Abstract
Cryo-electron microscopy of “single particles” is a powerful method to analyze structures of large macromolecular assemblies that are not amenable to investigation by traditional X-ray crystallographic methods. A key step in these studies is to obtain atomic interpretations of multiprotein complexes by fitting atomic structures of individual components into maps obtained from electron microscopic data. Here, we report the use of a “core-weighting” method, combined with a grid-threading Monte Carlo (GTMC) approach for this purpose. The “core” of an individual structure is defined to represent the part where the density distribution is least likely to be altered by other components that comprise the macromolecular assembly of interest. The performance of the method has been evaluated by its ability to determine the correct fit of (i) the α-chain of the T-cell receptor variable domain into a simulated map of the αβ complex at resolutions between 5 and 40 Å, and (ii) the E2 catalytic domain of the pyruvate dehydrogenase into an experimentally determined map, at 14 Å resolution, of the icosahedral complex formed by 60 copies of this enzyme. Using the X-ray structures of the two test cases as references, we demonstrate that, in contrast to more traditional methods, the combination of the core-weighting method and the grid-threading Monte Carlo approach can identify the correct fit reliably and rapidly from the low-resolution maps that are typical of structures determined with the use of single-particle electron microscopy. Published by Elsevier Science (USA).
Keywords: Conformational search, Correlation function, Cryo-electron microscopy, Docking, Monte Carlo, Multiprotein complex, Single-particle microscopy
1. Introduction
High-resolution electron microscopy of single particles is rapidly emerging as a method with tremendous potential to obtain molecular structures of complex macromolecular assemblies that function as dynamic cellular machines (e.g., Frank, 1996). Many of these complexes are too large to be studied by NMR methods and often do not form the three-dimensional crystals required for X-ray crystallographic analyses, despite the fact that the structure of individual complex components may be obtained using either or both of these methods. While maps derived from ordered twodimensional protein crystals at the highest resolutions approach atomic resolution (e.g., Subramaniam and Henderson, 2000), the maps derived in single particle microscopy are most often at much lower resolution, in the range of 10–30 A (Frank et al., 2000; Ranson et al., 2001; Stark et al., 2000). The use of reliable docking algorithms to position atomic coordinates of the individual components into these lower resolution maps holds the promise of significant biological insight into understanding the architectures of complex macromolecular machines. A variety of computational docking algorithms have been developed to perform reliable and reproducible fitting into low-resolution maps (Baker and Cheng, 1996; Chacon and Wriggers, 2002; Roseman, 2000; Rossmann, 2000; Rossmann et al., 2001; Wriggers et al., 1999; Wriggers and Chacon, 2001) and have been well reviewed (Chacon and Wriggers, 2002; Wriggers and Chacon, 2001).
Correlation coefficients have often been used as a criterion for fitting atomic structures into low-resolution EM maps (e.g., Jiang et al., 2001). Usually, the best results are obtained when the surface edges of individual components in a complex are well defined, and where there are only small regions of densities that cannot be assigned uniquely to a single component. However, the use of standard correlation coefficients is not particularly reliable, especially in the case of low-resolution maps from complex assemblies where each of the docked structures represents only a portion of the whole map, and the boundaries of individual components may not be clearly defined. Furthermore, the density distributions of adjacent components can overlap significantly with each other. Without taking this density alteration into account, the correlation function cannot correctly describe the fit between the map corresponding to an individual component and a map of the complex, and global optimization of the correlation coefficient may worsen the fit. Several methods have been proposed to overcome these difficulties. One approach involves the use of a mask to focus on the overlapping region between the densities arising from the individual docked components and the target map in calculations of the correlation coefficient (Roseman, 2000). Another approach involves altering the functional form of the compared densities by applying a filter that enhances detection of contours in the maps being compared (Chacon and Wriggers, 2002; Wriggers and Chacon, 2001). However, the significant levels of noise that are present in low-resolution maps derived from electron microscopy can be amplified by certain density filtering approaches, which may increase the likelihood of “false-positive” fits of density.
The docking of multiple atomic structures into low-resolution density maps is a many-body search problem. An ideal search should accurately position and orient each of the individual components so that a combined density map calculated from all of these components matches the experimentally determined map. In general, the conformational space of such a many-body system is prohibitively large for an exhaustive conformational search. This computationally expensive approach can be overcome if the many-body search problem is reduced to a series of single-body search problems. To do this, a target function must be defined that can recognize the correct fit despite the possible overlap between neighboring components. In this work, we present a “coreweighting” approach in which the construction of a complex structure from many components is simplified to a series of single component fitting procedures. The single component fitting is conducted using a grid-threading Monte Carlo (GTMC) method that identifies the global maximum state (best fit) among a series of local maximum states determined by short Monte Carlo searches originating at a variety of grid points. The details and performance of this approach are described in the following sections. As a brief comparison, we chose SITUS 2.0 (Chacon and Wriggers, 2002) from available public softwares for map fitting, e.g., COAN (Volkmann and Hanein, 1999), EMfit (Rossmann et al., 2001), EMAN (Jiang et al., 2001; Ludtke et al., 1999), to demonstrate the performance of our method.
2. Materials and methods
2.1. The core index
A map is described here as a distribution of a property, e.g., density, on grid points in a certain space. Molecules produce a high-density distribution at the place occupied by their structures. Due to the low resolution of a map, neighboring structures have significant density distributions overlapping with each other. We operationally define the “core” region of a structure as the part whose density distribution is unlikely to be altered by the presence of adjacent components. The “surface” region is the part that is accessible or can interact with other components. The region enclosed by the accessible surface thus belongs to the core region. We used a Laplacian filter (Russ, 1998), defined by the finite difference approximation as follows, to define the boundary of the surface,
| (1) |
where ρijk and ∇2ρijk represent the density and the Laplacian filtered density at grid point (i, j, k). The Laplacian filter produces an approximation of the secondary derivatives of the scalar density as respect to spatial positions, which changes from positive to negative when crossing the surface from the exterior to the interior.
It is then possible to define a core index, which describes the depth of a grid point located within this core as follows:
| (2) |
where fijk is the core index of grid point (i, j, k), ρc is a cutoff density, and min[fi±1jk, fij±1k, fijk±1] represents the minimum core index of the neighboring grid points around grid point (i, j, k). The core index is zero for grid points outside the core and increases progressively for grid points located deeper in the core. Eq. (2) implies that a grid point outside the core region must neighbor at least one grid point that is also outside the core. Similarly, a grid point within the core cannot neighbor a grid point outside the core unless it satisfies the condition ∇2ρijk ≤ 0 and ρijk > ρc. This definition indicates that within the core, the value of the core index of a grid point is one greater than the minimum core index of its neighboring grid points. Therefore, the core index is larger for a grid point deeper inside the core. To calculate the core index, we used the following iterative procedure:
- Initialize the core index according to Eq. (3) so that all core indices are 1 except the grid points at the boundary.
here, grid indices are from 1 to nx, 1 to ny, and 1 to nz for x, y, and z directions, respectively.(3) Loop over all grid points to calculate the core index of each grid point according to Eq. (2).
