Abstract
Globozoospermia has been reported to be a rare but severe causation of male infertility, which results from the failure of acrosome biogenesis and sperm head shaping. Variants of dpy-19-like 2 (DPY19L2) are highly related to globozoospermia, but related investigations have been mainly performed in patients from Western countries. Here, we performed a screening of DPY19L2 variants in a cohort of Chinese globozoospermic patients and found that five of nine patients carried DPY19L2 deletions and the other four patients contained novel DPY19L2 point mutations, as revealed by whole-exome sequencing. Patient 3 (P3) contained a heterozygous variant (c.2126+5G>A), P6 contained a homozygous nonsense mutation (c.1720C>T, p.Arg574*), P8 contained compound heterozygous variants (c.1182-1184delATC, p.Leu394_Ser395delinsPhe; c.368A>T, p.His123Arg), and P9 contained a heterozygous variant (c.1182-1184delATCTT, frameshift). We also reported intracytoplasmic sperm injection (ICSI) outcomes in the related patients, finding that ICSI followed by assisted oocyte activation (AOA) with calcium ionophore achieved high rates of live births. In summary, the infertility of these patients results from DPY19L2 dysfunction and can be treated by ICSI together with AOA.
Keywords: DPY19L2, globozoospermia, male infertility, point mutation, whole-exome sequencing
INTRODUCTION
Globozoospermia (MIM 102530) is a major subtype of teratozoospermia and is a rare but severe causation of male infertility characterized by round-headed spermatozoa.1 Because acrosomes are impaired or completely lost in globozoospermia, spermatozoa fail to fertilize oocytes.2 Normal spermatozoa are shaped through spermatid elongation, which is coordinated with the biogenesis of acrosomes. During spermatid elongation or differentiation, Golgi apparatus-derived pro-acrosome vesicles are gathered and attached to the anterior face of the spermatozoa, so failure in either sperm-head elongation or acrosome biogenesis can result in globozoospermia.3,4,5,6 From animal models and genome sequencing of related patients, many genetic factors have been revealed to be associated with globozoospermia and have been found to be involved in acrosome biogenesis or attachment to the nuclear membranes.4,7,8,9,10,11,12
DPY19L2 is highly expressed in the testis and it is a globozoospermia pathogenic gene, which has been identified in globozoospermic patients and validated in animal models. The first case of a DPY19L2 variant in globozoospermia was a homozygous deletion of 200 kb on chromosome 12, which contains the DPY19L2 locus. The large-scale deletion led to the complete loss of the DPY19L2 gene, and the affected sperms were round headed without acrosomes.13 The DPY19L2 protein is localized on the inner membrane of the sperm nuclear envelope (NE), facing the acrosome and with both its N- and C-terminuses embedded in the nucleoplasm. The inner NE-localized DPY19L2 facilitates the attachment of the acrosome to the NE. In Dpy19l2-deficient mouse spermatids, the development of the acrosome has been found to be hampered as early as step 4 spermatids.14 Without Dpy19l2, the acrosome matrix is no longer bound tightly to the acroplaxome, and the acrosome extension along the nucleus stops. All these defects result in round-headed spermatozoa.14
In addition to the loss of DPY19L2 protein, substitution of certain amino acids in the protein can also lead to globozoospermia. More than ten point variants of DPY19L2 have been reported in globozoospermic patients, with DPY19L2 variants showing a relatively high frequency in different cohorts.15,16,17,18,19 Reported DPY19L2 variants include nonsense mutations, missense mutations, and deletions of certain nucleotides, resulting in mutated or truncated DPY19L2 proteins. However, most of the studies were performed in European, North African, and Middle Eastern populations, and only one cohort of Chinese patients has been reported.18 Here, we conducted a systematic survey on a new Chinese cohort of globozoospermic patients and found complete or partial deletion of the DPY19L2 gene and six additional new point variants. We also found that most of the infertility of the patients could be overcome by intracytoplasmic sperm injection (ICSI) with assisted oocyte activation (AOA). Our results provide new insight into DPY19L2 variant-associated globozoospermia, which have very important implications for the diagnosis and treatment of this severe male infertility.
PATIENTS AND METHODS
Patients
Globozoospermic patients were recruited from Peking University First Hospital (Beijing, China), Peking University Third Hospital (Beijing China), and The First Affiliated Hospital of Anhui Medical University (Hefei, China). Informed consent was obtained from all individual participants included in the study. Related examinations of their samples are listed in Table 1. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees of three above mentioned hospitals and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Table 1.
