Abstract
We demonstrated the effects of exosomes secreted by cardiac mesenchymal stem cells (C-MSC-Exo) in protecting acute ischemic myocardium from reperfusion injury. To investigate the effect of exosomes from C-MSC on angiogenesis. We injected C-MSC-Exo or PBS intramuscularly into ischemic hind limb. Blood perfusion of limb was evaluated by laser Doppler Imaging. We observed that ischemic limb treated with C-MSC-Exo exhibit improved blood perfusion compared to ischemic limb treated with PBS at 2 weeks and 1 month after induction of limb ischemia. To explore the potential mechanisms underlying C-MSC-Exo’s angiogenetic effect, we performed microRNA array analysis, and identify mmu-miR-7116-5p as the most abundant enriched miRNA detected in C-MSC-Exo. Bioinformatics’ analysis shows that miR-7116-5p negatively regulates of protein polyubiquitination. In conclusion, our study demonstrated that intramuscular delivery of C-MSC-Exo after limb ischemia improves blood perfusion, and we identified most abundant miRNAs that are preferentially enriched in C-MSC-Exo.
Keywords: Cardiac Mesenchymal Stem cells, Exosomes, hind limb ischemia, Angiogenesis, miR-7116-5p
Introduction
Ischemic heart disease is the leading cause of death in the developed and developing countries[1]. An acute myocardial infarction can lead to rapid loss of billions of adult cardiomyocytes[2], however, adult hearts have very limited capacity for regeneration[3], therefore, replacement of dead cardiomyocytes with scar tissue leads to left ventricular remodeling and heart failure[4, 5]. While the revascularization treatments, such as percutaneous transluminal coronary angioplasty (PTCA) and coronary artery bypass grafting (CABG) surgery, can reopen occluded coronary arteries in patients with coronary artery disease[6], current research shows that these treatments are inefficient in solving microvascular dysfunction (CMD)[7], so the risk myocardium is still in a microenvironment with insufficient blood supply, which trig cell apoptosis[8]. The progressive apoptosis of cardiomyocytes exacerbates adverse left ventricular remodeling and ultimately promotes the progress of chronic heart failure[5].
Stem cells have the potential for promoting heart repair. Endothelial progenitor cells (EPCs) have been shown to protect ischemic myocardium via angiogenesis and vasculogenesis[9]. There are cardiac mesenchymal stem cells (C-MSCs) in the adult heart[10]. C-MSCs express different levels of GATA4 (early heart transcription factor) and mesenchymal stem cell markers, such as Sca-1, CD105, and CD44 on their cell surface, indicating that C-MSCs are population of cardiac specific mesenchymal stem cells[10-14]. MSCs are supporting cells in tissues and play a very important role in maintenance of tissue homeostasis[15]. Our previous studies showed that transplantation of bone marrow derived MSCs promotes cardiac angiogenesis through paracrine effects [16, 17]. Since cultured stem cells are at risk of tumorigenesis for clinical application [18], we want to determine if the non-cellular, paracrine fraction of C- MSCs can be used for angiogenesis.
We have reported that C-MSC secretes exosomes that have paracrine effects to protect ischemic myocardium from apoptosis induced by acute myocardial ischemia/reperfusion [19]. Other group also reported that serum exosomes attenuate H2O2-induced apoptosis in cultured cardiomyocytes [20]. Exosomes are nano-sized cargoes with lipid layers protect the integrity of microRNAs (miRNAs), which play a key role in cell-to-cell communication [21-24]. Exosome-derived miRNAs can also be used as biomarkers for diagnosis and therapy [25].
The purpose of this study was to determine whether the delivery of cardiac MSC- derived exosomes (C-MSC-Exo) can improve limb blood perfusion in ischemic hind limbs, and identifies the most enriched, abundant miRNA in C-MSC-Exo. Our results show that intramuscular injection of C-MSC-Exo can improve blood perfusion in the hindlimb, while miR-7116-5p are the most enriched, abundant miRNAs in C-MSC-Exo.
Methods
C-MSC isolation and culture
Mouse C-MSCs were isolated from 2 to 3 months old mouse hearts (C57BL/6, The Jackson Laboratory, Bar Harbor, Maine) by a 2-step procedure as previously described with modification [11, 12, 26]. Briefly, in step 1, ventricular heart tissues were minced into 1 mm3 size, and then digested with 0.1% collagenase IV and 1 U/mL Dispase in DMEM/F-12. The digested heart tissue was seeded into 6 well plate coated with fibronectin/gelatin (0.5 mg fibronectin in 100 mL 0.1% gelatin). After two weeks, the migrated round, phase-bright cells migrated from adherent explants were collected, and undergone hematopoietic cell depletion using the mouse hematopoietic lineage depletion cocktail kit (Stem Cell Technologies) by magnetic activated cell sorting (MACS) followed by cell enrichment with Sca-1 magnetic beads (Miltenyi Biotec Inc., Auburn, CA) according to the manufacturer's protocol. The sorted Sca-1 cells were cultured in complete medium (DMEM/F12 containing 10% fetal bovine serum (FBS), 200mmol/L L-Glutamine, 55nmol/L β-mercaptoethanol and 1% MEM non-essential amino acid).
Flow cytometry
The surface markers of cultured C-MSC are characterized by flow cytometry analyses with a BD LSRII flow cytometer and BD FACSDiva™ software as previously described[27]. Briefly, C-MSCs were blocked with 5% rat serum and stained with a panel of conjugated antibodies, including anti-CD105-APC (BioLegend), anti-CD140-PE (eBioscience) and anti-CD117-FITC (BD Biosciences Pharmingen).