Repeat step (b) until all grid points satisfy Eq. (2).
Upon forming a complex, the density distribution of each component is expected to remain the same for regions with a high core index. This may or may not hold true for regions near the surface of the core with a low core index depending upon whether the surface contacts other components. Therefore, even in the case of an exact fit, one cannot always expect a perfect one-to-one correlation between the density distributions of a component in its isolated and complexed forms.
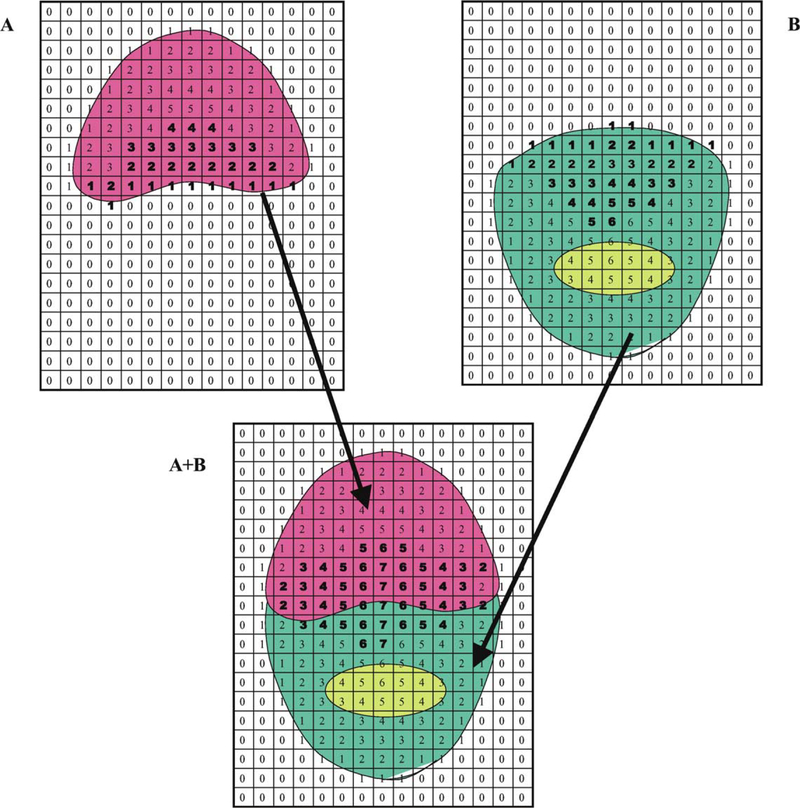
Fig. 1 shows the distribution of core indices for two individual proteins, A and B, and their complex. For each map, the core index is zero outside the domains, 1 at the outer edge and becomes larger for the grid points that are located more deeply in the core region. Since the core region does not necessarily need to correspond to the region with high density, it is possible that the index can have a high value for internal cavities that are buried well below the surface of the structure (e.g., the cavity in protein B). When proteins A and B interact, the core indices of their interaction surfaces dramatically increase, especially in regions where the surfaces become deeply buried in the AB complex.
Fig.1.
The core indices of schematic two-dimensional maps of proteins A and B and their complex. Regions of protein density are colored red and green, respectively, and a region of protein B containing an inaccessible cavity is shown in light green. Regions outside of the protein are white. The numerical value of the core index for each grid point is indicated. Bold numbers indicate the core indices of proteins A and B that change upon formation of the AB complex.
2.2. The core-weighted correlation function
The match in density between two maps is often described with a cross-correlation function as shown below, which we refer to as the “density correlation” function (DC):
| (4) |
Here, subscripts m and n refer to the two maps being compared,
and
represent the average and fluctuation of the density distribution, respectively.
An alternative measure of the correlation is the Laplacian correlation (LC), as used in the work of Chacon and Wriggers,
| (5) |
where the Laplacian filtered density, ∇2ρ, is calculated using Eq. (1).
We expect the following features when we consider the match between the map of an individual component and the map of a multicomponent assembly:
If the core region (high core index) of an individual component matches the core region (high core index) of the complex, the distribution property of this core region should not change appreciably for the correct fit.
If the surface (low core index) of an individual component matches the surface (low core index) of the complex, the distribution property of the surface region should not change appreciably for the correct fit.
If the surface (low core index) of an individual component matches the core (high core index) of the complex, the distribution property of the surface region should change significantly for the correct fit.
If the core (high core index) of an individual component matches the surface (low core index) of the complex, it cannot be a correct fit.
For scenarios 1, 2, and 4, a correlation function works fine to distinguish the correct fit from wrong fits. But for scenario 3, the distribution property is altered by overlap from neighboring components in a complex map and a correlation function is likely to fail. To overcome the effect of overlap, and properly describe the correct fit, we need to minimize the contribution from scenario 3 in the correlation function calculation. This can be achieved by “down-weighting” the match between a region with low core index in the map of individual components and a region with high core index in the complex map. We have chosen the following weighting function to implement this idea,
| (6) |
where wmn is the core-weighting function for the individual component, m, to the complex, n. Three parameters, a, b, and c, control the dependence of the function on the core indices. Typically, we chose a = 2 and b = 1, and set c to be a very small constant (e.g., 10−6) to ensure wmn = 0 when fm = 0 and fn = 0. We call this function the core-weighting function because it is based on the core index. Introducing this core-weighting function leads to the core-weighted correlation function,
| (7) |
where represents a core-weighted average of property X:
| (8) |
and
| (9) |
If we choose densities for the calculation, Eq. (7) results in the core-weighted density correlation (CWDC),
| (7a) |
and if we choose to apply the Laplacian filter, Eq. (7) results in the core-weighted Laplacian correlation (CWLC):
| (7b) |
These core-weighted correlation functions are designed to down-weight the regions overlapping with other components, while emphasizing the regions with no overlap. As explained above, the regions with significant overlap have small fm and large fn, and thus a small weighting function. By down-weighting the overlapping regions, the core-weighted correlation functions can minimize the overlap effect in predicting the correct fit.
