Abstract
Background and objectives
Quantitative MRI (qMRI) permits the quantification of brain changes compatible with inflammation, degeneration and repair in multiple sclerosis (MS) patients. In this study, we propose a new method to provide personalized maps of tissue alterations and longitudinal brain changes based on different qMRI metrics, which provide complementary information about brain pathology.
Methods
We performed baseline and two-years follow-up on (i) 13 relapsing-remitting MS patients and (ii) four healthy controls. A group consisting of up to 65 healthy controls was used to compute the reference distribution of qMRI metrics in healthy tissue. All subjects underwent 3T MRI examinations including T1, T2, T2* relaxation and Magnetization Transfer Ratio (MTR) imaging. We used a recent partial volume estimation algorithm to estimate the concentration of different brain tissue types on T1 maps; then, we computed a deviation map (z-score map) for each contrast at both time-points. Finally, we subtracted those deviation maps only for voxels showing a significant difference with healthy tissue in one of the time points, to obtain a difference map for each subject.
Results and conclusion
Control subjects did not show any significant z-score deviations or longitudinal z-score changes. On the other hand, MS patients showed brain regions with cross-sectional and longitudinal concomitant increase in T1, T2, T2* z-scores and decrease of MTR z-scores, suggesting brain tissue degeneration/loss. In the lesion periphery, we observed areas with cross-sectional and longitudinal decreased T1/T2 and slight decrease in T2* most likely related to iron accumulation. Moreover, we measured longitudinal decrease in T1, T2 - and to a lesser extent in T2* - as well as a concomitant increase in MTR, suggesting remyelination/repair.
In summary, we have developed a method that provides whole-brain personalized maps of cross-sectional and longitudinal changes in MS patients, which are computed in patient space. These maps may open new perspectives to complement and support radiological evaluation of brain damage for a given patient.
Keywords: Multi-parametric MRI, Quantitative MRI, Personalized MRI, Multiple sclerosis, Deviation maps, MRI pathology
1. Introduction
Neuroinflammatory and neurodegenerative diseases induce variable damage to the central nervous system, which can be quantified using magnetic resonance imaging (MRI). New advanced MRI techniques exhibit a level of sensitivity and specificity to focal and diffuse pathology that are not available through conventional MRI (Enzinger et al., 2015). Relaxometry and magnetization transfer imaging (MTI), for example, provide measures of biophysical parameters that show different sensitivity to the amount of free water, the amount of water bound to micro- and macromolecules and the amount of paramagnetic substances (i.e. iron), for review see (Helms, 2015). The combination of these MRI contrasts are sensitive and specific to the consequences of neuroinflammation and neurodegeneration, thus they are invaluable assets to perform disease diagnosis and monitoring.
Recent advances in qMRI mainly focus on how to accelerate (Bilgic et al., 2015; Hilbert et al., 2018; Metere et al., 2017; Sumpf et al., 2011) and to standardize (Deoni et al., 2008; Droby et al., 2015; Helms, 2015) qMRI acquisitions, in order to improve their translation into clinical applications. Yet, how to perform individual evaluation of qMRI changes remains an open question.
In contrast to studies based on the comparison between groups, single-subject predictions (i.e. the classification of one subject as being part of one of the groups of the study, e.g. healthy vs. patient) are challenging and prone to the biases induced by the analysis method and the bias inherent to the data, which is increased in subjects with neurological pathologies (Arbabshirani et al., 2017).
In recent years, machine-learning based methods have been proposed to classify patients with pathology and healthy subjects, which provide the probability of a dataset to belong to either one of the categories. Yet, these approaches suffer from the intrinsic limitations related to (i) how feature selection is performed to differentiate the targeted classes (i.e. classical machine methods) and/or (ii) the risk of overfitting when very complex models are applied to relatively small amount of data (classical and modern machine learning methods, (Arbabshirani et al., 2017; Meyer et al., 2017; Stephan et al., 2017).
A more traditional approach to compare single-patient data to a cohort of healthy individuals is based on a voxel basis by using voxel-based morphometry (VBM) analysis (Beeson et al., 2011; Colliot et al., 2006; Kruggel et al., 2017; Muhlau et al., 2009; Sajjadi et al., 2013). Voxel-based methods rely on the non-rigid registration of the patient brain volume to a group of healthy control volumes in a common space prior to the voxel-wise comparison. The non-rigid registration can be then combined with a deformation field, which is encoded for each subject as tissue density changes in the normal space.
Despite the undeniable utility of voxel-based methods, they suffer from important limitations arising from (i) the large topological structural differences among individual brains, (ii) the necessary averaging resulting in image blurring introduced to achieve noise reduction and spatial coherence – and (iii) partial volume effects (PVE). PVE occurs when an imaging voxel contains more that one tissue type, yielding a signal equal to the weighted average of its components. The main caveat is that in regions with spatially varying intensities such as edge structures, the aforementioned effects can shift the apparent position of abnormalities detected by several voxels (Bookstein, 2001). Moreover, the interpretation of the results using single-to-group VBM analysis is problematic; especially if we consider that any statistical difference between a single subject and a group might either just reflect individual variability in neuroanatomy (instead of any alteration due to the disease under study) or designate a false positive as result of the data being sampled from a non-normally distributed population (Scarpazza et al., 2013).
In this study, we propose a new method to perform an individual analysis of qMRI maps that provides complementary information about changes in brain tissue on a single-subject basis. Different than voxel-based methods, our methodology allows a voxel-wise comparison in the subject space without the need of inter-patient registration. In short, for a given voxel in a qMRI map, we compare its scalar value with the distribution of the voxels belonging to the same tissue class and region in a group of healthy subjects and provide deviation maps from its “normative” range. To assess our method and its ability to detect subtle microstructural pathological changes within the brain, we applied our approach to a longitudinal multi-contrast dataset of relapsing-remitting multiple sclerosis (RRMS) patients, which included T1, T2, T2* relaxometry and magnetization transfer imaging.
2. Methods
2.1. Population
Fifteen (15) RRMS patients (10 women/5 men, age = 32 ± 4 years [mean ± standard deviation-SD], month since diagnosis = 33 ± 22 [mean ± SD], EDSS = 1.5 [1–2] [median [range]]) diagnosed using the revised McDonald criteria (Polman et al., 2011) and twenty (20) healthy controls (HC) (12 women/8 men, age = 31 ± 7 years) were enrolled at the Lausanne University Hospital (CHUV), Lausanne, Switzerland. A larger cohort of 65 healthy controls was used as a reference for qT1 maps, which were acquired at the CHUV in the context of another study, with the same protocol and in the same scanner applied for the current study (see Table 1).
Table 1.
Characteristics of the study participants. Data are presented as mean (standard deviation) for continuous variables and count for categorical variables. NA stands for not applicable. Lesion load corresponds to the percentage of the total lesion volume over the total intracranial volume (TIV).
| RRMS (n = 15) | HC (n = 4) | HD (n = 16) | HD2 (n = 65) | |
|---|---|---|---|---|
| Age [years] | 32 (4.6) | 34.5 (7.5) | 32.5 (10) | 51.68 (3.46) |
| Gender (male/female) | 5/10 | 2/2 | 6/10 | 23/42 |
| Time since diagnosis [months] | 33.2 (22.4) | NA | NA | NA |
| Time since first relapse [months] | 33.5 (25.8) | NA | NA | NA |
| Lesion load [%] | 0.55 (0.6) | NA | NA | NA |
Inclusion criteria were: RRMS diagnosis and less than five year disease duration from diagnosis; for HC: absence of previous neurological and psychiatric disease, no history of smoke and alcohol abuse and no current medication. Patients were followed up at two years (21.9 ± 2 months). At study enrollment, no patient had received corticosteroid therapy for at least three months. Immunomodulatory treatment consisted of high dosage of either interferon beta or fingolimod. Fingolimod is used as a first line therapy in Switzerland.
The ethics committee of the Lausanne University Hospital (CHUV) approved the study and all the participants gave written informed consent.
After preprocessing and qualitative evaluation of acquired MR images, 2 RRMS subjects were excluded due to motion, distortion and other artifacts seen in at least one of the contrasts used in the study (see Table 1).
The baseline and longitudinal personalized maps were computed for the 13 RRMS patients and four healthy subjects (2 women/2 men; age 34.5 ± 7.5 years), who were randomly chosen out of the control group (HC).
The healthy distribution (HD) of each contrast's parametric map used for reference was computed from the remaining 16 healthy controls (10 women/6 men; age 32 ± 10 years). An additional healthy distribution from 65 controls (42 women/23 men; age 51.68 ± 3.46 years) was used to calculate a reference for qT1 maps as stated above (HD2, Table 1). The four healthy subjects selected as a control test group for the computation of the personalized deviation map were not part of the group of controls used to calculate the healthy distribution.
2.2. Image acquisition and reconstruction
All subjects underwent MRI examinations in a 3T Magnetom Trio (Siemens Healthcare, Erlangen, Germany) using the following protocol: (1) High-resolution magnetization-prepared acquisition with gradient echo (MPRAGE); (2) 3D fluid attenuated inversion recovery (3D FLAIR); (3) 3D double inversion recovery (3D DIR); (4) Magnetization-Prepared 2 Rapid Acquisition Gradient Echoes MP2RAGE (Marques et al., 2010) for lesion identification and T1 relaxometry maps; (5) T2* relaxation using a prototype Gradient-Recalled-Echo (GRE) sequence with and without magnetization transfer (MT) preparation pulse (flip-angle 220°, duration = 4000 ms; pulse offset = 2000 Hz; spoiler moment 25,000 μs × mT/m (Helms and Hagberg, 2009)). MTR maps were derived from the T2* data using the following formula:
With M0 and MT the images acquired without and with MT pulse, respectively. MTR was computed for each echo and averaged over all echoes. Finally, (6) T2 relaxation obtained using a prototype 2D Carr-Purcell-Meiboom-Gill (CPMG) sequence with a nonlinear inverse reconstruction algorithm that directly estimates a T2 and spin-density map from a train of undersampled spin echoes (Sumpf et al., 2011). The acquisition of T2 relaxometry maps was performed with a lower spatial resolution than the one achieved for the other MRI contrasts.