Histology
For cell staining, C-MSC were plated on 8-well chamber slides (Millipore, Billerica, MA) and fixed with 4% paraformaldehyde. After blocking with 5% goat serum, cells were incubated with rabbit anti-GATA4 antibody (1:100; Aviva System Biology, San Diego, CA) or rabbit anti-Ki67 antibody (1:400; Cell Signaling Technology, Danvers, MA) at 4 °C overnight. Primary antibodies were resolved via secondary staining with goat anti-rabbit Alexa Fluor 555-conjugated (1:400, Life Technologies, Carlsbad, CA). Slides were mounted using VECTASHIELD HardSet Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA).
Purification of C-MSC-Exo
Exosomes/microvesicles secreted by C-MSCs were purified from conditioned media by polymer-based exosome precipitation protocol as we described previously [19, 28, 29]. Briefly, C-MSC were cultured in culture medium containing 10% exosome-depleted FBS for 48 hours, collected media was centrifuged at 1000 rpm for 10 minutes, and then filtered through a 0.22 μm filters. C-MSC-Exo were precipitated with 5x polyethylene glycol 4000 (PEG 4000, 8.5% final concentration) and 10x NaCl (0.4 mol/L final concentration) overnight at 4 °C followed by centrifugation at 3,000 rpm for 30 min. The pellets were resuspended with PBS and stored at −80°C until use. The size of exosome particle was measured with nanoparticle tracking analysis (NTA) with ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany) at 23°C and corresponding software ZetaView 8.02.28 as we described previously[30]. The ZetaView system was calibrated using 100 nm polystyrene particles.
Immuno-electron microscopy imaging
Standard immunoelectron staining with anti-CD63 antibody was performed as previously described [12]. The fixed exosomal preparations were placed on a carbon Formvar-coated 200-mesh nickel grid and incubated for 30 minutes. The grid was then quenched with 1 M ammonium chloride for 30 min and blocked with 0.4% BSA in PBS for 2 hrs. The grid was washed with PBS and incubated with primary rabbit anti-CD63 (1:100 Santa Cruz Biotechnology, Inc., Santa Cruz, CA) for 1 hr. The grid was then washed with ddH2O and PBS, and 1.4 nm anti-rabbit nanogold (1:1000, Nanoprobes, Inc.) were dropped in blocking buffer for 1 hr. After enhancement with HQ Silver (gold enhancement reagent, Nanoprobes, Inc.), the samples were dried prior to observation in a transmission electron microscope (JEOL JEM 1230, Peabody, MA). TEM sample preparation and imaging were performed at the Electron Microscopy and Histology Core Laboratory at Augusta University (www.augusta.edu/mcg/cba/emhisto/).
Western blotting assay
Exosomes were lysed in RIPA buffer with Triton X-100 (Alfa Aesar, Ward Hill, MA). The proteins from C-MSC-Exo were resolved on 10% SDS gel and transferred to nitrocellulose membrane (LI-COR Biosciences, Lincoln, NE). The membranes were blocked with Odyssey blocking buffer (LICOR Biosciences, Lincoln, NE), exposed to rabbit anti-Tsg101 (1:1000, Thermo Scientific), and rabbit anti-CD63 (1:250, Santa Cruz Biotechnology, Inc.) overnight at 4°C. Then membranes were incubated with IRDye 680 goat anti-rabbit IgG (LI-COR Biosciences) at 1:10,000 for 1 hr at room temperature. Probed blots were scanned using Odyssey infrared imager.
Murine hind limb ischemia model and intramuscular C-MSC-Exo delivery
Wide type C57BL/6 mice were anesthetized with intraperitoneal injection of 100mg/kg ketamine and 10mg/kg Xylazine. A ligation was made around the left femoral artery, and all arterial branches were removed. A small segment of the artery was then dissected free. Mice were randomly assigned to receive intramuscular injections of PBS or C-MSC-Exo after induction of ischemia. The ischemic hindlimb were intramuscularly injected with in a 30 μL PBS or 30 μL C-MSC-Exo (50μg) at 4 different locations immediately after the surgery. Animals were treated according to approved protocols and animal welfare regulations of the Institutional Animal Care and Use Committee of the Medical College of Georgia, Augusta University.
Laser Doppler Perfusion Imaging
Mice were anesthetized with isoflurane (2%), and subjected to laser Doppler Perfusion imaging measurements before induction of hind limb ischemia (HLI) (baseline), 1 day, 2 weeks, and 1 month after HLI. At each time point, blood flow in ischemic and non-ischemic limbs was measured, and results were reported as a ratio of these 2 measurements.
MicroRNA microarray assay
To identify the most enriched, abundant miRNAs in C-MSC-Exo, we extracted total RNAs from C-MSC and C-MSC-Exo using RNAzol® (Molecular Research Center, Cincinnati, OH). The RNA samples were subjected to microarray analysis using GeneChip miRNA 4.0 Array (Affymetrix, Santa Clara, CA, USA). The RNA labeling, microarray hybridization, washing, and scanning were performed based on the manufacturer’s standard protocols in Integrated Genomics Core in the Georgia Cancer Center, Augusta University.