2.3. Grid-threading Monte Carlo search
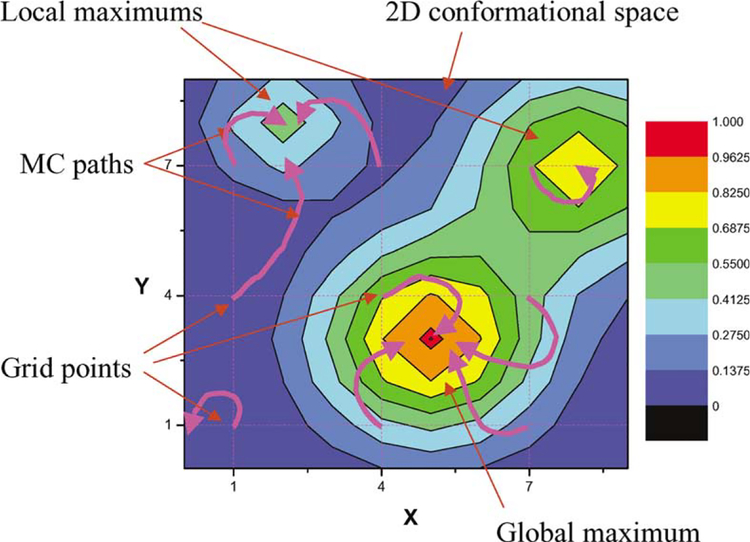
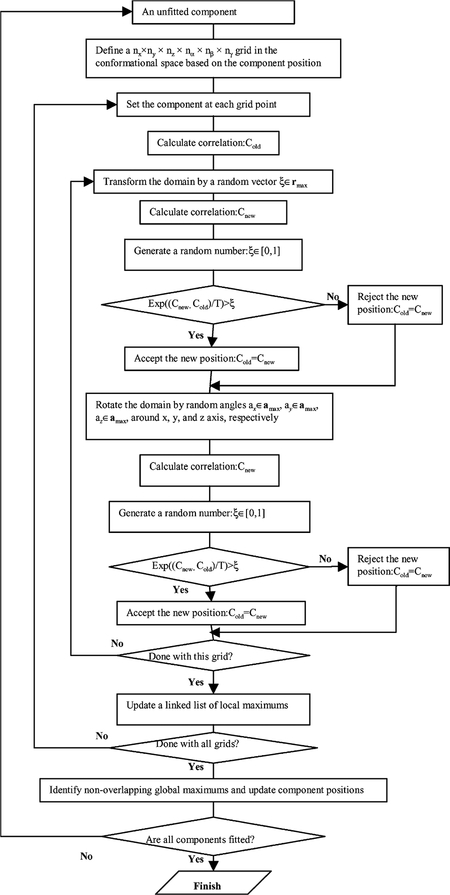
The grid-threading Monte Carlo search is a combination of a grid search and Monte Carlo sampling (Allen and Tildesley, 1987). The conformational space is split into grid points and short Monte Carlo searches are performed to identify local maxima close to the grid points. The global maximum is then identified from among the local maxima. This procedure is illustrated in Fig. 2 for the simple case of a search in two dimensions, where the conformational space is divided into a 3 × 3 grid. The local maximum at (5,3) has the highest correlation of all of the local maxima and thus is the global maximum. Fig. 3 shows the overall flow chart of the grid-threading Monte Carlo search algorithm, which is carried out in the following sequence:
Fig. 2.
The grid-threading Monte Carlo search in two-dimensional space. The conformational space is divided into a 3 × 3 grid. From each of the 9 grid points, short MC searches (shown as purple curves) are performed to locate a nearby local maximum. The global maximum is identified from among these local maxima. Only conformations along the 9 Monte Carlo paths are searched.
Fig. 3.
Schematic flow diagram of the grid-threading Monte Carlo search method to fit individual components into density maps.
For a protein component, the six-dimensional search space (three dimensions for translation and three dimensions for orientation) is divided to provide initial conformational states covering the whole space. The translational space is divided into an nx × ny × nz grid and at each translational grid point the orientational space is divided into an nα × nβ×nγ grid.
- A Monte Carlo search is performed from each grid point to identify a local maximum in the vicinity. The MC search lasts NMC steps. At each step the component is translated along a random vector (xr, yr, zr) and then rotated around x, y, z axes for random angles (αr, βr, γr), where xr, yr, zr are random numbers within (−δmax, δmax), αr, βr, γr are random numbers within (−θmax, θmax), and δmax, θmax are the maximum translation and rotation step sizes, respectively. A trial movement is accepted if
or is rejected otherwise. Here, Cold and Cnew are correlations before and after the movement, T is a reduced temperature which controls the sampling distribution. A larger T corresponds to a “flatter” sampling distribution and to a stronger ability to cross barriers during sampling. ξ is a random number between 0 and 1. Typically, we chose δmax = 15 Å, θmax = 30°, and T = 0.01. Nonoverlapping local maxima are stored in a sorted, linked list. Step 2 is repeated until all grid points are searched.
The global maximum is identified from the linked list and assigned to the component. When there are multiple copies of a given component, multiple global maxima may be identified and assigned to components, depending on how finely the grid-threading Monte Carlo search is carried out. All local maxima with Cmax — Ci ≤ ΔC are identified as global maxima. Here, Cmax and Ci are the correlation coefficients of the true global maximum among the linked list and a local maximum, respectively. ΔC is the threshold of correlation coefficient difference. Typically, we set ΔC = 0.01.
Steps 1 to 4 are repeated until all components have been fitted into the density map.
2.4. Maps of TCR variable domain and the E2 catalytic domain complex
The map of the TCR variable domain was generated from the X-ray structure (PDB code: 1A7N) using the program “pdblur” in the SITUS package (Wriggers et al., 1999). The atomic coordinates were interpolated to a 3D lattice with a voxel space of 3 A, with each lattice point convolved with a Gaussian function to lower the resolution to the indicated values (Wriggers and Birmanns, 2001). When fitting an atomic structure to a target map, a theoretical map is generated from the atomic structure at the same resolution as the target map for correlation function calculation.
The experimental density map of the E2 icosahedral core of pyruvate dehydrogenase was obtained as follows. The icosahedral complex of the E2 catalytic domain was prepared as described by Allen and Perham (1997) and was kindly provided by Dr. Richard Perham (University of Cambridge, Cambridge, UK). Images were recorded from frozen-hydrated specimens on SO163 film using a Tecnai F30 microscope operating at 300 kV at about 3 μm underfocus, at a nominal magnification of 39,000 ×. Films displaying low drift and negligible astigmatism were scanned on a flatbed Zeiss SCAI scanner using a pixel size of 7 μm. Pixels were binned to obtain a final pixel size of 14 μm corresponding to a distance of 3.59 Å in the specimen plane. Estimates for underfocus values of each image were determined computationally using algorithms in the MRC package of image processing programs (Crowther et al., 1996) for subsequent correction of the contrast transfer function. Individual molecular images (total of 4346) were selected automatically using the program BOXER within the EMAN image processing package (Ludtke et al., 1999) and refined using the program FREALIGN (Grigorieff, 1998) to the model of the E2 icosahedral core presented in Milne et al. (2002). The resolution of the resultant model was estimated to be ~14 Å, corresponding to the resolution at which the Fourier Shell Correlation between random halves of the data set is 0.5.
