1) and CRTC1 (CREB-regulated transcription coactivator 1) to trigger macrophage pro-tumorigenic activation.
Implications of all the available evidence
Our findings suggest LAMP2a as a potential candidate in TAM-targeting tumor immunotherapy, and partly illustrate the molecular mechanism of TAMs' multi-functional and bi-directional activation.
Alt-text: Unlabelled Box
1. Introduction
Macrophages are main component of immune cell in tumor microenvironment (TME), and immunosuppressive tumor associated-macrophages (TAMs) frequently aggregate in tumor lesions even at early stage [[1], [2], [3]]. Recent findings indicated TAMs derive from dual origins. Firstly, through cytokines and chemokines, like CCL2 or CSF-1, whether released from tumor cells, immune cells and stromal cells, can recruit abundance of monocytic precursors into TME [[4], [5], [6], [7]]. Secondly, TAMs can also maintain their population through self-proliferation in some tumor models [8,9]. As a complex inflammatory milieu, TME can not only make circulating precursors differentiate into anti-tumorigenic macrophage in the beginning [10,11], but also “re-educate” them subsequently towards immunosuppressive phenotype to favor tumor growth, like promoting angiogenesis, producing growth factor and increasing metastasis [[12], [13], [14], [15], [16]]. TAMs population is heterogeneous even in same tumor lesion, such as TAMs from the core or the periphery can exhibit different phenotypes [17]. Evidence shows that extreme M1/M2 classification do not fit TAMs in complex multistage immune response in tumor process, and TAMs represent unique phenotypes in different tumors [[18], [19], [20], [21]].
High numbers of TAMs infiltration is correlated with poor prognosis in various solid tumor types [22,23]. Therefore, considering TAMs as accessory cells of tumor lesions, they are promising target to restore the anti-tumorigenic immune response [[24], [25], [26]]. To date, the main strategies to target macrophage are recruit inhibition and polarization revision, and there have been several approaches of TAM targeting under evaluation in clinical trials [27]. Like CSF-1/CSF-1R axis [[28], [29], [30]] and PI3kγ inhibitor [31,32]. Another therapeutic strategy is to enhance phagocytosis and immune-stimulatory of TAMs, like CD47/SIRPα antibody and class II HDAC inhibitor [33,34]. These TAM-targeting therapies can reshape tumor immune microenvironment, promote CD8+ cytotoxic-T-cell activation, and when combined with other treatments can significantly benefit overall survival. A recent study revealed that TAMs also express PD-1, and further confirmed PD-1 inhibition in TAMs reduces tumor growth [35].
A major challenge for developing TAM-targeted therapies is to investigate the molecular mechanisms of TAMs activation and function. In this study, we reveal LAMP2a (lysosome-associated membrane protein type 2A) as a novel regulator for TAMs activation. LAMP2a locates in the lysosomal membrane, and plays essential role in chaperone-mediated autophagy (CMA) [36]. The main difference between CMA and other types of autophagy, like macro- and micro-autophagy, is to selectively target and degrade specific substrate proteins, without affecting organelles or neighboring proteins [37]. LAMP2a binds to target proteins in co-operated with its chaperonin, HSC70, and then forms a homomultimer to transport target proteins into lysosome [38]. Previous researches about LAMP2a-concerned physiology or diseases focus on aging [39], renal hypertrophy [40], neurodegeneration like Parkinson's disease [41,42] and Alzheimer's disease [43,44]. Recently, there are reports revealing LAMP2a's pro-tumorigenic role in tumor cells [45,46]. Beside, LAMP2a also contributes to MHC-II function in antigen presenting cells [47] and T cell activation [48]. These findings introduce LAMP2a into cancer and immunology fields.
In this study, we set out to describe LAMP2a function in TAMs, assess its potential in TAM-targeting therapy, and investigate the molecular mechanism under its effects.
2. Materials and methods
2.1. Cell lines and primary cells culture
RAW264.7 was cultured in DMEM (Dulbecco's Modified Eagle Medium). CT26, 4 T1 and mouse primary cells were cultured in RPMI-1640. All mediums were supplemented with 10% FBS, penicillin (100 mg/mL) and streptomycin (100 mg/mL). Sorted mouse hematopoietic cells were cultured in SFEM (STEMCELL, 09600) containing 10% FBS at 37 °C humidified chamber with 7.5% CO2. All cell lines were obtained from ATCC (American Tissue Type Culture Collection) and conducted following guidelines of SKLB (State/National Key Laboratory of Biotherapy), Sichuan University.
2.2. Mice
All mice were housed in the Animal Facility at State/National Key laboratory of Biotherapy, Sichuan University, under standard pathogen-free conditions. All animal experiments were approved by the institutional Animal Care and Use Committee.
For subcutaneous tumor models, female Balb/c mice age 6–8 weeks (obtained from Beijing Vital River Laboratory) were subcutaneously injected in the right upper abdomen with 1 × 106 4T1 or CT26 cells suspended in 100 μL normal saline per mouse. After tumorigenesis, mice that carried approximately same tumor volumes were randomly grouped and tail intravenously (i.v.) or intratumorally (i.t.) injected with 2 OD GHOSTs loading with shRNA targeting LAMP2a (sh-L2a) or non-coding vectors (sh-NC) suspended in 100 μL normal saline, with same volume normal saline (NS) as control, respectively. The treatments were performed every three days for four times in total.
For spontaneous breast cancer studies, MMTV-PyMT (PyMT) mice were obtained from Jackson Laboratory (Stock No: 002374). All experimental mice were born approximately at same time and grouped randomly. To ensure a same tumor growth state among all experimental groups, treatments were initiated about four days after tumorigenesis (palpable tumors appearing) and performed by tail intravenously injected with 2 OD GHOSTs loading with sh-L2a or sh-NC suspended in 100 μL normal saline, with same volume normal saline (NS) as control, every three days.
2.3. Clinical samples
All patient samples concerned studies were approved by West China Hospital IRB committee. All human subjects (except for tissue microarray) were obtained from patients who were receiving treatment in West China Hospital, with all patients fully informed consent.
2.4. Antibodies, critical commercial assays and oligonucleotides sequence
See Supplementary Tables 1 and 2.
2.5. Immunohistochemistry and immunofluorescence
For immunofluorescence staining of OCT frozen tumor tissue sections, serum and catalase blocking was performed by goat serum (ZSGB-Bio Company) and 0.3% H2O2 respectively. Then sections were incubated with anti-LAMP2a (Abcam ab18528), anti-F4/80 (Abcam ab16911), anti-human CD68 (Biolegend 333805), anti-human CD163 (Biolegend 333605), and corresponding secondary antibodies (Invitrogen, Abcam). After washing, DAPI (Beyotime Biotechnology) was stained and coverslips were mounted by Pro-Long Gold antifade reagent (Invitrogen). The staining sections were imaged by Leica TCS SP5 II microscope.
For staining of cell sliders, mouse primary cells were seeded in well-plates with glass slides placed in advance. Exfoliated cells from patients or sorted mouse cells from FACS were spotted in slides by Thermo Scientific Cytospin 4 Cytocentrifuge. The immunofluorescence staining procedures were consistent with tumor tissue sections above. The Giemsa staining was performed by commercial assay (BASO BA4007), imaged by Leica DM2500 microscope.
For TUNEL assays, the formalin treated mouse tumor tissues were sectioned by paraffin embedding. And the staining procedures followed manufacturer's protocol (Promega G7130), with images obtained by Leica DM2500 microscope.
2.6. Flow cytometry
For samples from mouse tumors, tumor tissues about 2 g per samples were cut up and digested in 10 mL RPMI-1640 medium containing 10 mg collagenase (Gibco) and 5 μg/mL DNase I (Sigma) in 37 °C with periodic vortex and inversion for 2 h. For tumor microenvironment analyses, the suspensions further flew through 70 μm cell strainers, and the filtrated cells were resuspended in 30% and 70% Percoll (Sigma, P1644) gradient and centrifugated to isolate immune cells. Before antibodies labelling, cells were incubated with live/dead staining dye (Invitrogen, L34974) and FcR blocking (Invitrogen, 31205) for 15 min in 4 °C. Then cells were stained by antibodies following per corresponding protocol. For intracellular staining, the surface antigens-labeled cells were fixed and permeabilized either by 4% paraformaldehyde/1% Triton X-100 or intracellular staining buffers (eBioscience, 00-5523), and subsequent staining was performed following specific antibody protocols. Samples were analyzed or sorted by using BD FACSAria III, and data was analyzed by FlowJo V10 (FlowJo, LLC).
For cultured cell sample preparation, CT26, RAW264.7, and mouse primary macrophages were dissociated by trypsin (Gibco), mechanical manners, and Cell Dissociation Buffer (Gibco, 13151014) respectively. The staining procedures were similar to description above.
For other mouse tissues sample, like peripheral blood and peritoneal lavage fluids, harvested cells were stained following red blood cell lysis. Spleens were mechanically dissociated by grinding through a 70 μm cell strainer. Livers and lungs were digested following mechanical disruption. These cells were stained as described above after red blood cell lysis.
2.7. Tissue microarray analyses
The breast cancer and matched adjacent tissues microarray was obtained from Shanghai Outdo Biotech (National Engineering Center for Biochip at Shanghai). The specimens were stained by anti-LAMP2a (Abcam, ab125068) and H&E respectively. For analyses of staining outcomes, two observers who were blinded to patients' information, independently scored and grouped these specimens by LAMP2a expression in tumor cells and stromal cells.
2.8. Tumor cells-supernatant medium preparation
CT26 or 4 T1 cells were cultured in complete RPMI-1640 medium at 100 mm dishes, and when cell density reached about 80%, the medium was replaced by new medium. After 24 h, new conditioned medium was harvested, centrifuged in 2000 rpm for 5 min, filtrated by 0·22 μm filters, and restored at −80 °C in aliquots.
2.9. Western blot
For total cellular protein extraction, the harvested cells were washed twice with cold PBS buffer, and lysed in RIPA buffer (Radio Immunoprecipitation Assay buffer, Beyotime, P0013B) containing protease inhibitor cocktail (Millipore). The protein concentrations were determined by Bradford dye (BIO-RAD, 5000205) in Eppendorf Bio-photometer Plus (Eppendorf). Afterwards, the protein extracts were separated in NuPAGE Bis-Tris gels (Invitrogen), and transferred to PVDF (polyvinylidene difluoride) membranes (Millipore). The membranes were probed by specific antibodies.
2.10. Mouse primary cells extraction and stimulation
For mouse bone marrow cells extraction, the bilateral tibias, femurs and hip bones were detached in sterile condition, with bone marrow flushed out by cold normal saline containing 5% FBS. After centrifuging, total bone marrow cells were resuspended by ACK lysis buffer in volume of 5 mL/per mouse, and incubated on ice for 10 min. After a washing, the bone marrow cells were cultured in complete RPMI-1640 medium containing 20 ng/mL M-CSF (Novoprotein, C756) for 48 h. Afterwards, the suspension cells were removed by washing and medium replacement, and the remaining adherent cells were stimulated according to specific experiments. For tumor-supernatant (TS) treatment, TS conditioned medium was added into cultured medium in about 1/3 volume, for 3 to 5 days. For macrophage classical and alternative activation, 20 ng/mL INF-γ (PeproTech) + 100 ng/mL LPS (Sigma) or 20 ng/mL IL-4 (PeproTech) were added in medium, respectively, with incubation for 3 days. For bafilomycin A1 treatment, 10 nM final concentration of bafilomycin (InvivoGen) was added with TS in cultured medium.
