Abstract
Antiphospholipid syndrome (APS) is a systemic autoimmune disease characterized by thromboembolic events and pregnancy loss. We sought to characterize the DNA methylation profile of primary APS in comparison to healthy controls and individuals with SLE. In primary APS neutrophils compared to controls, 17 hypomethylated and 25 hypermethylated CpG sites were identified. Notable hypomethylated genes included ETS1, a genetic risk locus for SLE, and PTPN2, a genetic risk locus for other autoimmune diseases. Gene ontology analysis of hypomethylated genes revealed enrichment of genes involved in pregnancy. None of the differentially methylated sites in primary APS were differentially methylated in SLE neutrophils, and there was no demethylation of interferon signature genes in primary APS as is seen in SLE. Hypomethylation within a single probe in the IFI44L promoter (cg06872964) was able to distinguish SLE from primary APS with a sensitivity of 93.3% and specificity of 80.0% at a methylation fraction of 0.329.
Keywords: Antiphospholipid syndrome, Autoimmunity, Epigenetics, Lupus, Methylation, Neutrophil
1. Introduction
Antiphospholipid syndrome (APS) is a systemic autoimmune disorder characterized by the occurrence of thromboembolic events and/or pregnancy loss in the setting of antiphospholipid autoantibodies [1]. In roughly half of individuals with APS, the disease occurs as an isolated syndrome, in which case it is classified as primary APS [2]. In the majority of remaining cases, however, APS occurs as a secondary manifestation of systemic lupus erythematosus (SLE) or other connective tissue disease. As such, the diagnosis of APS often prompts clinical evaluation for possible underlying SLE, which can be challenging to diagnose especially in its early stages. Among those who initially present with primary APS, 13–23% may later develop SLE or lupus-like disease within the next decade of their life based on retrospective studies [3,4].
The pathophysiology of APS remains incompletely understood, especially with regard to its mechanisms of autoimmunity. Indeed, APS is generally treated with anticoagulation rather than immunosuppression given the lack of clear immunologic target pathways and lack of clinical benefit when immunosuppressive strategies have been trialed empirically [5]. The close association between APS and SLE raises the obvious question of whether these disorders might share common underlying pathogenic mechanisms. Differential DNA methylation is a known epigenetic feature of SLE that not only differentiates SLE from healthy individuals, but also correlates with various organ-specific manifestations, and may play a dynamic role in disease activity via mediation of T helper cell response, among other pathophysiologic mechanisms [6]. We have previously demonstrated epigenetic aberrancies in SLE neutrophils, with robust demethylation in interferon-regulated genes [7]. More recently, we demonstrated a pro-inflammatory transcriptional signature in APS neutrophils, suggesting a role for neutrophils in the pathogenesis of APS [8]. The DNA methylation profile of APS remains to be characterized, however.
In this study, we examined the genome-wide DNA methylation signatures in neutrophils from individuals with primary APS as compared to healthy controls and compared to lupus neutrophils from the cohort which our group previously reported on [7], with the intent to inform our understanding of the pathophysiology and differentiation of these two closely interrelated diseases. We also specifically examined the methylation status of the IFI44L promoter in primary APS versus SLE. In whole blood, the methylation level of this promoter has been shown to distinguish individuals with SLE from healthy controls with >90% sensitivity and specificity in a Chinese population, with more modest results (sensitivity and specificity ranging roughly 70–90% – superior to currently available biomarkers) in those of European descent and in distinguishing SLE from rheumatoid arthritis and primary Sjögren’s syndrome [9].
2. Methods
2.1. Primary APS participants and controls
Initially, 12 participants with primary APS were recruited from the University of Michigan rheumatology clinics. All met the Sydney APS Classification Criteria [10]. From this group, 10 were successfully run on the DNA methylation array, and as such the final primary APS cohort was comprised of these 10 individuals. Their demographic and clinical characteristics are summarized in Table 1. Healthy controls were recruited by advertisement at the University of Michigan. A total of 12 controls (not shown) were matched to the original 12 primary APS participants by age (± 5 years), sex, and ethnicity.
Table 1.
Demographic and clinical characteristics of the primary APS cohort. All participants were of European-American ethnicity.
| Number | Age | Sex | Disease duration (yrs) | Antibody profile | Clinical characteristics |
|---|---|---|---|---|---|
| 1 | 33 | F | 5 | aCL | Pregnancy morbidity |
| 2 | 67 | M | 16 | aCL, LA | Venous and arterial thrombosis |
| 3 | 40 | F | 7 | aCL, LA | Arterial thrombosis, livedo reticularis, thrombocytopenia |
| 4 | 61 | M | 1 | aCL | Venous thrombosis |
| 5 | 37 | M | 12 | aB2G, aCL, LA | Arterial thrombosis, livedo reticularis |
| 6 | 36 | M | 15 | aB2G, aCL, LA | Venous and arterial thrombosis |
| 7 | 28 | F | 7 | aCL, LA | Pregnancy morbidity, venous thrombosis |
| 8 | 52 | M | 5 | aB2G, aCL, LA | Venous thrombosis |
| 9 | 40 | F | 1 | aB2G, aCL | Pregnancy morbidity, livedo reticularis, thrombocytopenia |
| 10 | 42 | F | 16 | aB2G, aCL, LA | Venous thrombosis |
aB2G, anti-beta2 glycoprotein I antibody; aCL, anti-cardiolipin antibody; LA, lupus anticoagulant.
The average age of the primary APS and control group was 44 ± 13 and 43 ± 13 years old, respectively. There was no statistically significant demographic difference between the two groups. All participants in both groups were of European-American ethnicity.
2.2. SLE participants and controls
DNA methylation data from neutrophils isolated from SLE patients and matched healthy controls were extracted from our previous study [7]. In total, 15 participants with SLE and 15 matched controls were initially included. Within the control group, two samples were found to have methylation patterns incongruous with their reported sex and were subsequently excluded from later analysis in this study. As such, the control cohort was ultimately comprised of 13 healthy individuals (not shown).
The average age of the SLE and control groups was 37 ± 10 and 40 ± 9 years old, respectively. There was no statistically significant difference between the two groups with regard to age or ethnicity. All participants were female in both groups.