3. Results
3.1. The performance of the core-weighted correlation functions
To test the performance of the core-weighted correlation functions relative to the standard density correlation and to the contour-based Laplacian correlation (Chacon and Wriggers, 2002), we used the α-chain of the T-cell receptor (TCR) variable domain to examine how well these correlations describe the correct fit. Fig. 4a shows the map of the TCR variable domain with the atomic model of the TCR superposed at the correct position. For a correlation function to predict the correct fit, it is essential that it has a global maximum near the correct fit. However, due to the overlap from neighboring components, the global maximum may not occur at the correct fit. Indeed, this was observed here when the α-chain of TCR variable domain was fitted into the 15 Å TCR map using simple density correlation; as illustrated in Fig. 4b, the global maximum was far from the correct position. Here, we operationally define a local maximum with root-mean-square deviation (rmsd) of the Cα backbone from the true structure less than 10 Å as the “near” maximum, and the highest local maximum far from the correct fit, with rmsd >10 Å, as the “far” maximum. Table 1 lists the near maxima and far maxima for the fit of the TCR α-chain with each of the four types of correlation functions at various resolutions. Clearly, the near maximum needs to be higher than the far maximum for a correlation function to predict the correct fit. As can be seen, density correlation only predicted the correct fit at a map resolution of 5 Å, but failed at map resolutions of 10 Å or worse because the near maximum is not higher than the far maximum. Laplacian correlation extended the useful resolution limit to 15 Å, but failed at resolutions of 20 Å or worse. Coreweighted density correlation identified the correct fit at resolutions up to 20 Å, while core-weighted Laplacian correlation was successful at map resolutions up to 30 Å. Thus, for these noise-free maps, the use of core-weighted correlation functions facilitates correct fitting at significantly lower resolutions and is likely to be of special utility when fitting X-ray structures of individual components into electron microscopic density maps, which are often determined in the resolution range of 10–30 Å.
Fig. 4.
(a) The X-ray structure of TCR variable domain (PDB code: 1A7N) and a 15 Å map generated from the structure using the program pdblur (Wriggers et al., 1999). The α- and β-chains are colored red and green, respectively. (b) The α-chain at the maximum density correlation position. The β-chain is at its X-ray position for reference. The map is generated from the X-ray complex structure at resolution 15 Å.
Table 1.
Comparison of the four types of correlation functions for fitting the α-chain of the TCR variable domain into simulated maps of the TCR αβ complex at various resolutions
| Types | Resolution, Å | Near maximum* | Far maximum | ||
|---|---|---|---|---|---|
| rms, Å | Correlation | rms, Å | Correlation | ||
| DC | 5 | 0.71 | 0.638 | 21.8 | 0.598 |
| 10 | 5.1 | 0.738 | 22.9 | 0.819 | |
| 15 | 0 | 0.716 | 23.6 | 0.886 | |
| 20 | 0 | 0.737 | 24.3 | 0.929 | |
| 30 | 0 | 0.801 | 22.9 | 0.970 | |
| 40 | 0 | 0.838 | 23.8 | 0.985 | |
| LC | 5 | 1.2 | 0.400 | 25.1 | 0.147 |
| 10 | 1.0 | 0.586 | 24.4 | 0.334 | |
| 15 | 1.6 | 0.605 | 23.9 | 0.533 | |
| 20 | 2.1 | 0.623 | 24.4 | 0.674 | |
| 30 | 0 | 0.642 | 23.9 | 0.871 | |
| 40 | 0 | 0.687 | 23.4 | 0.946 | |
| CWDC | 5 | 1.2 | 0.711 | 21.7 | 0.401 |
| 10 | 1.8 | 0.875 | 22.0 | 0.784 | |
| 15 | 2.8 | 0.928 | 22.2 | 0.910 | |
| 20 | 5.3 | 0.928 | 22.9 | 0.925 | |
| 30 | 8.4 | 0.949 | 23.2 | 0.962 | |
| 40 | 12.6 | 0.971 | 23.2 | 0.984 | |
| CWLC | 5 | 0.8 | 0.426 | 23.0 | 0.287 |
| 10 | 1.2 | 0.771 | 25.2 | 0.333 | |
| 15 | 1.9 | 0.892 | 23.0 | 0.676 | |
| 20 | 1.8 | 0.936 | 20.7 | 0.824 | |
| 30 | 4.9 | 0.930 | 22.8 | 0.907 | |
| 40 | 9.1 | 0.925 | 23.1 | 0.961 | |
rms = 0 Å indicates no near maximum (rms < 20 Å) was detected. The X-ray position of the α-chain was used to calculate the correlations.
3.2. Conformational search with the grid-threading Monte Carlo method
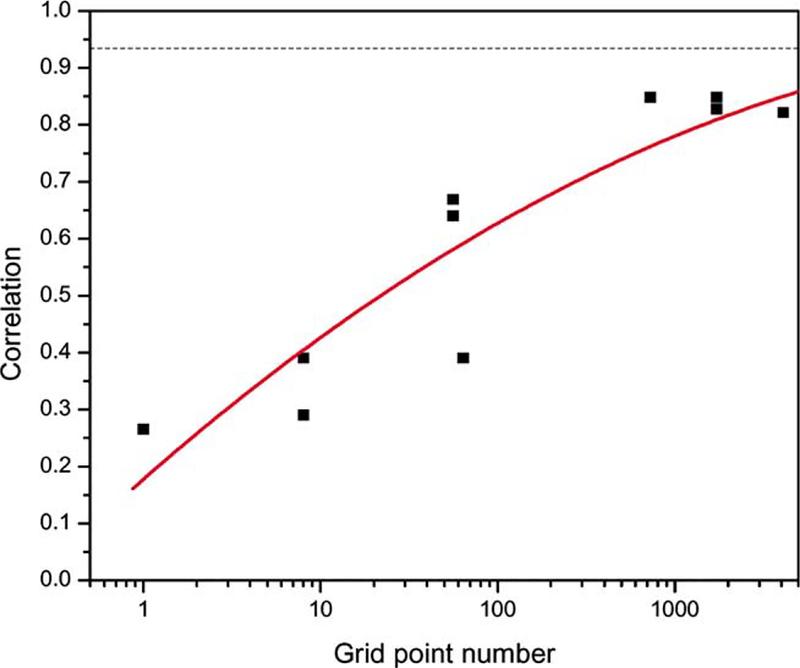
For simple systems with a limited number of degrees of freedom, it is possible to identify the best fit from a grid search of conformational space. For a single rigid body, six independent degrees of freedom need to be searched: three translational variables (x, y, z) and three orientational variables (α, β, γ). Since the accuracy of the fit should depend upon the grid size of these parameters, we fit the α-chain of the TCR variable domain into the simulated 15 Å map of the entire complex using coreweighted density correlation and a range of grid sampling sizes. The highest correlation values obtained at each of the grid sizes is shown in Fig. 5. As can be seen, an exponential increase in grid sampling size was required to improve the correlation values, indicating that a grid search is computationally inefficient.
Fig. 5.
The maximum core-weighted density correlations between the map of TCR α-chain and the map of TCR αβ complex identified from grid searches. These maps are generated at a resolution of 15 Å. The black dashed line represents the correlation value at the position corresponding to the X-ray coordinates. The six-dimensional conformational space was divided into a n × n × n × n × n × n grid with n6 grid points for grid search.