Sequence's parameters are summarized in Table 2. Visual inspection of image quality was performed in all cases.
Table 2.
MRI protocol. TR: Repetition time [ms]; TE: Echo time [ms]; TI: Inversion time [ms]; FoV [mm]; Field of view [mm3]; TA [min:sec]; Acquisition time; Misc. stands for parameters specific to a sequence.
| TR/TE | Voxel size | FoV | TA | Misc | |
|---|---|---|---|---|---|
| MPRAGE | 2300/2.98 | 1.0 × 1.0 × 1.2 | 256 × 240 × 192 | 5:12 | GRAPPA factor = 2 |
| 3D FLAIR | 5000/394 | 1.0 × 1.0 × 1.2 | 256 × 240 × 212 | 6:27 | TI = 1800 |
| 3D DIR | 10,000/218 | 1.1 × 1.2 × 1.2 | 256 × 240 × 192 | 12:52 | TI1/TI2 = 450/3652 |
| MP2RAGE | 5000/2.89 | 1 × 1 × 1.2 | 256 × 240 × 212 | 8:22 | TI1/TI2 = 700/2500 |
| CMPG | 5850/9 | 1 × 1 × 4 | 210 × 175 × 120 | 3 | 21 echoes - 30 slices |
| GRE | 47/1.23 | 1.6 × 1.6 × 1.6 | 217 × 217 × 179 | 11:16 | 32 gradient echoes w/wo MT |
2.3. Personalized qMRI analysis
The proposed method relies on a voxel-to-region comparison, calculating a deviation score (i.e. a z-score) between (i) the MR metric of the input image in each voxel and (ii) the MR metric distribution of the corresponding healthy tissue of the same brain region. To test the sensitivity of each parametric map to detect neuroinflammatory or neurodegenerative processes, the personalized maps were computed over the T1, T2 and T2* relaxation and MTR maps.
The condition that should be fulfilled to minimize biases is that at least one contrast should provide sufficient detailed structural information in order to quantify the concentration of each brain tissue-type, which is done to minimize PVE.
The overall methodology can be summarized as follows:
-
(1)
data preprocessing.
-
(2)
brain segmentation into white matter (WM), gray matter (GM), cerebrospinal fluid (CSF) and a segmentation into four main lobes and deep gray matter nuclei (DGMN);
-
(3)
tissue concentration's estimation for partial volume correction and tissue's intensity characterization at each voxel;
-
(4)
segmentation and identification of lesions as WM, GM or mixed lesions;
-
(5)
computation of the data statistics (mean, variances and co-variances) of the healthy tissue intensity distribution for each tissue and region of interest;
-
(6)
computation of the deviation score for each voxel in the patient MRI based on the healthy tissue distribution previously calculated, for each qMRI contrast.
Steps (2) and (3) were computed using only the uniform (UNI) image from MP2RAGE. Step (4) was computed on the UNI (MP2RAGE), 3D FLAIR and DIR contrasts. An additional affine transformation to UNI image was added for the T2, T2* and MTR contrasts in order to map segmentation masks and tissue concentration maps (the masks and tissue concentration maps were computed only over the T1 maps, since they are more sensitive to differences between brain tissue types than other maps; see Section 2.3.2).
All data outputs were inspected at all processing steps to assure the accuracy of volume estimation, alignment of multi-modal MRI and fusion with lesion maps. Note that no registration is needed if only one contrast is used and the tissue characterization using the PVE is done over the same contrast. The following paragraphs provide a comprehensive description of these steps.
2.3.1. Data preprocessing
UNI MP2RAGE images were skull-stripped using the Morphobox prototype (see next section). T2* and MTR MR scans were corrected for gradient distortions and B1 inhomogeneities
2.3.2. Brain segmentation
Tissue segmentation was performed over the UNI MP2RAGE image (in patient space) using the Morphobox prototype, a software based on variational expectation-maximization tissue classification (Roche et al., 2014; Schmitter et al., 2015). The same algorithm parcellated the brain in four main lobes: parietal, frontal, occipital and temporal (from now our regions-of-interest, ROIs), as well as the DGMN (thalamus, caudate, putamen and globus pallidus, see Supplementary Information section, SI.1, for an example of ROIs segmentation).
2.3.3. Tissue concentration estimation for partial volume correction
In many brain regions, especially at the border between different brain tissues, MRI intensities suffer from the so-called PVE, i.e. the imaging voxels contains more than one tissue type, yielding a signal equal to the weighted average of its components. To account for this effect in our analysis, we estimated the concentration of GM, WM and CSF in each of these neighboring voxels, assigning value computed as a mixture of tissues instead of assigning them a single tissue type. This was done by applying a PVE algorithm that was proposed by Roche and Forbes (Roche et al., 2014), which relies on continuous Markov Random Field (MRF) models, with the tissue concentration estimation formulated as a Bayesian maximum a posterior (MAP). This algorithm specifically uses a model that describes the intensity of each voxel (y) as the sum of GM, WM and CSF characteristic intensities in a given region, weighted by their respective local concentrations, with additive Gaussian noise:
where μGM, μWM, μCSF are the characteristic intensities of each tissue and CGM, CWM, CCSF are the concentrations of GM, WM, and CSF respectively. ε is the additive Gaussian noise (with zero mean and σ standard deviation).
In order to solve this ill-posed problem, we set three hyper-parameters to regularize the following assumptions on brain tissue intensities: (i) voxels with mild partial volume are more frequent than those with strong partial volume; (ii) tissue concentration maps are spatially smooth; and (iii) mean tissue intensities are bounded (see SI.2 section for more details).
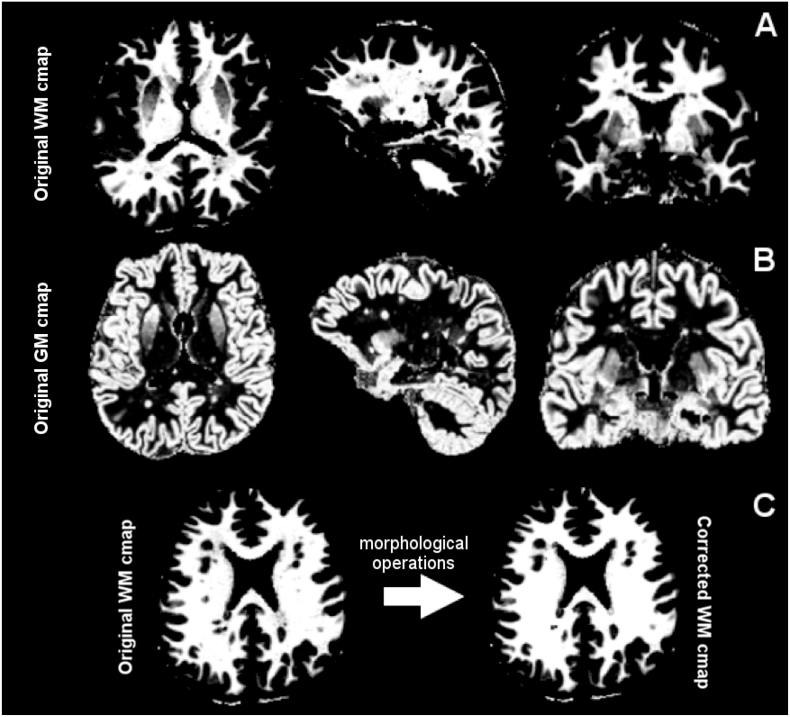
The algorithm proceeds iteratively until convergence. Once the characteristic intensities μGM, μWM, μCSF are initialized, the algorithm estimates the tissue concentrations CGM, CWM, CCSF (step 1) and the characteristic intensities μGM, μWM, μCSF (step 2) as well as the noise standard deviation, σ. These steps are repeated until the cost function of the algorithm converges below a defined threshold (see SI, for summary description of the PVE method). The algorithm outputs three concentration maps, one for each tissue (GM, WM and CSF) and the characteristic intensity values for GM, WM and CSF (see Fig. 1A and B for WM and GM concentration maps [cmap] examples).
Fig. 1.
Example of WM and GM concentration maps (cmaps) as computed from an exemplary T1 map. From left to right, top row (A): original white matter (WM) cmap in axial, sagittal and coronal view; middle row (B), original gray matter (GM) cmap in axial, sagittal and coronal view. On the bottom row (C), example of a WM cmap correction. From left to right, original WM cmap and corrected cmap.
Since T1 maps are more sensitive to differences between brain tissue types than other maps, we performed the tissue concentration estimation on quantitative T1 maps: we initialized the mean intensities at 1350 ms for the GM and 850 ms for WM, which are the characteristic mean relaxation times according to (Marques et al., 2010). In addition, we applied the algorithm on each ROI separately to consider the variability of T1 signal over the brain (see Section 2.3.6.). We then registered the T2, T2* and MTR maps to the subjects' T1 map space (intra-subject registration) to use tissue concentrations maps estimated from the T1 contrast. To do this, we estimated an affine transformation from the T2 and T2* map space (which is the same than the MTR space) to the T1 map space using Elastix c++ library (Klein et al., 2010).