For mouse peritoneal cells extraction, the mice were humanely sacrificed and injected intraperitoneally with 10 mL cold normal saline containing 5% FBS. After a gentle massage on abdomen, the ascites was extracted carefully to avoid wounding the organs. The cells suspension should be clear without blood or intestinal contents. After centrifuging, the peritoneal cells were cultured in complete RPMI-1640 medium for 2 h to allow the adherence of macrophages, following suspension cells removal.
2.11. RT-PCR (qPCR)
Total RNA was extracted by using RNA Extraction Kit (TIANGEN BIOTECH, DP419), and reverse transcribed to cDNA by using Prime Script RT Kit (Takara, RR036), according to per manufacturer's protocol. qPCR was performed by using SYBR Select Master Mix (Invitrogen, 4472908), with specific gene primers, in StepOnePlus PCR System (Thermo). Each sample was run in triplicate, and target genes expression levels were normalized to β-actin and analyzed by using ∆∆Ct method.
2.12. shRNA knockdown of LAMP2a
The specific sequences targeting the regions in mouse Lamp2a exon 9 were designed following previous studies [45,48,49], and three parallel clones were synthesized. All these sequences were respectively constructed into shRNA vector pENTR/U6 (Invitrogen), with a non-coding vector (sh-NC) as control. Afterwards, these shRNA vectors were loaded in GHOSTs to perform LAMP2a knockdown.
2.13. RNA sequencing
For RNA samples preparation, TS-primed mouse BMDMs were treated by sh-NC, sh-L2a or not, with three biological duplicates for each condition. Before RNA extraction, cells were lysed in TRIzol reagent and stored at −80 °C. The integrity and concentration of RNA extracts was determined by Agilent 2100 Bioanalyzer and RNA Nano 6000 Assay Kit (Agilent Technologies), and RNA integrity numbers ranged between 8·3 and 9·7. To prepare RNA-seq library, total RNA was purified by oligo (dT) beads and fragmented, followed by synthesis of first and second strand, 3′ ends adenylation and adapter ligation. Afterwards, samples were amplified by PCR subsequently to gel extraction. Libraries were analyzed on Illumina HiSeq 2500 (Illumina) following PE150 sequencing strategy.
2.14. CRISPR/Cas9-mediated deletion in mouse hematopoietic stem cells (HSCs)
The oligo sequences for guide RNA targeting Lamp2a, Prdx1 and Crtc1 were designed by DNA 2·0, with three to five candidates of highest scores obtained. After the synthesis of these oligonucleotides, they were respectively constructed into 12-2 CRISPR vector followed by lentiviral transduction to test work efficiency. Next, the cassettes with workable sgRNAs were transferred into a retroviral CRISPR vector which contains GFP expression cassettes. In multiple-CRISPR experiments, the guide RNAs either targeted Lamp2a, Prdx1 and Crtc1 were conjoined into three combinations as sg-L+P, sg-L+C, sg-L+P+C, and transferred into CRISPR vector respectively. All vectors used in CRISPR/Cas9 experiments were generously provided by Prof. Chong Chen.
For detection of protein level of LAMP2a, PRDX1, CRTC1 and mRNA expression, the genetically modified mouse HSCs were treated by M-CSF (20 ng/mL) and TS to enable macrophage differentiation and activation.
2.15. Mouse HSCs transplantation
The HSCs from FVB mice bone marrow were isolated by EasySep Mouse Hematopoietic Cell Isolation Kit (STEMCELL, 19856) following manufacturer's protocol. After transfection by retrovirus that loading with sg-L2a or sg-SCRAMBLE (sg-SCR) control vectors, the injection amounts were determined by GFP and living cell properties measured by flow cytometry.
Before HSCs transplantation, the recipient PyMT mice with 7–8 weeks age were irradiated with 5 Gy. To minimize the irradiation effect on tumor formation and exclude the mice failed in tumorigenesis, the irradiation was performed after palpable tumors appeared. Two hours after irradiation, sg-L2a or sg-SCR transfected HSCs (2 × 106 cells/mouse) were injected by tail vein. Afterwards, the recipient mice were fed in standard condition with monitoring for tumor progress.
2.16. Immunoprecipitation and mass spectrometry
The proteins samples used for immunoprecipitation (IP) were extracted from mouse BMDMs treated by tumor-supernatant (TS) alone or with bafilomycin (TS + Bafilo). Antibody immobilization was performed by incubating anti-LAMP2a (Hangzhou HUAAN Biotechnology, ET1601–24) with Dynabeads Streptavidin magnetic beads (Invitrogen, 65801D) in PBS at 4 °C for 4 h. After separating the antibody-coated beads by a magnetic rack (Bio-Rad) and 4–5 times washing, the coated beads were resuspended with protein extracts at 4 °C with continuous inversion for 8 h. Next, the IP products were separated and washed in a magnetic rack, with magnetic beads releasing by incubating in 0·1% SDS at 95 °C for 10 min and magnetic separation. The final products without beads were quantified by Bradford dye and analyzed by Western blot or mass spectrometry.
For mass spectrometry, the samples were subjected into NuPAGE Bis-Tris gels, followed by Coomassie Blue staining. Then gels were de-stained and cut into slices for subsequent reduction, alkylation and trypsin digestion. The extracted peptides were analyzed in Q Exactive Plus mass spectrometer (Thermo) and identified by database on Uniprot following standard procedures.
2.17. Protein affinity measurements
The affinities of LAMP2a binding to PRDX1, CRTC1 and IRG1 were measured by Surface Plasmon Resonance (SPR) in Biacore T200 (GE Healthcare). LAMP2a was immobilized on Sensor Chip CM5, while PRDX1, CRTC1 and IRG1 were double diluted to concentrations ranging from 7·8125 nM to 1000 nM, flowed through the chip. The dissociation constants (KDs) were fitted by Biacore T200 Evaluation Software.
2.18. Cell products analyses
Nitric oxide (NO) and lactate productions were detected in cell culture supernatant. Mouse BMDMs were cultured by normal medium, TS alone, TS + sh-NC or TS + sh-L2a respectively, and by IFN-γ + LPS or IL-4 as control. After stimulation, the medium supernatant was collected and filtrated. The products were detected by NO Griess Reagent System (Promega, G2903) and Lactate Colorimetric Assay Kit (BioVision, K607), following per manufacturer's protocol. For ROS detection, mouse BMDMs were cultured in 96-well plates with same treatments as described above, and measured for total ROS production by using ROS-Glo H2O2 Assay (Promega, G8820). All the outcomes were analyzed by luminescence determination in a luminometer.
2.19. Cytotoxicity assay
The macrophages' tumor cytotoxicity assay was performed by referring Current Protocols in Immunology [50,51]. To determine in vitro macrophages tumor cytotoxicity, LDH release was detected in supernatant medium from tumor cells co-cultured with genetically modified mouse HSC-derived macrophages. Before co-culture, genetically modified HSCs were counted and planted at required number (4 × 104, 2 × 104, 104, 0.5 × 104 cells/well, as E:T = 40:1, 20:1, 10:1, 5:1) in 96-well plates, cultured in complete RPMI-1640 containing 20 ng/mL M-CSF for 4 days to ensure macrophage differentiation. Then these HSCs-derived macrophages were washed to remove suspending cells and co-cultured with 4 T1, CT26 or LL/2 cells at 1000 cells/well respectively. After 12 h and 24 h, the supernatant medium was transferred to new 96-well plates to determine cytotoxicity as LDH release by using CytoTox 96 Non-Radioactive Cytotoxicity Assay Kit (Promega). The percentage of macrophages tumor cytotoxicity was calculated according manufacturer's instruction as
The cell viability of these co-cultured cells was measured by Alamar Blue (Thermo Fisher), after four days of co-culture, followed by manufacturer's protocol.
2.20. Statistical analysis
All statistical analyses were performed by Graph Pad Prism 7 and 8. All data are shown as mean ± SD. A two-tailed Student's t-test was used to analyze difference between two groups, and one-way ANOVA was used for multiple groups comparison. Log-rank (Mantel-Cox) test was used for survival curves analyses. The statistical tests used are also stated in the Figure legends. The statistic differences are shown as * p < 0·05, ** p < 0·01, *** p < 0·001, ns, no significance, and p < 0·05 was considered as statistic significant.
3. Results
3.1. LAMP2a is upregulated in TAMs and indicates poor prognosis in breast cancer
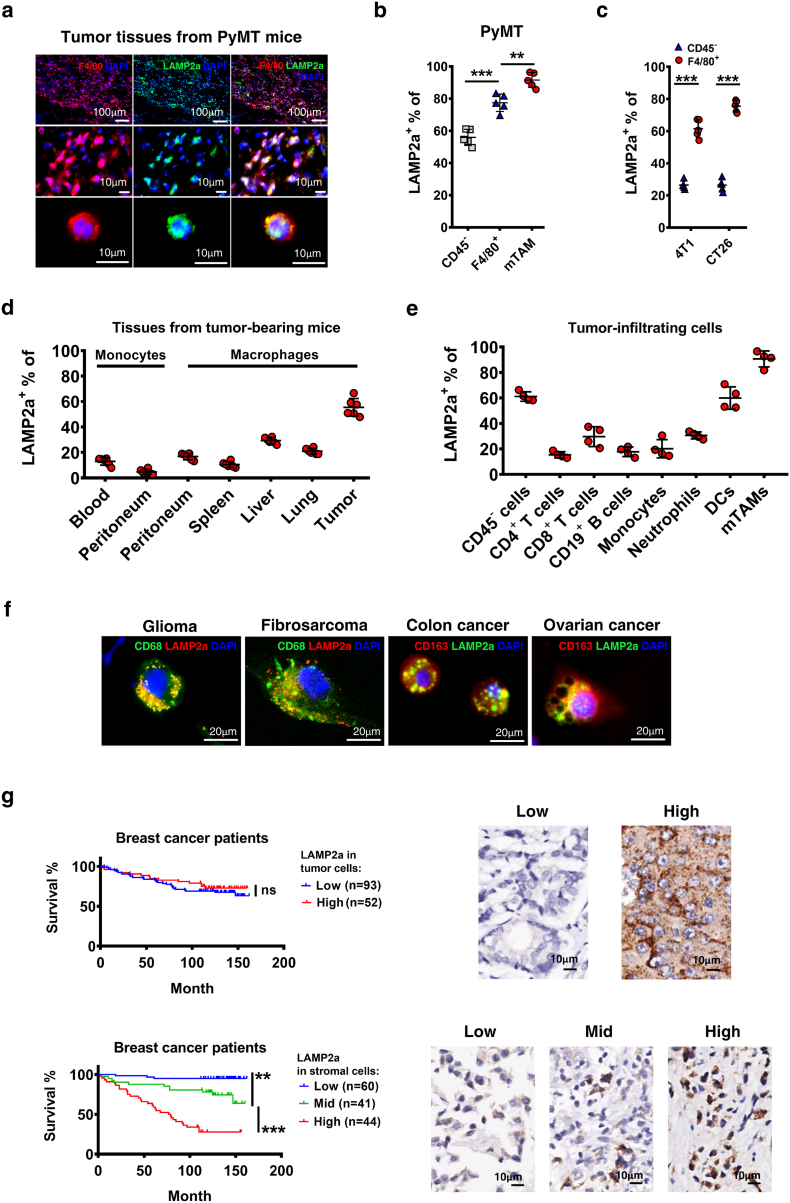
LAMP2a mostly locates in lysosome membrane and assembles when chaperone-mediated autophagy occurs, and usually remains at a low level in non-autophagic cells [52]. Current studies about LAMP2a in cancers mostly focus on its expression in tumor cells [45,46], here we tried to illustrate its role in macrophages. To validate LAMP2a activation in TAMs, LAMP2a expression was detected in F4/80+ cells in tumor tissues from MMTV-PyMT (PyMT) mice (Fig. 1a). Then, LAMP2a expression was measured by flow cytometry in tumor cells (CD45−), tumor-infiltrating macrophages (TIMs, CD11b+ F4/80+), and a specific TAM subpopulation (MHC-IIhi CD11blo) defined by Ruth A. Franklin and colleagues in PyMT mouse model [9]. To distinguish this TAMs subpopulation from generalized TIMs, we named it as mTAM (mammary-TAM) in this paper. The results showed that LAMP2a was extensively expressed in TIMs and mTAMs (Fig. 1b, Supplementary Fig. S1a and b). Although tumor cells represent most of tumor mass, in our study we found their average LAMP2a level was not comparable with TAMs'. To confirm this, two more mouse subcutaneous inoculation tumor models of 4 T1 (mouse breast cancer cell line) and CT26 (mouse colon cancer cell line) were detected for LAMP2a expression in TAMs. Consistent with PyMT mice, TAMs (CD11b+ F4/80+) in 4 T1 and CT26 inoculated mice also broadly expressed LAMP2a (Fig. 1c). Next, we tried to determine whether macrophages are the predominant source for LAMP2a expression in tumor stroma. LAMP2a expression was assessed in macrophages and monocytes from different tissues of tumor-bearing PyMT mice, and we found that TAMs expressed higher level of LAMP2a than other tissue macrophages (Fig. 1d and Supplementary Fig. S1a). Additionally, within tumor milieu mTAMs still expressed highest level of LAMP2a than other tumor-infiltrating immune cells (Fig. 1e and Supplementary Fig. S1b).