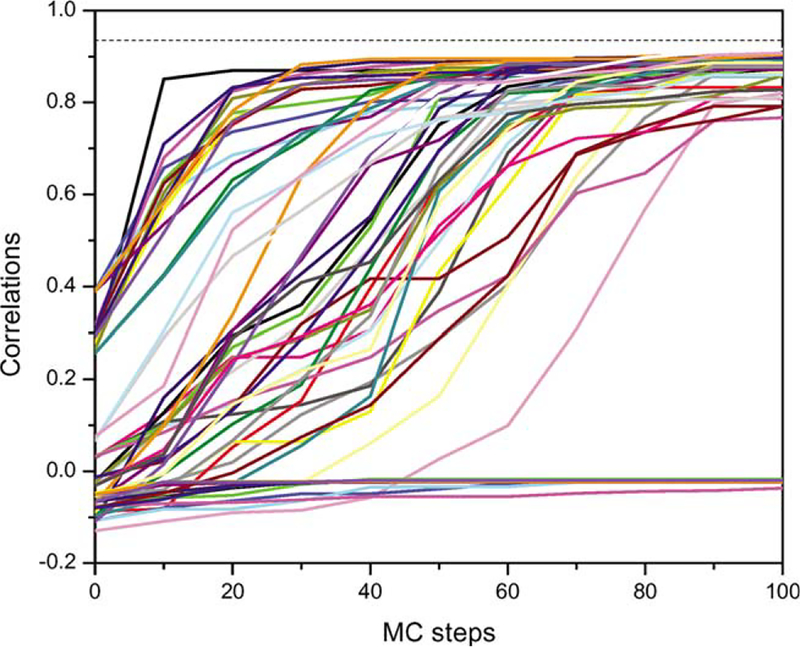
An alternative approach to identify the correct fit is to use the Monte Carlo (MC) search algorithm, which is widely used in sampling multidimensional conformational space (Allen and Tildesley, 1987) in a variety of applications. This method works efficiently for homogeneous systems, but less well for systems containing large energy barriers. In the latter, the result of an MC search often depends upon the starting position and the simulation length. To test the applicability of this method to low-resolution density maps, we again fit the α-chain of the TCR variable domain into a 15 Å simulated map of the complex. We performed 64 MC searches with different starting positions. These starting positions are the 64 grid points after splitting the sixdimensional conformational space into a 2 × 2 × 2 × 2 × 2 × 2 grid. Fig. 6 illustrates the core-weighted density correlation coefficients between the maps of the α-chain and the αβ complex measured throughout each of these searches. The ability to converge to the correct fit and the speed of convergence depended significantly upon the starting position. Certain initial positions led to convergence with only 10 MC steps, others required up to 90 steps, and yet others never displayed positive correlation values even after 100 steps. Thus, a useful strategy is to identify the best local fits through short MC searches starting from discrete grid points covering conformational space, and to select the global best fit among these candidate fits, which is the basis of our proposed grid-threading Monte Carlo search.
Fig. 6.
The core-weighted density correlation function between the map of TCR α-chain and the map of the TCR αβ complex during Monte Carlo searches starting from each of the 2 × 2 × 2 × 2 × 2 × 2 grid points. The Monte Carlo searches were performed with δmax = 15 Å, θmax = 30°, and T = 0.01. Each line represents one Monte Carlo search procedure.
3.3. Fitting of the α-chain of the TCR variable domain
To assess how well the grid-threading Monte Carlo method works in conjunction with various correlation functions, 1000 step MC searches were conducted from each of the 64 (2 × 2 × 2 × 2 × 2 × 2) grids in conformational space, using an initial displacement of 15 AA and an initial rotational step size of 30°. Table 2 lists these fitting results, together with the results obtained using the Situs 2.0 package (Chacon and Wriggers, 2002). An rmsd value of larger than 20 Å indicates that the search converged to a far maximum. MC searches undertaken with density correlation alone did not converge to the correct fit of the α-chain (Table 2). This is an expected result since all test map resolutions were 15 Å or worse, where density correlation does not have a global maximum near the correct fit (see Table 1). Laplacian correlation, core-weighted density correlation, and core-weighted Laplacian correlation all found best fits close to the X-ray position at resolution 15 Å. Laplacian correlation failed to generate the correct fit at resolutions worse than 15 Å. Core-weighted density correlation produced a correct fit up to 20 Å resolution, and core-weighted Laplacian correlation succeeded even at resolutions of 30 Å. The results of SITUS 2.0 are comparable to those obtained using Laplacian correlation; i.e, it worked at resolutions of 15 Å or better.
Table 2.
The rms deviations of the best fits from the X-ray structure using different correlation functions
| Resolution | 15 Å | 20 Å | 30 Å | 40 Å |
|---|---|---|---|---|
| Situs 2.0* | 0.4 | 24.2 | 23.9 | 23.9 |
| DCa | 24.3 | - | - | - |
| LCa | 1.8 | 24.3 | - | - |
| CWDCa | 1.3 | 1.2 | 20.7 | - |
| CWLCa | 1.5 | 2.2 | 1.9 | 21.9 |
The SITUS 2.0 program colores is used with degree = 20, cut-off = 0.0.
The grid-threading MC is performed with a 2 × 2 × 2 × 2 × 2 × 2 grid, Nmc = 1000, δmax = 15 Å, θmax = 30°, and T = 0.01.
3.4. Fitting of the E2 catalytic domain of pyruvate dehydrogenase
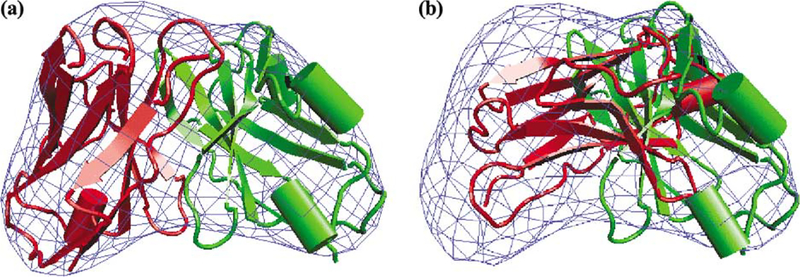
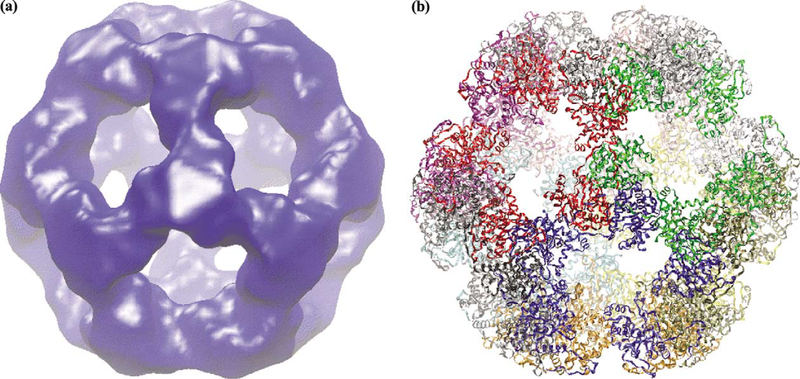
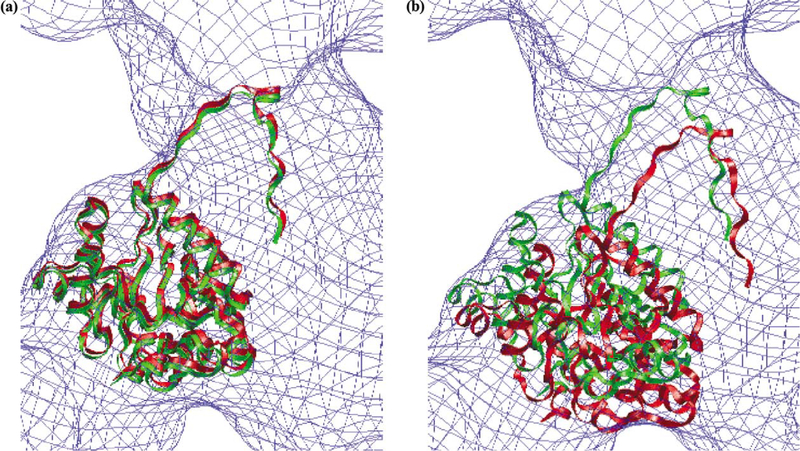
The applicability of the grid-threading Monte Carlo method to fit components into experimental low-resolution density maps was tested using a 14 Å electron microscopic map of the icosahedral core of pyruvate dehydrogenase (Fig. 7a), an 1.8-MDa complex comprised of 60 copies of the E2 catalytic domain, whose structure (Fig. 7b) has been determined using X-ray crystallographic methods. We performed grid-threading Monte Carlo searches with core-weighted Laplacian correlation; Table 3 lists the rms deviations from the X-ray structure and total cpu times of these searches. When 64 (2 × 2 × 2 × 2 × 2 × 2) grids were searched, only 34 of 60 correct fits were identified, likely because the grid size is too coarse for the short MC searches (NMC = 5000 steps) to search out all 60 global maximum positions. When 729 (3 × 3 × 3 × 3 × 3 × 3) or 4096 (4 × 4 × 4 × 4 × 4 × 4) grids were searched, all 60 monomers could be correctly fit into the experimental density map. Note that using finer grids did not result in a proportional increase in cpu time. This is because with finer grids, more best fits can be identified in each loop over all grid points, and fewer loops are need to search out all 60 best fits. Fig. 8 shows the best and worst fits among the 60 fitting results identified with the (4 × 4 × 4 × 4 × 4 × 4) grid, which are both very close to the positions of the corresponding monomer in the X-ray structure.