2.3.4. Lesion segmentation and lesion tissue identification
Brain lesions were manually counted by consensus between an expert neurologist and radiologist the in the 3DFLAIR, DIR and MP2RAGE uniform images as previously performed (Bonnier et al., 2014; Bonnier et al., 2015; Kober et al., 2012; Romascano et al., 2015). A trained technician generated manual contours for each lesion in the three different contrasts. In order to maximize the sensitivity of lesion count and volume, the masks of segmented lesions in each contrast (3D FLAIR, DIR and MP2RAGE) were then merged in a single “union lesion mask” as in (Bonnier et al., 2014; Bonnier et al., 2015; Kober et al., 2012; Romascano et al., 2015). Lesion volumes were computed and normalized by total intracranial volume as obtained using an in-house software and classified as WM, GM or mixed lesions. The lesion union mask and the ROIs masks were then registered to the T1, T2, T2*, and MTR maps.
The method used to estimate tissue concentration is robust to subtle signal intensity variations, but fails when tissue damage drastically changes MR signal like in brain lesions and their vicinity. In that case, the algorithm identifies altered WM as a mix of WM and GM (or even CSF). Thus, we a posteriori modified the concentration maps inside brain lesions, based on a single hypothesis: there is no GM inside WM. Specifically, we attributed (i) 100% WM concentration to a voxel belonging to a pure WM lesion, (ii) 100% GM concentration to a voxel belonging to a GM lesion and (iii) 50% WM and 50% GM concentration to a voxel belonging to a mixed WM-GM lesion. While in our work lesions were manually segmented, we have also integrated a software for automated lesion detection in the pipeline (Fartaria MJ, 2017a, b).
Additionally, we performed morphological operations on the white matter concentration map to apply the a-priori knowledge that there is no GM in the WM: first, we binarized the WM concentration map by thresholding it (we use a high threshold in order to keep voxels with partial volume (GM/WM border) out of the process). Then we filled the holes inside the WM binary map and finally combine the binary map with original WM concentration map (see Fig. 1C).
2.3.5. Computation of healthy tissue distribution for each qMRI map
T1, T2, T2* and MTR metrics vary depending on the underlying microstructure in the subjects and therefore exhibit different values across the brain (Aubert-Broche et al., 2009; Wansapura et al., 1999). However, these parameters are considered relatively homogeneous within each cerebral lobe (Georgiades et al., 2001; Hasan et al., 2012).
For each qMRI contrast, we estimated a healthy tissue distribution for each brain lobe (frontal, parietal, occipital and temporal). We used a cohort of 65 healthy controls to compute the reference healthy tissue distribution for the T1 parametric maps and 15 controls for the other parametric maps in each brain tissue (WM, GM, and CSF) and ROI.
For each control, we first segmented the brain into WM, GM and CSF tissues as well as in four different lobes (ROIs) (Section 2.3.1) and then estimated their tissue concentrations separately (Section 2.3.2). For each control subject and ROI, only voxels with at least 95% concentration of the same tissue (WM or GM) were considered. To estimate the data's statistics (mean, standard deviation and covariances) considering intra- and inter-subjects variability, for each ROI we performed a bootstrapping analysis on the entire healthy controls cohort. In this context, we consider the data statistics (mean and standard deviation values) derived from the healthy distribution as being reasonably representative of the distribution, so that the z-score (used as a measure of deviation) shows abnormal brain tissue microstructure (see SI.3).
2.3.6. Deviation maps
The personalized deviation maps were computed by comparing the intensity of a single voxel in the patient scalar map with the corresponding tissue type (WM, GM or mixed) and brain ROI (frontal, parietal, temporal or occipital) value in the reference healthy distribution. The mean and standard deviation values derived from the healthy distribution of each single parameter in each ROI was considered as being reasonably representative of the distribution, so that each deviation from “healthy tissue properties” may be calculated using the z-score formula. Formally, we estimated the deviation as:
where ZV is the z-score for a single voxel, Iv is the intensity of voxel v, μ is the mean of the healthy distribution over the 15 controls, and σ its standard deviation. This expression of the z-score can be extended to a more general approach taking inn consideration the different tissue concentrations. In this case, the z-core per each voxel can be re-written as:
with
where Iv is the intensity of the voxel v in image and CGM, CWM, CCSF are the concentration of GM, WM and CSF in the voxel v, respectively. μGM, μWM, μCSF and σGM, σWM, σCSF are, respectively, the characteristic intensities and variances of the GM, WM and CSF tissues in the ROI where the voxel v belongs, while CovGM/WM, CovGM/CSF, CovWM/CSF are respectively the covariances of the GM and WM, GM and CSF, WM and CSF in the same region (see Fig. 2)
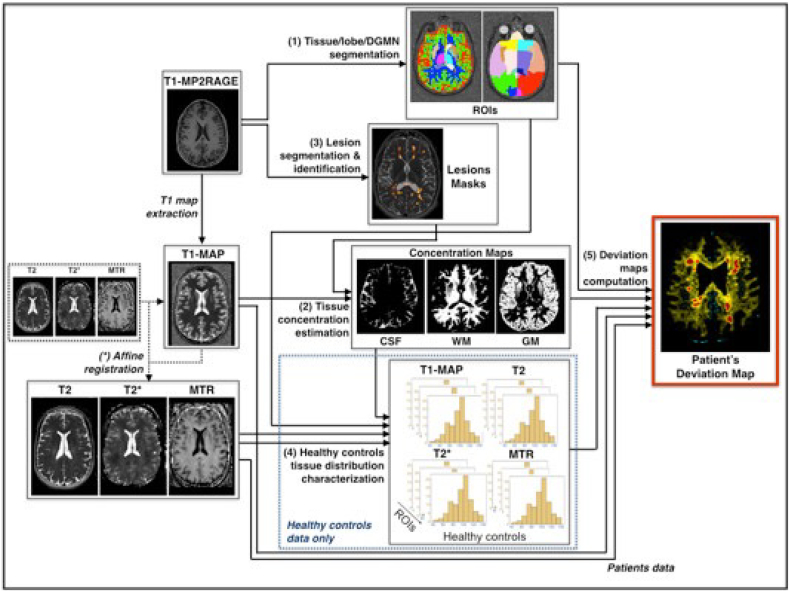
Fig. 2.
Processing pipeline for the computation of the deviation map for the T1, T2, T2* and MTR acquisitions.
For the deep gray matter nuclei, we used the original z-score formula (without concentrations, ) as we considered that the deep gray nuclei are composed by a single tissue characterized by a mean and standard deviation values (μDGM and σDGM) calculated over all voxels belonging to the deep gray nuclei of the group of controls.
2.4. Statistical analysis
Each deviation map quantifies the parametric variation representative of the changes occurring in an individual brain compared to a distribution of healthy controls. Thus, in order to highlight both focal and diffuse brain changes at baseline and two-years follow up, we first computed the deviation maps over the four quantitative MR contrasts (T1, T2, T2* and MTR) for both RRMS subjects and the HC group. Then, we analyzed the mean z-scores inside all voxels in all patients (i) within the lesions, (ii) in the lesions vicinity as well as (iii) in the normal appearing white matter (NAWM) and gray matter (NAGM) for all parametric maps.
The classification of “pathological” z-scores was performed based on a previous work (G. Bonnier, A. Roche and Romascano, et al. 2015), where we MS lesions were classified into 3 groups as follows: (i) z very low (z < −2), (ii) z very high (z > 2), and (iii) z close to the HC distribution (−2 ≤ z ≤ 2), considering the continuous distribution (without distinct cluster) of lesions z-scores in each contrast: these thresholds were chosen based on the fact that in a normal distribution >95% of the z-scores belongs to the interval [−2, 2] and that values beyond this interval reflect significant differences in patients compared to controls (P < .05).
For the longitudinal analysis, we computed difference maps (between the z-score maps obtained from the T1 map acquired at baseline and the ones obtained at 2-years follow-up) to show focal and diffuse brain changes.
3. Results
3.1. Healthy tissue distribution
The assessment of the distribution of the T1, T2, T2* and MTR maps in our group of healthy controls validate the hypothesis of a reasonably normal distribution of MRI intensities in all four lobes (see Supplementary information, Section SI.3). Interestingly, the standard deviation (σ) of the intensities in the voxels within the different lobes of the healthy tissue distribution appear higher in GM than WM for all qMRI metrics (σGM,T1 = 160 ms; σGM,T2 = 16 ms; σGM,T2⁎ = 22 ms; σGM,MTR = 0.04 ms; σWM,T1 = 60 ms; σWM,T2 = 8 ms; σWM,T2⁎ = 10 ms; σWM,MTR = 0.02 ms).