2.3. Neutrophil isolation, DNA extraction, and bisulfite conversion
Peripheral blood samples were collected from each participant, and Ficoll density gradient centrifugation was used to isolate peripheral blood mononuclear cells (PBMCs). Neutrophils were further isolated from the granulocyte layer per previously described protocols [11]. DNA was extracted from the neutrophils using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) then bisulfite converted for DNA methylation studies using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA) using the provided protocol and thermocycler settings recommended by Zymo for Illumina DNA methylation array samples included in the product literature.
2.4. DNA methylation profiling
Genome-wide DNA methylation analysis was performed on extracted neutrophil DNA using the Infinium HumanMethylation450 BeadChip Kit as previously outlined [12]. This array allows for the interrogation of over 485,000 methylation sites (primarily CpG dinucleotides) across the entire genome. It covers 99% of RefSeq genes and 96% of CpG islands. Multiple other methylation sites are also covered, including microRNA promoter regions and several thousand non-CpG sites identified in human stem cells. All array handling, sample hybridization, and array scanning was performed at the University of Michigan DNA Sequencing Core.
2.5. Methylation data processing
The R software package minfi [13] was used to perform probe normalization and extract M and beta (β) values for each methylation site, the latter of which represents the fraction of methylated cytosines at a given site. Probes located on sex chromosomes, cross-reactive probes [14], probes targeting CpG sites within 10 base pairs of a SNP with a minor allele frequency >5% (as reported by Illumina’s probe annotation documentation), and probes that failed (detection p-value >0.05 compared to baseline) in greater than 20% of samples were all excluded.
2.6. Statistical and bioinformatics analysis
Delta beta (Δβ) was defined as the difference in average β value in either the primary APS or SLE participants versus their respective controls at each CpG site. Probes were filtered for effect size, and we retained only those for which the Δβ was greater than 10%. Multiple linear regression was used to evaluate for statistically significant differences in methylation patterns (per Benjamini-Hochberg procedure with a false discovery rate of 0.05) in either primary APS or SLE versus corresponding controls while controlling for age, sex, ethnicity, and chip placement. Regression analyses were performed using M values.
For the primary APS cohort, differentially methylated loci were mapped to genes or gene regions and subjected to gene ontology and pathway analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [15,16]. An Expression Analysis Systematic Explorer (EASE) Score threshold of <0.1 was used to assign statistical significance, with results additionally filtered for fold enrichment ≥1.5 and false discovery rate <10% as multiple testing correction.
In comparing the methylation status of the IFI44L promoter between cohorts, probe cg06872964 was specifically examined as this was one of the exact same methylation probes evaluated by Zhao et al. in the aforementioned study of IFI44L methylation status in SLE [9]. As DNA methylation status at this probe is not normally distributed, the Mann-Whitney U test was used to assess for a statistically significant difference between the two disease cohorts. Receiver operating characteristic (ROC) curves and associated statistics were generated using the R package pROC [17]. The Youden index was used to determine the optimal threshold point.
3. Results
3.1. DNA methylation signatures
In the primary APS group, a total of 42 differentially methylated sites were identified with 17 (40%) being hypomethylated and 25 (60%) being hypermethylated in primary APS neutrophils as compared to neutrophils from the matched control cohort (Table 2). Within the SLE group, a total of 179 differentially methylated sites were identified (Supplementary Table 1), similar to our previous analysis of this same cohort showing robust hypomethylation of interferon-regulated genes [7]. Notably, no differentially methylated sites were shared between the primary APS and SLE signatures.
Table 2.
Differentially methylated CpG sites in primary APS neutrophils compared to healthy controls. Results are arranged in order of absolute change in methylation fraction (Δβ), where negative values represent hypomethylation, and positive values represent hypermethylation.
| CGID | Chr | Position (HG19) |
Gene | Mean β | Δβ | Adjusted p | |
|---|---|---|---|---|---|---|---|
| APS | Controls | ||||||
| cg13341454 | 18 | 12884064 | PTPN2 | 0.059 | 0.435 | −0.376 | 4.3E–04 |