C-MSC-Exo treatment and qRT-PCR of muscle tissue
To determine whether local delivery of C-MSC-Exo can increase level of miR-7116-5p in treated muscle, we intramuscularly injected 50μg C-MSC-Exo into left tibialis anterior (TA) muscle, and used right TA as non-treated control. Left TA and right TA muscles were dissected after 24hrs. Total RNA was isolated by RNAzol RT (Molecular Research Center, Inc., Cincinnati, OH) following the manufacturer's instructions. cDNAs were synthesized from total RNA by using Mir-X™ miRNA First-Strand Synthesis kit (Clontech Laboratories, Inc.). The cDNA synthesized was used to perform quantitative PCR on CFX96 Touch Real-Time PCR Detection System (Bio-Rad) using PowerUp SYBR® Green Master Mix (ThermoFisher). Amplification was performed at 50°C for 2min, 95 °C for 2 min, followed by 51 cycles of 95 °C for 15 s, and 60 °C for 1 min with the following primes: miR-7116-5p Forward primers: 5’ GCAGCGTGTGAAGACATCAGGA 3’ Reverse primer (mRQ 3’) and U6 Forward and Reverse Prime were provided in Mir-X™ miRNA First-Strand Synthesis kit.
Statistical Analysis
Results are presented as the mean ± standard error of the mean (SEM). Comparisons between two groups were made by two-tailed Student’s t test. Differences were considered statistically significant at p < 0.05.
Results:
Characterization of C-MSC
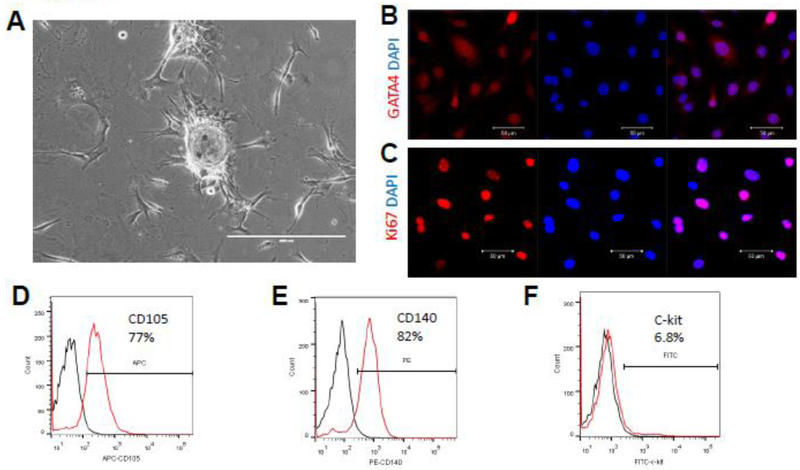
As mentioned previously, we isolated C-MSC from the adult mouse heart using a two- step protocol (Fig. 1A) [31, 32]. Immunofluorescent staining showed that C-MSC express GATA4, a marker for early cardiac transcription factor (Fig. 1B), and Ki67, a cellular marker for proliferation (Fig. 1C). Flow cytometry showed that C-MSCs express high levels of mesenchymal stem cell markers CD105 (77%) and CD140 (82%) (Fig. 1D-E), and about 6.8% C-MSC are c-kit positive, a controversial cardiac stem cell marker (Fig. 1E). Taken together, these data indicate that C-MSCs represent a subpopulation of cardiac-derived mesenchymal stem cells.
Figure 1:
Phenotypic characterization of C-MSC cells. (A) Phase contrast picture of cultured C-MSC; (B) Immunofluorescent staining of C-MSC cells for expression of the cardiac transcription factors GATA4 (red); cell nuclei were counterstained with DAPI (blue); (C) Immunofluorescent staining of C-MSC cells for expression of the proliferative marker Ki67 (red); cell nuclei were counterstained with DAPI (blue); (D-F). Flow cytometric analyses of C-MSC cells for expression of CD105, CD140, and c-Kit.
Characterization of C-MSC derived Exosomes
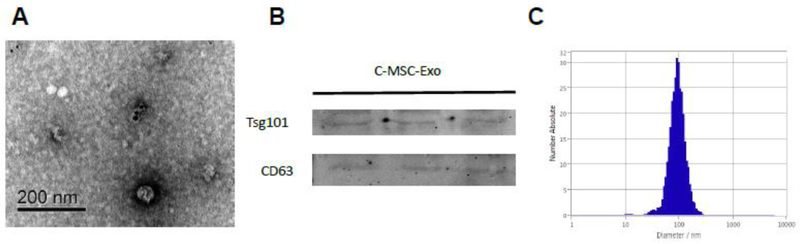
Morphological analysis of C-MSC-Exo using electron micrography demonstrated the typical appearance of microvesicles positive for exosomal marker CD63(Fig. 2A). Western blot analysis confirmed the presence of exosome markers, including TSG101, and CD63 (Fig. 2B). ZetaView®, a nanoparticle tracking analyzer that uses Brownian motion, was employed to measure the size of the microvesicles. The particles exhibited an average diameter around 100 nm, consistent with the characteristic size range of exosomes (Fig. 2C).
Figure 2:
Characterization of C-MSC derived exosomes. (A) Transmission Electron micrograph image of C-MSC-derived exosomes after immunoelectron labeling with anti- CD63 antibody. Scale bar = 200 nm; (B) Western blot results demonstrate the expression of Tsg101, and CD63 in exosomes derived from C-MSC; (C) Particle size distribution in purified pellets consistent with size range of exosomes (average size around 100 nm), measured by ZetaView® Particle Tracking Analyzer.