Clinical information of the globozoospermic patients
| Patient | Age (year) | Semen volume (ml) | Sperm concentration (×106 per ml) | Total sperm count (×106) | Motility PR/NP/IM (%) | Sperm morphology (%) | ||
|---|---|---|---|---|---|---|---|---|
| Normally formed | Round headed | Acrosomeless | ||||||
| P1 | 35 | 4 | 53 | 212 | 19/4/77 | 0 | 100 | 100 |
| P2 | 35 | 3 | 37 | 111 | 8/1/91 | 0 | 100 | 100 |
| P3 | 31 | 2 | 61 | 122 | 11/9/79 | 0 | 100 | 100 |
| P4 | 35 | 3 | 36 | 108 | 34/15/51 | 0 | 100 | 100 |
| P5 | 35 | 1 | 72 | 72 | 17/16/72 | 0 | 100 | 100 |
| P6 | 30 | 2 | 33 | 66 | 49/22/30 | 0 | 100 | 100 |
| P7 | 24 | 6 | 7 | 42 | 12/20/68 | 0 | 100 | 100 |
| P8 | 25 | 2 | 55 | 110 | 17/19/64 | 0 | 100 | 100 |
| P9 | 33 | 2 | 35 | 70 | 14/10/77 | 0 | 100 | 100 |
| Median | 33 | 2 | 37 | 108 | 16/15/70 | 0 | 100 | 100 |
The sperm motility and the percentages of morphologically normal and abnormal spermatozoa were evaluated according to the guidelines (WHO, 2010). PR: progressive motility; NP: nonprogressive motility; IM: immotility; WHO: World Health Organization
Morphological examinations of human spermatozoa
Semen spermatozoa were spread onto polylysine-coated slides and dried in air, and then fixed by 4% (w/v) polyformaldehyde (PFA, Solarbio, Beijing, China). Acrosomes were stained by fluorescein isothiocyanate (FITC)-labeled Peanut agglutinin (PNA, ENZO, Farmingdale, NY, USA), and nuclei were stained by 4’, 6-diamidino-2-phenylindole (DAPI, Roche, Basel, Switzerland).
DNA extraction and DPY19L2 deletion screening
Genomic DNA was extracted from blood with DNA extraction kits (DP304), according to the instructions of the manufacturers (TIANGEN, Beijing, China). DPY19L2 deletion screening primers were designed as previously described.13 Fifty nanograms genomic DNA, 10 pmol forward and reverse primers, and PCR premix (Yeasen, Shanghai, China) were used in each PCR reaction, and the annealing temperature was the same as that described previously.13 Each PCR reaction was performed for 35 cycles, and the same amount of products were analyzed with 1.5% (w/v) agarose gel (Amresco, Solon, OH, USA).
Whole-exome sequencing
The qualified genomic DNA samples were randomly fragmented by Covaris technology,20 with the size of the library fragments being mainly distributed between 150 bp and 250 bp. The end repair of DNA fragments was performed and an “A” base was added to the 3’ end of each strand. Adapters were then ligated to the A-tailed DNA fragments for amplification and sequencing using Truseq DNA sample preparation (Illumina Inc., San Diego, CA, USA). Size-selected DNA fragments were amplified by ligation-mediated PCR (LM-PCR), purified, and hybridized to the exome array for enrichment using Sure SelectXT Custom kit (Agilent, Santa Clara, CA, USA). Nonhybridized fragments were then washed away and captured products circularized. Rolling circle amplification (RCA) was performed to produce DNA Nanoballs (DNBs). Each resulting qualified captured library was then loaded on a BGISEQ-500 sequencing platform (BGI, Shenzhen, China), and we performed high-throughput sequencing for each captured library to ensure that each sample met the desired average sequencing coverage (globally over 95%) with a depth of ×30. Sequencing-derived raw image files were processed by BGISEQ-500 base-calling software (BGI, Shenzhen, China), using Confirm-Show-All base calling with default parameters. The sequence data of each individual were generated as a paired-end read, which was defined as “raw data” and stored in FASTQ format.