| cg24586205 | 1 | 222162028 | - | 0.550 | 0.234 | 0.315 | 4.6E–02 |
| cg23174406 | 12 | 97304436 | NEDD1 | 0.248 | 0.561 | −0.312 | 3.6E–02 |
| cg20927656 | 12 | 7863229 | DPPA3 | 0.387 | 0.665 | −0.278 | 4.5E–02 |
| cg08820231 | 12 | 58013687 | SLC26A10 | 0.581 | 0.324 | 0.257 | 3.6E–02 |
| cg07792871 | 6 | 29942706 | HCG9 | 0.299 | 0.526 | −0.226 | 3.6E–02 |
| cgl5415945 | 6 | 31627678 | C6orf47 | 0.839 | 0.613 | 0.226 | 3.6E–02 |
| cg27333952 | 16 | 1509206 | CLCN7 | 0.490 | 0.307 | 0.183 | 3.4E–02 |
| cg25528199 | 19 | 7505344 | ARHGEF18 | 0.696 | 0.867 | −0.171 | 3.6E–02 |
| cg01962183 | 16 | 1507866 | CLCN7 | 0.659 | 0.491 | 0.168 | 3.6E–02 |
| cg22325292 | 17 | 80708367 | FN3K | 0.800 | 0.647 | 0.154 | 3.8E–02 |
| cg07644321 | 4 | 2847213 | ADD1 | 0.272 | 0.124 | 0.148 | 3.6E–02 |
| cg22813794 | 7 | 75677469 |
STYXL1; MDH2; MDH2 |
0.236 | 0.096 | 0.140 | 3.4E–02 |
| cg23773946 | 20 | 62327968 | TNFRSF6B | 0.213 | 0.352 | −0.139 | 3.6E–02 |
| cg00893875 | 10 | 91597593 | - | 0.389 | 0.251 | 0.138 | 3.6E–02 |
| cg05731801 | 16 | 10647200 | EMP2 | 0.154 | 0.292 | −0.138 | 3.6E–02 |
| cg14964336 | 4 | 1523275 | - | 0.104 | 0.238 | −0.135 | 3.6E–02 |
| cg23758309 | 12 | 97273300 | - | 0.608 | 0.476 | 0.132 | 3.6E–02 |
| cg04811706 | 16 | 55686715 | - | 0.597 | 0.728 | −0.130 | 3.6E–02 |
| cg04385631 | 12 | 46385625 | SFRS2IP | 0.161 | 0.032 | 0.129 | 2.0E–07 |
| cg10196532 | 13 | 50134640 | RCBTB1 | 0.586 | 0.458 | 0.128 | 3.4E–02 |
| cg05200811 | 21 | 47581042 | - | 0.702 | 0.575 | 0.127 | 2.7E–02 |
| cg22129323 | 12 | 6572482 | VAMP1 | 0.201 | 0.326 | −0.125 | 3.6E–02 |
| cg18505691 | 6 | 29723320 | - | 0.591 | 0.469 | 0.122 | 3.4E–02 |
| cg01775802 | 14 | 72945461 | RGS6 | 0.573 | 0.454 | 0.119 | 8.9E–07 |
| cg17910899 | 13 | 100218552 | - | 0.648 | 0.531 | 0.117 | 4.1E–02 |
| cg11345693 | 17 | 79170810 | AZI1 | 0.665 | 0.552 | 0.113 | 3.6E–02 |
| cg03084983 | 2 | 180294010 | - | 0.641 | 0.529 | 0.112 | 2.4E–02 |
| cg03100639 | 16 | 31483137 |
TGFB1I1 SNORD115–15 |
0.458 | 0.569 | −0.111 | 3.8E–02 |
| cg01393705 | 15 | 25457603 |
SNORD115–21; HBII–52–24 |
0.527 | 0.638 | −0.111 | 3.6E–02 |
| cg10692528 | 12 | 6605071 | NCAPD2 | 0.458 | 0.568 | −0.110 | 3.6E–02 |
| cg06484123 | 19 | 53107200 | - | 0.425 | 0.315 | 0.110 | 4.6E–02 |
| cg15555017 | 11 | 128455958 | ETS1 | 0.528 | 0.637 | −0.109 | 2.7E–02 |
| cg07747220 | 20 | 3052115 | OXT | 0.567 | 0.675 | −0.108 | 3.3E–02 |
| cg12578536 | 5 | 43003251 | - | 0.554 | 0.449 | 0.105 | 1.1E–07 |
| cg14789911 | 21 | 47582049 | C21orf56 | 0.777 | 0.674 | 0.103 | 2.1E–02 |
| cg24354818 | 20 | 62328094 | TNFRSF6B | 0.186 | 0.289 | −0.103 | 3.6E–02 |
| cg16591159 | 16 | 31487813 | TGFB1I1 | 0.435 | 0.333 | 0.102 | 3.6E–02 |
| cg01675596 | 8 | 145911897 | - | 0.448 | 0.346 | 0.102 | 3.6E–02 |
| cg07187855 | 6 | 30854161 | DDR1 | 0.652 | 0.551 | 0.102 | 3.8E–02 |
| cg13752184 | 13 | 100219013 | - | 0.680 | 0.578 | 0.102 | 3.8E–02 |
| cg 16702083 | 20 | 62328427 | TNFRSF6B | 0.298 | 0.399 | −0.101 | 4.4E–02 |
APS, antiphospholipid syndrome.
The single most hypomethylated gene in primary APS neutrophils was PTPN2, which encodes a widely expressed protein tyrosine phosphatase (protein tyrosine phosphatase non-receptor type 2) that acts as a negative regular of T cell activation [18]. Another notably hypomethylated gene was DPPA3, also known as PGC7 or STELLA. In mice, the expressed protein from this gene acts is a maternal factor that plays a critical role in normal early embryogenesis by protecting the DNA methylation state of several imprinted loci on the maternal genome [19]. In the APS neutrophils, several differentially methylated genes were located in major histocompatibility complex (MHC) regions, including hypomethylated HCG9, a non-coding gene within the MHC class I region on chromosome 6, and hypermethylated C6orf47, an MHC class III gene [20].
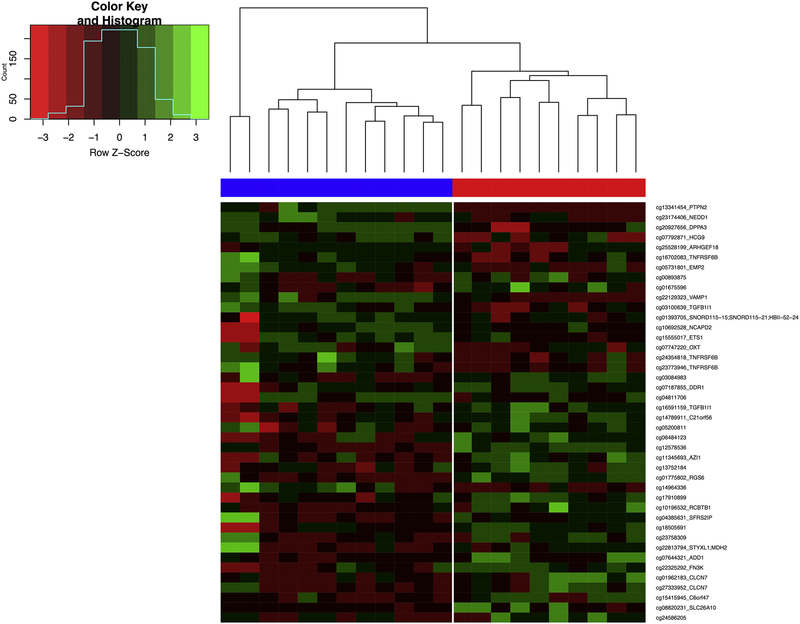
Hierarchical clustering of primary APS and healthy control samples shows that these 42 CpG sites can cluster patient and control methylation profiles accurately and reflects their disease status (Figure 1).
Figure 1.
Heatmap of the 42 differentially methylated CpG sites in the primary APS group (red) compared to healthy controls (blue). A Z-score for each sample was calculated as the difference of the row mean beta value and sample beta value divided by the sample standard deviation. Negative Z-scores (red) are lower than the row mean and represent hypomethylation, and positive Z-scores (green) are higher than the row mean and represent hypermethylation. A dendrogram was drawn by calculating Euclidean distances between sample rows and columns and hierarchical clustering using complete linkage. Rows are sorted from the most hypomethylated CpG sites in the primary APS group (top) to most hypermethylated sites (bottom).