Effect of C-MSC-Exo on blood perfusion of hind limb by laser Doppler imaging
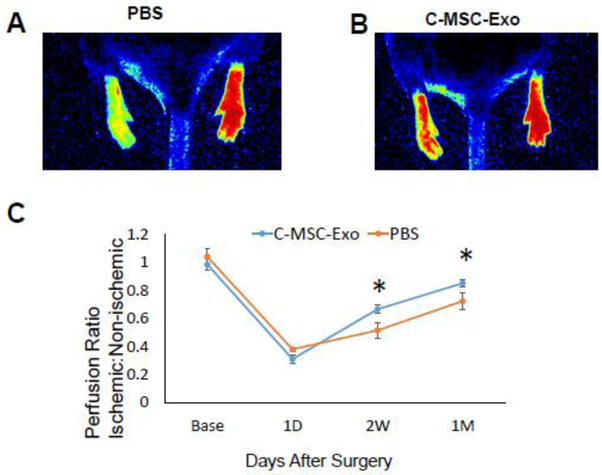
We developed ischemia in the mouse hindlimb by ligating the femoral artery. After intramuscular injection of C-MSC-Exo or PBS, we observed that C-MSC-Exo treatment restored approximately 85% of perfusion relative to non-ischemic limbs after 1 month (Fig. 3A-C) while PBS-treated control has about 72% perfusion restoration relative to the non-ischemic limb, indicating C-MSC-Exo treatment can improve blood perfusion in limb ischemia.
Figure 3:
Laser Doppler Perfusion imaging measurements before induction of hind limb ischemia (HLI) (baseline), 1 day, 2 weeks, and 1 month after HLI. (A-B) Representative Laser Doppler images of mice treated with PBS or C-MSC-Exo at 1 month post HLI; (C) Time course of perfusion ratio (Ischemic: non-ischemic paw) at baseline, 1 day, 2 weeks, and 1 month after PBS and C-MSC-Exo treatment in mice (*, P<0.05, n=5 for PBS group, n=7 for C-MSC-Exo group).
Identify the most enriched, abundant miRNAs from C-MSC-Exo
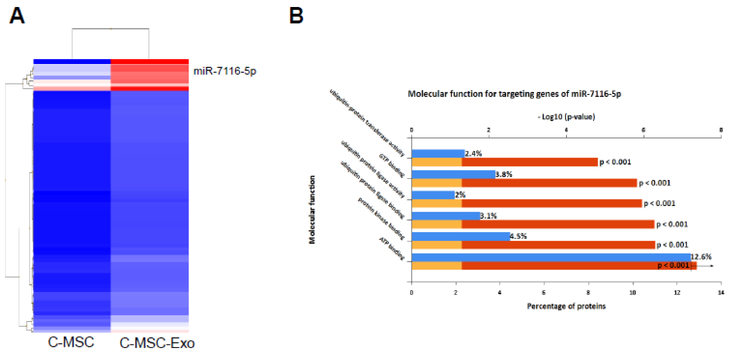
Exosomal miRNAs play an important role in stimulating angiogenesis[33], to identify the microRNA profile of the C-MSC-Exo, we analyzed microRNA expression profiles of C-MSC-Exo and C-MSC by microarray based miRNA profiling. There were 72 miRNAs enriched in C-MSC-Exo compared with C-MSC (Fold change ≥2, Fig. 4A, Table 1), among them, miR-7116 is the most enriched, high abundant miRNA with 32-fold enrichment in C-MSC-Exo.
Figure 4:
The enriched microRNAs in C-MSC-Exo comparing with C-MSC. (A) Enriched miRNAs in C-MSC were identified by differential expression analysis of miRNAs in C-MSC-Exo versus C-MSC when setting the Fold change ≥2, and miR-7116-5p is the most enriched, abundant miRNA in C-MSC-Exo; (B) FunRich functional enrichment analysis of predicted, targeted genes of miR-7116-5p. MiR-7116-5p is supposed to participate in regulation of protein ubiquitination.
Table 1.
Summary of enriched miRNAs (Fold change >2) in C-MSC-Exo compared with C-MSC by microRNA array
| miRNAs | C-MSC Avg (log2) |
C-MSC-Exo Avg (log2) |
Fold Change |
Accession |
|---|---|---|---|---|
| mmu-miR-7116-5p | 2.86 | 7.87 | 32.28 | MIMAT0028129 |
| mmu-mir-465c-1 | 3.94 | 8.45 | 22.65 | MI0005500 |
| mmu-mir-465c-2 | 3.94 | 8.45 | 22.65 | MI0005501 |
| mmu-miR-7025-5p | 4.23 | 7.8 | 11.82 | MIMAT0027954 |
| mmu-miR-5128 | 6.68 | 9.71 | 8.12 | MIMAT0020639 |
| mmu-miR-195a-3p | 1.64 | 4.52 | 7.35 | MIMAT0017000 |
| mmu-miR-6968-5p | 5.37 | 8.05 | 6.42 | MIMAT0027838 |
| mmu-miR-7001-5p | 2.63 | 4.87 | 4.74 | MIMAT0027904 |
| mmu-miR-7030-5p | 1.48 | 3.56 | 4.21 | MIMAT0027964 |
| mmu-miR-1931 | 3.51 | 5.57 | 4.15 | MIMAT0009394 |
| mmu-miR-126b-5p | 0.42 | 2.18 | 3.37 | MIMAT0029894 |
| mmu-miR-16-1-3p | 0.48 | 2.23 | 3.37 | MIMAT0004625 |
| mmu-miR-6347 | 1.85 | 3.53 | 3.22 | MIMAT0025090 |
| mmu-miR-6379 | 0.04 | 1.5 | 2.75 | MIMAT0025125 |
| mmu-miR-871-5p | 0.18 | 1.64 | 2.75 | MIMAT0004841 |
| mmu-mir-541 | 0.35 | 1.81 | 2.73 | MI0003521 |
| mmu-miR-340-3p | 0.2 | 1.65 | 2.73 | MIMAT0000586 |
| mmu-mir-452 | 0.45 | 1.83 | 2.61 | MI0001734 |
| mmu-miR-599 | −0.09 | 1.3 | 2.61 | MIMAT0012772 |
| mmu-mir-2137 | 0.92 | 2.29 | 2.58 | MI0010750 |