The bioinformatics analysis began with the sequencing data (raw data from the BGISEQ platform). First, clean data were produced by filtering the raw data, and the clean data of each sample were mapped to the human reference genome (GRCh37). Burrows–Wheeler Aligner (BWA) software (https://github.com/lh3/bwa/archive/v0.7.17.tar.gz) was used to do the alignment.21,22 To ensure accurate variant calling, we followed the recommended best practices for variant analysis with the Genome Analysis Toolkit (GATK, https://github.com/broadinstitute/gatk/archive/4.0.1.1.tar.gz).23,24 Local realignment around InDels and base quality score recalibration were performed by GATK, with duplicate reads removed by Picard tools (https://github.com/broadinstitute/picard/releases/download/2.18.11/picard.jar). The sequencing depth and coverage for each individual were calculated from the alignments.
In addition, the strict data analysis quality control system (QC) of the whole pipeline was built to guarantee qualified sequencing data. All genomic variations, including single-nucleotide polymorphisms (SNPs) and InDels, were detected by state-of-the-art software, such as Haplotype Caller of GATK (v3.8; Broad Institute, Cambridge, MA, USA). After that, hard-filtering methods were applied to get high-confident variant calls. Next, the Intervar tool (https://github.com/WGLab/InterVar/archive/v2.0.1.tar.gz) was applied to perform a series of annotations for variants.25 The final variants and annotation results were used in the downstream advanced analysis.
Oocyte activation procedure
In vitro fertilization (IVF) medium (COOK Medical, Brisbane, Australia) containing 10 mol l−1 calcium ionophore (A23187, Sigma-Aldrich, St. Louis, MO, USA) was prepared before ICSI. Thirty minutes after ICSI, oocytes were activated in the prepared IVF medium for 5 min. These oocytes were washed three times in fresh IVF medium without calcium ionophore and then were individually transferred into a drop of IVF medium under mineral oil in disc for incubation at 37°C under 5% (v/v) CO2 air atmosphere.26 IVF was considered successful if zygotes presented two pronuclei (2PN) on day 1 (16–18 h after ICSI). On days 3, 5, and 6, the number and quality of embryos were checked and then were classified into different grades.27,28 On day 5 or 6, fresh or frozen embryos were transferred in the following cycle, respectively.
RESULTS
Confirmation of the globozoospermia of the patients
Patients were referred to our center for semen analysis after trying unsuccessfully for a pregnancy for at least 3 years. Semen samples, obtained after 3 days of sexual abstinence, showed nine cases of total globozoospermia (100%). These nine genetically independent patients ranged from 24 to 35 years of age (Table 1), and the median progressive motility of the spermatozoa was 16% with seven of the nine patients below the WHO 2010 normal threshold.18 DNA and acrosomes were stained during morphological analysis, with results showing that all the spermatozoa were round headed and lacked normal acrosomes. No other defects in sperm tails were found in the patients, indicating total globozoospermia. Representative images of the spermatozoa are shown in Figure 1.
Figure 1.
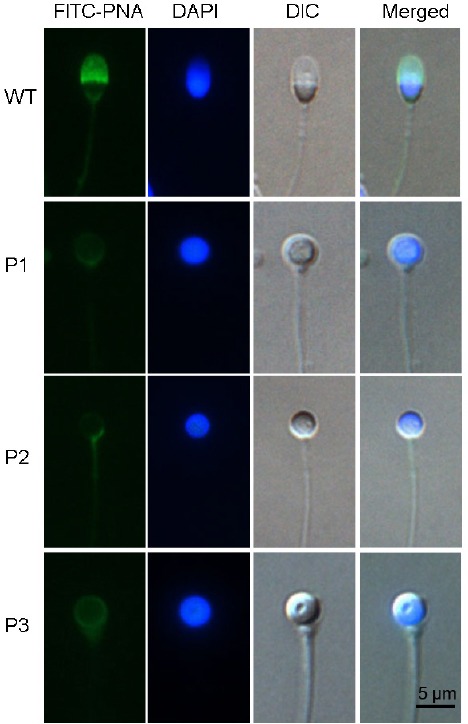
Representative images of sperm from globozoospermic patients. All the spermatozoa from affected individuals and control individuals were stained with PNA for the acrosome and DAPI for the nucleus. Scale bars = 5 μm. PNA: peanut agglutinin; DAPI: 4’, 6-diamidino-2-phenylindole; FITC: fluorescein isothiocyanate; DIC: differential interference contrast microscope; WT: wild type.