3.2. Gene ontology and pathway analysis in primary APS neutrophils
Gene ontology analysis of hypomethylated genes in the primary APS cohort using category GOTERM_BP_FAT revealed significant association with female pregnancy (p = 0.005), with fold enrichment of 24.3 and a false discovery rate of 8%. Hypomethylated genes associated with this term include ETS1 (E26 transformation-specific proto-oncogene 1, transcription factor), EMP2 (epithelial membrane protein 2), and OXT (oxytocin/neurophysin I prepropeptide). No other terms were both statistically significant and with a false discovery rate <10%. No gene ontology terms were associated with hypermethylated genes in the primary APS group.
Pathway analysis using the KEGG database (via DAVID) was also performed, but did not reveal any canonical pathways associated with the DNA methylation profile of neutrophils in primary APS.
3.3. IFI44L promoter methylation status in primary APS versus SLE
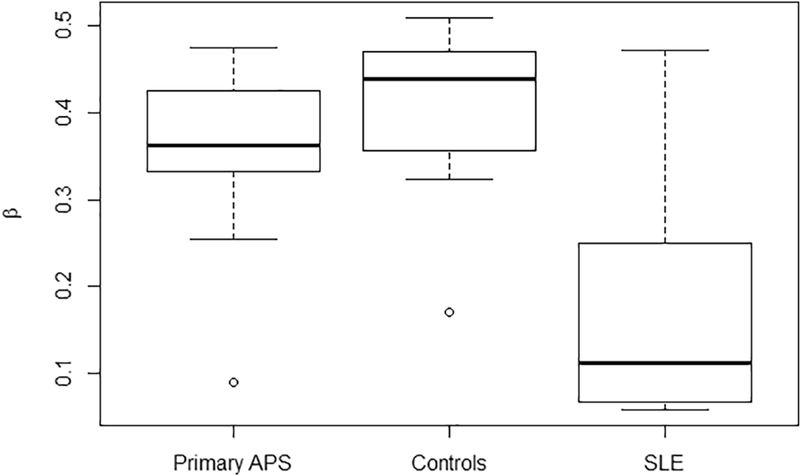
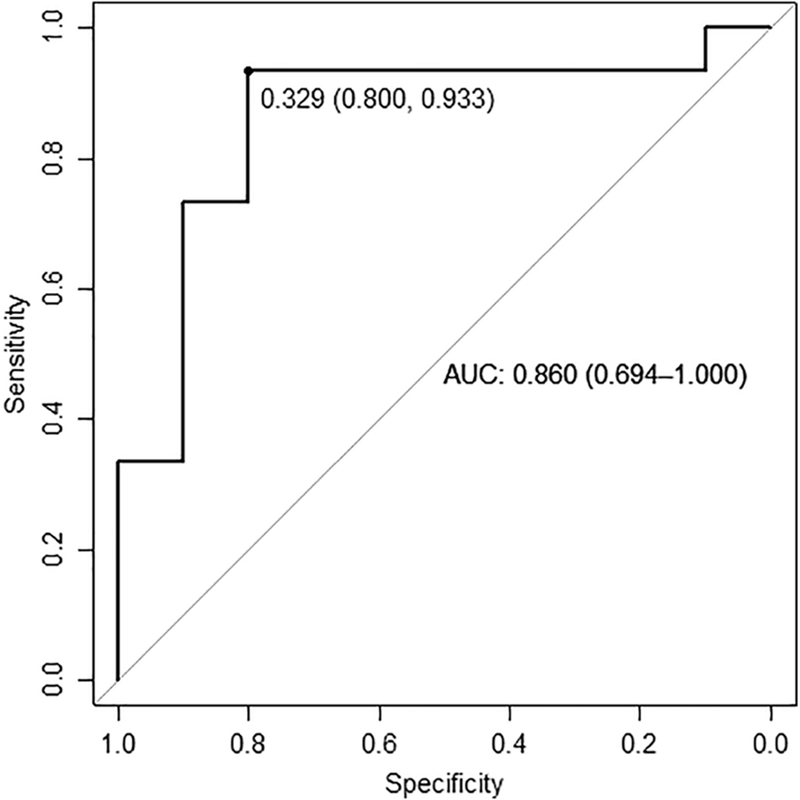
DNA methylation status at probe cg06872964 within the IFI44L promoter was examined and found to be markedly reduced in SLE neutrophils compared to neutrophils from those with primary APS or healthy controls (Figure 2). Specifically, the average ± SD of the methylation fraction β for the SLE cohort was 0.171 ± 0.126 compared to 0.346 ± 0.109 in the primary APS cohort (p = 0.002). Methylation in the primary APS cohort was comparable to the combined control group, with average β 0.418 ± 0.077 (p = 0.05). ROC curve analysis comparing the primary APS cohort to the SLE cohort is shown in Figure 3. Area under the curve was high at 0.860 (95% CI 0.694–1.000). The sensitivity and specificity for distinguishing SLE from primary APS at an optimal methylation cutoff level of 0.329 were 93.3% and 80.0% respectively.
Figure 2.
Methylation status of probe cg06872964 within the IFI44L promoter in the primary APS, SLE, and combined control cohorts. Beta (β) represents the fraction of methylated cytosines at this particular probe.
Figure 3.
ROC curves of DNA methylation level at probe cg06872964 within the IFI44L promoter in the primary APS cohort as compared to the SLE cohort. Optimal threshold per Youden index is shown as a point along the curve, with specificity and sensitivity in parentheses respectively. Area under the curve is also shown with 95% confidence interval.
4. Discussion
We performed a genome-wide DNA methylation analysis of neutrophils from individuals with primary APS versus healthy controls, and compared the differential methylation profile of primary APS to that of SLE. To the authors’ knowledge, this is the first genome-wide DNA methylation study of primary APS in neutrophils to be described in the literature. Arguably the most significant result of this work is that the DNA methylation profile of primary APS is entirely distinct from that of SLE, at least in neutrophils. Robust demethylation of interferon signature genes, an established feature of SLE which has been found in all immune cells studied to date [7,12,21], was not observed in the primary APS methylation signature. Furthermore, no other differentially methylated genes or gene regions were shared between the two disease methylation profiles.