3.2. T1 deviation maps
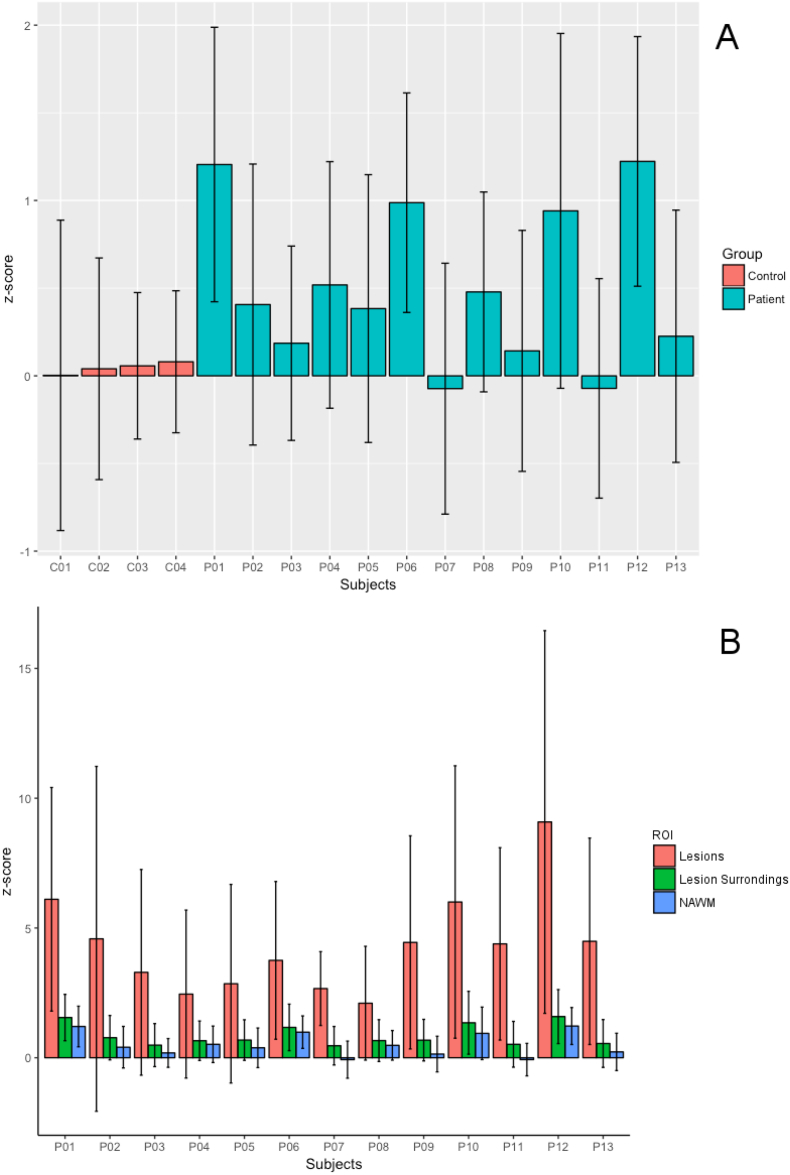
The 4 healthy controls exhibited a much lower T1 deviation in WM than patients in NAWM tissue (controls: zT1 0.04 ± 0.20 vs. RRMS patients: zT1 0.50 ± 0.10 [mean ± standard deviation, SD]), Fig. 3A.
Fig. 3.
Error bar plots for T1 mean z-score in (A) NAWM in the RRMS patients (blue bars) and HC WM (red bars) and in (B) lesions (red bars), lesion surroundings (green) and NAWM (blue) in RRMS patients. Whisker extends to the most extreme data points. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Compared to controls, the T1 deviation maps of RRMS patients showed: (i) a strong increase in T1 z-score in lesions (zT1: 4.30 ± 1.60 [mean ± SD], Figs. 3B and 4), (ii) a moderate increase in T1 z-score in the region surrounding the lesions (zT1: 0.90 ± 1.30 [mean ± SD]), Figs. 3B and 6; and (iii) a lower increase in NAWM (zT1: 0.50 ± 0.10 [mean ± SD]) and NAGM tissue (zT1: 0.04 ± 0.09 [mean ± SD]) (Fig. 3A-B and 5).
Fig. 4.
Axial view of one T1 z-score map of five RRMS patients (top row) and the four healthy controls (bottom row).
Fig. 6.
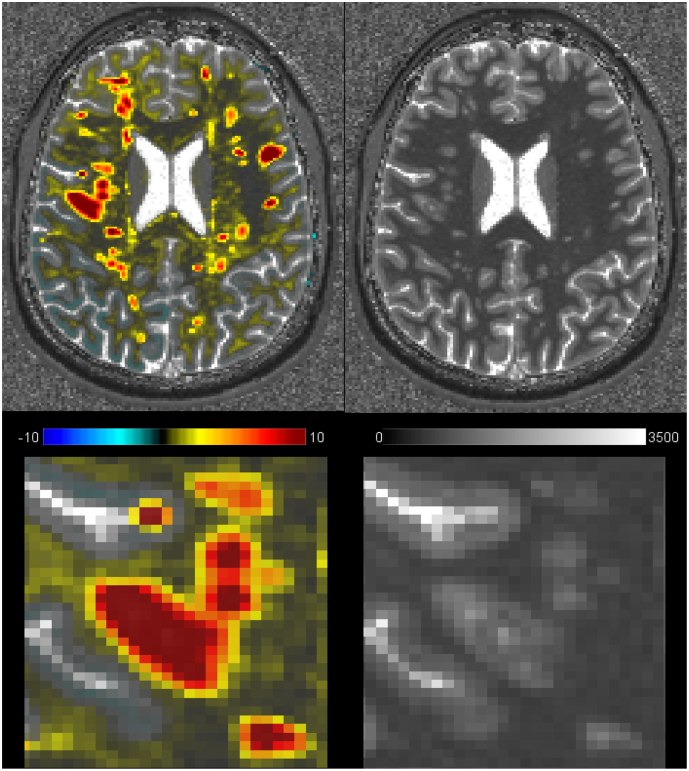
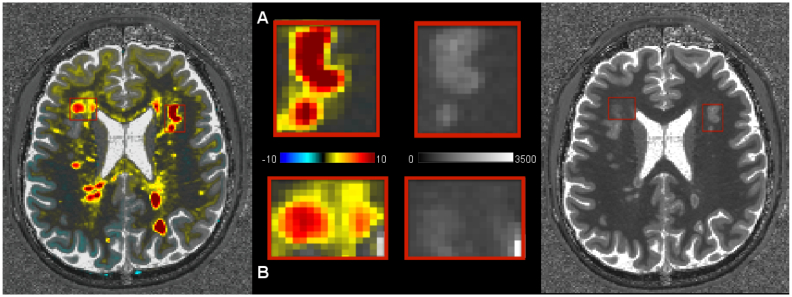
Axial view of a T1 z-score map and a T1 map in RRMS patient 1 (top row). Zoomed area showing the T1 z-score in one exemplary WM lesion (left) and the corresponding T1 map (right). The lesion appears larger in the deviation map with larger T1 z-scores in the center of the lesion and lower but still ≥3T1 z-scores in the lesion periphery/surrounding tissue.
Regarding the NA tissue, patients 1, 6 and 10 with a high LL but also patient 12 with a low LL (LL12 = 0.40), showed a higher positive T1 z-score in NA white matter (NAWM) (zT1 > 0.80) than other patients. All patients showed a low z-scores in NAGM, with patients 1 and 13 have the highest deviation (zT1 > 0.40) and patients 3 and 4 have the lowest deviation (zT1 < 0.10). The highest z-scores were observed around and between lesions (Fig. 5).
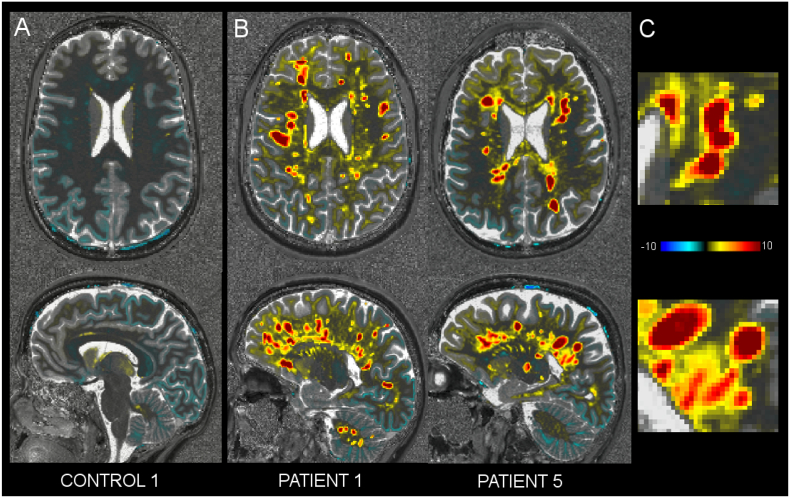
Fig. 5.
Axial (top) and sagittal (bottom) view of a T1 z-score map (over the corresponding T1 map) in control 1 (A) and patients 1 and 5, who exhibit high lesion load (B). (C) The zoomed area from patient's maps shows a diffuse pattern between the lesions.
Also, MS lesions in the T1 deviation maps often appeared to be larger than the manually segmented lesions (Fig. 6). This was most probably due to areas of increased T1 relaxation times surrounding MS lesions that were not perceived as lesion tissue by our neurologist and radiologist experts.
Finally, the z-score map inside some large lesions revealed presence of “core” of increased T1 relaxation time where the z-score is even higher than in the rest of the lesion (Fig. 7).
Fig. 7.
Axial view of a T1-zscore map over the corresponding T1 map (left) in patient 6 showing (A) a strong gradient in T1 z-scores within some WM lesions (T1 deviation z-score ranges from 10 in the core of the lesions to 2 in the edges) and (B) a moderate or minimal gradient in others (T1 z-score range left lesion: [2–4] and right lesion: [2–3]). On the right side, the axial view of the corresponding T1 map.