New variants of DPY19L2
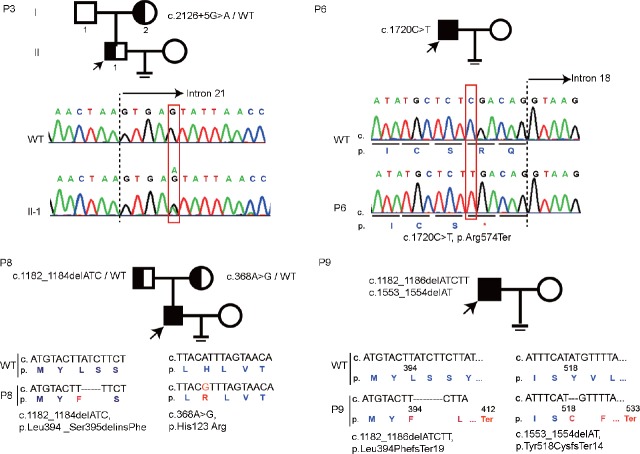
We first screened for DPY19L2 deletions according to the previously established strategy13 and amplified seven loci around the DPY19L2 gene (a–g) from genomic DNA (Supplementary Figure 1a (407KB, tif) ). The seven loci were successfully amplified in control fertile individuals, and four globozoospermia patients also yielded complete amplification of the loci (P3, P6, P8, and P9 in Supplementary Figure 1b (407KB, tif) ). To identify novel variants from globozoospermic patients, we next applied whole-exome sequencing (WES) to the four DPY19L2 nondeleterious globozoospermic patients. To establish the family pedigree of the potential variants, we collected genomic DNA of some patients’ parents (P3 and P8). P3 carried a heterozygous nucleotide substitution in intron 21 next to exon 21. Pedigree analysis of the affected family revealed that this variant was maternally inherited (Figure 2). The WES results indicated that P6 carried a homozygous variant on exon 18- c.1720C>T.
Figure 2.
Novel DPY19L2 variants in globozoospermic patients without gene deletion revealed by whole-exome sequencing. P3 carried a heterozygous variant c.2126+5G>A, which was maternally inherited. P6 carried a homozygous variant c.1720C>T leading to PTC in DPY19L2 mRNA. P8 contained a compound heterozygous DPY19L2 variant, paternally inherited c.1182-1184delATC and maternally inherited c.368A>G. P9 contained two heterozygous variants c.1182-1186delATCTT and c.1553-1554delAT, which are probable compound heterozygous variants. WT: wild type; DPY19L2: dpy-19-like 2; PTC: premature termination codon.
DPY19L2 deletion screening of the globozoospermic patients. (a) The distribution of a-g loci on and around DPY19L2. (b) Genomic DNA was extracted from the blood of the patients and the reported seven loci of DPY19L2 were amplified. P1, P2, P4, P5, and P7 show complete or partial deletion of the DPY19L2 gene, while P3, P6, P8, and P9 show no deletion in the DPY19L2 gene.
P8 contained compound heterozygous DPY19L2 variants: one was a three-nucleotide deletion from c.1182-1184 (c.1182-1184delATC), which was paternally inherited, and the other was a nucleotide substitution c.368A>G, which was maternally inherited. The variant c.1182-1184delATC disrupted the code for Leu394 and Ser395, and the new frame encoded for a Phe without influencing the following coding frame. The final result of this variant was Leu394_Ser395delinsPhe. The A to G substitution at nucleotide 368 resulted in an amino acid change from His to Arg in the protein sequence. P9 also contained a deletion near c.1182, a five-nucleotide deletion from 1182 to 1186 (c.1182-1186delATCTT) that led to a frameshift after Met392 and resulted in a stop code at position 413. P9 also contained a two-nucleotide deletion (c.1553-1554delAT) that led to a frameshift after Tyr518 and resulted in a stop code at position 523. Blood samples of the parents of P9 were unavailable, so it was difficult to determine whether P9's two deletions are compound heterozygous variants or not. In summary, the new DPY19L2 variants found in our cohort led to loss of function in the DPY19L2 protein but not any other new genes. All the newly identified variants are summarized in Table 2.
Table 2.