Fig. 1.
LAMP2a is upregulated in TAMs and correlates with overall survive in breast cancer.
(a) Representative immunofluorescence images of tumor sections from MMTV-PyMT mice, labeled by F4/80 (red), LAMP2a (green) and DAPI (blue). Scale bars are marked in individual images.
(b) LAMP2a expression in tumor cells (CD45−), TIMs (F4/80+) and mTAMs was detected by flow cytometry in PyMT mice. n = 5/group.
(c) LAMP2a expression in tumor cells (CD45−) and TAMs (F4/80+) was detected in 4 T1 and CT26 inoculation mouse models by flow cytometry. n = 5/group.
(d) and (e) LAMP2a expression in tissue macrophages (d) and tumor stroma cells (e) was detected by flow cytometry in PyMT mice. n = 4–6/group.
(f) Representative immunofluorescence images of tumor tissues or exfoliated cells from clinical samples. Sections were labeled with CD68 (green), LAMP2a (red), DAPI (blue) or CD163 (red), LAMP2a (green), DAPI (blue) as marked in images. Scale bar, 20 μm.
(g) Survival curves of breast cancer patients grouped by LAMP2a expression either in tumor cells (upper panel) or stromal cells (bottom panel), analyzed by Log-rank (Mantel-Cox) test. Immunohistochemistry images with labeled LAMP2a were representative regions of LAMP2a expression in tumor cells (upper panel) and stromal cells (bottom panel).
Data are represented as mean ± SD, with Student's t-tests unless noted. *p < 0·05, **p < 0·01, ***p < 0·001. Data represent 2–3 independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To examine whether cancer patients' TAMs also express LAMP2a, we detected patient tumor tissues or exfoliated cells from different cancer types for LAMP2a expression. In these samples, LAMP2a was also broadly expressed in macrophages (Fig. 1f and Supplementary Fig. S1c). Considering that previous studies about LAMP2a in cancer mainly focused on its expression in tumor cells, in which LAMP2a was regarded as an oncoprotein [45,46], we wondered whether TAMs-expressed LAMP2a might correlate with cancer prognosis. To explore this, tumor tissues microarray of breast cancer patients was labeled with anti-LAMP2a, and all 145 patients were sorted and graded by LAMP2a expression in tumor cells or in stromal cells that are mainly consisted of macrophages [3]. The results showed that, patients carried high or low amounts of LAMP2a+ tumor cells had barely difference in survival time (Fig. 1g upper panel), that is to say, tumor cells-expressed LAMP2a might not be a noteworthy prognostic variable for breast cancer, which has already been demonstrated [53]. However, if these samples were re-graded by LAMP2a expression in stromal cells, we found that elevated amounts of LAMP2a+ stromal cells infiltration implicated significantly shorter survival time of patients (Fig. 1g bottom panel). To further confirm this, these two variables that LAMP2a expression in tumor cells/stromal cells were combined to assess survival time difference. The results revealed that stromal cells-expressed LAMP2a positively correlated worse breast cancer prognosis in both high/low LAMP2a+ tumor cells groups (Supplementary Fig. S1d). In contrast, significant correlation between tumor cells-expressed LAMP2a and worse prognosis only presented in LAMP2amid stromal cells group (Supplementary Fig. S1e).
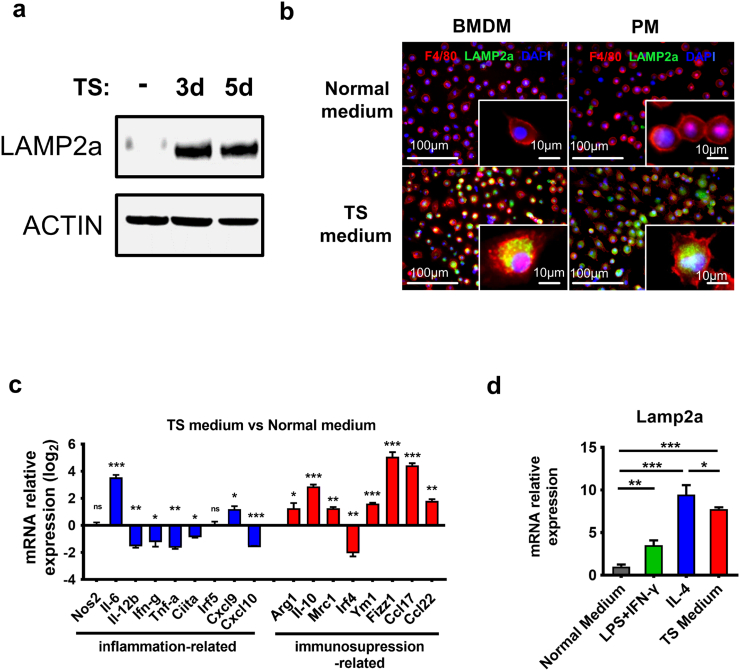
3.2. Tumor-supernatant upregulates LAMP2a expression and promotes macrophages activation
As the key factor of chaperone-mediated autophagy (CMA), LAMP2a responds to various stressors in TME, especially to autophagy-inducing stressors, including growth and inflammatory factors, tumor cells metabolites, hypoxia and nutrient deprivation [54,55], which also involve in TAMs activation. To mimic tumor cells' in vivo effect on macrophages, we utilized conditioned medium containing tumor cells-supernatant (TS) to stimulate mouse primary macrophages in vitro. The results showed that TS effectively elevated LAMP2a expression in mouse BMDMs (bone marrow-derived macrophages), and three days of TS-stimulation was enough for LAMP2a upregulation (Fig. 2a). Immunofluorescent staining further confirmed that TS activated LAMP2a in both mouse BMDMs and peritoneal macrophages (PMs) (Fig. 2b). Considering that BMDMs are the predominant replenishment for mammary gland macrophages and particularly for breast cancer TAMs [9,20], we focused on BMDMs in subsequent in vitro experiments. Next, we measured a series of genes that involved in inflammatory or immunosuppressive macrophages polarization in TS-stimulated BMDMs. The results exhibited a general amplification of immunosuppression-related genes expression and a concomitant reduction of inflammation-related genes expression in TS-stimulated BMDMs compared with normal medium treatment (Fig. 2c). These results suggested a positive correlation between LAMP2a expression and immunosuppressive phenotype of macrophages. To further validate this, LAMP2a expression in TS-stimulated BMDMs was compared with BMDMs treated by LPS+IFN-γ or IL-4, and we found that TS and IL-4 both induced higher level of LAMP2a than LPS+IFN-γ (Fig. 2d and Supplementary Fig. S2a). Interestingly, although LAMP2a expression in IL-4-primed BMDMs resembled that in TS-stimulated BMDMs, Giemsa staining revealed a unique multi-vacuolar cytoplasm in TS-stimulated BMDMs (Supplementary Fig. S2b). Coincidentally, this multi-vacuolar cytoplasm has been also observed in TAMs by other researchers [35]. Besides, surface markers of these BMDMs were also different (Supplementary Fig. S2c). Therefore, although IL-4 was a stronger inducer than TS in LAMP2a activation, we utilized TS medium for its ability to stimulate phenotypical heterogeneity of macrophages and mimic tumor cells' effect in vitro.
Fig. 2.
Tumor-supernatant (TS) induces LAMP2a expression and macrophage activation.
(a) Mouse BMDMs (bone marrow-derived macrophages) were cultured with TS conditioned medium for three days (lane 2) and five days (lane 3), with normal medium as control (lane 1), followed by LAMP2a protein level detection.
(b) Immunofluorescence staining of F4/80 (red), LAMP2a (green) and DAPI (blue) was performed in mouse BMDMs and PMs (peritoneal macrophages), which were treated by normal medium or TS conditioned medium for three days. Representative images, scale bars are marked in individual images.
(c) mRNA expression of genes related to inflammatory (blue) or immunosuppressive (red) macrophage activation was detected by qPCR in TS-primed BMDMs, data were normalized to normal medium treatment, and represented as log2 scale, with β-actin as control.
(d)Lamp2a mRNA expression was detected in mouse BMDMs cultured with normal medium or LPS + IFN-γ, IL-4, TS conditioned medium respectively. Data were normalized to normal medium treatment (first column), with β-actin as control.
Data are represented as mean ± SD, with one-way ANOVA tests. *p < 0·05, **p < 0·01, ***p < 0·001. Data represent 2–4 independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
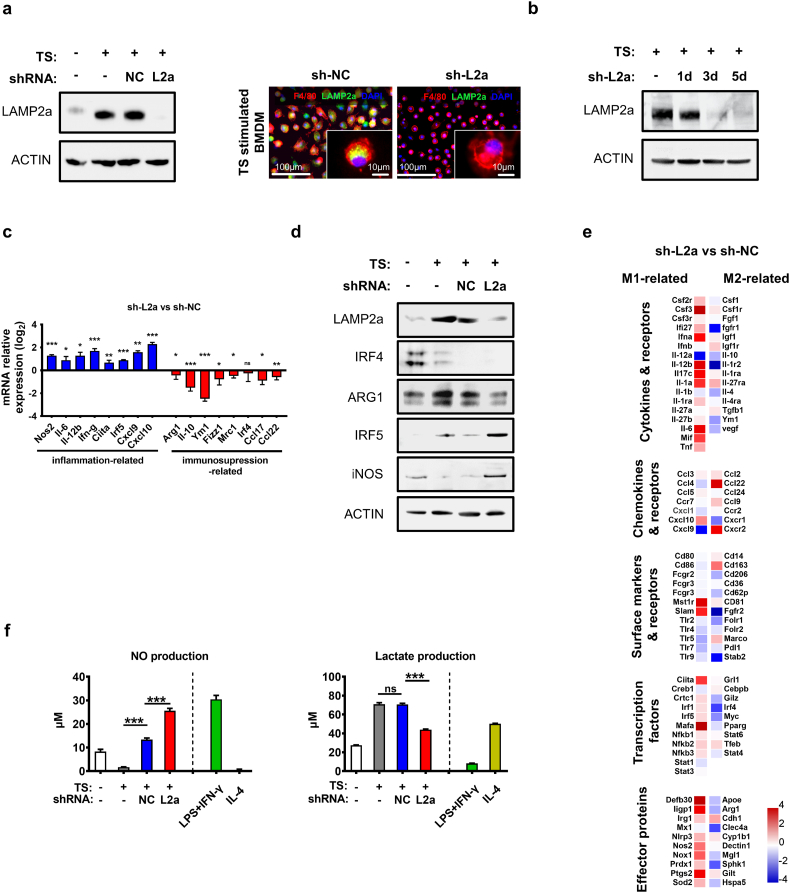
3.3. LAMP2a knockdown in vitro alters macrophages activation
To further assess LAMP2a's role in macrophage activation, we aimed to interfere with LAMP2a expression in macrophages. The Lamp2a mRNA-targeting shRNA (short hairpin RNA) design and selection was referred to previous studies [45,48,49]. Considering that tumor cells and other immune cells also express LAMP2a, bacterial GHOSTs were used as shRNA-encoding plasmids vehicles to specifically and effectively target macrophages [56,57]. It is reported that LAMP2a has a basic expression on lysosome membrane, which would exhaust in three to five days after RNA interference [58], and TS-stimulation also needs at least three days to reach a sufficient effect, mouse BMDMs were transfected by GHOSTs loading-with three different Lamp2a mRNA-targeting shRNA (sh-L2a c1, c2 and c3) followed by an immediate TS-stimulation for three days. And the results showed that all three shRNA clones reduced LAMP2a protein level in these BMDMs (Supplementary Fig. S3a). Next, to exclude the possible nonspecific effects from GHOSTs or prokaryotic plasmids on LAMP2a expression, the non-targeting shRNA vector was also loaded into GHOSTs (sh-NC) as a control group throughout all shRNA-involved experiments. Although sh-NC induced a modest decrease of LAMP2a mRNA level, which might due to the inflammatory activation of macrophages induced by bacterial GHOSTs' immunogenicity, sh-L2a treatment was still significantly comparable with sh-NC (Supplementary Fig. S3b), and sh-NC treatment did not affect LAMP2a protein level (Fig. 3a). And as we assumed, the pre-existing LAMP2a in macrophages required at least three days for significant reduction of LAMP2a protein level after sh-L2a transfection (Fig. 3b). These interference effects of sh-L2a were also observed in IL-4-stimulated BMDMs (Supplementary Fig. S3c).
