Abstract
Reported here are DNA and RNA-like systems built from eight (hachi-) nucleotide letters (-moji) that form four orthogonal pairs. This synthetic genetic biopolymer meets the structural requirements needed to support Darwinism, including a polyelectrolyte backbone, predictable thermodynamic stability, and stereoregular building blocks that fit a Schrödinger aperiodic crystal. Measured thermodynamic parameters predict the stability of hachimoji duplexes, allowing hachimoji DNA to double the information density of natural terran DNA. Three crystal structures show that the synthetic building blocks do not perturb the aperiodic crystal seen in the DNA double helix. Hachimoji DNA was then transcribed to give hachimoji RNA in the form of a functioning fluorescent hachimoji aptamer. These results expand the scope of molecular structures that might support life, including life throughout the cosmos.
One Sentence Summary:
DNA with eight nucleotide letters forms four orthogonal pairs with predictable thermodynamics, supports transcription to give a functioning 8-letter fluorescent RNA aptamer, and creates double helices that fit the Schrödinger aperiodic crystal needed for Darwinism, presumed to be universally necessary for life in the cosmos.
No behaviors are more central to biology than the storage, transmission, and evolution of genetic information. In modern terran biology, this is achieved by DNA double helices whose strands are joined by regularly sized nucleobase pairs with hydrogen bond complementarity (1). Schrödinger theorized that such regularity in size was necessary for the pairs to fit into an aperiodic crystal, which he proposed to be necessary for reliable molecular information storage, and faithful information transfer (2). This feature is also essential for any biopolymer that might support Darwinian evolution, as it ensures that changes in the sequence of the informational building blocks do not damage the performance of the biopolymer, including its interactions with enzymes that replicate it.
Complementary inter-base hydrogen bonding has been proposed to be dispensable in Darwinian molecules, provided that size complementarity is retained (3). Thus, hydrophobic nucleotide analogs have been incorporated into duplexes (4), aptamers (5), and living cells (6). These analogs increase the number of genetic letters from four to six. However, pairs lacking inter-base hydrogen bonds evidently must be flanked by pairs joined by hydrogen bonds. Further, unless they are constrained by an enzyme active site, hydrophobic pairs can slip atop each other (7), shortening the rung in the DNA ladder, distorting the double helix, and damaging the aperiodic crystal uniformity of the duplex.
When hydrogen bonding is used to give a third pair, behaviors characteristic of natural DNA are also reproduced (8). Thus, 6-letter DNA alphabets with inter-pair hydrogen bonds can be copied (9), PCR-amplified and sequenced (10, 11), transcribed to 6-letter RNA and back to 6-letter DNA (12), and used to encode proteins with added amino acids (13). Six-letter alphabets with all pairs joined by inter-base hydrogen bonds also support Darwinian selection, evolution and adaptation (14), all hallmarks of the living state.
Here, we test the limits of molecular information storage that combines Watson-Crick hydrogen bonding with Schrödinger’s requirement for crystal-like uniformity, building an alien genetic system from eight (“hachi-”) building block “letters” (-”moji”). This required the design of two sets of heterocycles that implement two additional hydrogen bonding patterns that join two additional pairs (Fig. 1).
Figure 1.
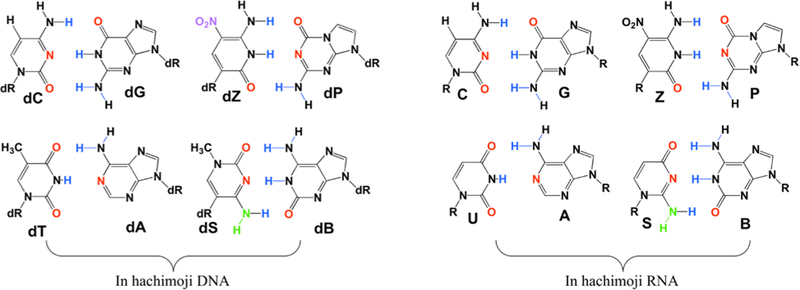
The eight nucleotides of hachimoji DNA (left) and hachimoji RNA (right) are designed to form four size- and hydrogen bond-complementary pairs. Hydrogen bond donor atoms involved in pairing are blue; hydrogen bond acceptor atoms are red. The left two pairs in each set are formed from the four standard nucleotides (note missing hydrogen bonding group in the A:T pair, a peculiarity of standard terran DNA/RNA). The right two pairs are formed from the four new non-standard nucleotides. Notice the absence of electron density in the minor groove of S, which has a NH (green) moiety.