Summary of the new DPY19L2 variants found in the investigated globozoospermic patients
| Patients | mRNA change | Protein change | Variant type | Max allele frequency | GERP (prediction of conservation) | Zygosity |
|---|---|---|---|---|---|---|
| P3 | c.2126+5G>A | - | Splice region variant | 0 | 3.78 | Heterozygous |
| P6 | c.1720C>T | p.Arg574Ter | Nonsense | 0 | 2.51 | Homozygous |
| P8 | c.1182_1184delATC | p.Leu394_Ser395delinsPhe | Inframe deletion | <0.0001 | 1.37 | Compound heterozygous |
| c.368A>G | p.His123Arg | Missense | 0 | 2.5 | ||
| P9 | c.1182_1186delATCTT | p.Leu394PhefsTer19 | Frame shift | 0 | 2.71 | Compound heterozygous |
| c.1553_1554delAT | p.Tyr518CysfsTer14 | Frame shift | <0.0001 | 3.18 |
The max allele frequency of the variants was from gnomAD and ExAC databases. Polyphen scores were only suitable to assess the pathogenicity of missense mutations, and the Polyphen score of c.368A>G is 0.998, which means likely pathogenic. GERP: Genomic Evolutionary Rate Profiling
Effects of single-nucleotide substitution on the properties of conserved amino acid in the DPY19L2 protein
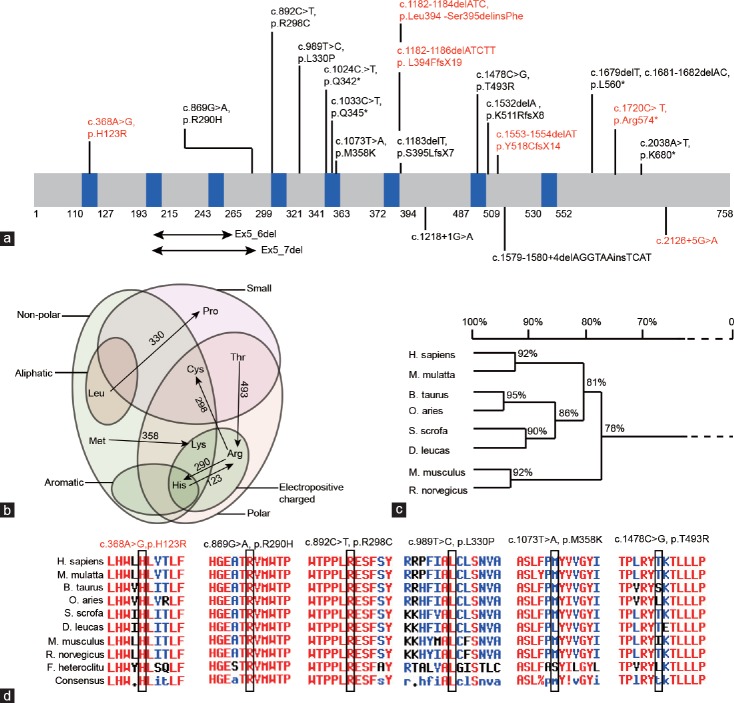
In addition to gene deletions, numerous point mutations of DPY19L2 have been identified in globozoospermic patients. All the reported mutations and the novel variants found in our study are summarized in Figure 3a, which includes 19 mutations. These mutations include nine nonsense mutations, seven missense mutations, and three intronic mutations. We then analyzed the amino acid properties before and after mutation, finding that all the mutations resulted in amino acid property changes, either from polar to nonpolar or from noncharged to charged (Figure 3b). Since DPY19L2 is highly conserved among species, we then investigated whether the mutated amino acids were conserved or not (Figure 3c). We found that all the affected amino acids were highly conserved among species, with only T493 being less conserved among all species yet still conserved in humans, monkeys, and sheep (Figure 3d). These analyses revealed that single-nucleotide substitutions might change the important properties of the conserved amino acids, thus disrupting the function of DPY19L2 protein.
Figure 3.
The mutated DPY19L2 sites were highly conserved among species. (a) The distribution of reported and newly identified variants on DPY19L2 protein. The blue boxes show the predicted transmembrane domains. The variants in black are reported variants, and those in red are newly found variants in our study. (b) The amino acid property changes before and after mutations. (c) DPY19L2 is highly conserved among species; the sequences were aligned with DNAMAN. (d) The mutated amino acids are conserved among species. The sequences were aligned with Multalign; amino acids in red show high identity among species, those in blue show neutral identity, and those in black show low identity. DPY19L2: dpy-19-like 2.