3.3. Deviation map of T2, T2* and MTR
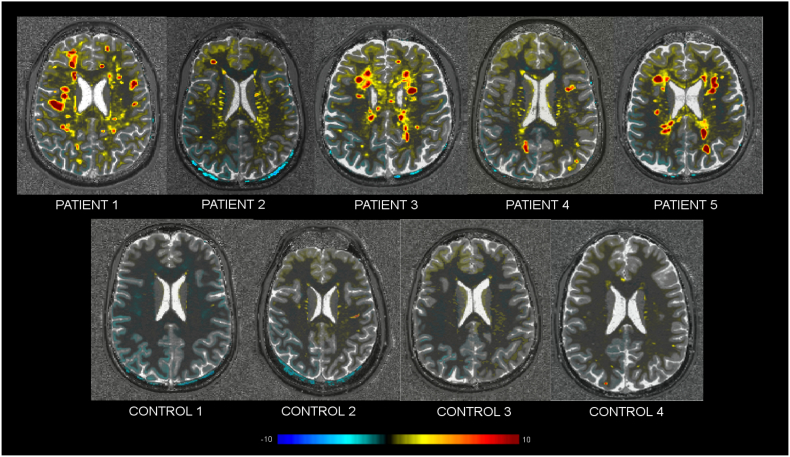
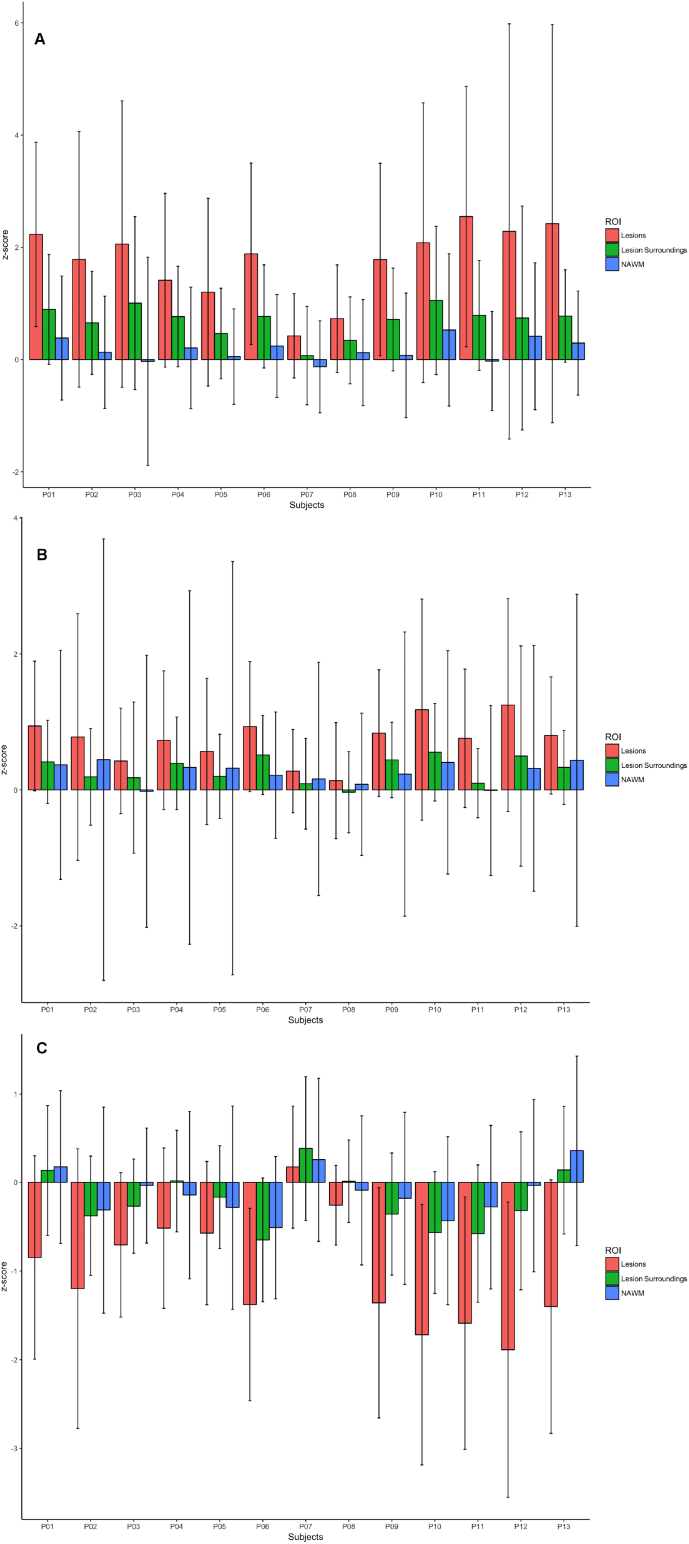
Compared to controls, patient's T2 deviation maps revealed a moderate T2 relaxation time increase inside lesions (zT2 = 1.76 ± 0.60 [mean ± standard deviation, SD]) and mild diffuse increases around lesions (zT2 = 0.70 ± 0.30 [mean ± SD]). T2* maps revealed also slight increases inside lesions although lower than T2 (lesions: zT2⁎ = 0.74 ± 0.36 [mean ± SD]) and no changes in lesion surroundings (zT2⁎ = 0.30 ± 0.20 [mean ± SD]), while the MTR maps showed a slight decrease inside the lesions (zMTR = −1.02 ± 0.60 [mean ± SD]) and no apparent diffuse changes around them (zMTR = −0.20 ± 0.30 [mean ± SD]) (Fig. 8, Fig. 9). No differences were observed between patients with high and low lesion loads.
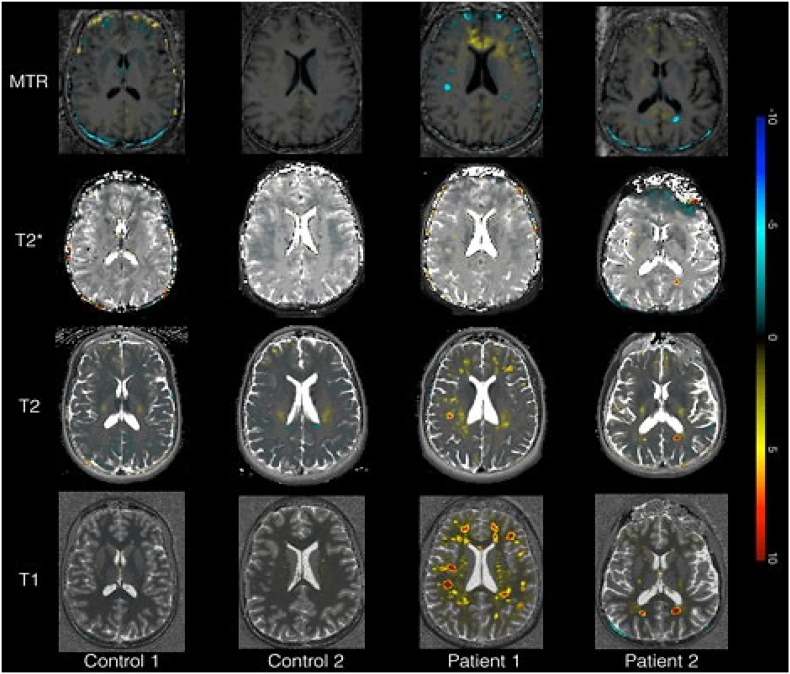
Fig. 8.
Axial view of MTR, T2*, T2 and MTR z-score maps in controls 1 and 2 and patient 1 (high lesion load) and 2 (low lesion load).
Fig. 9.
Barplots for mean (A) T2, (B) T2* and (C) MTR z-scores in RRMS NAWM (blue bar), lesions (red bar) and lesion surroundings (green bar). Whiskers extend to the most extreme data points.
The 4 healthy controls globally exhibited a similar T2, T2* and MTR deviation than in NAWM and NAGM tissue in patients (T2 controls vs patients NAWM [mean ± SD]: zT2–0.28 ± 0.07 vs zT2 0.21 ± 0.27; NAGM [mean ± SD] zT2 – −0.18 ± 0.02; zT2 0.15 ± 0.; T2* controls vs patients NAWM [mean ± SD]: zT2⁎- 0.14 ± 0.11 vs zT2⁎ 0.22 ± 0.08; NAGM [mean ± SD]: zT2⁎– −0.11 ± 0.08; zT2⁎ 0.04 ± 0.006; MTR controls vs patients NAWM [mean ± SD]: zMTR −0.12 ± 0.17 vs zMTR 0.10 ± 0.21; NAGM [mean ± SD]: zMTR 0.03 ± 0.08; zMTR 0.06 ± 0.07).
3.4. Longitudinal analysis
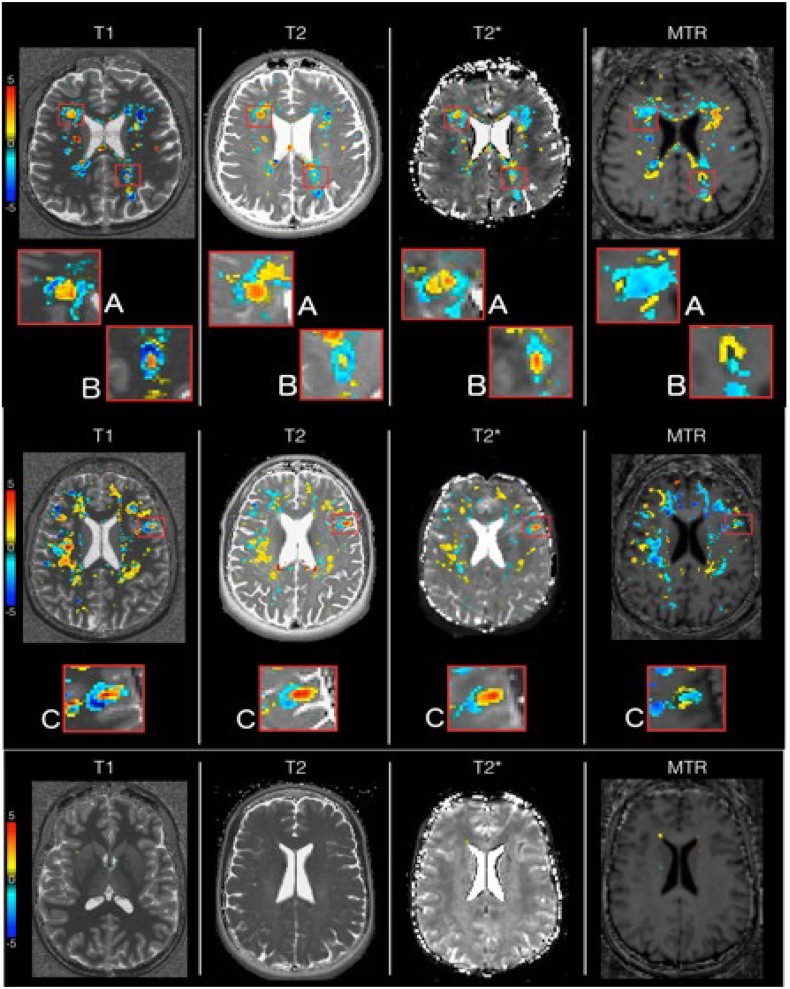
In patients, some subcortical areas, predominant in the fronto-parietal lobes, showed a concomitant increase in T1, T2, T2* z-scores and decrease of MTR z-scores over two years (Fig. 10A and C). In the lesion periphery, some areas e exhibited a decrease in T1/T2 and slight decrease of T2* z-scores (Fig. 10A and B). Besides, other areas showed a decrease in T1, T2 (T2*) and a concomitant increase in MTR (Fig. 10B).