Fig. 7.
(a) Surface representation of the experimental map (at 14 Å resolution) of the icosahedral complex formed from 60 copies of the E2 catalytic domain of the pyruvate dehydrogenase. (b) The X-ray structure of the same complex (PDB code: 1B5S).
Table 3.
The fitting results of the 60 E2 catalytic domains of the pyruvate dehydrogenase in the icosahedral complex using the grid-threading Monte Carlo search algorithm (GTMC) with the core-weighted Laplacian correlation function and the colores program from SITUS 2.0*
| Monomer index |
GTMC | SITUS 2.0 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Ngrid = 26 | Ngrid = 36 | Ngrid = 46 | Deg = 20° Nex = 60 |
Deg = 20° Nex = 100 |
Deg = 10° Nex = 60 |
Deg = 10° Nex = 100 |
Deg = 5° Nex = 60 |
Deg = 5° Nex = 100 |
|
| 1 | 2.28 | 1.68 | 2.13 | 2.17 | 2.17 | 2.18 | 2.18 | 2.18 | 2.18 |
| 2 | 2.53 | 2.07 | 2.15 | 2.30 | 2.30 | 2.30 | 2.30 | 2.30 | 2.30 |
| 3 | 2.57 | 2.25 | 2.28 | 2.41 | 2.41 | 2.40 | 2.40 | 2.43 | 2.43 |
| 4 | 2.58 | 2.55 | 2.38 | 2.58 | 2.46 | 2.46 | 2.46 | 2.46 | 2.46 |
| 5 | 2.59 | 2.69 | 2.43 | 2.65 | 2.58 | 2.59 | 2.59 | 2.59 | 2.59 |
| 6 | 2.87 | 2.78 | 2.53 | 2.86 | 2.65 | 2.87 | 2.65 | 2.86 | 2.65 |
| 7 | 2.94 | 2.85 | 2.71 | 2.94 | 2.86 | 2.93 | 2.87 | 2.93 | 2.86 |
| 8 | 3.13 | 2.88 | 2.75 | 2.94 | 2.94 | 2.98 | 2.93 | 2.98 | 2.93 |
| 9 | 3.24 | 3.03 | 2.82 | 2.98 | 2.94 | 3.02 | 2.93 | 3.01 | 2.94 |
| 10 | 3.52 | 3.07 | 2.84 | 3.02 | 2.97 | 3.18 | 2.98 | 3.11 | 2.99 |
| 11 | 3.61 | 3.07 | 2.86 | 3.11 | 3.02 | 3.21 | 3.02 | 3.18 | 3.02 |
| 12 | 3.72 | 3.13 | 2.98 | 3.18 | 3.11 | 3.21 | 3.02 | 3.20 | 3.02 |
| 13 | 3.72 | 3.15 | 3.01 | 3.20 | 3.18 | 3.41 | 3.12 | 3.25 | 3.11 |
| 14 | 3.74 | 3.19 | 3.04 | 3.22 | 3.20 | 3.43 | 3.18 | 3.42 | 3.18 |
| 15 | 3.77 | 3.19 | 3.07 | 3.41 | 3.22 | 3.52 | 3.21 | 3.52 | 3.20 |
| 16 | 3.79 | 3.22 | 3.14 | 3.53 | 3.41 | 3.52 | 3.23 | 3.54 | 3.25 |
| 17 | 3.81 | 3.23 | 3.17 | 3.55 | 3.44 | 3.54 | 3.41 | 3.60 | 3.42 |
| 18 | 3.87 | 3.33 | 3.24 | 3.61 | 3.52 | 3.60 | 3.44 | 3.63 | 3.44 |
| 19 | 3.90 | 3.52 | 3.32 | 3.64 | 3.53 | 3.60 | 3.52 | 3.64 | 3.52 |
| 20 | 3.96 | 3.53 | 3.34 | 3.66 | 3.55 | 3.64 | 3.53 | 3.67 | 3.52 |
| 21 | 3.98 | 3.55 | 3.35 | 3.67 | 3.61 | 3.64 | 3.54 | 3.67 | 3.54 |
| 22 | 4.02 | 3.57 | 3.38 | 3.67 | 3.61 | 3.66 | 3.60 | 3.76 | 3.60 |
| 23 | 4.26 | 3.57 | 3.38 | 3.67 | 3.64 | 3.76 | 3.61 | 3.82 | 3.61 |
| 24 | 4.34 | 3.62 | 3.41 | 3.76 | 3.66 | 3.82 | 3.64 | 3.83 | 3.64 |
| 25 | 4.43 | 3.62 | 3.44 | 3.77 | 3.67 | 3.83 | 3.64 | 3.88 | 3.66 |
| 26 | 4.49 | 3.63 | 3.49 | 3.78 | 3.67 | 3.88 | 3.66 | 3.92 | 3.67 |
| 27 | 4.52 | 3.70 | 3.50 | 3.83 | 3.67 | 3.92 | 3.68 | 3.95 | 3.67 |
| 28 | 4.63 | 3.79 | 3.61 | 3.83 | 3.76 (~#19) |
3.95 | 3.68 | 3.97 | 3.68 |
| 29 | 4.82 | 3.81 | 3.65 | 3.92 | 3.77 | 3.96 | 3.76 | 4.05 | 3.76 |
| 30 | 4.95 | 3.84 | 3.66 | 3.94 | 3.78 | 4.04 | 3.78 (~#21) |
4.14 | 3.82 |
| 31 | 5.08 | 3.87 | 3.73 | 3.96 | 3.83 | 4.13 | 3.78 | 4.20 | 3.83 |
| 32 | 5.51 | 3.89 | 3.84 | 4.21 | 3.83 | 4.20 | 3.82 | 4.21 | 3.88 |
| 33 | 5.63 | 3.91 | 3.87 | 4.23 | 3.92 | 4.22 | 3.83 | 4.24 | 3.92 |
| 34 | 5.87 | 3.94 | 3.91 | 4.23 | 3.94 (~#29) |
4.23 | 3.88 (~#31) |
4.24 | 3.95 |
| 35 | 15.15 | 3.95 | 3.94 | 4.24 | 3.96 | 4.24 | 3.92 | 4.26 | 3.97 |
| 36 | 15.52 | 3.98 | 3.95 | 4.26 | 4.02 | 4.25 | 3.96 | 4.27 | 4.05 |