We first assessed the regularity and predictability of the thermodynamics of interaction between hachimoji DNA strands. With standard DNA, the energy of duplex formation is not accurately modeled by a single parameter for each base pair. Instead, to make usefully accurate predictions of duplex stability, predictive tools must account for sequence context (15). With standard DNA, this is done by obtaining nearest-neighbor (NN) thermodynamic parameters for all base pair dimers (BPDs) (15). Parameters are often added to account for the decrease in translational degrees of freedom when two strands become one duplex, and to specially treat the distinctively weak A:T pair at the ends of duplexes.
If context dependence is similar in hachimoji DNA, tools that make usefully accurate predictions should also require parameters for all BPDs for an 8-letter alphabet. Of course, with eight building blocks instead of four, hachimoji DNA has many more BPDs to parameterize. After accounting for symmetry (e.g. AC/TG is equivalent to GT/CA), 40 parameters are required (28 more than the 12 required for standard DNA). These comprise 36 added BPDs plus four for pairs initiated with terminal G:C and terminal effects for A:T, S:B, and Z:P, where S is 3- methyl-6-amino-5-(1’-β-D-2’-deoxyribofuranosyl)-pyrimidin-2-one, B is 6-amino-9-(1’-β-D-2’- deoxyribofuranosyl)-4-hydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-purin-2-one, Z is 6-amino- 3-(1’-β-D-2’-deoxyribofuranosyl)-5-nitro-1H-pyridin-2-one, and P is 2-amino-8-(1’-β-D-2’- deoxyribofuranosyl)-imidazo-[1,2a]-1,3,5-triazin-[8H]-4-one.
To obtain these additional parameters, 94 hachimoji duplexes (Table S1) were designed with standard A, T, G, and C, purine analogs “P” and “B”, and pyrimidine analogs “Z” and “S” (Fig. 1). If the design is successful, these duplexes should be joined by P:Z and B:S pairs in addition to standard G:C and A:T pairs. The paired hachimoji DNA oligonucleotides were synthesized by solid phase chemistry from synthetic phosphoramidites, assembled, and melted to collect thermodynamic data. These data were processed with Meltwin v.3.5 (16) to obtain a parameter set using both the (Tm-1 vs. Ln(Ct)) method and the Marquardt non-linear curve fit method. The error-weighted average of the values from the two methods yielded the ΔG°37 and ΔH° values for the 94 duplexes (17).
This analysis allowed us to determine the 28 additional parameters for the 8-letter genetic system using singular value decomposition methods (SI Tables S4, S7, and S10, Fig. S1, S3 and S5). As this number of measurements over-determines the unknown parameters by a factor of 3.3, we could test the applicability of the BPD model, using error propagation to derive standard deviations in the derived parameters (17). The parameters and standard deviations are in Figs. S1, S3, and S5. A cross-validation approach gave the same result, as expected given the over-determination.
The resulting parameters proved to usefully predict melting temperatures for hachimoji DNA. Plots of experimental vs. predicted free-energy changes and experimental and predicted melting temperatures (Fig 2) show that on average, Tm is predicted within 2.1 °C for the 94 GACTZPSB hachimoji duplexes, and ΔG°37 is predicted to within 0.39 kcal/mol (Tables S3, S6, and S9). These errors are similar to those observed with nearest-neighbor parameters for standard DNA:DNA duplexes (15). Thus, GACTZPSB hachimoji DNA reproduces, in expanded form, the molecular recognition behavior of standard 4-letter DNA. It is an informational system.
Figure 2.