| mmu-mir-497 | 0.92 | 2.29 | 2.58 | MI0004636 |
| mmu-miR-185-3p | 0.4 | 1.76 | 2.57 | MIMAT0016996 |
| mmu-miR-433-5p | 0.4 | 1.76 | 2.57 | MIMAT0001419 |
| mmu-miR-3106-3p | 0.3 | 1.66 | 2.57 | MIMAT0014818 |
| mmu-mir-297b | 0.54 | 1.9 | 2.57 | MI0004674 |
| mmu-mir-1943 | 0.24 | 1.6 | 2.57 | MI0009932 |
| mmu-miR-466m-3p | 0.73 | 2.09 | 2.57 | MIMAT0014883 |
| mmu-mir-3086 | 0.26 | 1.58 | 2.5 | MI0014049 |
| mmu-miR-370-5p | 0.08 | 1.4 | 2.5 | MIMAT0017174 |
| mmu-miR-411-5p | 0.02 | 1.27 | 2.37 | MIMAT0004747 |
| mmu-mir-7032 | 1.24 | 2.36 | 2.18 | MI0022881 |
| mmu-miR-686 | 0.19 | 1.31 | 2.18 | MIMAT0003464 |
| mmu-miR-5103 | 0.19 | 1.31 | 2.18 | MIMAT0020610 |
| mmu-mir-7655 | 0.19 | 1.31 | 2.18 | MI0024995 |
| mmu-miR-878-3p | 0.36 | 1.49 | 2.18 | MIMAT0004933 |
| mmu-mir-6394 | 0.35 | 1.48 | 2.18 | MI0021928 |
| mmu-mir-6948 | 0.36 | 1.49 | 2.18 | MI0022795 |
| mmu-mir-7084 | 0.49 | 1.62 | 2.18 | MI0022934 |
| mmu-mir-30d | 0.54 | 1.67 | 2.18 | MI0000549 |
| mmu-mir-3966 | 0.17 | 1.3 | 2.18 | MI0016975 |
| mmu-mir-412 | 0.02 | 1.12 | 2.14 | MI0001164 |
| mmu-mir-5112 | 1.22 | 2.31 | 2.14 | MI0018021 |
| mmu-miR-7050-3p | 0.36 | 1.46 | 2.14 | MIMAT0028005 |
| mmu-miR-3062-3p | 1.01 | 2.1 | 2.14 | MIMAT0014831 |
| mmu-miR-18b-3p | 0.72 | 1.82 | 2.14 | MIMAT0017270 |
| mmu-miR-6950-3p | 0.37 | 1.47 | 2.14 | MIMAT0027801 |
| mmu-mir-150 | 0.53 | 1.63 | 2.14 | MI0000172 |
| mmu-mir-433 | 0.53 | 1.63 | 2.14 | MI0001525 |
| mmu-mir-1948 | 0.53 | 1.63 | 2.14 | MI0009939 |
| mmu-miR-3099-5p | 0.19 | 1.28 | 2.14 | MIMAT0014815 |
| mmu-miR-3110-5p | 0.56 | 1.65 | 2.14 | MIMAT0014951 |
| mmu-miR-6365 | 0.37 | 1.47 | 2.14 | MIMAT0025109 |
| mmu-miR-7688-3p | 0.37 | 1.47 | 2.14 | MIMAT0029907 |
| mmu-mir-384 | 0.37 | 1.47 | 2.14 | MI0001146 |
| mmu-mir-544 | 0.19 | 1.28 | 2.14 | MI0005555 |
| mmu-mir-466k | 0.19 | 1.28 | 2.14 | MI0006292 |
| mmu-miR-551b-5p | 0.23 | 1.31 | 2.11 | MIMAT0017236 |
| mmu-miR-5620-3p | 0.38 | 1.46 | 2.11 | MIMAT0022368 |
| mmu-miR-6993-3p | 0.38 | 1.46 | 2.11 | MIMAT0027889 |
| mmu-miR-7008-5p | 0.38 | 1.46 | 2.11 | MIMAT0027920 |
| mmu-mir-208b | 0.38 | 1.46 | 2.11 | MI0005552 |
| mmu-mir-6349 | 0.73 | 1.81 | 2.11 | MI0021877 |
| mmu-miR-741-3p | 0.54 | 1.62 | 2.11 | MIMAT0004236 |
| mmu-miR-190b-3p | 0.2 | 1.27 | 2.11 | MIMAT0017267 |
| mmu-miR-7012-3p | 0.54 | 1.62 | 2.11 | MIMAT0027929 |
| mmu-miR-7236-5p | 0.2 | 1.27 | 2.11 | MIMAT0028440 |
| mmu-miR-18b-5p | 0.52 | 1.59 | 2.11 | MIMAT0004858 |
| mmu-miR-3075-5p | 0.21 | 1.29 | 2.11 | MIMAT0014858 |
| mmu-miR-3547-5p | 5.09 | 6.16 | 2.09 | MIMAT0027832 |
| mmu-miR-1928 | 0.42 | 1.42 | 2 | MIMAT0009391 |
| mmu-mir-1947 | 0.25 | 1.25 | 2 | MI0009937 |
| mmu-mir-7010 | 0.77 | 1.77 | 2 | MI0022859 |
To predict the potential molecular function of miR-7116-5p, we use TargetScan, a popular bioinformatics tool to analyze 5795 predicted gene targets of miR-7116-5p, and then use Funrich software for molecular function enrichment analysis [34]. Figure 4B shows the percentage of genes predicted to be involved in molecular function of miR-7116-5p, including regulation of protein polyubiquitination, GTP binding, and ATP binding. This result suggests that miR-7116-5p may play an important role in C-MSC- Exo mediated paracrine effect via inhibiting protein polyubiquitination.