Fig. 10.
Axial difference between baseline (time point 1) and 2 years follow up (time point 2) of T1, T2, T2* and MTR z-score maps in patient 6 (top), patient 1 (middle) and control 2 (bottom). The difference maps give an overview of the changes occurring in the brain tissue of patients 6 and 1, and control 2.
4. Discussion
We have developed a novel approach to provide a personalized quantification of brain damage using a voxel-wise comparison between quantitative parameters of tissue integrity in a patient and the distribution of the same parameters in the same region and tissue in a population of healthy controls.
To test the sensitivity and robustness of our method in the detection of subtle pathological brain changes, we have applied it to a longitudinal dataset of early-stage RRMS patients, who were imaged using four quantitative and semi-quantitative contrasts (T1, T2, T2*, MTR). Those contrasts/metrics were chosen because their combination has been previously proven to increase sensitivity and specificity to pathological brain changes related to inflammation and degeneration compared to single contrast/metric approaches (Bonnier et al., 2014; Bonnier et al., 2015; Granziera et al., 2015; Romascano et al., 2015).
When both RRMS and control subjects were analyzed at baseline, the proposed method was able to detect deviation from “normative ranges” (computed as healthy tissue characteristic intensity obtained from healthy subjects) for RRMS subjects, while no significant deviations were found for our control test group. These differences were especially remarkable within MS lesions and their vicinity but were also sometimes evident in the remaining normal-appearing tissue.
As expected from previous group studies (Bonnier et al., 2014; Bonnier et al., 2015; Deoni et al., 2008; Neema et al., 2007), T1 relaxation deviation maps computed in RRMS patients showed a global increase in T1 relaxation time compared to the healthy control population, which suggests the loss of tissue integrity (i.e. axonal, myelin and cell damage) and/or extracellular water accumulation.
In MS lesions, T1 z-scores were in general increased and some lesions showed the peculiar microstructure reported in postmortem studies i.e. a gradient of damage from a central core with more pronounced tissue loss (i.e. more prolonged T1 relaxation times compared to healthy tissue) and areas surrounding it with less prolonged T1 relaxation times (Fig. 6, Fig. 7) (Bruck et al., 2002; Lassmann et al., 2001; Lucchinetti and Bruck, 2004; Metz et al., 2014).
In the normal appearing tissue, the highest T1 z-scores were measured in the regions surrounding MS lesions, as previously reported (Bonnier et al., 2014; Davies et al., 2007; Parry et al., 2002; Seewann et al., 2009; Vrenken et al., 2010; West et al., 2014), (Fig. 3, Fig. 5). Increased T1 relaxation times point at a loss of tissue microstructural damage and/or to an increase in iron content in the tissue (Bonnier et al., 2017; Bonnier et al., 2014; Bonnier et al., 2015; Granziera C., 2015), which are visible also in the absence of macroscopic global or localized changes in brain volume (Bonnier et al., 2017; Bonnier et al., 2014).
The remaining normal appearing tissue showed mild T1 increases compared to the healthy population of references and interestingly, NAGM showed lower deviation in T1 relaxation times compared to healthy tissue than NAWM. This may be due to the presence of a lower degree of tissue damage in GM than WM at early disease stages, which is consistent with previous studies (Davies et al., 2007; Klaver et al., 2013; Muhlau et al., 2013). Yet, this may also be due to higher PVE, which induces a higher variability when computing the healthy GM distribution compared to the WM.
As well, and as expected from previous literature (Parry et al., 2002, Parry et al., 2003; Rovira et al., 2013), the evaluation of T1 z-scores deviations in MS patients showed a diffuse microstructural pathology that is not related to lesion load. Therefore, the personalized maps of tissue damage – obtained with the proposed methodology- may help to better depict the global damage provoked by MS disease, beyond the areas of focal lesions.
On the other hand, the deviation maps obtained from T2, T2* and MTR maps showed brain tissue alterations, which were located in the same regions as the ones obtained from T1 maps (Fig. 8). Yet, while the T2 and T2* deviation maps highlighted more diffuse changes in patient white matter and gray matter tissue, the MTR deviation maps mainly revealed focal changes in lesions (Fig. 8). Lesion and normal-appearing tissue z-score deviations were positive for T2, T2* maps and negative for MTR maps (Fig. 9), which is compatible with (i) tissue degeneration/loss (in the presence of concomitant and strong increase in T1) or (ii) inflammatory phenomena (in the presence of less pronounced increase in T1 and/or concomitant increase in T2/T2*).
Nonetheless, the z-scores measured in T2, T2* and MTR maps were less important than the ones revealed by T1 maps: this may be due to the lower spatial resolution of the T2, T2* and MTR maps, which translates in an increase of partial volume effect, but also to a higher variability of the healthy distribution compared to the pathological effect on the T2 and T2* relaxation times.
Longitudinal analysis of parametric deviation maps showed no changes in control subjects but areas of z-score alterations in patients. Specifically, in RRMS patients, we have evidenced regions with a concomitant increase in T1, T2, T2* z-scores and decrease of MTR z-scores, a behavior suggesting brain tissue loss over time (Bonnier et al., 2017; Bonnier et al., 2014; Deoni, 2011; Filippi and Agosta, 2007). Also, in the lesion periphery, we could observe areas with decreased T1/T2 and slight decrease of T2* indicating iron accumulation (Dusek et al., 2013). Moreover, in other areas, we assessed longitudinal decrease in T1, T2 (T2*) and a concomitant increase in MTR occurs, suggesting remyelination/repair (Mallik et al., 2014), Fig. 10.
In summary, we have developed and assessed a method that provides a whole-brain personalized information of brain tissue damage and showed its sensitivity to diffuse and focal changes in MS patients, both cross-sectionally and longitudinally. One advantage of the proposed method compared to conventional voxel-based morphometry is that the maps are computed in patient space, therefore bypassing all limitations of inter-subject registration (Klein et al., 2010). Another advantage is the fact that it may be easily extended to other type of images, provided that a large number of healthy subjects are also imaged with the same settings. Also, the proposed approach may be easily extended to other neurological diseases since the applied metrics are sensitive to neurodegenerative and neuroinflammatory changes also in other neurological disorders such as mild cognitive impairment (Granziera et al., 2015), neuroHIV infection (Granziera et al., 2013) and migraine (Granziera et al., 2014).
The proposed method may still benefit of a number of targeted improvements. Indeed, the healthy cohort of reference presently consists of 65 and 15 subjects, who are reasonably but not perfectly representative of the healthy distribution of each metric. Because of this, the presented results require confirmation using a larger reference cohort of healthy controls, which may reduce the variability of the intensity distribution, increase the statistical power and improve sensitivity of the comparison.
In addition, the T2 mapping method that we have applied in this work models a mono-exponential T2 signal decay (Sumpft et al., 2011), which may be violated by the presence of stimulated echoes. To minimize this issue, we have discarded the first acquired echo (at TE = 9 ms). Despite that, however, the signal sampled with the remaining echoes (in the range from 20 to 170 ms) may be still subjected to model violations by the different T2 contributions in individual brain voxels (MacKay, Whittal, et al. 1994, MacKay, Laule, et al. 2006, Barta, et al. 2015). Indeed, for a precise measurement and quantification of the different T2 components of the brain tissue, a much longer acquisition and a more complex data modeling are necessary (MacKay, Whittal, et al. 1994, MacKay, Laule, et al. 2006, Barta, et al. 2015). Nevertheless, the applied T2 mapping measurement has shown high sensitivity to brain pathological changes in neuroinflammatory and neurodegenerative diseases (Bonnier et al., 2017; Bonnier et al., 2014; Bonnier et al., 2015; Granziera C., 2015) as well as high precision and reproducibility (T2 variation of <0.5 ms were reported in scan-rescan measurements and < 4 ms across subjects) (Hilbert, et al. 2018) and relatively short scan times, all characteristics that render it suitable for clinical studies. Furthermore, the approach we propose uses the information provided by different contrasts and metrics (i.e. T1, T2, T2* and MTR) to identify pathological changes in brain tissue microstructure and this is complementing - to a certain extent - the missing information derived from a mono-exponential fitting of the T2 decay.
Lastly, future studies should also further explore the optimal cut-off or range to detect neurodegenerative and inflammatory processes.
In conclusion, we have provided a method to calculate personalized deviation maps in MRI, which may open new perspectives to complement and support clinical decisions. Future work will focus on increasing the healthy control population, integrating automatic lesion detection tools in the pipeline and in the identification of the optimal threshold for pathological changes in each contrast.
Acknowledgements
This study was supported by the Swiss National Science Foundation, Switzerland (under grants P2LAP3_164894 to GB, P2ELP2_172286 to EF and PZ00P3 131914/1 to CG), the Swiss MS Society and the Societé Académique Vaudoise in Switzerland. The funding sources had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report or in the decision to submit the paper for publication.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nicl.2018.11.017.