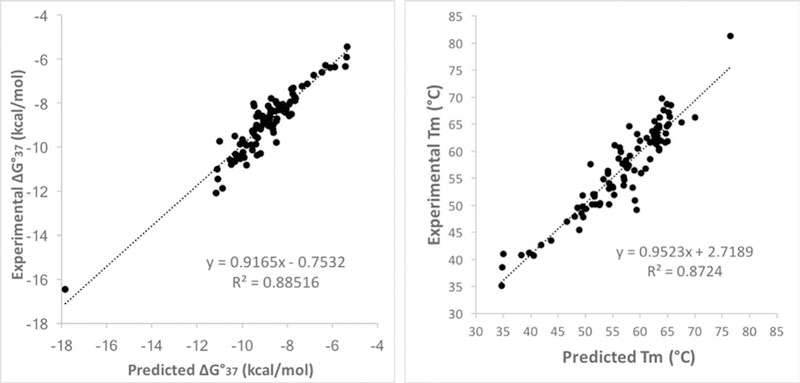
(left) Plot of experimental vs. predicted free energy changes (ΔG°37) for 94 SBZP- containing hachimoji DNA duplexes (Tables S3, S6, and S9). (right) Plot of experimental vs. predicted melting temperatures of 94 SBZP-containing hachimoji DNA duplexes (Tables S3, S6, and S9). The outlier is a sequence embedded in the PP guest (Fig. 3E).
We then asked whether hachimoji DNA might be mutable without damaging the Schrödinger aperiodic crystal required to support mutability and Darwinian evolution. High-resolution crystal structures were determined for three different hachimoji duplexes assembled from three self-complementary hachimoji 16-mers: 5’-CTTATPBTASZATAAG (“PB”, 1.7 Å; PDB ID 6MIG), 5’-CTTAPCBTASGZTAAG (“PC”, 1.6 Å; PDB ID 6MIH), and 5’-CTTATPPSBZZATAAG (“PP”, 1.7 Å; PDB ID 6MIK). These duplexes were crystallized with Moloney murine leukemia virus reverse transcriptase to give a “host-guest” complex with two protein molecules (host) bound to each end of a 16mer duplex (guest) (Fig. 3) (18). With interactions between the host and guest limited to the ends, the intervening 10 bp are free to adopt a sequence-dependent structure (Fig. 3A) (19).
Figure 3.
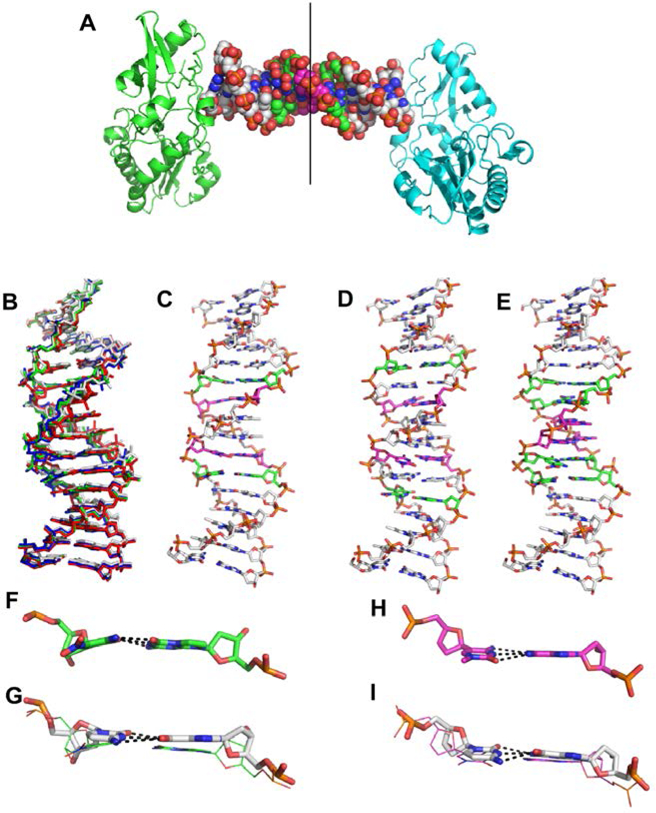
Crystal structures of PB, PC, and PP hachimoji DNA. (A) The host-guest complex with two N-terminal fragments from Moloney murine leukemia virus reverse transcriptase in green and cyan bound to a 16mer PP hachimoji DNA; in the duplex sphere model, Z:P pairs are green, S:B pairs magenta. The asymmetric unit includes one protein molecule and half of the 16mer DNA as indicated by the line. (B) Hachimoji DNA structures PB (green), PC (red), PP (blue) are superimposed with GC DNA (gray). (C) Structure of hachimoji DNA with self- complementary duplex CTTATPBTASZATAAG (“PB”). (D) Structure of hachimoji DNA with self-complementary duplex 5’-CTTAPCBTASGZTAAG, (“PC”). (E) Structure of hachimoji DNA with self-complementary duplex with six consecutive non-standard components 5’-CTTATPPSBZZATAAG (PP). DNA structures are shown as stick models with P:Z pairs (C, green), B:S pairs (C, magenta), and natural pairs (C, gray). Examples of largest differences in detailed structures: (F) The Z:P pair (from the PB structure) is more buckled than corresponding G:C pair in (G). (H) The S:B pair (from the PB structure) exhibits a propeller angle similar to that in the corresponding G:C pair shown in (I).