Fig. 3.
Macrophage activation is reversed by LAMP2a inhibition.
(a) Left panel: LAMP2a protein level was detected in mouse BMDMs cultured with normal medium or TS for three days prior to shRNA transfection. Right panel: Immunofluorescence staining of F4/80 (red), LAMP2a (green) and DAPI (blue) in TS-stimulated BMDMs. Representative images. Scale bars are marked in individual images.
(b) TS-stimulated BMDMs were treated by sh-L2a for one day, three days or five days respectively, with LAMP2a protein level detection. The TS medium was replaced by normal medium when TS treatment was already done for three days but cells were not prepared for protein detection.
(c) mRNA expression of genes related to inflammatory (blue) or immunosuppressive (red) macrophage activation was detected by qPCR in TS-primed BMDMs which were subsequently treated by sh-NC, sh-L2a or not. Data were normalized to sh-NC treatment, and represented as log2 scale, with β-actin as control.
(d) Protein level of IRF4, ARG1, IRF5 and iNOS was detected in normal medium- or TS-cultured BMDMs which were then transfected by shRNA or not.
(e) Heatmap of relative mRNA expression in TS-primed BMDMs which were transfected by sh-NC, sh-L2a or not. 110 macrophage activation-related genes were picked from total 17,983 genes analyzed by RNA sequencing. Data were normalized to sh-NC treatment, represented as log2 scale, and grouped by functional features.
(f) Nitric oxide (NO) and Lactate production was measured in BMDMs which were transfected by shRNA after TS or normal medium culture, with LPS + IFN-γ- or IL-4-primed BMDMs as controls.
Data are represented as mean ± SD, with one-way ANOVA tests. *p < 0·05, **p < 0·01, ***p < 0·001, ns, no significance. Data represent 2–3 independent experiments except for RNA-seq. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Next, to examine the effects of LAMP2a knockdown on macrophages activation, TS-stimulated BMDMs were transfected by sh-NC, sh-L2a or not, and detected for polarization-related genes expression as in Fig. 2c. The results showed that LAMP2a knockdown in TS-stimulated BMDMs enhanced inflammation-related genes expression and suppressed immunosuppression-related genes expression (Fig. 3c). These changes indicated that LAMP2a-inhibition might reverse TS-primed immunosuppressive macrophages activation. To further confirm these results, we detected IRF4, IRF5, ARG1 and iNOS protein levels in those BMDMs, and found that LAMP2a knockdown elevated IRF5 and iNOS protein level, with a decrease in IRF4 and ARG1 level, although IRF4 was not upregulated by TS treatment (Fig. 3d). Meanwhile, RNA-seq was performed in these BMDMs to test a broader range of genes related to macrophage activation. Consistently, LAMP2a knockdown in TS-stimulated BMDMs leaded to a global upregulation of inflammatory-related genes expression and downregulation of immunosuppression-related genes expression (Fig. 3e and Supplementary Fig. S3d). Next, these BMDMs were detected for extracellular secretions of nitric oxide (NO) and lactate, which respectively indicate inflammatory and immunosuppressive macrophage activation. The outcomes from both NO and lactate productions consistently revealed a re-programming of TS-stimulated BMDMs in consequence of LAMP2a knockdown (Fig. 3f). Similar to Lamp2a mRNA results in Fig. S3b, although there was an obvious raise of NO production in sh-NC treatment compared with TS-stimulation, the increase in sh-L2a treatment was more intense and closer to LPS + IFN-γ treatment. Additionally, to explore whether LAMP2a knockdown in BMDMs would impact the lysosomal biogenesis and function, we detected TFEB (transcription factor EB) expression, which also activates LAMP2a expression [36], in LAMP2a-inactivating BMDMs. And the results showed that LAMP2a knockdown had no comparable effect on TS-elevated TFEB expression (Supplementary Fig. S3e).
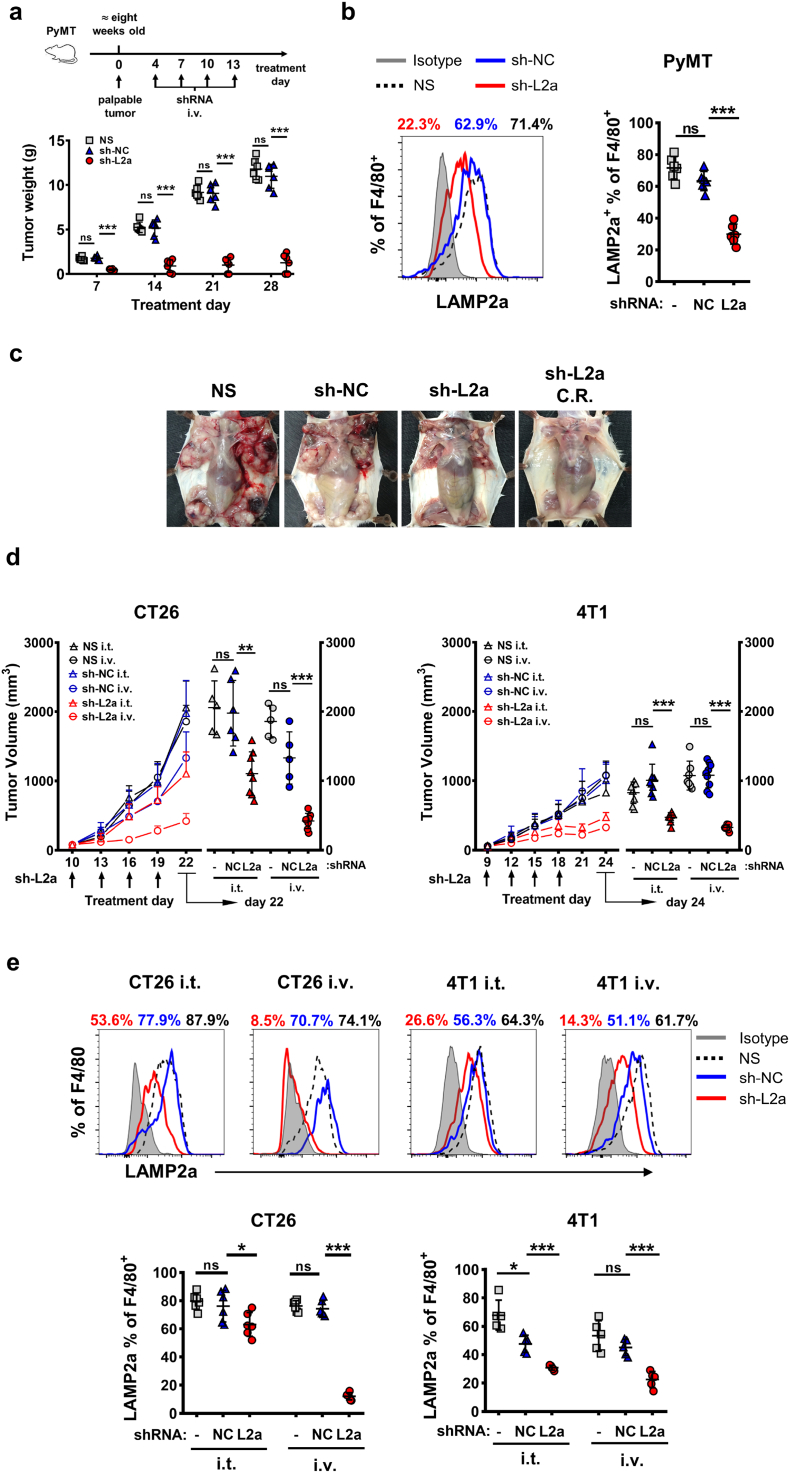
3.4. Inhibiting LAMP2a in TAMs represses tumor growth in vivo
Next, we tried to examine whether LAMP2a knockdown in TAMs in vivo would impact tumor progression. Tumor-bearing PyMT mice were injected intravenously with GHOSTs loading with sh-NC or sh-L2a, or normal saline (NS) with same volume as control. To exclude the mice failed in tumorigenesis, the treatments started at least four days after the tumors becoming palpable and reaching an approximate same diameter among all experimental mice. As a result, sh-L2a treatment exhibited a comparable tumor suppression effect since early time points, while sh-NC treatment showed no obvious effects compared with NS (Fig. 4a). Additionally, flow cytometric results confirmed that sh-L2a treatment effectively decreased LAMP2a expression in TAMs compared with sh-NC and NS treatment (Fig. 4b). And the decrease of LAMP2a expression did not present in other tumor-infiltrating immune cells nor tumor cells (Supplementary Fig. S4a). Notably, at end time points of treatment, about one-third sh-L2a-receiving mice showed complete response (Fig. 4c).
Fig. 4.
LAMP2a inhibition in TAMs suppresses tumor progression.
(a) PyMT mice were tail intravenously injected with NS or bacterial GHOSTs loading with sh-NC/sh-L2a every three days for up to four times total, since at least four days after palpable tumors appearing. At each time points, mice were euthanized to weigh tumors. n = 4–6/group.
(b) LAMP2a expression was analyzed by flow cytometry in TAMs from PyMT mice treated by NS, sh-NC or sh-L2a. n = 4–6/group.
(c) Representative images of PyMT mice which received NS, sh-NC or sh-L2a treatment. C.R. refers to complete response.
(d) Tumor growth curves and scatters of CT26 and 4T1 subcutaneous inoculation mice. Mice were tail intravenously (i.v.) or intratumorally (i.t.) injected with NS or bacterial GHOSTs loading with sh-NC/sh-L2a. Tumor volumes were measured by digital calipers, calculated by V = (longer diameter) × (shorter diameter)2/2. n = 5–8/group.