Appendix A. Supplementary data
Supplementary material
References
- Arbabshirani M.R., Plis S., Sui J., Calhoun V.D. Single subject prediction of brain disorders in neuroimaging: promises and pitfalls. NeuroImage. 2017;145:137–165. doi: 10.1016/j.neuroimage.2016.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert-Broche B., Grova C., Pike G.B., Collins D.L. Clustering of atlas-defined cortical regions based on relaxation times and proton density. NeuroImage. 2009;47:523–532. doi: 10.1016/j.neuroimage.2009.04.079. [DOI] [PubMed] [Google Scholar]
- Barta R., Kalantari S., Laule C., Vavasour I.M., MacKay A.L., Michal C.A. Modeling T(1) and T(2) relaxation in bovine white matter. J. Magn. Reson. 2015 Oct;259:56–67. doi: 10.1016/j.jmr.2015.08.001. Epub 2015 Aug 6. [DOI] [PubMed] [Google Scholar]
- Beeson P.M., King R.M., Bonakdarpour B., Henry M.L., Cho H., Rapcsak S.Z. Positive effects of language treatment for the logopenic variant of primary progressive aphasia. J. Mol. Neurosci. 2011;45:724–736. doi: 10.1007/s12031-011-9579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgic B., Gagoski B.A., Cauley S.F., Fan A.P., Polimeni J.R., Grant P.E., Wald L.L., Setsompop K. Wave-CAIPI for highly accelerated 3D imaging. Magn. Reson. Med. 2015;73:2152–2162. doi: 10.1002/mrm.25347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnier G., Roche A., Romascano D., Simioni S., Meskaldji D., Rotzinger D., Lin Y.C., Menegaz G., Schluep M., Du Pasquier R., Sumpf T.J., Frahm J., Thiran J.P., Krueger G., Granziera C. Advanced MRI unravels the nature of tissue alterations in early multiple sclerosis. Ann. Clin. Transl. Neurol. 2014;1:423–432. doi: 10.1002/acn3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnier G., Roche A., Romascano D., Simioni S., Meskaldji D.E., Rotzinger D., Lin Y.C., Menegaz G., Schluep M., Du Pasquier R., Sumpf T.J., Frahm J., Thiran J.P., Krueger G., Granziera C. Multicontrast MRI quantification of focal inflammation and degeneration in Multiple Sclerosis. Biomed. Res. Int. 2015;2015 doi: 10.1155/2015/569123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnier G., Marechal B., Fartaria M.J., Falkowskiy P., Marques J.P., Simioni S., Schluep M., Du Pasquier R., Thiran J.P., Krueger G., Granziera C. The combined quantification and interpretation of multiple quantitative magnetic Resonance imaging metrics enlightens longitudinal changes compatible with brain repair in relapsing-remitting Multiple Sclerosis patients. Front. Neurol. 2017;8:506. doi: 10.3389/fneur.2017.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein F.L. "Voxel-based morphometry" should not be used with imperfectly registered images. NeuroImage. 2001;14:1454–1462. doi: 10.1006/nimg.2001.0770. [DOI] [PubMed] [Google Scholar]
- Bruck W., Lucchinetti C., Lassmann H. The pathology of primary progressive multiple sclerosis. Mult. Scler. 2002;8:93–97. doi: 10.1191/1352458502ms785rr. [DOI] [PubMed] [Google Scholar]
- Colliot O., Bernasconi N., Khalili N., Antel S.B., Naessens V., Bernasconi A. Individual voxel-based analysis of gray matter in focal cortical dysplasia. NeuroImage. 2006;29:162–171. doi: 10.1016/j.neuroimage.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Davies G.R., Hadjiprocopis A., Altmann D.R., Chard D.T., Griffin C.M., Rashid W., Parker G.J., Tofts P.S., Kapoor R., Thompson A.J., Miller D.H. Normal-appearing grey and white matter T1 abnormality in early relapsing-remitting multiple sclerosis: a longitudinal study. Mult. Scler. 2007;13:169–177. doi: 10.1177/1352458506070726. [DOI] [PubMed] [Google Scholar]
- Deoni S.C. Magnetic resonance relaxation and quantitative measurement in the brain. Methods Mol. Biol. 2011;711:65–108. doi: 10.1007/978-1-61737-992-5_4. [DOI] [PubMed] [Google Scholar]
- Deoni S.C., Williams S.C., Jezzard P., Suckling J., Murphy D.G., Jones D.K. Standardized structural magnetic resonance imaging in multicentre studies using quantitative T1 and T2 imaging at 1.5 T. NeuroImage. 2008;40:662–671. doi: 10.1016/j.neuroimage.2007.11.052. [DOI] [PubMed] [Google Scholar]
- Droby A., Lukas C., Schanzer A., Spiwoks-Becker I., Giorgio A., Gold R., De Stefano N., Kugel H., Deppe M., Wiendl H., Meuth S.G., Acker T., Zipp F., Deichmann R. A human post-mortem brain model for the standardization of multi-centre MRI studies. NeuroImage. 2015;110:11–21. doi: 10.1016/j.neuroimage.2015.01.028. [DOI] [PubMed] [Google Scholar]
- Dusek P., Dezortova M., Wuerfel J. Imaging of iron. Int. Rev. Neurobiol. 2013;110:195–239. doi: 10.1016/B978-0-12-410502-7.00010-7. [DOI] [PubMed] [Google Scholar]
- Enzinger C., Barkhof F., Ciccarelli O., Filippi M., Kappos L., Rocca M.A., Ropele S., Rovira A., Schneider T., de Stefano N., Vrenken H., Wheeler-Kingshott C., Wuerfel J., Fazekas F., Group, M.S. Nonconventional MRI and microstructural cerebral changes in multiple sclerosis. Nat. Rev. Neurol. 2015;11:676–686. doi: 10.1038/nrneurol.2015.194. [DOI] [PubMed] [Google Scholar]
- Fartaria M.J.R.A., Şorega A., O'Brien K., Krueger G., Maréchal B., Sati P., Reich D.S., Kober T., Bach Cuadra M., Granziera C. International Society for Magnetic Resonance Imaging; Honolulu, Hawaii: 2017. Automated Detection of White Matter and Cortical Lesions in MP2RAGE at Ultra-High Fied using a Single Scan. [Google Scholar]
- Fartaria M.J.R.A., Şorega A., O'Brien K., Krueger G., Maréchal B., Sati P., Reich D.S., Kober T., Bach Cuadra M., Granziera C. ECTRIMS; 2017. Automated Validation of Automated Cortical and Subcortical Multiple Sclerosis Lesion Detection Using 7T MP2RAGE Imaging. [Google Scholar]
- Filippi M., Agosta F. Magnetization transfer MRI in multiple sclerosis. J. Neuroimaging. 2007;17(Suppl. 1):22S–26S. doi: 10.1111/j.1552-6569.2007.00132.x. [DOI] [PubMed] [Google Scholar]
- Georgiades C.S., Itoh R., Golay X., van Zijl P.C., Melhem E.R. MR imaging of the human brain at 1.5 T: regional variations in transverse relaxation rates in the cerebral cortex. AJNR Am. J. Neuroradiol. 2001;22:1732–1737. [PMC free article] [PubMed] [Google Scholar]
- Granziera C.S.T. Brain Inflammation, degeneration and plasticity in Multiple Sclerosis. In: Toga A.W., editor. Brain Mapping: An Encyclopedic Reference. 2015. pp. 917–928. [Google Scholar]
- Granziera C., Daducci A., Simioni S., Cavassini M., Roche A., Meskaldji D., Kober T., Metral M., Calmy A., Helms G., Hirschel B., Lazeyras F., Meuli R., Krueger G., Du Pasquier R.A. Micro-structural brain alterations in aviremic HIV+ patients with minor neurocognitive disorders: a multi-contrast study at high field. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granziera C., Daducci A., Romascano D., Roche A., Helms G., Krueger G., Hadjikhani N. Structural abnormalities in the thalamus of migraineurs with aura: a multiparametric study at 3 T. Hum. Brain Mapp. 2014;35:1461–1468. doi: 10.1002/hbm.22266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granziera C., Daducci A., Donati A., Bonnier G., Romascano D., Roche A., Bach Cuadra M., Schmitter D., Kloppel S., Meuli R., von Gunten A., Krueger G. A multi-contrast MRI study of microstructural brain damage in patients with mild cognitive impairment. Neuroimage Clin. 2015;8:631–639. doi: 10.1016/j.nicl.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan K.M., Walimuni I.S., Abid H., Datta S., Wolinsky J.S., Narayana P.A. Human brain atlas-based multimodal MRI analysis of volumetry, diffusimetry, relaxometry and lesion distribution in multiple sclerosis patients and healthy adult controls: implications for understanding the pathogenesis of multiple sclerosis and consolidation of quantitative MRI results in MS. J. Neurol. Sci. 2012;313:99–109. doi: 10.1016/j.jns.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms G. Tissue properties from quantitative MRI. In: Toga A.W., editor. Brain Mapping: An Encyclopedic Reference. Elsevier; 2015. pp. 287–294. [Google Scholar]
- Helms G., Hagberg G.E. In vivo quantification of the bound pool T1 in human white matter using the binary spin-bath model of progressive magnetization transfer saturation. Phys. Med. Biol. 2009;54:N529–N540. doi: 10.1088/0031-9155/54/23/N01. [DOI] [PubMed] [Google Scholar]
- Hilbert T., Sumpf T.J., Weiland E., Frahm J., Thiran J.P., Meuli R., Kober T., Krueger G. Accelerated T2 mapping combining parallel MRI and model-based reconstruction: GRAPPATINI. J. Magn. Reson. Imaging. 2018;48(2):359–368. doi: 10.1002/jmri.25972. [DOI] [PubMed] [Google Scholar]