Local delivery of C-MSC-Exo transfers miR-7116-5p in muscle of mice
To determine whether miR-7116-5p can be transferred into muscle tissue by local C- MSC-Exo delivery, we intramuscularly injected 50ug C-MSC-Exo into left TA muscle, and used right TA muscle as non-treated control. To allow time for miRNA transfer, we harvested muscle 1 day after injection, we found that the level of miR-7116-5p increased about 20 fold by local C-MSC-Exo delivery in treated muscles compared with non-treated control muscle by qRT-PCR assay, suggesting that miR-7116-5p can be transferred into muscle by C-MSC-Exo.
Discussion:
In this study, we found that intramuscular injection of C-MSC-Exo into ischemic limb improves blood perfusion in ischemic limbs. Using microRNA array, we identified the most enriched, abundant miRNAs in C-MSC-Exo, which have not been reported yet. MiRNAs from C-MSC-Exo are among the most important molecular factors controlling beneficial paracrine effect of C-MSC. Therefore, we evaluated the miRNA expression profile in C-MSC and C-MSC-Exo through miRNA microArray. Our results showed that miR-7116-5p is the most enriched, abundant miRNA in C-MSC-Exo compared to C-MSC, A report has shown that inhibition of miR-7116-5p level sin microglia enhances TNF-α production and inflammation, which lead to neuron damage [35], however, there is no report about the function of miR-7116-5p in cardiovascular and muscle systems. Bioinformatics pathway analysis shows that miR-7116-5p negatively regulates protein polyubiquitination. Certainly, we need to verify the exact targets of the miRNAs and define the pathways in future studies in order to better understand the role miR-7116-5p in functional benefit of C-MSC-Exo in ischemic muscle.
Exosome mediated local and distal cell-to-cell communication, and stem cell-derived exosomes have potential for treating cardiovascular diseases[36]. Exosomes can carry miRNAs to modify the molecular signals of recipient cells. Therefore, miRNAs play a key role for exosome- mediated communication [11, 37]. Sorting miRNAs into exosomes is cell-specific and related to cytopathophysiology. Exosomes have a unique mechanism for enriching miRNAs, Pigati L et al. [38] found the existence of a cellular selection mechanism determining the difference of extracellular and cellular miRNA profile. Chen L. et al [19] also found that CPC-Exo have high level expression of GATA4-reactive miR-451, but not miR-144, the asymmetric distribution of miR-451 is also reported by other studies in normal and malignant cells, and their derived exosomes[38]. The mechanisms by which miRNAs were loaded from their derived cells into exosomes remain elusive. Villarroya-Beltri C et al [39] show that the SUMOylated form of heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) specifically binds to exosomal miRNAs by recognizing these motifs and controls their loading into exosomes. Recently, Santangelo L et al.[40] demonstrated that the SUMOylated RNA binding protein SYNCRIP, which binds to GGCU seed sequence of specific miRNAs, was involved in hepatocyte exosomal miRNA sorting. Liu X et al[41] also reported the phenomenon of the selective packing of miRNA cargo into exosomes under hypertensive status. In this study, we analyzed the miRNA transcriptome of C-MSC and their secreted exosomes, and identified selectively enriched miRNAs in mouse C-MSC-Exo, such as miR-7116-5p, the function of miR-7116-5p has not been well evaluated in the cardiovascular system yet.
In summary, we have shown that C-MSC-derived exosomes have the potential to act as therapeutic agents for angiogenesis. Our study supports that MSC-derived exosomes improve blood perfusion in ischemic hindlimb, and miR-7116-5p is the most enriched, high abundant miRNAs inside of C-MSC-Exo, which might play a critical role in regulating protein ubiquitination. The roles and mechanism of miR-7116-5p in cell protection deserves further study.
Figure 5:
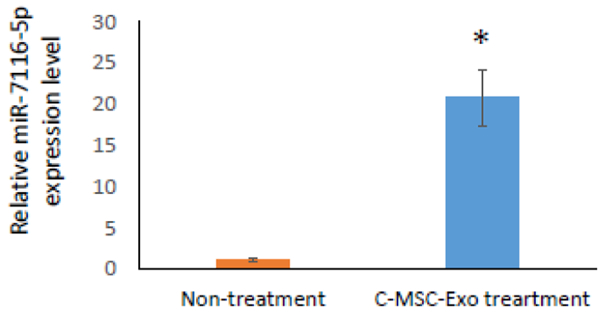
Relative miR-7116-5p levels in non-treated or C-MSC-Exo treated TA muscles as quantified by qRT-PCR (*, P<0.05, n=3).