(e) LAMP2a expression was analyzed in TAMs from CT26 and 4 T1 subcutaneous inoculation mice, which received NS, sh-NC or sh-L2a transfection. n = 5–8/group.
Data are represented as mean ± SD, with one-way ANOVA tests. *p < 0·05, **p < 0·01, ***p < 0·001, ns, no significance. Data represent 2–4 independent experiments.
Next, we tested this LAMP2a knockdown effect in mouse CT26 and 4 T1 subcutaneous inoculation models. For there is only one tumor node in subcutaneous inoculation model, additional intratumor injection (i.t.) groups were added to compare with intravenous injection (i.v.). The results showed that in both CT26 and 4 T1 models, sh-L2a i.v. treatment exhibited obviously comparable tumor suppression with NS and sh-NC treatment, on tumor volume and weight, while sh-L2a i.t. treatment only worked effectively in 4 T1 model (Fig. 4d and Supplementary Fig. S4b). LAMP2a expression in TAMs of these two models showed similar trends (Fig. 4e). The slight decrease of LAMP2a expression in sh-L2a i.t. group might due to the weak fluidity of subcutaneous solid tumors for GHOSTs transfection.
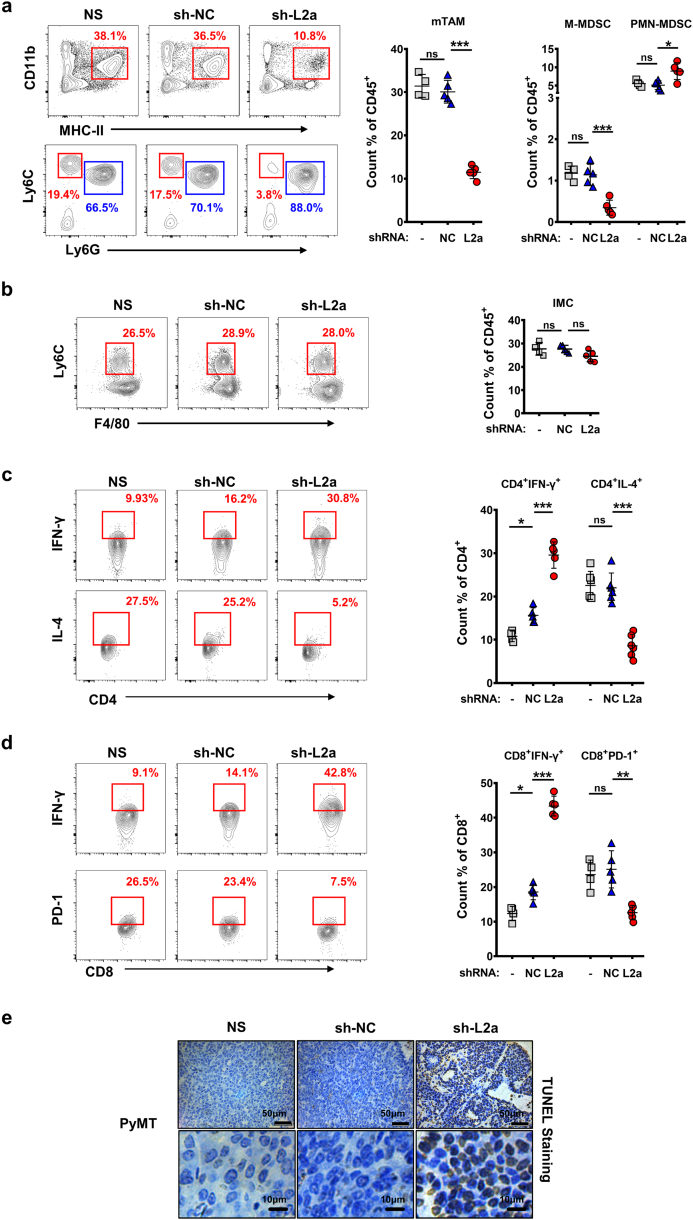
3.5. LAMP2a knockdown in TAMs restores tumor immune microenvironment
Since we have found LAMP2a knockdown alters macrophage activation in vitro, we next investigated whether LAMP2a knockdown in TAMs in vivo would modulate tumor immune microenvironment (TIME). Firstly, we analyzed the population of mTAMs in PyMT mice, which is positively associated with tumor burden [9]. Consistent with tumor suppression, mTAM population in sh-L2a-receiving mice suffered a significant decrease, while sh-NC treatment showed no considerable effect (Fig. 5a). Next, tumor-infiltrating MDSCs (myeloid-derived suppressor cells) population was also measured. Given that although polymorphonuclear MDSCs (PMN-MDSCs) represents the major population of MDSC, monocytic MDSCs (M-MDSCs) is more immunosuppressive [59], we assessed both of them (Supplementary Fig. S5a). The results showed that M-MDSCs population significantly declined in sh-L2a-receiving mice compared with NS or sh-NC treatment, while PMN-MDSCs population exhibited inconsequential changes (Fig. 5a). Additionally, a reduction of M-MDSC/PMN-MDSC ratio was also observed in consequence of LAMP2a knockdown (Supplementary Fig. S5b), which indicated the descending immunosuppression of intratumoral MDSCs [59]. To examine whether the decrease of mTAMs and MDSCs population came from restrained recruiting or infiltrating of monocytic precursors, we further measured the population of tumor-infiltrating inflammatory monocytes (IMCs) (Supplementary Fig. S5c), which was recognized as monocytic precursor of TAMs [9,11], and total populations of tumor-infiltrating F4/80+ cells and Ly6C+ cells were also assessed. Interestingly, LAMP2a knockdown had no comparable effects on the population of intratumoral IMCs, F4/80+ cells or Ly6C+ cells (Fig. 5b and Supplementary Fig. S5d). Besides, LAMP2a knockdown in vitro in mouse BMDMs did not induce observable changes in cell viability, death or density (Supplementary Fig. S5e and S5f). Next, the mTAMs population was isolated by using flow cytometry from PyMT mice which were treated by NS, sh-NC or sh-L2a. Afterwards, these sorted TAMs were analyzed for genes expression. The results further showed that sh-L2a treatment in vivo partly neutralized these TAMs' immunosuppression function, although these changes were not comparable with in vitro treatment (Supplementary Fig. S5 g). These results suggested that LAMP2a knockdown in vivo did not cause amount losses of TAMs, but remodeled them from tumor-promoting phenotypes into undefined phenotypes, whose markers spectrum is still elusive and might be characterized in future.
Fig. 5.
Tumor immuno-environment is restored by LAMP2a inhibition.
(a) and (b) Cell population of mTAMs, MDSCs (a) and IMCs (inflammatory monocytes) (b) from PyMT mice. Flow cytometric graphs were representative results, red and blue gates in MDSC graphs indicate M-MDSC and PMN-MDSC respectively. n = 4–6/group.
(c) and (d) Cell population of CD4+ IFN-γ+, CD4+ IL-4+(c) and CD8+ IFN-γ+, CD8+ PD-1+ T cells (d) from PyMT mice. Flow cytometric graphs were representative results. n = 4–6/group.
(e) TUNEL staining of tumor tissue sections from PyMT mice which received NS, sh-NC or sh-L2a transfection. Scale bars are marked in individual images.
Data are represented as mean ± SD, with one-way ANOVA tests. *p < 0·05, **p < 0·01, ***p < 0·001, ns, no significance. Data represent 2–3 independent experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In addition to immunosuppressive myeloid cells, effector T cell non-responsiveness is also an essential factor for TIME and largely induced by TAMs and M-MDSCs in tumor milieu [59]. So we next assessed tumor-infiltrating T cell response in consequence of LAMP2a knockdown in TAMs. As a result, sh-L2a-receiving mice exhibited significantly elevated proportions of CD4+ IFN-γ+ T cells and decreased CD4+ IL-4+ T cells (Fig. 5c), which indicated Th1-dominant responses. Meanwhile, CD8+ IFN-γ+ T cells population was also expanded in sh-L2a treatment, with reduced CD8+ PD-1+ T cells population (Fig. 5d). However, the proportions of tumor-infiltrating CD4+ or CD8+ cells showed no considerable differences, as well as CD4+ Foxp3+, CD4+ PD-1+ or CD8+ GZMB+ T cells (Supplementary Fig. S5h and i). In summary, we found that LAMP2a knockdown in TAMs restored TIME, enhanced effector T cell responses, and leaded tumor cells death (Fig. 5e).
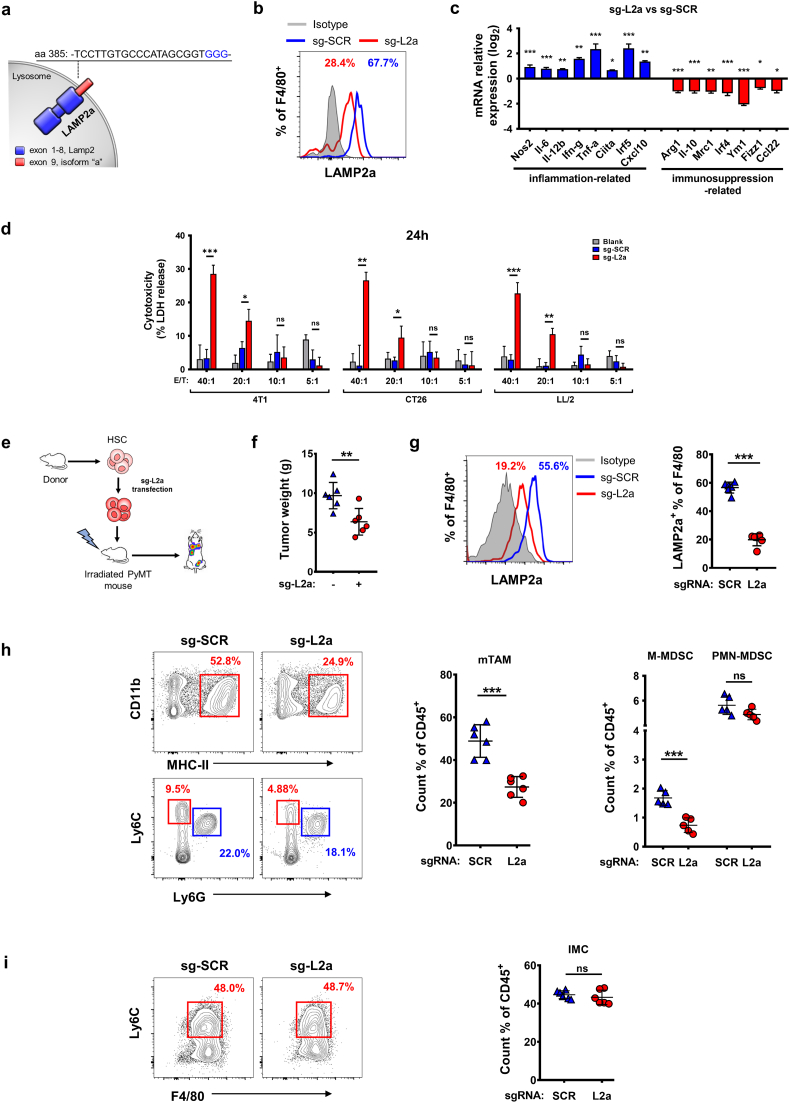
3.6. CRISPR/Cas9-induced LAMP2a-inactivation induces macrophage tumor cytotoxicity enhancement and in vivo tumor suppression
Next, we utilized CRISPR/Cas9 to induce LAMP2a inactivation in macrophages to complement shRNA-involved experiments. To ensure the maximum functional losses of LAMP2a, without effecting its whole structure, the CRISPR-targeted sequences were restricted in the last exon (exon 9, isoform Lamp2a specificity) of Lamp2a gene (Fig. 6a and Supplementary Fig. S6a), for the cytoplasmic tail of LAMP2a encoded by exon 9 is essential for LAMP2a specifically binding to its substrate proteins [52]. Then, isolated mouse HSCs were transfected by retrovirus carrying non-specific scrambled CRISPR vector (sg-SCR) or Lamp2a-targeting CRISPR vector (sg-L2a), and measured for their subsequent differentiation in vitro (Supplementary Fig. S6b). In the meantime, we utilized M-CSF and TS to induce these genetically modified HSCs to differentiate into macrophages and express LAMP2a. As a result, sg-L2a-treated HSCs-derived macrophages showed obviously decreased protein level of LAMP2a compared with sg-SCR transfection (Fig. 6b). Then, these genetically modified HSCs-derived macrophages were further analyzed for phenotypic differences. Similar to shRNA experiments, sg-L2a-treated HSCs-derived macrophages showed elevated inflammation-related genes expression and decreased immunosuppression-related genes expression (Fig. 6c). Next, we set out to determine whether CRISPR/Cas9-induced LAMP2a-inactivation would affect tumor cytotoxicity of these macrophages. Firstly, a wild type HSCs group was added as control to ensure that these HSCs' differentiation into macrophages was not impacted (Supplementary Fig. S6c). Afterwards, 4 T1, CT26 and LL/2 cells were co-cultured with these wild type or genetically modified HSCs-derived macrophages respectively, with gradient ratios of E/T referring to previous studies [50,51]. At 12 h and 24 h after co-culture, LDH release was measured to calculate tumor cytolysis. As a result, sg-L2a treatment exhibited significantly comparable enhanced tumor cytotoxicity compared with sg-SRC or wild type group at high ratios (Fig. 6d and Supplementary Fig. S6d).