The hachimoji DNA in all three structures adopts a B-form (Fig. 3B-E) with 10.2–10.4 bp/turn, as analyzed by 3DNA (20). The major and minor groove widths for hachimoji DNA are similar to one another and to GC DNA (5’-CTTATGGGCCCATAAG), but not AT DNA (5’- CTTATAAATTTATAAG) (Fig. S16). For nucleotide pair parameters, the S:B pairs at position 7 in both the PC and PB structures exhibit very similar propeller angles but slightly greater opening angles as compared to G:C in the same position. The P:Z pairs adjacent to natural pairs exhibit larger buckle angles compared to G:C pairs (Fig. 3F-I).
Even with these differences, the structural parameters for the individual pairs and the dinucleotide steps of the hachimoji DNA fall well within the ranges observed for natural four letter DNA. Thus, it appears that hachimoji DNA meets the Schrödinger requirement for a Darwinian system, forming essentially the same “aperiodic crystal” regardless of the sequences. It is a mutable information storage system.
With the information storage and mutability properties shown for hachimoji DNA, we asked whether hachimoji information could also be transmitted, here to give hachimoji GACUZPSB RNA, where S is 2-amino-1-(1’-β-D-ribofuranosyl)-4(1H)-pyrimidinone, B is 6-amino-9-(1’-β-D-ribofuranosyl)-4-hydroxy-5-(hydroxymethyl)-oxolan-2-yl]-1H-purin-2-one, Z is 6-amino-3- (1’-β-D-ribofuranosyl)-5-nitro-1H-pyridin-2-one, and P is 2-amino-8-(1’-β-D-ribofuranosyl)- imidazo-[1,2a]-1,3,5-triazin-[8H]-4-one. To develop RNA polymerases able to transcribe hachimoji DNA, we started with four model sequences that each contained a single nonstandard hachimoji component, B, P, S, and Z, each followed by a single C nucleotide. To analyze hachimoji RNA products, transcripts were labeled with alpha-32P-CTP; digestion with ribonuclease T2 then generated the corresponding hachimoji 3’-phosphates (21). These were resolved in TLC systems and compared with synthetic authentic nonstandard 3’-phosphates.
These experiments showed that native T7 RNA polymerase incorporates riboZTP opposite template dP, riboPTP opposite template dZ, and riboBTP opposite template dS (Figs. S11 and S12). However, incorporation of riboSTP opposite template dB was not seen with native RNA polymerase (22). This was attributed to an absence of electron density in the minor groove from the aminopyridone heterocycle on riboSTP (Fig. 1); polymerases are believed to recognize such density, which is presented by all other triphosphate substrates.