Along similar lines, methylation status of the IFI44L promoter (at probe cg06872964) was able to distinguish individuals with SLE from those with primary APS with a high sensitivity and moderately high specificity. As compared to the results of Zhao et al. in whole blood, hypomethylation of this probe in neutrophils was able to discriminate SLE from primary APS with greater sensitivity and specificity than hypomethylation of this same probe for SLE versus primary Sjögren’s syndrome and SLE versus controls in a European cohort, and with comparable performance in distinguishing SLE from RA. Though we make this comparison with caution, given that this study was performed in neutrophils rather than whole blood, it does provide proof of principle that DNA methylation status might prove useful as a clinical tool in detecting underlying SLE or similar connective disease in individuals who present with APS. Notably, one individual in the primary APS cohort (APS #3 per Table 1) had a DNA methylation profile that was overall more similar to SLE, including significant hypomethylation of IFI44L as illustrated graphically by the lone outlier in the primary APS group in Figure 2. Interestingly, this participant was the only one in the primary APS cohort to have autoimmune hemolytic anemia, which refers to a group of hematologic disorders that can be associated with SLE, especially in conjunction with anti-cardiolipin IgG and thrombocytopenia (both of which this participant also had) [22]. The significance of this finding is not clear. Perhaps such individuals with SLE-like methylation patterns might be at a higher risk for progression to SLE or other connective tissue disease in the future, or these SLE-like methylation changes have some distinct relation to autoimmune hemolytic anemia itself, either as a primary means of pathogenesis or as some secondary epiphenomenon.
Compared to controls, the differential methylation profile of primary APS is relatively modest, in contrast to the marked global changes seen in the SLE methylome. Two of the most significantly hypomethylated genes were PTPN2 and DPPA3. In rheumatoid arthritis (RA), PTPN2 is overexpressed in synovial fibroblasts and contributes to disease pathogenesis via regulation of IL-6 production, cell death, and autophagy [23]. Genetic variants of PTPN2 have been associated with an increased risk for rheumatoid arthritis, as well as other autoimmune diseases including inflammatory bowel disease and type 1 diabetes mellitus [24–27]. DPPA3, as mentioned in Section 3.1, plays a role in early embryogenesis in mice by protecting the DNA methylation state of the maternal genome [19]. Its function in humans has not been characterized. It has been shown, however, to perform a similar epigenetic regulatory role in bovine embryos, suggesting that this behavior may be preserved across multiple mammalian species [28].
An association between differential methylation in APS and genetic regions regulating pregnancy is a notable finding, given that pregnancy loss is a defining feature of APS. Beyond the hypomethylation of DPPA3, gene ontology analysis revealed enrichment of hypomethylated genes known to be associated with human pregnancy, namely ETS1, EMP2, and OXT. Genome-wide association studies have identified ETS1 as a susceptibility locus for SLE and other autoimmune diseases [29,30]. The associated protein, Ets1, is a transcription factor which regulates numerous cellular processes including stem cell development and cell senescence [31]. It is highly expressed in lymphocytes, though it is present at reduced levels in PBMCs and regulatory T cells from individuals with SLE [32,33]. Indeed, this altered expression is postulated to contribute to the pathophysiology of SLE, in part because Ets1 plays an important genetic regulatory role in preventing plasma cell differentiation and maintaining B cell self-tolerance [34]. ETS1 knock-out mice develop an autoimmune syndrome similar to SLE, complete with serum autoantibodies against double-stranded DNA and histones (as seen in SLE in humans) as well as anti-cardiolipin (an antiphospholipid antibody) [35]. With regard to its role in pregnancy, Ets1 is expressed in the human placenta in the first and third trimesters [36], and was later discovered to induce differentiation of the trophoblast into its multiple cell layers. Specifically, overexpression of Ets1 in primary trophoblast cells promotes a multinucleated syncytiotrophoblast cell phenotype.
The protein encoded by EMP2 also plays a critical role in human pregnancy, specifically as a protective factor that mediates placental vascularization and maintenance. EMP2 is a transmembrane protein that is expressed on the epithelial cell surface of the uterus as well as on the trophoblast of implanting embryos. Knockdown of EMP2 in mice impairs successful implantation [37]. In vitro investigation of human trophoblast cell lines has shown that EMP2 similarly regulates angiogenesis and trophoblast invasion, and that its deficiency is associated with intrauterine growth restriction [38]. In this same study, upregulation of EMP2 correlated with increased trophoblast cell migration.
Finally, OXT encodes a precursor protein to oxytocin and its carrier protein, neurophysin I. Oxytocin is well-known for its many important function in sexual reproduction both during and after childbirth, including contraction of the uterus during labor and mediating the milk letdown reflex during lactation [20].
With regard to the role of neutrophils in pregnancy, there is increasing evidence that neutrophil extracellular traps (NETs) may be of critical importance in mediating fetal loss [39]. Early studies using murine models demonstrated that antiphospholipid antibodies can promote pregnancy loss via activation of the complement cascade, which in turn leads to polymorphonuclear neutrophil activation via tissue factor [40–42]. It was proposed that the resulting production of reactive oxygen species would then cause irreparable damage to the trophoblast [43,44]. More recently, Erpenbeck and colleagues examined antiangiogenic factor-mediated pregnancy loss in mice that are deficient in peptidylarginine deiminase 4 (PAD4) and thereby not able to form NETs [45]. Overexpression of soluble fms-like tyrosine kinase 1, an antiangiogenic protein, lead to excessive accumulation of neutrophils and NETs in the placentas of wild type mice and promoted pregnancy loss. In the PAD4 deficient mice, however, overexpression of this same protein resulted in a significantly dampened inflammatory and thrombotic response with a correspondingly lower rate of pregnancy loss. Similarly, Mizugishi and Yamashita demonstrated that NETs also appear to play a role in sphingosine kinase-mediated pregnancy loss [46]. In their murine model, inhibition of the activated sphingolipid metabolic pathway by way of sphingosine kinase gene disruption resulted in increased NET formation specifically at the fetomaternal interface and associated early fetal death. Furthermore, use of a PAD4 inhibitor to block NET formation partially restored normal embryonic development and protected against pregnancy loss.