| 37 | 16.36 | 4.01 | 3.95 | 4.27 | 4.04 | 4.26 | 3.97 | 4.27 | 4.14 |
| 38 | 17.28 | 4.04 | 3.99 | 4.30 | 4.18 | 4.27 | 4.04 | 4.41 | 4.20 |
| 39 | 18.15 | 4.09 | 4.03 | 4.42 | 4.21 | 4.27 | 4.13 | 4.52 | 4.21 |
| 40 | 19.25 | 4.13 | 4.10 | 4.51 | 4.23 | 4.39 | 4.18 (~#39) |
4.58 | 4.24 |
| 41 | 19.30 | 4.17 | 4.17 | 4.60 | 4.24 | 4.42 | 4.20 | 4.79 | 4.24 |
| 42 | 19.39 | 4.21 | 4.19 | 4.79 | 4.24 | 4.43 | 4.22 | 4.84 | 4.25 |
| 43 | 19.88 | 4.23 | 4.21 | 4.86 | 4.26 | 4.52 | 4.23 | 5.17 | 4.26 |
| 44 | 20.43 | 4.24 | 4.26 | 5.20 | 4.27 | 4.60 | 4.24 | 5.23 | 4.27 |
| 45 | 20.64 | 4.25 | 4.28 | 5.21 | 4.30 | 4.65 | 4.25 | 5.28 | 4.27 |
| 46 | 22.30 | 4.27 | 4.39 | 5.23 | 4.39 | 4.80 | 4.26 | 5.30 | 4.39 |
| 47 | 29.28 | 4.28 | 4.42 | 5.29 | 4.42 | 4.84 | 4.27 | 5.30 | 4.41 |
| 48 | 30.15 | 4.29 | 4.44 | 5.30 | 4.51 | 5.17 | 4.27 | 5.32 | 4.42 |
| 49 | 31.20 | 4.30 | 4.47 | 5.30 | 4.63 | 5.23 | 4.39 | - | 4.52 |
| 50 | 31.90 | 4.35 | 4.50 | 5.34 | 4.80 | 5.29 | 4.42 | - | 4.58 |
| 51 | 32.14 | 4.37 | 4.50 | - | 4.86 | 5.29 | 4.42 | - | 4.65 |
| 52 | 32.29 | 4.37 | 4.54 | - | 5.20 | 5.30 | 4.52 | - | 4.79 |
| 53 | 32.34 | 4.43 | 4.58 | - | 5.21 | - | 4.58 | - | 4.84 |
| 54 | 33.09 | 4.59 | 4.63 | - | 5.23 | - | 4.65 | - | 5.17 |
| 55 | 33.79 | 4.76 | 4.65 | - | 5.28 | - | 4.79 | - | 5.23 |
| 56 | 33.83 | 4.84 | 4.69 | - | 5.30 | - | 4.84 | - | 5.24 |
| 57 | 33.96 | 5.01 | 5.26 | - | 5.30 | - | 5.17 | - | 5.28 |
| 58 | 34.77 | 5.02 | 5.48 | - | 5.34 | - | 5.23 | - | 5.30 |
| 59 | 35.00 | 5.29 | 5.51 | - | 12.52 (~#26) |
- | 5.23 | - | 5.30 |
| 60 | 37.02 | 5.63 | 6.52 | - | 12.62 (~#50) |
- | 5.29 | - | 5.31 |
| - | - | - | - | - | 13.86 (~#15) |
- | 5.29 | - | - |
| - | - | - | - | - | 14.47 (~#53) |
- | 5.30 | - | - |
| - | - | - | - | - | 14.91 (~#6) |
- | 5.34 | - | - |
| - | - | - | - | - | 15.38 (~#47) |
- | - | - | - |
| - | - | - | - | - | 15.54 (~#8) |
- | - | - | - |
| - | - | - | - | - | 15.80 (~#49) |
- | - | - | - |
| Average, Åa |
3.90 | 3.76 | 3.73 | 3.85 | 3.83 | 3.87 | 3.85 | 3.85 | 3.85 |
| Time, hours |
1.84 | 8.61 | 19.43 | 9.05 | 11.53 | 19.31 | 22.00 | 130.78 | 133.80 |
Ngrid = M6 indicates a M × M × M × M × M × M grid is used in a GTMC search with Nmc = 5000, δmax = 30 Å, θmas = 30°, and T = 0.01. For SITUS fitting, “Deg” represents the angular grid size and “Nex” represents the number of the candidate solutions to be refined.
This table lists the rms deviations of all solutions obtained in the fittings. Some solutions overlapping with other solutions are labeled out with “~#XX”, where XX is the overlapping solution number.
The averages are calculated over those solutions with correct fit (rms < 10Å).
Fig. 8.
Comparison of the location of the E2 catalytic domain obtained using a GTMC search (green) with that of the corresponding domain from the X-ray structure (red). The experimental map obtained by electron microscopy is shown in blue. (a) The best fit obtained, rms = 2.13 Å; (b). The worst fit obtained, rms = 6.52 Å. The grid-threading Monte Carlo search was conducted with a 4 × 4 × 4 × 4 × 4 × 4 grid, Nmc = 5000, δmax = 30 Å, θmax = 30°, and T = 0.01. The core-weighted Laplacian correlation function was used. The average root mean square deviation of the Cα backbone (averaged over all 60 copies) between the X-ray structure and the fitted coordinates is 3.73 Å.