Acknowledgments
Funding: Y. Tang were partially supported by the American Heart Association: GRNT31430008, NIH-AR070029, NIH-HL086555, NIH-HL134354.
Footnotes
Conflict of Interest: All authors declares that he/she has no conflict of interest.
Ethical approval: Animals were treated according to approved protocols and animal welfare regulations of the Institutional Animal Care and Use Committee of the Medical College of Georgia, Augusta University. This article does not contain any studies with human participants performed by any of the authors.
Literature cited
- [1].Gaziano TA, Bitton A, Anand S, Abrahams-Gessel S, Murphy A, Growing epidemic of coronary heart disease in low- and middle-income countries, Curr Probl Cardiol, 35 (2010) 72–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Xin M, Olson EN, Bassel-Duby R, Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair, Nat Rev Mol Cell Biol, 14 (2013) 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Steinhauser ML, Lee RT, Regeneration of the heart, EMBO Mol Med, 3 (2011) 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Domenech M, Polo-Corrales L, Ramirez-Vick JE, Freytes DO, Tissue Engineering Strategies for Myocardial Regeneration: Acellular Versus Cellular Scaffolds?, Tissue Eng Part B Rev, 22 (2016) 438–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Z, Su X, Ashraf M, Kim IM, Weintraub NL, Jiang M, Tang Y, Regenerative Therapy for Cardiomyopathies, Journal of cardiovascular translational research, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Foussas SG, Tsiaousis GZ, Revascularization treatment in patients with coronary artery disease, Hippokratia, 12 (2008) 3–10. [PMC free article] [PubMed] [Google Scholar]
- [7].Loffler AI, Bourque JM, Coronary Microvascular Dysfunction, Microvascular Angina, and Management, Curr Cardiol Rep, 18 (2016) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Awada HK, Hwang MP, Wang Y, Towards comprehensive cardiac repair and regeneration after myocardial infarction: Aspects to consider and proteins to deliver, Biomaterials, 82 (2016) 94–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Djohan AH, Sia CH, Lee PS, Poh KK, Endothelial Progenitor Cells in Heart Failure: an Authentic Expectation for Potential Future Use and a Lack of Universal Definition, Journal of cardiovascular translational research, (2018). [DOI] [PubMed] [Google Scholar]
- [10].Tang YL, Wang YJ, Chen LJ, Pan YH, Zhang L, Weintraub NL, Cardiac-derived stem cell-based therapy for heart failure: progress and clinical applications, Experimental biology and medicine (Maywood, N.J.), 238 (2013) 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ruan XF, Li YJ, Ju CW, Shen Y, Lei W, Chen C, Li Y, Yu H, Liu YT, Kim IM, Wang XL, Weintraub NL, Tang Y, Exosomes from Suxiao Jiuxin pill-treated cardiac mesenchymal stem cells decrease H3K27 demethylase UTX expression in mouse cardiomyocytes in vitro, Acta pharmacologica Sinica, 39 (2018) 579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ruan XF, Ju CW, Shen Y, Liu YT, Kim IM, Yu H, Weintraub N, Wang XL, Tang Y, Suxiao Jiuxin pill promotes exosome secretion from mouse cardiac mesenchymal stem cells in vitro, Acta pharmacologica Sinica, 39 (2018) 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen L, Ashraf M, Wang Y, Zhou M, Zhang J, Qin G, Rubinstein J, Weintraub NL, Tang Y, The role of notch 1 activation in cardiosphere derived cell differentiation, Stem cells and development, 21 (2012) 2122–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen L, Phillips MI, Miao HL, Zeng R, Qin G, Kim IM, Weintraub NL, Tang Y, Infrared fluorescent protein 1.4 genetic labeling tracks engrafted cardiac progenitor cells in mouse ischemic hearts, PloS one, 9 (2014) e107841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Klimczak A, Kozlowska U, Mesenchymal Stromal Cells and Tissue-Specific Progenitor Cells: Their Role in Tissue Homeostasis, Stem Cells Int, 2016 (2016) 4285215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tang YL, Zhao Q, Zhang YC, Cheng L, Liu M, Shi J, Yang YZ, Pan C, Ge J, Phillips MI, Autologous mesenchymal stem cell transplantation induce VEGF and neovascularization in ischemic myocardium, Regulatory peptides, 117 (2004) 3–10. [DOI] [PubMed] [Google Scholar]
- [17].Tang YL, Zhao Q, Qin X, Shen L, Cheng L, Ge J, Phillips MI, Paracrine action enhances the effects of autologous mesenchymal stem cell transplantation on vascular regeneration in rat model of myocardial infarction, The Annals of thoracic surgery, 80 (2005) 229–236; discussion 236-227. [DOI] [PubMed] [Google Scholar]
- [18].Zhang L, Pan Y, Qin G, Chen L, Chatterjee TK, Weintraub NL, Tang Y, Inhibition of stearoyl-coA desaturase selectively eliminates tumorigenic Nanog-positive cells: improving the safety of iPS cell transplantation to myocardium, Cell Cycle, 13 (2014) 762–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen L, Wang Y, Pan Y, Zhang L, Shen C, Qin G, Ashraf M, Weintraub N, Ma G, Tang Y, Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury, Biochemical and biophysical research communications, 431 (2013) 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li P, Liu Z, Xie Y, Gu H, Dai Q, Yao J, Zhou L, Serum Exosomes Attenuate H2O2-Induced Apoptosis in Rat H9C2 Cardiomyocytes via ERK1/2, Journal of cardiovascular translational research, (2018). [DOI] [PubMed] [Google Scholar]