- Klaver R., De Vries H.E., Schenk G.J., Geurts J.J. Grey matter damage in multiple sclerosis: a pathology perspective. Prion. 2013;7:66–75. doi: 10.4161/pri.23499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging. 2010;29:196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- Kober T., Granziera C., Ribes D., Browaeys P., Schluep M., Meuli R., Frackowiak R., Gruetter R., Krueger G. MP2RAGE multiple sclerosis magnetic resonance imaging at 3 T. Investig. Radiol. 2012;47:346–352. doi: 10.1097/RLI.0b013e31824600e9. [DOI] [PubMed] [Google Scholar]
- Kruggel F., Masaki F., Solodkin A., Alzheimer's Disease Neuroimaging, I Analysis of longitudinal diffusion-weighted images in healthy and pathological aging: an ADNI study. J. Neurosci. Methods. 2017;278:101–115. doi: 10.1016/j.jneumeth.2016.12.020. [DOI] [PubMed] [Google Scholar]
- Lassmann H., Bruck W., Lucchinetti C. Heterogeneity of multiple sclerosis pathogenesis: implications for diagnosis and therapy. Trends Mol. Med. 2001;7:115–121. doi: 10.1016/s1471-4914(00)01909-2. [DOI] [PubMed] [Google Scholar]
- Lucchinetti C., Bruck W. The pathology of primary progressive multiple sclerosis. Mult. Scler. 2004;10(Suppl. 1):S23–S30. doi: 10.1191/1352458504ms1027oa. [DOI] [PubMed] [Google Scholar]
- MacKay A., Whittall K., Adler J., Li D., Paty D., Graeb D. In vivo visualization of myelin water in brain magnetic resonance. Magn. Reson. Med. 1994 Jun;31(6):673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- MacKay A., Laule C., Vavasour I., Bjarnason T., Kolind S., Mädler B. Insights into brain microstructure from the T2 distribution. Magn. Reson. Imaging. 2006 May;24(4):515–525. doi: 10.1016/j.mri.2005.12.037. Epub 2006 Mar 20. [DOI] [PubMed] [Google Scholar]
- Mallik S., Samson R.S., Wheeler-Kingshott C.A., Miller D.H. Imaging outcomes for trials of remyelination in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry. 2014;85:1396–1404. doi: 10.1136/jnnp-2014-307650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques J.P., Kober T., Krueger G., van der Zwaag W., Van de Moortele P.F., Gruetter R. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage. 2010;49:1271–1281. doi: 10.1016/j.neuroimage.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Metere R., Kober T., Moller H.E., Schafer A. Simultaneous quantitative MRI mapping of T1, T2* and magnetic susceptibility with multi-echo MP2RAGE. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz I., Weigand S.D., Popescu B.F., Frischer J.M., Parisi J.E., Guo Y., Lassmann H., Bruck W., Lucchinetti C.F. Pathologic heterogeneity persists in early active multiple sclerosis lesions. Ann. Neurol. 2014;75:728–738. doi: 10.1002/ana.24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S., Mueller K., Stuke K., Bisenius S., Diehl-Schmid J., Jessen F., Kassubek J., Kornhuber J., Ludolph A.C., Prudlo J., Schneider A., Schuemberg K., Yakushev I., Otto M., Schroeter M.L., Group, F.T.S Predicting behavioral variant frontotemporal dementia with pattern classification in multi-center structural MRI data. Neuroimage Clin. 2017;14:656–662. doi: 10.1016/j.nicl.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlau M., Wohlschlager A.M., Gaser C., Valet M., Weindl A., Nunnemann S., Peinemann A., Etgen T., Ilg R. Voxel-based morphometry in individual patients: a pilot study in early Huntington disease. AJNR Am. J. Neuroradiol. 2009;30:539–543. doi: 10.3174/ajnr.A1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlau M., Buck D., Forschler A., Boucard C.C., Arsic M., Schmidt P., Gaser C., Berthele A., Hoshi M., Jochim A., Kronsbein H., Zimmer C., Hemmer B., Ilg R. White-matter lesions drive deep gray-matter atrophy in early multiple sclerosis: support from structural MRI. Mult. Scler. 2013;19:1485–1492. doi: 10.1177/1352458513478673. [DOI] [PubMed] [Google Scholar]
- Neema M., Stankiewicz J., Arora A., Dandamudi V.S., Batt C.E., Guss Z.D., Al-Sabbagh A., Bakshi R. T1- and T2-based MRI measures of diffuse gray matter and white matter damage in patients with multiple sclerosis. J. Neuroimaging. 2007;17(Suppl. 1):16S–21S. doi: 10.1111/j.1552-6569.2007.00131.x. [DOI] [PubMed] [Google Scholar]
- Parry A., Clare S., Jenkinson M., Smith S., Palace J., Matthews P.M. White matter and lesion T1 relaxation times increase in parallel and correlate with disability in Multiple sclerosis. J. Neurol. 2002;249:1279–1286. doi: 10.1007/s00415-002-0837-7. [DOI] [PubMed] [Google Scholar]
- Parry A., Clare S., Jenkinson M., Smith S., Palace J., Matthews P.M. MRI brain T1 relaxation time changes in MS patients increase over time in both the white matter and the cortex. J. Neuroimaging. 2003;13:234–239. [PubMed] [Google Scholar]
- Polman C., Reingold S., Banwell B., Clanet M., Cohen J., Filippi M., Fujihara H., Havrdova E., Hutchinsom M., Kappos L., Lublin F., Montalban X., O'Connor P., Sandberg-Wollheim M., Thompson A., Waubant E., Weinshenker B., Wolinsky J. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche A., Forbes F., Alzheimer's Disease Neuroimaging, I Partial volume estimation in brain MRI revisited. Med. Image Comput. Comput. Assist. Interv. 2014;17:771–778. doi: 10.1007/978-3-319-10404-1_96. [DOI] [PubMed] [Google Scholar]
- Romascano D., Meskaldji D.E., Bonnier G., Simioni S., Rotzinger D., Lin Y.C., Menegaz G., Roche A., Schluep M., Pasquier R.D., Richiardi J., Van De Ville D., Daducci A., Sumpf T., Fraham J., Thiran J.P., Krueger G., Granziera C. Multicontrast connectometry: a new tool to assess cerebellum alterations in early relapsing-remitting multiple sclerosis. Hum. Brain Mapp. 2015;36:1609–1619. doi: 10.1002/hbm.22698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A., Auger C., Alonso J. Magnetic resonance monitoring of lesion evolution in multiple sclerosis. Ther. Adv. Neurol. Disord. 2013;6:298–310. doi: 10.1177/1756285613484079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjadi S.A., Acosta-Cabronero J., Patterson K., Diaz-De-Grenu L.Z., Williams G.B., Nestor P.J. Diffusion tensor magnetic resonance imaging for single subject diagnosis in neurodegenerative diseases. Brain. 2013;136:2253–2261. doi: 10.1093/brain/awt118. [DOI] [PubMed] [Google Scholar]
- Scarpazza C., Sartori G., De Simone M.S., Mechelli A. When the single matters more than the group: very high false positive rates in single case Voxel based Morphometry. NeuroImage. 2013;70:175–188. doi: 10.1016/j.neuroimage.2012.12.045. [DOI] [PubMed] [Google Scholar]
- Schmitter D., Roche A., Marechal B., Ribes D., Abdulkadir A., Bach-Cuadra M., Daducci A., Granziera C., Kloppel S., Maeder P., Meuli R., Krueger G., Alzheimer's Disease Neuroimaging, I An evaluation of volume-based morphometry for prediction of mild cognitive impairment and Alzheimer's disease. Neuroimage Clin. 2015;7:7–17. doi: 10.1016/j.nicl.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewann A., Vrenken H., van der Valk P., Blezer E.L., Knol D.L., Castelijns J.A., Polman C.H., Pouwels P.J., Barkhof F., Geurts J.J. Diffusely abnormal white matter in chronic Multiple sclerosis: imaging and histopathologic analysis. Arch. Neurol. 2009;66:601–609. doi: 10.1001/archneurol.2009.57. [DOI] [PubMed] [Google Scholar]
- Stephan K.E., Schlagenhauf F., Huys Q.J.M., Raman S., Aponte E.A., Brodersen K.H., Rigoux L., Moran R.J., Daunizeau J., Dolan R.J., Friston K.J., Heinz A. Computational neuroimaging strategies for single patient predictions. NeuroImage. 2017;145:180–199. doi: 10.1016/j.neuroimage.2016.06.038. [DOI] [PubMed] [Google Scholar]
- Sumpf T.J., Uecker M., Boretius S., Frahm J. Model-based nonlinear inverse reconstruction for T2 mapping using highly undersampled spin-echo MRI. J. Magn. Reson. Imaging. 2011;34:420–428. doi: 10.1002/jmri.22634. [DOI] [PubMed] [Google Scholar]
- Vrenken H., Seewann A., Knol D.L., Polman C.H., Barkhof F., Geurts J.J. Diffusely abnormal white matter in progressive multiple sclerosis: in vivo quantitative MR imaging characterization and comparison between disease types. AJNR Am. J. Neuroradiol. 2010;31:541–548. doi: 10.3174/ajnr.A1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansapura J.P., Holland S.K., Dunn R.S., Ball W.S., Jr. NMR relaxation times in the human brain at 30 tesla. J. Magn. Reson. Imaging. 1999;9:531–538. doi: 10.1002/(sici)1522-2586(199904)9:4<531::aid-jmri4>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- West J., Aalto A., Tisell A., Leinhard O.D., Landtblom A.M., Smedby O., Lundberg P. Normal appearing and diffusely abnormal white matter in patients with multiple sclerosis assessed with quantitative MR. PLoS One. 2014;9 doi: 10.1371/journal.pone.0095161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material