Fig. 6.
LAMP2a-inactivation in HSCs-derived macrophage alters macrophage phenotypes and represses tumor progression.
(a) Schematic diagram describing the design of mouse Lamp2a-targeting CRISPR guideRNAs. PAM sites are shown in blue.
(b) LAMP2a expression in sg-SCR or sg-L2a transfected HSCs-derived macrophages.
(c) mRNA expression of genes related to inflammatory (blue) or immunosuppressive (red) macrophage activation was detected by qPCR in sg-SCR or sg-L2a transfected HSCs-derived macrophages. Results were represented as log2 scale, with β-actin as control.
(d) LDH release from 4T1, CT26, LL/2 cells after 24 h co-culture with gradient ratios of sg-SCR or sg-L2a transfected HSCs-derived macrophages. The percentage of cytotoxicity was calculated by maximum tumor cells LDH release control. Data were analyzed with one-way ANOVA tests.
(e) The procedures of LAMP2a-inactivating HSCs chimera establishment.
(f) and (g) Tumor weights (f) and LAMP2a expression in TAMs (g) of LAMP2a-inactivating or wild type bone marrow chimera PyMT mice.
(h) and (i) Cell population of mTAMs, MDSCs (h) and IMCs (i) from LAMP2a-inactivating or wild type bone marrow chimera PyMT mice.
Data are represented as mean ± SD, with Student's t-tests unless noted. *p < 0·05, **p < 0·01, ***p < 0·001, ns, no significance. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To assess the tumor suppression effect of these LAMP2a-inactivating macrophages in vivo, PyMT mice with initial palpable tumors were irradiated by X-ray and then tail intravenously injected with sg-SCR or sg-L2a transfected HSCs (Fig. 6e). The results showed that sg-L2a treatment had obviously suppression effects on tumor growth and LAMP2a expression in TAMs compared with sg-SCR (Fig. 6f and g). The populations of mTAMs, intratumoral MDSCs and IMCs were also detected in these chimera mice. Consistent with shRNA experiments, the populations of mTAMs and M-MDSCs decreased significantly with a concomitant reduction of M-MDSC/PMN-MDSC ratio in sg-L2a group (Fig. 6h and Supplementary Fig. S6e). While the tumor-infiltrating IMCs population showed negligible differences (Fig. 6i).
3.7. PRDX1 and CRTC1 are critical for LAMP2a-modulated macrophages activation and tumor promotion
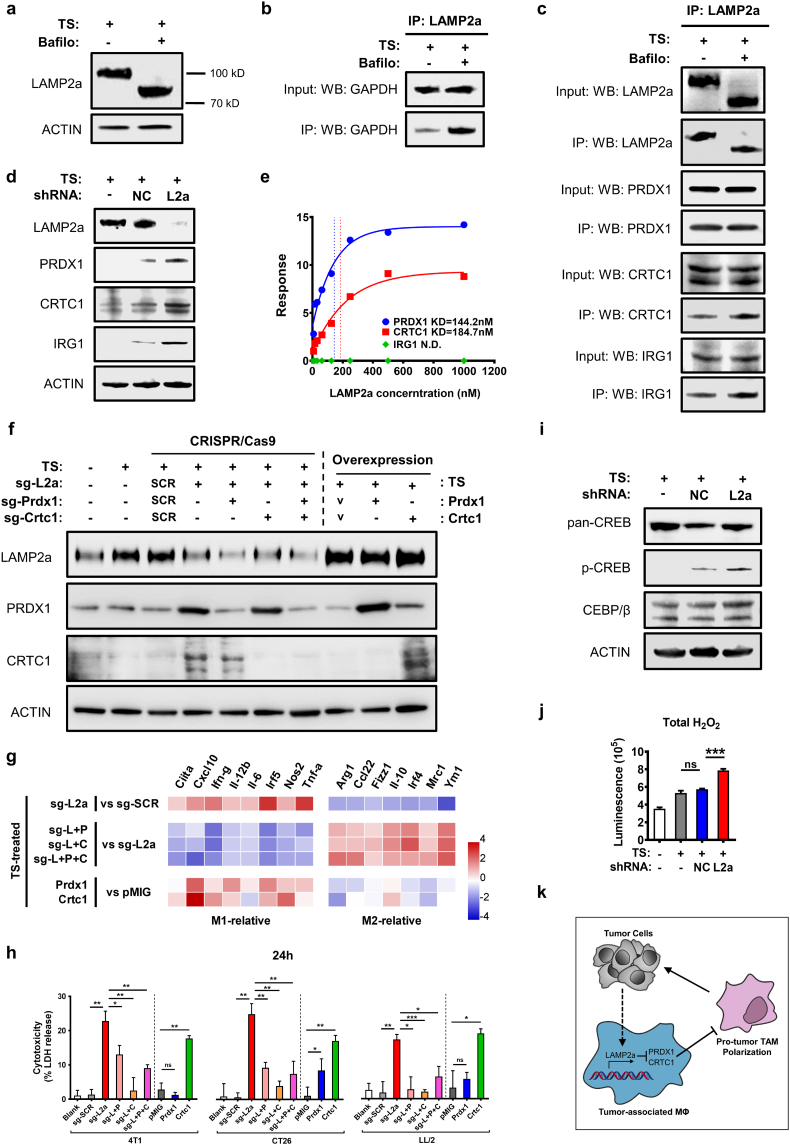
To investigate the underlying mechanisms of LAMP2a-modulated macrophage activation, we attempted to identify LAMP2a's substrate proteins in macrophages. The vacuolar-type H+ ATPase inhibitor bafilomycin A1 was utilized to prevent the lysosomal degradation of target proteins binding to LAMP2a [60]. Interestingly, although bafilomycin treatment in TS-stimulated BMDMs did not affect LAMP2a protein level, which is consistent with previous studies [60], but the molecular weight of LAMP2a was decreased from about 100 kD to 80 kD (Fig. 7a). Considering that the antibodies used in this study specifically binds to LAMP2a's unique peptides (aa 380–410), we speculated that LAMP2a's specific domain, as well as its function, was unperturbed. To validate this, we detected the protein level of LAMP2a-binding GAPDH, which was reported as a substrate protein of LAMP2a [61], in TS-stimulated BMDMs treated by bafilomycin (Bafilo +) or not (Bafilo -). As a result, in Bafilo + macrophages, GAPDH still bound to LAMP2a and obviously accumulated compared with Bafilo - (Fig. 7b). These results suggested that bafilomycin treatment did not impact LAMP2a binding to its substrates in macrophages. The decreased molecular weight of LAMP2a might come from the altered protein modification, for the predicted MW of LAMP2a is only 55kD. Next, all LAMP2a-binding proteins were collected in Bafilo +/− BMDMs, in which Bafilo - group served as a control to exclude the possibility that bafilomycin-induced lysosomal inactivation might increase non-specific binding of LAMP2a. Then, these substrate proteins were analyzed by mass spectrometry and western blotting simultaneously. As a result, three most probable substrate proteins were identified, as PRDX1 (peroxiredoxin 1), CRTC1 (CREB-regulated transcription coactivator 1) and IRG1 (immune-responsive gene 1 protein) (Fig. 7c). In parallel WB experiments, PRDX1, CRTC1 and IRG1 all showed increased protein level in consequence of LAMP2a knockdown (Fig. 7d). To exclude the indirect binding, PRDX1, CRTC1, IRG1 and LAMP2a were synthesized and assessed for their respective binding kinetics in Biacore system. As a result, the KD (dissociation constant) values of PRDX1 and CRTC1 indicated their direct binding to LAMP2a, even in low concentrations, while IRG1 failed in KD fitting (Fig. 7e and Supplementary Fig. S7a).
Fig. 7.
LAMP2a targets PRDX1 and CRTC1 to modulate macrophages activation.
(a) LAMP2a expression in TS-stimulated BMDMs which were treated by bafilomycin (Bafilo) or not.
(b) Protein level of GPADH binding to LAMP2a in TS-stimulated BMDMs treated by bafilomycin (Bafilo) or not.
(c) LAMP2a, PRDX1, CRTC1 and IRG1 expression in co-IP experiments of TS-stimulated BMDMs treated by bafilomycin (Bafilo) or not. All the IP-proteins were immunoprecipitated by anti-LAMP2a antibody and detected by respective antibodies.
(d) Protein level of PRDX1, CRTC1 and IRG1 in TS-stimulated BMDMs which were transfected by sh-NC, sh-L2a or not.
(e) The surface plasmon resonances (SPR) of LAMP2a-bound PRDX1, CRTC1 and IRG proteins. The results were fitted as dissociation constants (KDs).
(f) Protein level of LAMP2a, PRDX1 and CRTC1 in TS-stimulated, genetically modified mouse HSCs-derived macrophages. “SCR” represents sg-SRC vector and “V” represents overexpression control vector.
(g) Heatmap of relative mRNA expression of macrophage activation-related genes in mouse HSCs-derived macrophages described as (f). “pMIG” stands for overexpression control vector. Data were measured by qPCR, normalized to corresponding groups as noted, represented as log2 scale, with β-actin as control.
(h) LDH release of 4 T1, CT26, LL/2 cells after 24 h co-culture with genetically modified HSCs-derived macrophages at a ratio of 40:1 as E:T. The percentage of cytotoxicity was calculated by maximum tumor cells LDH release control.
(i) CREB and CEBP/β expression in TS-stimulated BMDMs treated by sh-NC, sh-L2a or not. Total CREB protein was shown as pan-CREB, and S133-phosphorylated CREB was shown as p-CREB.
(j) Luminescence assays of H2O2 production in BMDMs which were transfected by shRNA after TS or normal medium culture.
(k) Illustration of mechanism for LAMP2a-PRDX1/CRTC1 axis.