We therefore searched for T7 RNA polymerase variants able to transcribe a complete set of hachimoji nucleotides (Table S11). One variant (Y639F H784A P266L, “FAL”) was especially effective at incorporating riboSTP opposite template dB (Figs. S13 and S14). FAL was originally developed as a thermostable polymerase to accept 2’-O-methyl triphosphates (23). HPLC analysis of its transcripts showed that 1.2 ± 0.4 riboSTP nucleotides were incorporated opposite a single template dB (Fig. S9). FAL also incorporated the other non-standard components of the hachimoji system into transcripts (Figs. S13-S14).
We then designed a hachimoji variant of the spinach fluorescent RNA aptamer (24). In its standard form, spinach folds and binds 3,5-difluoro-4-hydroxybenzylidene imidazolinone which, when bound, fluoresces green. An analogous hachimoji aptamer with non-standard nucleotides placed strategically to avoid disrupting its fold (Fig. 4) was prepared by transcribing hachimoji DNA using the FAL variant of RNA polymerase. The aptamer’s sequence was confirmed by label transfer experiments; incorporation of riboZ was further confirmed by HPLC and UV spectroscopy.
Figure 4.
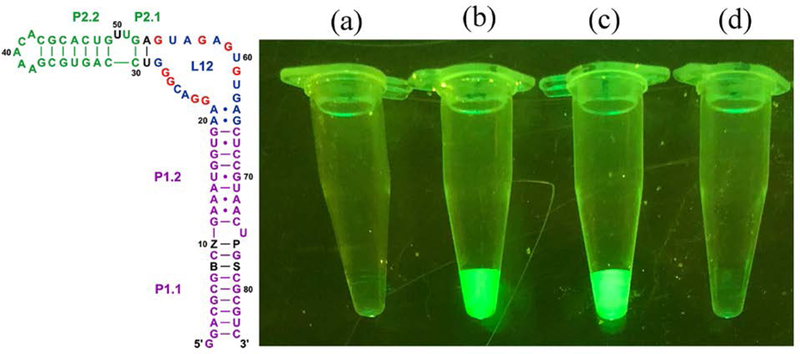
(Left) Schematic showing the full hachimoji spinach variant aptamer; additional nucleotide components of the hachimoji system are shown as black letters at positions 8, 10,76, and 78 (B, Z, P, and S respectively). The fluor binds in loop L12 (25). (Right) Fluorescence of various species in equal amounts as determined by UV. Fluorescence was visualized under a blue light (470 nm) with an amber (580 nm) filter. From left to right: (a) Control with fluor only, lacking RNA, (b) hachimoji spinach with the sequence shown in the left panel (c) native spinach aptamer with fluor, and (d) fluor and spinach aptamer containing Z at position 50, replacing A:U pair at positions 53:29 with G:C to restore the triple observed in the crystal structure. This places the quenching Z chromophore near the fluor; CD spectra suggest that this variant had the same fold as native spinach (Fig. S8).
The hachimoji spinach fluoresced green (Fig. 4). As a control, a spinach variant was prepared with a Z incorporated at position 50, near enough to the bound fluor to quench its fluorescence. That variant did not fluoresce, even though circular dichroism (CD) experiments (Fig. S8) suggested that its overall fold was undisturbed. These results precluded the possibility that non-standard hachimoji components are generally misincorporated throughout the aptamer, as these would disrupt the fold needed for the fluor to bind or place a quenching riboZ nucleotide near enough to loop L12 to eliminate its fluorescence.
Concluding, this synthetic biology makes available a mutable genetic system built from eight different building blocks. With double the information density of standard DNA and predictable duplex stability across (evidently) all 8n sequences of length n, hachimoji DNA has potential applications in bar-coding and combinatorial tagging, retrievable information storage, and self-assembling nano-structures. The structural differences between three different hachimoji duplexes are not larger than the differences between various standard DNA duplexes, making this system potentially able to support molecular evolution. Further, the ability to have structural regularity independent of sequence shows the importance of inter-base hydrogen bonding in such mutable informational systems. Thus, in addition to its technical applications, this work provides a specifically expanded scope of the structures that we might encounter as we search the cosmos.