Epigenetic regulation of NETosis remains incompletely understood, as do the direct mechanisms by which NETs might cause abnormal placentation and fetal loss. Nevertheless, it is apparent that ongoing investigation into the neutrophil-mediated inflammatory responses in APS may yield important insights into the pathophysiology of this disease. Based on our study, hypomethylation of ETS1 and EMP2 in primary APS neutrophils may indicate dysregulation of trophoblast differentiation and migration which could certainly contribute to increased fetal morbidity. DPPA3, also hypomethylated in primary APS as previously discussed, plays an important role in embryogenesis in murine and bovine studies, though its role in humans is less clear. Functional studies of how DNA methylation affects these genes and their associated cellular functions may elucidate the mechanisms by which APS causes pregnancy morbidity and potentially guide future treatment strategies. In particular, ETS1 is also a susceptibility locus for SLE, though we again note that it is not differentially methylated in SLE neutrophils. While it is known that Ets1 plays an important role in SLE pathogenesis, this DNA methylation study appears to be the first to suggest some possible unifying connection between Ets1, SLE, APS, and pregnancy morbidity.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health grants number R01AI097134 and U19AI110502, and the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health Intramural Research Program ZIA AR041199. Dr. Knight was supported by a career development award from the Burroughs Wellcome Fund.
Abbreviations:
- APS
antiphospholipid syndrome
- C6orf47
chromosome 6 open reading frame 47
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- DNA
deoxyribonucleic acid
- DPPA3
developmental pluripotency associated 3
- EASE
Expression Analysis Systematic Explorer
- EMP2
epithelial membrane protein 2
- ETS1
E26 proto-oncogene 1
- HCG9
human leukocyte antigen complex group 9
- IFI44L
interferon induced protein 44 like
- MHC
major histocompatibility complex
- NET
neutrophil extracellular trap
- OXT
oxytocin/neurophysin I prepropeptide
- PAD4
peptidylarginine deiminase 4
- PMBC
peripheral blood mononuclear cell
- PTPN2
protein tyrosine phosphatase non-receptor type 2
- ROC
receiver operating characteristic
- SD
standard deviation
- SLE
systemic lupus erythematosus
- SNP
single nucleotide polymorphism
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interest: None of the authors have any financial conflict of interest to declare with the work presented. AHS is listed as inventor on a patent application to use IFI44L methylation as a diagnostic tool for lupus.
References
- [1].Gómez-Puerta JA, Cervera R, Diagnosis and classification of the antiphospholipid syndrome., J. Autoimmun 48–49 (n.d.) 20–5. doi: 10.1016/j.jaut.2014.01.006. [DOI] [PubMed] [Google Scholar]
- [2].Cervera R, Piette J-C, Font J, Khamashta MA, Shoenfeld Y, Camps MT, Jacobsen S, Lakos G, Tincani A, Kontopoulou-Griva I, Galeazzi M, Meroni PL, Derksen RHWM, de Groot PG, Gromnica-Ihle E, Baleva M, Mosca M, Bombardieri S, Houssiau F, Gris J-C, Quéré I, Hachulla E, Vasconcelos C, Roch B, Fernández-Nebro A, Boffa M-C, V Hughes GR, Ingelmo M, Euro-Phospholipid Project Group, Antiphospholipid syndrome: clinical and immunologic manifestations and patterns of disease expression in a cohort of 1,000 patients., Arthritis Rheum. 46 (2002) 1019–27. [DOI] [PubMed] [Google Scholar]
- [3].Gómez-Puerta JA, Martín H, Amigo M-C, Aguirre MA, Camps MT, Cuadrado MJ, V Hughes GR, Khamashta MA, Long-term follow-up in 128 patients with primary antiphospholipid syndrome: do they develop lupus?, Medicine (Baltimore). 84 (2005) 225–30. [DOI] [PubMed] [Google Scholar]
- [4].Veres K, Szodoray P, Szekanecz Z, Lakos G, Kiss E, Laczik R, Sipka S, Bodolay E, Zeher M, Muszbek L, Szegedi G, Soltész P, Clinical and immunoserological characteristics of the transition from primary to overlap antiphospholipid syndrome., Lupus. 19 (2010) 1520–6. doi: 10.1177/0961203310374336. [DOI] [PubMed] [Google Scholar]
- [5].Myones BL, McCurdy D, The antiphospholipid syndrome: immunologic and clinical aspects. Clinical spectrum and treatment., J. Rheumatol. Suppl 58 (2000) 20–8. [PubMed] [Google Scholar]
- [6].Weeding E, Sawalha AH, Deoxyribonucleic Acid Methylation in Systemic Lupus Erythematosus: Implications for Future Clinical Practice., Front. Immunol 9 (2018) 875. doi: 10.3389/fimmu.2018.00875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Coit P, Yalavarthi S, Ognenovski M, Zhao W, Hasni S, Wren JD, Kaplan MJ, Sawalha AH, Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils., J. Autoimmun 58 (2015) 59–66. doi: 10.1016/j.jaut.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Knight JS, Meng H, Coit P, Yalavarthi S, Sule G, Gandhi AA, Grenn RC, Mazza LF, Ali RA, Renauer P, Wren JD, Bockenstedt PL, Wang H, Eitzman DT, Sawalha AH, Activated signature of antiphospholipid syndrome neutrophils reveals potential therapeutic target, JCI Insight. 2 (2017). doi: 10.1172/jci.insight.93897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q, Liu Y, Jiang J, Luo S, Tan Y, Wu H, Renauer P, Del M Gutiérrez Mar Ayala, Palma M.J. Castillo, Castro R. Ortega, Fernández-Roldán C, Raya E, Faria R, Carvalho C, Alarcón-Riquelme ME, Xiang Z, Chen J, Li F, Ling G, Zhao H, Liao X, Lin Y, Sawalha AH, Lu Q, IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus., Ann. Rheum. Dis 75 (2016) 1998–2006. doi: 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Miyakis S, Lockshin MD, Atsumi T, Branch DW, Brey RL, Cervera R, Derksen RHWM, DE Groot PG, Koike T, Meroni PL, Reber G, Shoenfeld Y, Tincani A, Vlachoyiannopoulos PG, Krilis SA, International consensus statement on an update of the classification criteria for definite antiphospholipid syndrome (APS), J. Thromb. Haemost. JTH 4 (2006) 295–306. doi: 10.1111/j.1538-7836.2006.01753.x. [DOI] [PubMed] [Google Scholar]