- [21].Sahoo S, Klychko E, Thorne T, Misener S, Schultz KM, Millay M, Ito A, Liu T, Kamide C, Agrawal H, Perlman H, Qin G, Kishore R, Losordo DW, Exosomes from human CD34(+) stem cells mediate their proangiogenic paracrine activity, Circulation research, 109 (2011) 724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Khan M, Nickoloff E, Abramova T, Johnson J, Verma SK, Krishnamurthy P, Mackie AR, Vaughan E, Garikipati VN, Benedict C, Ramirez V, Lambers E, Ito A, Gao E, Misener S, Luongo T, Elrod J, Qin G, Houser SR, Koch WJ, Kishore R, Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction, Circulation research, 117 (2015) 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kishore R, Khan M, More Than Tiny Sacks: Stem Cell Exosomes as Cell-Free Modality for Cardiac Repair, Circulation research, 118 (2016) 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kishore R, Khan M, Cardiac cell-derived exosomes: changing face of regenerative biology, Eur Heart J, 38 (2017) 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lu M, Yuan S, Li S, Li L, Liu M, Wan S, The Exosome-Derived Biomarker in Atherosclerosis and Its Clinical Application, Journal of cardiovascular translational research, (2018). [DOI] [PubMed] [Google Scholar]
- [26].Chen L, Pan Y, Zhang L, Wang Y, Weintraub N, Tang Y, Two-step protocol for isolation and culture of cardiospheres, Methods in molecular biology (Clifton, N.J.), 1036 (2013) 75–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tang YL, et al. (2009). Hypoxic preconditioning enhances the benefit of cardiac progenitor cell therapy for treatment of myocardial infarction by inducing CXCR4 expression. Circulation Research, 104, 1209–1216. 10.1161/CIRCRESAHA.109.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Y, Zhang L, Li Y, Chen L, Wang X, Guo W, Zhang X, Qin G, He SH, Zimmerman A, Liu Y, Kim IM, Weintraub NL, Tang Y, Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium, International journal of cardiology, 192 (2015) 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen Z, Li Y, Yu H, Shen Y, Ju C, Ma G, Liu Y, Kim IM, Weintraub NL, Tang Y, Isolation of Extracellular Vesicles from Stem Cells, Methods in molecular biology (Clifton, N.J.), 1660 (2017) 389–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y, A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents, PloS one, 12 (2017) e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ruan XF, Ju CW, Shen Y, Liu YT, Kim IM, Yu H, Weintraub N, Wang XL, Tang YL, Suxiao Jiuxin pill promotes exosome secretion from mouse cardiac mesenchymal stem cells in vitro, Acta pharmacologica Sinica, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ruan XF, Li YJ, Ju CW, Shen Y, Lei W, Chen C, Li Y, Yu H, Liu YT, Kim IM, Wang XL, Weintraub NL, Tang YL, Exosomes from Suxiao Jiuxin Pill-treated cardiac mesenchymal stem cells decrease H3K27 demethylase UTX expression in mouse cardiomyocytes in vitro, Acta pharmacologica Sinica, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S, Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function, Genomics, Proteomics & Bioinformatics, 13 (2015) 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pathan M, Keerthikumar S, Chisanga D, Alessandro R, Ang CS, Askenase P, Batagov AO, Benito-Martin A, Camussi G, Clayton A, Collino F, Di Vizio D, Falcon-Perez JM, Fonseca P, Fonseka P, Fontana S, Gho YS, Hendrix A, Hoen EN, Iraci N, Kastaniegaard K, Kislinger T, Kowal J, Kurochkin IV, Leonardi T, Liang Y, Llorente A, Lunavat TR, Maji S, Monteleone F, Overbye A, Panaretakis T, Patel T, Peinado H, Pluchino S, Principe S, Ronquist G, Royo F, Sahoo S, Spinelli C, Stensballe A, Thery C, van Herwijnen MJC, Wauben M, Welton JL, Zhao K, Mathivanan S, A novel community driven software for functional enrichment analysis of extracellular vesicles data, Journal of extracellular vesicles, 6(2017)1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].He Q, Wang Q, Yuan C, Wang Y, Downregulation of miR-7116-5p in microglia by MPP(+) sensitizes TNF-alpha production to induce dopaminergic neuron damage, Glia, 65 (2017) 1251–1263. [DOI] [PubMed] [Google Scholar]
- [36].Ni J, Sun Y, Liu Z, The Potential of Stem Cells and Stem Cell-Derived Exosomes in Treating Cardiovascular Diseases, Journal of cardiovascular translational research, (2018). [DOI] [PubMed] [Google Scholar]
- [37].Campbell CR, Berman AE, Weintraub NL, Tang YL, Electrical stimulation to optimize cardioprotective exosomes from cardiac stem cells, Medical hypotheses, 88 (2016) 6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM, Selective release of microRNA species from normal and malignant mammary epithelial cells, PloS one, 5 (2010) e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sanchez-Madrid F, Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs, Nature communications, 4 (2013) 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M, The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting, Cell reports, 17 (2016) 799–808. [DOI] [PubMed] [Google Scholar]
- [41].Liu X, Yuan W, Yang L, Li J, Cai J, miRNA Profiling of Exosomes from Spontaneous Hypertensive Rats Using Next-Generation Sequencing, Journal of cardiovascular translational research, (2018). [DOI] [PubMed] [Google Scholar]