Supplementary Material
Acknowledgments:
Funding: This work was supported by grants from the National Aeronautics and Space Administration under award NNX15AF46G, the National Institute of General Medical Science at the NIH (R41GM119494, R01GM128186, R01GM102489), and the Defense Threat Reduction Agency under award HDTRA1–13-1–0004. This publication also was made possible through the support of a grant from the John Templeton Foundation 54466 and the Templeton World Charity Foundation, Inc. 0092/AB57. Use of the Stanford Synchrotron Radiation Lightsource (SLAC National Accelerator Laboratory) was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02–76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (including P41GM103393). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the NIH, DOE, NASA, DTRA, JTF, or TWCF. We thank Accelero Biostructures, Inc. for their rapid, high quality X-ray diffraction data collection services.
Footnotes
Competing interests: S.A.B., N.A.L., S.H., and the Foundation for Applied Molecular Evolution have a financial interest in the intellectual property in the area of expanded genetic alphabets. Firebird Biomolecular Sciences, which makes various hachimoji reagents available for sale, is owned by S.A.B. N.E.W., H.A.S., and J.S.L. are owners of DNA Software, Inc., which owns intellectual property and software unrelated to this paper that makes thermodynamic predictions concerning nucleotide binding. A.D.E. has a financial interest in the T7 RNA polymerase variants used here. A. D. E. and A. J. M. are inventors on patent application US 15/127,617 held by The Board of Regents of The University of Texas System which covers “T7 RNA polymerase variants with expanded substrate range and enhanced transcriptional yield.” S.A.B. and N.A.L filed patent application 16226963 “Enzymatic Processes for Synthesizing RNA Containing Certain Non-Standard Nucleotides”, on 20 December 2018.
Data and materials availability: Coordinates are deposited as PDB ID 6MIG, PDB ID 6MIH, and PDB ID 6MIK. All reagents can be purchased from Firebird Biomolecular Sciences LLC.
References and Notes:
- 1.Watson JD, Crick FHC, Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. Nature 171, 737–738 (1953). [DOI] [PubMed] [Google Scholar]
- 2.Schrödinger E, Was ist Leben Serie Piper, Vol. 1134 (1943). [Google Scholar]
- 3.Goodman MF, On the wagon. DNA polymerase joins “H-bonds anonymous”. Nature Biotechnol 17, 640–640 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Moran S, Ren RX-F, Rumney S IV, Kool ET, Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc 119, 2056–2057 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimoto M, Yamashige R, Matsunaga KI, Yokoyama S, Hirao I, Generation of high- affinity DNA aptamers using an expanded genetic alphabet. Nature Biotechnol 31, 453–457 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Ptacin JL, Fischer EC, Aerni HR, Caffaro CE, Jose KS, et al. , A semi-synthetic organism that stores and retrieves increased genetic information. Nature 551, 644–647 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, et al. , KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry. Nat. Chem. Biol 8, 612–614 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benner SA, Karalkar NB, Hoshika S, Laos R, Shaw RW, Matsuura M, et al. , Alternative Watson-Crick synthetic genetic systems. Synthetic Biology Cold Spring Harbor Perspectives in Biology, Cold Spring Harbor Press; (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Sismour AM, Sheng P, Puskar NL, Benner SA, Enzymatic incorporation of a third nucleobase pair. Nucl. Acids Res 35, 4238–4249 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sismour AM, Lutz S, Park J-H, Lutz MJ, Boyer PL, Hughes SH, et al. , PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from human immunodeficiency virus-1. Nucl. Acids. Res 32, 728–735 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Chen F, Alvarado JB, Benner SA, Amplification, mutation, and sequencing of a six-letter synthetic genetic system. J. Am. Chem. Soc 133, 15105–15112 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leal NA, Kim H-J, Hoshika S, Kim M-J, Carrigan MA, Benner SA, Transcription, reverse transcription, and analysis of RNA containing artificial genetic components. ACS Synthetic Biol 4, 407–413 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Bain JD, Chamberlin AR, Switzer CY, Benner SA, Ribosome-mediated incorporation of non-standard amino acids into a peptide through expansion of the genetic code. Nature 