- [11].Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM, Rubin CJ, Zhao W, Olsen SH, Klinker M, Shealy D, Denny MF, Plumas J, Chaperot L, Kretzler M, Bruce AT, Kaplan MJ, Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus., J. Immunol. Baltim. Md 1950 187 (2011) 538–52. doi: 10.4049/jimmunol.1100450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, Merrill JT, McCune WJ, Sawalha AH, Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naïve CD4+ T cells from lupus patients., J. Autoimmun 43 (2013) 78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays., Bioinforma. Oxf. Engl 30 (2014) 1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray., Epigenetics. 8 (2013) 203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Huang DW, Sherman BT, Lempicki RA, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists., Nucleic Acids Res. 37 (2009) 1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources., Nat. Protoc 4 (2009) 44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- [17].Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M, pROC: an open-source package for R and S+ to analyze and compare ROC curves., BMC Bioinformatics. 12 (2011) 77–77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wiede F, Shields BJ, Chew SH, Kyparissoudis K, van Vliet C, Galic S, Tremblay ML, Russell SM, Godfrey DI, Tiganis T, T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice, J. Clin. Invest 121 (2011) 4758–4774. doi: 10.1172/JCI59492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, Nakano T, PGC7/Stella protects against DNA demethylation in early embryogenesis, Nat. Cell Biol 9 (2007) 64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- [20].O’Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, Astashyn A, Badretdin A, Bao Y, Blinkova O, Brover V, Chetvernin V, Choi J, Cox E, Ermolaeva O, Farrell CM, Goldfarb T, Gupta T, Haft D, Hatcher E, Hlavina W, Joardar VS, Kodali VK, Li W, Maglott D, Masterson P, McGarvey KM, Murphy MR, O’Neill K, Pujar S, Rangwala SH, Rausch D, Riddick LD, Schoch C, Shkeda A, Storz SS, Sun H, Thibaud-Nissen F, Tolstoy I, Tully RE, Vatsan AR, Wallin C, Webb D, Wu W, Landrum MJ, Kimchi A, Tatusova T, DiCuccio M, Kitts P, Murphy TD, Pruitt KD, Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation, Nucleic Acids Res. 44 (2016) D733–745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP, Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations., PLoS Genet. 9 (2013) e1003678–e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kokori SI, Ioannidis JP, Voulgarelis M, Tzioufas AG, Moutsopoulos HM, Autoimmune hemolytic anemia in patients with systemic lupus erythematosus., Am. J. Med 108 (2000) 198–204. [DOI] [PubMed] [Google Scholar]
- [23].Aradi B, Kato M, Filkova M, Karouzakis E, Klein K, Scharl M, Kolling C, Michel BA, Gay RE, Buzas EI, Gay S, Jüngel A, Protein tyrosine phosphatase nonreceptor type 2: an important regulator of lnterleukin-6 production in rheumatoid arthritis synovial fibroblasts, Arthritis Rheumatol. Hoboken NJ 67 (2015) 2624–2633. doi: 10.1002/art.39256. [DOI] [PubMed] [Google Scholar]
- [24].Wellcome Trust Case Control Consortium, Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls, Nature. 447 (2007) 661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Glas J, Wagner J, Seiderer J, Olszak T, Wetzke M, Beigel F, Tillack C, Stallhofer J, Friedrich M, Steib C, Göke B, Ochsenkühn T, Karbalai N, Diegelmann J, Czamara D, Brand S, PTPN2 gene variants are associated with susceptibility to both Crohn’s disease and ulcerative colitis supporting a common genetic disease background, PloS One. 7 (2012) e33682. doi: 10.1371/journal.pone.0033682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Kawaguchi T, Stahl EA, Kurreeman FAS, Nishida N, Ohmiya H, Myouzen K, Takahashi M, Sawada T, Nishioka Y, Yukioka M, Matsubara T, Wakitani S, Teshima R, Tohma S, Takasugi K, Shimada K, Murasawa A, Honjo S, Matsuo K, Tanaka H, Tajima K, Suzuki T, Iwamoto T, Kawamura Y, Tanii H, Okazaki Y, Sasaki T, Gregersen PK, Padyukov L, Worthington J, Siminovitch KA, Lathrop M, Taniguchi A, Takahashi A, Tokunaga K, Kubo M, Nakamura Y, Kamatani N, Mimori T, Plenge RM, Yamanaka H, Momohara S, Yamada R, Matsuda F, Yamamoto K, Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population, Nat. Genet 44 (2012) 511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- [27].Cobb JE, Plant D, Flynn E, Tadjeddine M, Dieudé P, Cornélis F, Ärlestig L, Dahlqvist SR, Goulielmos G, Boumpas DT, Sidiropoulos P, Krintel SB, Ørnbjerg LM, Hetland ML, Klareskog L, Haeupl T, Filer A, Buckley CD, Raza K, Witte T, Schmidt RE, FitzGerald O, Veale D, Eyre S, Worthington J, Identification of the tyrosine-protein phosphatase non-receptor type 2 as a rheumatoid arthritis susceptibility locus in europeans, PloS One. 8 (2013) e66456. doi: 10.1371/journal.pone.0066456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bakhtari A, Ross PJ, DPPA3 prevents cytosine hydroxymethylation of the maternal pronucleus and is required for normal development in bovine embryos, Epigenetics. 