Table 3 also compares the results from SITUS 2.0 package with different angular grid size and different solution numbers. The two most important parameters that bear on the results of automated fitting using SITUS 2.0 are the angular grid size and the number of solutions identified by the fitting algorithm. A small angular grid size results in a more detailed search in orientational space, but requires longer computational times. The number of solutions identified should be equal to or larger than the number of fits expected. If the search was restricted to identifying only 60 solutions, the algorithm in SITUS 2.0 did not arrive at all 60 correct fits, independent of whether the angular grid size is 20, 10, or 5°. This was because some of the best fits overlap with each other and therefore must be discarded. If the search criteria were relaxed to identify 100 solutions, the algorithm in SITUS 2.0 arrived at 66 best fits with an angular grid size of 20°, 58 of which represent correct fits (see rmsd values in Table 3). With a reduced angular grid size of 10°, SITUS 2.0 identified 63 best fits, 3 fits of which overlap with other fits. When the angular grid size was reduced to 5°, the SITUS 2.0 algorithm correctly placed all 60 monomers in the map. However, the grid-threading Monte Carlo search with 729 grid points was substantially quicker to arrive at the same result, taking about 8.6 h as compared to 133.8 h with SITUS 2.0 under similar computing conditions.
4. Discussion
In this work, we have described a core-weighting approach to overcome some of the problems that can arise in the fitting of atomic structures into low-resolution maps with multiple components. Using two model systems, we have demonstrated that coreweighted correlations have significantly improved sensitivity to distinguish the correct fit when compared with more traditional correlations. The construction of a molecular model for a complex macromolecular assembly is thus simplified from a many-body search problem to a series of single-body search problems, making the computational search for the correct fit much easier.
Like the core-weighting approach developed here, the Laplacian filter adopted by Chacon and Wriggers also extends the resolution limit significantly. But unlike the core-weighting approach, the use of a Laplacian filter alone does not adequately compensate for the overlapping effects resulted from neighboring components that are inherent in low-resolution maps. In addition, as pointed out by Chacon and Wriggers (2002), the Laplacian amplifies high-frequency noise in the map, which may cause the generation of false positives. In turn, the core-weighting correlation function has its own limits. It relies on the fit of the nonoverlap regions of the individual components, and therefore requires that these regions exhibit sufficiently distinct density distributions for obtaining a reliable fit. As shown in the two examples studied here, despite inheriting the noise amplification feature of Laplacian filtering, the combination of the Laplacian filter and the core-weighting function shows better performance at lower resolutions than the other correlation functions that were tested. It should be noted these results are based on the noise-free maps. The sensitivity of these correlation functions to the presence of noise must be tested further.
The use of the grid-threading Monte Carlo approach greatly enhances the speed of the calculation compared to exhaustive searches. This is obviously an important advantage for larger assemblies. The efficiency of the grid-threading Monte Carlo search comes from the fact that it only searches certain paths within the conformational space. Since an exhaustive search is not carried out, this method assumes that at least one of the paths starting from the grid points leads to the correct fit. However, because an exhaustive search is not performed, the grid size must be sufficiently fine, say, 3 × 3 × 3 × 3 × 3 × 3 or 4 × 4 × 4 × 4 × 4 × 4, to ensure that the correct fit is within the reach of a short Monte Carlo search initiated from nearby grid points.
For single-body searches, Fourier correlation theory and the fast Fourier transform (FFT) provide an attractive way to scan rapidly the correlation through the translation space. The SITUS 2.0 program (Chacon and Wriggers, 2002) uses this approach to achieve an efficient exhaustive search in translation space. To take advantage of this approach, we can modify the coreweighting function to the following form,
| (6a) |
where b is a nonzero constant. Any core-weighted summation can be calculated through the reverse Fourier transform of the product of two Fourier transforms:
| (10) |
Eq. (10) can be applied to all the core-weighted summations through Eqs. (7)–(9) to efficiently scan the core-weighted correlations in translation space. It should be noted that the introduction of the coreweighting function requires several more summations to be calculated through Eq. (10), as compared to the standard correlations.
One advantage of the grid-threading Monte Carlo search is that it can be extended to multibody systems without an exponential increase in the computational cost. Monte Carlo search methods are designed for multidimensional space sampling and have been widely used in many-body systems (Allen and Tildesley, 1987). Directly performing a multibody search does not require a target function to distinguish the correct fits of each individual domain to a complex map, because the overlap between neighboring components can be calculated directly from all components. Another advantage of this approach is that it is convenient for molecular modeling, including structure building, manipulation, and refinement based on low-resolution maps. It is also relatively easy to incorporate constraints to components during a search. The grid-threading MC method for construction of complex structures from EM maps has been implemented into CHARMM (Brooks et al., 1983) and will be available at its next release.
References
- Allen MD, Perham RN, 1997. The catalytic domain of dihydrolipoyl acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Expression, purification and reversible denaturation. FEBS Lett. 413, 339–343. [DOI] [PubMed] [Google Scholar]
- Allen MP, Tildesley DJ, 1987. Computer Simulations of Liquids. Clarendon, Oxford. [Google Scholar]
- Baker TS, Cheng RH, 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol 116, 120–130. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swamina-than S, Jaun B, Karplus M, 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem 4, 187–217. [Google Scholar]
- Chacon P, Wriggers W, 2002. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol 317, 375–384. [DOI] [PubMed] [Google Scholar]
- Crowther RA, Henderson R, Smith JM, 1996. MRC image processing programs. J. Struct. Biol 116, 9–16. [DOI] [PubMed] [Google Scholar]
- Frank J, 1996. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press, London. [Google Scholar]
- Frank J, Penczek P, Agrawal RK, Grassucci RA, Heagle AB, 2000. Three-dimensional cryoelectron microscopy of ribosomes. Methods Enzymol. 317, 276–291. [DOI] [PubMed] [Google Scholar]
- Grigorieff N, 1998. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J. Mol. Biol 277, 1033–1046. [DOI] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W, 2001. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol 308, 1033–1044. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W, 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol 128, 82–97. [DOI] [PubMed] [Google Scholar]
- Milne JLS, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, Brooks BR, Perham RN, Henderson R, Subramaniam S, 2002. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 21, 5587–5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, Horwich AL, Saibil HR, 2001. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell 107, 869–879. [DOI] [PubMed] [Google Scholar]
- Roseman AM, 2000. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr. D. Biol. Crystallogr 56, 1332–1340. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, 2000. Fitting atomic models into electron-microscopy maps. Acta Crystallogr. D. Biol. Crystallogr 56, 1341–1349. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Bernal R, Pletnev SV, 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol 136, 190–200. [DOI] [PubMed] [Google Scholar]
- Russ JC, 1998. The Image Processing Handbook. CRC Press, Boca Raton, FL. [Google Scholar]
- Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W, 2000. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100, 301–309. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Henderson R, 2000. Molecular mechanism of vectorial proton translocation by bacteriorhodopsin. Nature 406, 653–657. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Hanein D, 1999. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J. Struct. Biol 125, 176–184. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Birmanns S, 2001. Using situs for flexible and rigid-body fitting of multiresolution single-molecule data. J. Struct. Biol 133, 193–202. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Chacon P, 2001. Modeling tricks and fitting techniques for multiresolution structures. Structure 9, 779–788. [DOI] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA, 1999. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol 125, 185–195. [DOI] [PubMed] [Google Scholar]
- Yin Z, Zheng Y, Doerschuk PC, 2001. An ab initio algorithm for low-resolution 3-D reconstructions from cryoelectron microscopy images. J. Struct. Biol 133, 132–142. [DOI] [PubMed] [Google Scholar]