356, 537–539 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Yang Z, Sefah K, Bradley KM, Hoshika S, Kim M-J, et al. , Evolution of functional six-nucleotide DNA. J. Am. Chem. Soc 137, 6734–6737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.SantaLucia J Jr., A unified view of polymer, dumbbell, and oligonucleotide DNA nearest- neighbor thermodynamics. Proc. Natl Acad. Sci. USA 95, 1460–1465 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDowell JA, Turner DH, Investigation of the structural basis for thermodynamic stabilities of tandem GU mismatches: solution structure of (rGAGGUCUC)2 by two- dimensional NMR and simulated annealing. Biochemistry 35, 14077–14089 (1996). [DOI] [PubMed] [Google Scholar]
- 17.SantaLucia J Jr., Determination of nucleic acid thermodynamics by UV absorbance melting curves, in spectrophotometry and spectrofluorimetry: A practical approach (Gore MG, Ed.), Oxford U. Press; (2000). [Google Scholar]
- 18.Coté ML, Yohannon SJ, Georgiadis MM, Use of an N-terminal fragment from moloney murine leukemia virus reverse transcriptase to facilitate crystallization and analysis of a pseudo-16-mer DNA molecule containing G-A mispairs. Acta Crystallography Sect. D: Biol. Crystallography 56, 1120–1131 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Montano SP, Cote ML, Roth MJ, Georgiadis MM, Crystal structures of oligonucleotides including the integrase processing site of the Moloney murine leukemia virus. Nucleic Acids Res 34, 5353–5360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu XJ, Olson WK, 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res 31, 5108–5121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirao I, Ohtsuki T, Fujiwara T, Mitsui T, Yokogawa T, Okuni T, et al. , An unnatural base pair for incorporating amino acid analogs into proteins. Nature Biotechnol 20, 177–182 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Switzer CY, Moroney SE, Benner SA, Enzymatic recognition of the base pair between iso-cytidine and iso-guanosine. Biochemistry 32, 10489–10496 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Meyer AJ, Garry DJ, Hall B, Byrom MM, McDonald HG, Yang X, et al. , Transcription yield of fully 2′-modified RNA can be increased by the addition of thermostabilizing mutations to T7 RNA polymerase mutants. Nucleic Acids Res 43, 7480–7488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paige JS, Wu KY, Jaffrey SR, RNA mimics of green fluorescent protein. Science 333, 642–646 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, et al. , A G- quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat Chem Biol, 10, 686–691 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Hutter D, Sheng P, Sismour AM, Benner SA, Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. Nucleic Acids Res 34, 6095–6101 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim HJ, Leal NA, Benner SA, 2’-deoxy-1-methylpseudocytidine, a stable analog of 2’- deoxy-5-methylisocytidine. Bioorg. Med. Chem 17, 3728–3732 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allawi HT, SantaLucia J Jr., Thermodynamics and NMR of internal G•T mismatches in DNA. Biochemistry 36, 10581–10594 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Kim HJ, Leal NA, Hoshika S, Benner SA, Ribonucleosides for an Artificially Expanded Genetic Information System. J. Org. Chem 79, 3194–3199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh I, Laos R, Hoshika S, Benner SA, Georgiadis MM, Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide. Nucleic Acids Res 46, 7977–7988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DasGupta S, Shelke SA, Li N-S, Piccirilli JA, Spinach RNA detects lead (II) with high selectivity. Chem. Commun 51, 9034–9037 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun D, Jessen S, Liu C, Liu X, Najmudin S, Georgiadis MM, Cloning, expression, and purification of a catalytic fragment of Moloney murine leukemia virus reverse transcriptase: crystallization of nucleic acid complexes. Protein Sci 7, 1575–1582 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W, XDS. Acta Crystallogr. D Biol. Crystallogr 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN, How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ, Phaser crystallographic software. J. Appl. Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodel A, Kim S-H, Brunger AT Model bias in macromolecular crystal structures. Acta Crystallogr A Foundations and Advances, 48, 851–858 (1992). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