9 (2014) 1271–1279. doi: 10.4161/epi.32087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Han J-W, Zheng H-F, Cui Y, Sun L-D, Ye D-Q, Hu Z, Xu J-H, Cai Z-M, Huang W, Zhao G-P, Xie H-F, Fang H, Lu Q-J, Xu J-H, Li X-P, Pan Y-F, Deng D-Q, Zeng F-Q, Ye Z-Z, Zhang X-Y, Wang Q-W, Hao F, Ma L, Zuo X-B, Zhou F-S, Du W-H, Cheng Y-L, Yang J-Q, Shen S-K, Li J, Sheng Y-J, Zuo X-X, Zhu W-F, Gao F, Zhang P-L, Guo Q, Li B, Gao M, Xiao F-L, Quan C, Zhang C, Zhang Z, Zhu K-J, Li Y, Hu D-Y, Lu W-S, Huang J-L, Liu S-X, Li H, Ren Y-Q, Wang Z-X, Yang C-J, Wang P-G, Zhou W-M, Lv Y-M, Zhang A-P, Zhang S-Q, Lin D, Li Y, Low HQ, Shen M, Zhai Z-F, Wang Y, Zhang F-Y, Yang S, Liu J-J, Zhang X-J, Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus, Nat. Genet 41 (2009) 1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- [30].Yang W, Shen N, Ye D-Q, Liu Q, Zhang Y, Qian X-X, Hirankarn N, Ying D, Pan H-F, Mok CC, Chan TM, Wong RWS, Lee KW, Mok MY, Wong SN, Leung AMH, Li X-P, Avihingsanon Y, Wong C-M, Lee TL, Ho MHK, Lee PPW, Chang YK, Li PH, Li R-J, Zhang L, Wong WHS, Ng IOL, Lau CS, Sham PC, Lau YL, Asian Lupus Genetics Consortium, Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus, PLoS Genet. 6 (2010) e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Grund EM, Spyropoulos DD, Watson DK, Muise-Helmericks RC, Interleukins 2 and 15 regulate Ets1 expression via ERK1/2 and MNK1 in human natural killer cells, J. Biol. Chem 280 (2005) 4772–4778. doi: 10.1074/jbc.M408356200. [DOI] [PubMed] [Google Scholar]
- [32].Li Y, Sun L, Lu W, Hu W, Gao J, Cheng Y, Yu Z, Yao S, He C, Liu J, Cui Y, Yang S, Expression analysis of ETS1 gene in peripheral blood mononuclear cells with systemic lupus erythematosus by real-time reverse transcription PCR, Chin. Med. J. (Engl.) 123 (2010) 2287–2288. [PubMed] [Google Scholar]
- [33].Xiang N, Li X-P, Li X-M, Wang G-S, Tao J-H, Pan H-F, Fang X, Ma Q, Yu N, Expression of Ets-1 and FOXP3 mRNA in CD4(+)CD25 (+) T regulatory cells from patients with systemic lupus erythematosus, Clin. Exp. Med 14 (2014) 375–381. doi: 10.1007/s10238-013-0263-4. [DOI] [PubMed] [Google Scholar]
- [34].Garrett-Sinha LA, Kearly A, Satterthwaite AB, The Role of the Transcription Factor Ets1 in Lupus and Other Autoimmune Diseases, Crit. Rev. Immunol 36 (2016) 485–510. doi: 10.1615/CritRevImmunol.2017020284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Wang D, John SA, Clements JL, Percy DH, Barton KP, Garrett-Sinha LA, Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease, Int. Immunol 17 (2005) 1179–1191. doi: 10.1093/intimm/dxh295. [DOI] [PubMed] [Google Scholar]
- [36].Takai N, Ueda T, Narahara H, Miyakawa I, Expression of c-Ets1 protein in normal human placenta, Gynecol. Obstet. Invest 61 (2006) 15–20. doi: 10.1159/000087855. [DOI] [PubMed] [Google Scholar]
- [37].Wadehra M, Dayal M, Mainigi M, Ord T, Iyer R, Braun J, Williams CJ, Knockdown of the tetraspan protein epithelial membrane protein-2 inhibits implantation in the mouse, Dev. Biol 292 (2006) 430–441. doi: 10.1016/j.ydbio.2006.01.015. [DOI] [PubMed] [Google Scholar]
- [38].Williams CJ, Chu A, Jefferson WN, Casero D, Sudhakar D, Khurana N, Hogue CP, Aryasomayajula C, Patel P, Sullivan P, Padilla-Banks E, Mohandessi S, Janzen C, Wadehra M, Epithelial membrane protein 2 (EMP2) deficiency alters placental angiogenesis, mimicking features of human placental insufficiency, J. Pathol 242 (2017) 246–259. doi: 10.1002/path.4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hahn S, Giaglis S, Hoesli I, Hasler P, Neutrophil NETs in reproduction: from infertility to preeclampsia and the possibility of fetal loss, Front. Immunol 3 (2012) 362. doi: 10.3389/fimmu.2012.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Holers VM, Girardi G, Mo L, Guthridge JM, Molina H, Pierangeli SS, Espinola R, Xiaowei LE, Mao D, Vialpando CG, Salmon JE, Complement C3 activation is required for antiphospholipid antibody-induced fetal loss, J. Exp. Med 195 (2002) 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Girardi G, Berman J, Redecha P, Spruce L, Thurman JM, Kraus D, Hollmann TJ, Casali P, Caroll MC, Wetsel RA, Lambris JD, Holers VM, Salmon JE, Complement C5a receptors and neutrophils mediate fetal injury in the antiphospholipid syndrome, J. Clin. Invest 112 (2003) 1644–1654. doi: 10.1172/JCI18817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Redecha P, Tilley R, Tencati M, Salmon JE, Kirchhofer D, Mackman N, Girardi G, Tissue factor: a link between C5a and neutrophil activation in antiphospholipid antibody induced fetal injury, Blood. 110 (2007) 2423–2431. doi: 10.1182/blood-2007-01-070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Weiler H, Tracing the molecular pathogenesis of antiphospholipid syndrome, J. Clin. Invest 118 (2008) 3276–3278. doi: 10.1172/JCI37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Girardi G, Role of tissue factor in feto-maternal development: a xiphos, J. Thromb. Haemost. JTH 9 (2011) 250–256. doi: 10.1111/j.1538-7836.2010.04135.x. [DOI] [PubMed] [Google Scholar]
- [45].Erpenbeck L, Chowdhury CS, Zsengellér ZK, Gallant M, Burke SD, Cifuni S, Hahn S, Wagner DD, Karumanchi SA, PAD4 Deficiency Decreases Inflammation and Susceptibility to Pregnancy Loss in a Mouse Model, Biol. Reprod 95 (2016) 132. doi: 10.1095/biolreprod.116.140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mizugishi K, Yamashita K, Neutrophil extracellular traps are critical for pregnancy loss in sphingosine kinase-deficient mice on 129Sv/C57BL/6 background, FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 31 (2017) 5577–5591. doi: 10.1096/fj.201700399RR. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.