Next, to determine whether PRDX1 and CRTC1 is responsible for LAMP2a-modulated macrophage activation, PRDX1 and CRTC1 was further ablated in mouse HSCs by CRISPR/Cas9 (Supplementary Fig. S7b). Then, the sequences encoding Prdx1- or Crtc1-targeting sgRNAs were respectively or both transduced into Lamp2a-targeting CRISPR vector, to obtain three combined targeting vectors as sg-L + P, sg-L + C and sg-L + P + C (L: Lamp2a, P: Prdx1, C: Crtc1). Additionally, the expression plasmids of PRDX1 and CRTC1 were also constructed. Next, mouse HSCs were transfected by these vectors and induced to differentiate into macrophages to validate the effects of these vectors (Fig. 7f). To further assess the role of PRDX1 and CRTC1 in macrophage activation, a series of inflammation/immunosuppression-related gene expression was detected. As a result, either or both of PRDX1/CRTC1 inhibition reversed the sg-L2a-induced gene expression trends (Fig. 7g). Besides, PRDX1 and CRTC1 overexpression also neutralized LAMP2a-modulated genes expression (Fig. 7g). Afterwards, the tumor cytotoxicity of these genetically modified HSCs-derived macrophages was assessed. The results showed that in all three co-cultured tumor cells, PRDX1 and CRTC1 insufficiency greatly restrained sg-L2a-induced tumor cytotoxicity (Fig. 7h). Moreover, CRTC1 overexpression significantly restored LAMP2a-inhibited tumor cytotoxicity, while PRDX1 overexpression only exhibited a moderate increase of tumor cytotoxicity in co-cultured CT26 (Fig. 7h). Comparable results were also obtained in cell viability assay (Supplementary Fig. S7c).
Next, we tried to investigate how LAMP2a-PRDX1/CRTC1 axis modulates macrophage activation. Since CRTC1 is named as CREB-regulated transcription coactivator, and its downstream pathway factors, CREB and CEBP/β, are considered to promote M1-related and M2-related genes expression respectively in macrophages [[62], [63], [64]], we next detected CREB and CEBP/β expression. The results showed that LAMP2a knockdown notably increased p-CREB (Ser133 phosphorylated, transcriptionally activated form) expression, while CEBP/β expression was unperturbed (Fig. 7i). And this imbalance between CREB and CEBP/β expression is regarded as triggers for inflammatory activation of macrophages [62]. For PRDX1, whose main function is to interact with ROS, and considering that H2O2 is an ordinary form of ROS with longest half-life in cultured cells, we next detected H2O2 production as a downstream effect of PRDX1. As a result, sh-L2a-treated macrophages produced significantly higher level of H2O2 than sh-NC and control group (Fig. 7j). In this situation, the stabilized PRDX1 serves as a responder to activate ROS-responsive signal pathway [65,66], which directly contributes to macrophages inflammatory activation [67,68]. In summary, these results suggest that LAMP2a degrades PRDX1 and CRTC1 in macrophages to abrogate ROS and CREB-CEBP/β signal pathway, and PRDX1 and CRTC1 are responsible for LAMP2a-modulated macrophage activation (Fig. 7k).
4. Discussion
As a predominant cell type in TME, TAM exerts immunosuppressive function in most cancer types via producing growth factor, digesting extracellular matrix, promoting angiogenesis, and secreting immunosuppressive cytokines [[12], [13], [14], [15], [16]]. Although the negative prognostic significance of TAM in clinical therapy has been implicated [22,23], and immunotherapy strategies that targeting TAM have been validated a very promising approach in cancer therapy, with constantly developed new targets of TAM [24,25], the origins, surface markers and diversified features of TAMs still need further investigation [20,21]. Besides, the pivotal role of TAMs in tumor-infiltrating cells also largely relies on TAM-centered immune suppression network, in which tumor cells, Th2 cells, fibroblasts, B cells, and hypoxia drive TAMs towards “M2-like” phenotype [27]. And these “M2-like” TAMs further enhance TME immunosuppression, mainly via M-MDSCs and Treg cells [[69], [70], [71]]. Despite this knowledge, key pathways that inducing the immunosuppressive functions of TAMs still need to be further investigated.
In our research, we reveal a potential target LAMP2a in TAMs, find that LAMP2a is upregulated in multiple human cancers and experimental mouse tumor models, and verify the positive correlation of LAMP2a expression in TAMs with poor prognosis. Next, our data suggest that LAMP2a knockdown reverses macrophage activation, increases tumor cytotoxicity in vitro, suppresses cancer progression and restores immuno-environment in tumor milieu. Furthermore, we find CRTC1 and PRDX1 are responsible for LAMP2a-modulated macrophage activation.
TAM-targeting therapeutic approaches mainly focus on restricting cell population and reversing pro-tumorigenic phenotype [3], and we suggested LAMP2a as a potential target in re-programming macrophage activation. In our study, we used tumor-supernatant (TS) to induce immunosuppressive activation of BMDMs and HSC-derived macrophages, and LAMP2a knockdown reversed this process into an inflammatory trend. Besides, LAMP2a-inactivation also enhanced macrophage tumor cytotoxicity in vitro. The mouse tumor models further showed that LAMP2a knockdown in vivo significantly impact mTAMs and M-MDSCs population, and we speculated that this effect did not come from restrained recruiting or infiltrating of monocytic precursors, or simply amount losses, but due to TAMs' remodeling from known tumor-promoting phenotypes into unclear markers combinations, which might be defined in future.
Tumor milieu contains series of adverse environmental conditions to impede macrophages' anti-tumorigenic function, but the specific molecular mechanisms still remain to be defined. Besides, it is not clear that how TAMs “decide” their phenotypic trend when they simultaneously encounter diverse stressors that elicit both promoting and inhibiting effects on inflammation in TME. In exploring of these issues, the diversity or plasticity of macrophage, whether in phenotype or function level, is a critical factor [20,21,69,72]. The multi-functional and bi-directional polarization potencies of macrophages should require one or more “switch” to ensure their adaptation to different tissue microenvironments or respond to various signals. Considering the key feature of LAMP2a-represented CMA is selective removal of target proteins with neighboring proteins unperturbed [37], which allows LAMP2a to precisely modulate signal transduction, metabolism, transcription activation, and other cellular processes that directly involve in macrophage activation, we demonstrate that LAMP2a plausibly serves as a critical molecular switch in macrophage activation. Besides, the LAMP2a-mediated macrophages activation might also be an underlying mechanism for the phenotypical and functional heterogeneity of TAMs population, because various stressors in TME which involve in TAMs regulation also activate LAMP2a, like growth and inflammatory factors, tumor cells metabolites, hypoxia and nutrient deprivation [54,55].
Although current targets in TAMs are mostly membrane proteins like CSF-1R, CD47/SIRPα, class II HDAC, PD-1 and so on [[28], [29], [30],[33], [34], [35]], and intracellular targets have intrinsic difficulties on recognition, detection and blockage in basic research and clinical therapy, LAMP2a has its advantages in directly and rapidly participating in cell regulation. As in TAMs, LAMP2a has the capacity to rapidly and precisely control macrophage activation through regulating its specific substrate proteins. Besides, it is promising to utilize LAMP2a to explore underlying key factors participated in macrophage activation and differentiation, since our data suggest that macrophage activation can be reversed by LAMP2a regulation, and the key feature of LAMP2a is to selectively degrade target proteins [37]. As in this study, CRTC1 and PRDX1 are identified as substrates of LAMP2a in macrophage, and proved to be critical for LAMP2a-modulated macrophage activation.
There are still limitations in our work, and the foremost one is we do not identify the biological features of LAMP2a-inactivating macrophages in vivo, especially the specific cell marker spectrums. Besides, due to the lack of precise marker combinations or definitions to distinguish different types of macrophage activation in vivo [20,21,26], LAMP2a-PRDX1/CRTC1 axis is difficult to be confirmed in tumor milieu, where macrophages would encounter more complex conditions. To resolve these issues, we are still searching for more suitable marker spectrums in vivo. Moreover, there still might be other underlying substrates of LAMP2a in macrophage. To explore this, in the future we may utilize more precise approaches that only interfere with LAMP2a's recognizing or binding to substrates, without disrupting macrophage activation.
In summary, our results unravel a paradigm in which intracellular protein LAMP2a regulates TAMs activation to promote tumor progression. And this LAMP2a-dependent mechanism might partly illustrate how tumor cells utilize microenvironment to domesticate TAMs and suppress anti-tumor immunity.
Author contributions
R.W. and Y.L. designed and performed all experiments. X.W. and Y.W. supervised the study. L.L. and J.Y. assisted with most animal experiments. M.C. assisted with CRISPR/Cas9 experiments designing and operating. X.W. and Y.G. assisted with immunoprecipitation and mass spectrometry experiments. B.D. and X.W. assisted with experimental results conceptualizing and manuscript writing improvement. R.W., Y.L. and Y.W. wrote the manuscript.
Acknowledgments
Acknowledgments
We acknowledge Prof. Chong Chen for his guidance and generously sharing the vectors used in CRISPR/Cas9 experiments. We acknowledge Prof. Lunzhi Dai for his assistance with immunoprecipitation and mass spectrometry experiments. We acknowledge Prof. Yinglan Zhao, Dr. Yaqiong Li and Qianyi Deng for assistance with experimental animal management.
Funding
This work is supported by the National Natural Science Foundation of China (No. 81602492) and the National Key Research and Development Program of China (No. 2016YFA0201402).
Declaration of interests
All authors declare that they have no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.045.
Contributor Information
Yuquan Wei, Email: yqwei@scu.edu.cn.
Xiawei Wei, Email: xiaweiwei@scu.edu.cn.
Appendix A. Supplementary data
Supplementary material
References
- 1.Chevrier S., Levine J.H., Zanotelli V.R.T., Silina K., Schulz D., Bacac M. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749.e18. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavin Y., Kobayashi S., Leader A., Amir E. ad D., Elefant N., Bigenwald C. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cassetta L., Pollard J.W. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discov [Internet] 2018 doi: 10.1038/nrd.2018.169. Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 4.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 5.Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature [Internet] Nature Publishing Group. 2011;475:222–225. doi: 10.1038/nature10138. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin E.Y., Nguyen A.V., Russell R.G., Pollard J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med [Internet]. 2001;193:727–740. doi: 10.1084/jem.193.6.727. http://www.jem.org/lookup/doi/10.1084/jem.193.6.727 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shand F.H.W., Ueha S., Otsuji M., Koid S.S., Shichino S., Tsukui T. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc Natl Acad Sci U S A. 2014;111:7771–7776. doi: 10.1073/pnas.1402914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottazzi B., Erba E., Nobili N., Fazioli F., Rambaldi A., Mantovani A. A paracrine circuit in the regulation of the proliferation of macrophages infiltrating murine sarcomas. J Immunol. 1990;144:2409–2412. [PubMed] [Google Scholar]
- 9.Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K. The cellular and molecular origin of tumor-associated macrophages. Science (80- ) [Internet] 2014;344:921–925. doi: 10.1126/science.1252510. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4204732&tool=pmcentrez&rendertype=abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar V., Cheng P., Condamine T., Mony S., Languino L.R., McCaffrey J.C. CD45 phosphatase inhibits STAT3 transcription factor activity in myeloid cells and promotes tumor-associated macrophage differentiation. Immunity [Internet] 2016;44:303–315. doi: 10.1016/j.immuni.2016.01.014. Elsevier Inc.. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Movahedi K., Laoui D., Gysemans C., Baeten M., Stangé G., Van Den Bossche J. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 12.Hiratsuka S., Nakamura K., Iwai S., Murakami M., Itoh T., Kijima H. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/s1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 13.Du R., Lu K.V., Petritsch C., Liu P., Ganss R., Passegué E. HIF1α induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagemann T., Lawrence T., McNeish I., Charles K.A., Kulbe H., Thompson R.G. “Re-educating” tumor-associated macrophages by targeting NF-kappaB. J Exp Med. 2008;205:1261–1268. doi: 10.1084/jem.20080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mantovani A., Sica A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chittezhath M., Dhillon M.K., Lim J.Y., Laoui D., Shalova I.N., Teo Y.L. Molecular profiling reveals a tumor-promoting phenotype of monocytes and macrophages in human cancer progression. Immunity. 2014;41:815–829. doi: 10.1016/j.immuni.2014.09.014. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- 17.Laoui D., Van Overmeire E., Di Conza G., Aldeni C., Keirsse J., Morias Y. Tumor hypoxia does not drive differentiation of tumor-associated macrophages but rather fine-tunes the M2-like macrophage population. Cancer Res. 2014;74:24–30. doi: 10.1158/0008-5472.CAN-13-1196. [DOI] [PubMed] [Google Scholar]
- 18.Biswas S.K., Gangi L., Paul S., Schioppa T., Saccani A., Sironi M. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-κB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 19.Saccani A., Schioppa T., Porta C., Biswas S.K., Nebuloni M., Vago L. p50 nuclear factor-κB overexpression in tumor-associated macrophages inhibits M1 inflammatory responses and antitumor resistance. Cancer Res. 2006;66:11432–11440. doi: 10.1158/0008-5472.CAN-06-1867. [DOI] [PubMed] [Google Scholar]
- 20.Franklin R.A., Li M.O. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer [Internet] 2016;2:20–34. doi: 10.1016/j.trecan.2015.11.004. Elsevier Inc.. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity [Internet] 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. Elsevier. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Q., Liu L., Gong C., Shi H., Zeng Y., Wang X. Prognostic significance of tumor-associated macrophages in solid tumor: a meta-analysis of the literature. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis C.E., Pollard J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 24.Noy R., Pollard J.W. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruffell B., Coussens L.M. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. Elsevier Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med [Internet] 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. Springer US. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mantovani A., Marchesi F., Malesci A., Laghi L., Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol [Internet] 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. http://www.nature.com/doifinder/10.1038/nrclinonc.2016.217%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/28117416 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pyonteck S.M., Akkari L., Schuhmacher A.J., Bowman R.L., Sevenich L., Quail D.F. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med [Internet] 2013;19:1264–1272. doi: 10.1038/nm.3337. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quail D.F., Bowman R.L., Akkari L., Quick M.L., Schuhmacher A.J., Huse J.T. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science (80- ) [Internet] 2016;352 doi: 10.1126/science.aad3018. http://science.sciencemag.org/content/352/6288/aad3018.abstract aad3018. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ries C.H., Cannarile M.A., Hoves S., Benz J., Wartha K., Runza V. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25:846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 31.De Henau O., Rausch M., Winkler D., Campesato L.F., Liu C., Cymerman D.H. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature. 2016;539:1–16. doi: 10.1038/nature20554. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaneda M.M., Messer K.S., Ralainirina N., Li H., Leem C., Gorjestani S. PI3Kγ is a molecular switch that controls immune suppression. Nature. 2016;539:437–442. doi: 10.1038/nature19834. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guerriero J.L., Sotayo A., Ponichtera H.E., Castrillon J.A., Pourzia A.L., Schad S. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature [Internet] 2017;543:428–432. doi: 10.1038/nature21409. http://www.nature.com/doifinder/10.1038/nature21409 Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao M.P., Jaiswal S., Weissman-Tsukamoto R., Alizadeh A.A., Gentles A.J., Volkmer J. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001375. 63ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon S.R., Maute R.L., Dulken B.W., Hutter G., George B.M., McCracken M.N. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature [Internet] 2017;545:495–499. doi: 10.1038/nature22396. http://www.nature.com/doifinder/10.1038/nature22396%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/28514441 Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuervo A.M., Dice J.F. A receptor for the selective uptake and degradation of proteins by lysosomes. Science (80- ) [Internet] 1996;273:501–503. doi: 10.1126/science.273.5274.501. http://www.sciencemag.org/cgi/doi/10.1126/science.273.5274.501 Available from: [DOI] [PubMed] [Google Scholar]
- 37.Kiffin R. Activation of chaperone-mediated autophagy during oxidative stress [Internet] Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04-06-0477 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandyopadhyay U., Kaushik S., Varticovski L., Cuervo A.M. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol [Internet] 2008;28:5747–5763. doi: 10.1128/MCB.02070-07. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2546938&tool=pmcentrez&rendertype=abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuervo A.M., Dice J.F. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 40.Sooparb S., Price S.R., Shaoguang J., Franch H.A. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004;65:2135–2144. doi: 10.1111/j.1523-1755.2004.00639.x. [DOI] [PubMed] [Google Scholar]
- 41.Vogiatzi T., Xilouri M., Vekrellis K., Stefanis L. Wild type a-synuclein is degraded by chaperone-mediated autophagy and macroautophagy in neuronal cells. J Biol Chem. 2008;283:23542–23556. doi: 10.1074/jbc.M801992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-vicente M., Talloczy Z., Kaushik S., Massey A.C., Mazzulli J., Mosharov E.V. Dopamine-modified a-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777–788. doi: 10.1172/JCI32806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y., Martinez-Vicente M., Krüger U., Kaushik S., Wong E., Mandelkow E.M. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., Wang P., Song W., Sun X. Degradation of regulator of calcineurin 1 (RCAN1) is mediated by both chaperone-mediated autophagy and ubiquitin proteasome pathways. FASEB J [Internet] 2009;23:3383–3392. doi: 10.1096/fj.09-134296. http://www.ncbi.nlm.nih.gov/pubmed/19509306 Federation of American Societies for Experimental Biology. [cited 2017 Aug 20]. Available from. [DOI] [PubMed] [Google Scholar]
- 45.Kon M., Kiffin R., Koga H., Chapochnick J., Macian F., Varticovski L. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med [Internet] 2011;3 doi: 10.1126/scitranslmed.3003182. http://stm.sciencemag.org/cgi/doi/10.1126/scitranslmed.3003182 109a117-109ra117. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8:1643–1656. doi: 10.4161/auto.21654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou D., Li P., Lin Y., Lott J.M., Hislop A.D., Canaday D.H. Lamp-2a facilitates MHC class II presentation of cytoplasmic antigens. Immunity. 2005;22:571–581. doi: 10.1016/j.immuni.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 48.Valdor R., Mocholi E., Botbol Y., Guerrero-Ros I., Chandra D., Koga H. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol [Internet] 2014;15:1–11. doi: 10.1038/ni.3003. https://www.pubchase.com/article/25263126%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/25263126 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A [Internet] 2006;103:5805–5810. doi: 10.1073/pnas.0507436103. http://www.pnas.org/content/103/15/5805.long Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox G.W. Assay for macrophage-mediated anti-tumor cytotoxicity. Curr Protoc Immunol [Internet] 2018:14.7.1–14.7.10. doi: 10.1002/0471142735.im1407s12. Wiley-Blackwell. Available from: [DOI] [PubMed] [Google Scholar]
- 51.Escórcio-Correia M., Hagemann T. Measurement of tumor cytolysis by macrophages. Curr Protoc Immunol [Internet] 2011;92:14.18.1–14.18.11. doi: 10.1002/0471142735.im1418s92. Wiley-Blackwell. Available from: [DOI] [PubMed] [Google Scholar]
- 52.Cuervo A.M., Dice J.F. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci [Internet] 2000;113 Pt 24:4441–4450. doi: 10.1242/jcs.113.24.4441. http://www.ncbi.nlm.nih.gov/pubmed/11082038 Available from: [DOI] [PubMed] [Google Scholar]
- 53.Giatromanolaki A., Sivridis E., Kalamida D., Koukourakis M.I. Transcription factor EB expression in early breast cancer relates to lysosomal/autophagosomal markers and prognosis. Clin Breast Cancer [Internet] 2016:1–7. doi: 10.1016/j.clbc.2016.11.006. Elsevier Inc.. Available from: [DOI] [PubMed] [Google Scholar]
- 54.Settembre C., Di Malta C., Polito V.A., Arencibia M.G., Vetrini F., Erdin S. TFEB links autophagy to lysosomal biogenesis. Science (80- ) 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tasset I., Cuervo A.M. Role of chaperone-mediated autophagy in metabolism. FEBS J. 2016:2403–2413. doi: 10.1111/febs.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witte A., Lubitz W. Biochemical characterization of φX174-protein-E-mediated lysis of Escherichia coli. Eur J Biochem. 1989;180:393–398. doi: 10.1111/j.1432-1033.1989.tb14661.x. [DOI] [PubMed] [Google Scholar]
- 57.Szostak M., Wanner G., Lubitz W. Recombinant bacterial ghosts as vaccines. Res Microbiol [Internet] 1990;141:1005–1007. doi: 10.1016/0923-2508(90)90141-c. http://linkinghub.elsevier.com/retrieve/pii/092325089090141C [cited 2017 Aug 20]. Available from: [DOI] [PubMed] [Google Scholar]
- 58.Patel B., Cuervo A.M. Methods to study chaperone-mediated autophagy. Methods [Internet] 2015;75:133–140. doi: 10.1016/j.ymeth.2015.01.003. Elsevier Inc.. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar V., Patel S., Tcyganov E., Gabrilovich D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol [Internet] 2016;37:208–220. doi: 10.1016/j.it.2016.01.004. Elsevier Ltd. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hubbi M.E., Hu H., Kshitiz, Ahmed I., Levchenko A., Semenza G.L. Chaperone-mediated autophagy targets hypoxia-inducible factor-1a(HIF-1a) for lysosomal degradation. J Biol Chem. 2013;288:10703–10714. doi: 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuervo A.M., Terlecky S.R., Dice J.F., Knecht E. Selective binding and uptake of ribonuclease A and glyceraldehyde-3-phosphate dehydrogenase by isolated rat liver lysosomes. J Biol Chem. 1994;269:26374–26380. [PubMed] [Google Scholar]
- 62.Lawrence T., Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol [Internet] 2011;11:750–761. doi: 10.1038/nri3088. Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 63.Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R.G., Rosenthal N. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A [Internet] 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. http://www.pnas.org/content/106/41/17475.full.pdf Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Altarejos J.Y., Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol [Internet] 2011;12:141–151. doi: 10.1038/nrm3072. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4324555&tool=pmcentrez&rendertype=abstract Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morinaka A., Funato Y., Uesugi K., Miki H. Oligomeric peroxiredoxin-I is an essential intermediate for p53 to activate MST1 kinase and apoptosis. Oncogene [Internet] 2011;30:4208–4218. doi: 10.1038/onc.2011.139. http://www.ncbi.nlm.nih.gov/pubmed/21516123 Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 66.Jang H.H., Lee K.O., Chi Y.H., Jung B.G., Park S.K., Park J.H. Two enzymes in one: two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 67.Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E. Succinate dehydrogenase supports metabolic repurposing of mitochondria to drive inflammatory macrophages. Cell. 2016;167:457–470.e13. doi: 10.1016/j.cell.2016.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Neill L.A.J., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J Exp Med [Internet] 2016;213:15–23. doi: 10.1084/jem.20151570. http://jem.rupress.org/content/213/1/15 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Biswas S.K., Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol [Internet] 2010;11:889–896. doi: 10.1038/ni.1937%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/20856220. Nature Publishing Group. Available from: [DOI] [PubMed] [Google Scholar]
- 70.Bronte V., Brandau S., Chen S.-H., Colombo M.P., Frey A.B., Greten T.F. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun [Internet] 2016;7:12150. doi: 10.1038/ncomms12150. http://www.ncbi.nlm.nih.gov/pubmed/27381735%5Cnhttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC4935811 Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol [Internet] 2012;12:253–268. doi: 10.1038/nri3175. Nature Publishing Group. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mantovani A., Allavena P., Sica A. Tumour-associated macrophages as a prototypic type II polarised phagocyte population: role in tumour progression. Eur J Cancer. 2004:1660–1667. doi: 10.1016/j.ejca.2004.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material