DPY19L2 mutations are the major cause of globozoospermia
Although globozoospermia has been well documented, few pathogenic genes have been identified, so we analyzed the percentage of DPY19L2 mutations together with two other globozoospermia-associated genes spermatogenesis associated 16 (SPATA16) and protein interacting with C kinase 1 (PICK1) in all reported patients and our current Chinese cohort.7,8,15,16,17,18,19 We found that in 193 reported globozoospermic patients, over 57% of them carried DPY19L2 mutations, but less than 5% of the patients carried mutations in SPATA16 or PICK1. Nearly 40% of the patients were diagnosed with globozoospermia due to unknown reasons (Figure 4a). In all the patients with DPY19L2 variants, over 70% carried a gene deletion and about 23% carried point mutations (Figure 4b). By comparing reported genetic data from globozoospermic patients of Western countries with reported genetic data from those of China, we found that the frequency of DPY19L2 variants in Chinese patients is currently higher than that in patients of Western countries (Figure 4c and 4d).
Figure 4.
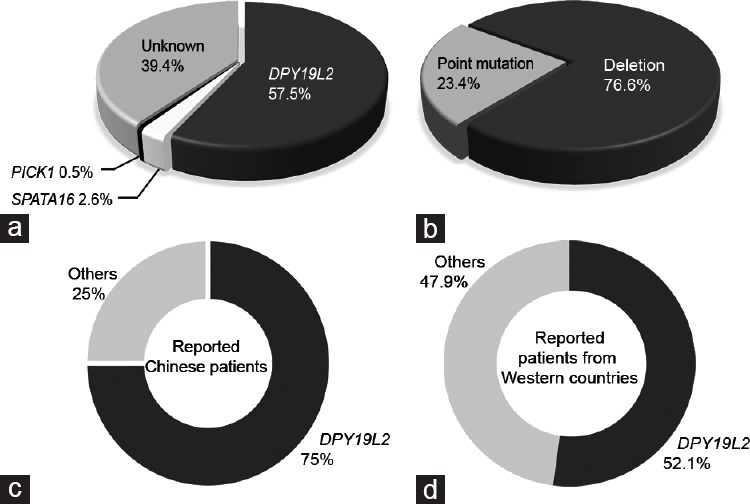
Variants in DPY19L2 are the major cause of globozoospermia. (a) The percentage of known and unknown gene variants in the reported globozoospermic patients from a total of 193 patients from reported cases and the current study was analyzed, among which 111 (57.5%) carried DPY19L2 variants, 5 (2.6%) carried SPATA16 variants, 1 (0.5%) carried PICK1 variant, and the remaining 76 (39.4%) patients carried unknown variants. (b) The percentage of gene deletion and point mutations in patients with DPY19L2 dysfunction in a total of 111 patients with DPY19L2 variants from reported cases and the current study was analyzed, among which 85 (76.6%) carried DPY19L2 deletions and 26 (23.4%) carried DPY19L2 point mutations. (c) The percentages of DPY19L2 and other known or unknown gene mutations in currently reported Chinese globozoospermic patients. A total of 24 patients were analyzed, among which 18 (75.0%) of them carried DPY19L2 mutations and 6 (25.0%) of them carried other mutations. (d) The percentages of DPY19L2 and other known or unknown gene mutations in currently reported globozoospermic patients from Western countries. A total of 169 patients were analyzed, and 88 (52.1%) of them carried DPY19L2 variants and 81 (47.9%) of them carried other variants. P = 0.034 as examined by Chi-squared test. The list of analyzed patients is included in the Supplementary Table 1. DPY19L2: dpy-19-like 2; SPATA16: spermatogenesis associated 16; PCK1: protein interacting with C kinase 1.
Supplementary Table 1.
The list of analyzed patients in Figure 4
| Genes | Nucleotide variants | Protein variants | Number of patients | Cohort size | References |
|---|---|---|---|---|---|
| DPY19L2 | c.869G>A | p.R290H | 3 | 34 | Hum Reprod 2012; 27: 2549–58 |
| c.1024C.>T | p.Q342* | ||||
| c.1073T.>A | p.M358K | ||||
| deletions | 23 | ||||
| c.1033C.>T | p.Q345X | 11 | 64 | Hum Mol Genet 2012; 21: 3695–702 | |
| c.1478C.>G | p.T493R | ||||
| c.2038A.T | p.K680X | ||||
| c.1183delT | p.S395LfsX7 | ||||
| deletion | |||||
| c.892C>T | p.R298C | ||||
| c.1218+1G>A | - | ||||
| deletions | 20 | ||||
| c.1532delA | p.K511RfsX8 | 5 | 15 | Mol Hum Reprod 2013; 19: 395–404 | |
| c.1679delT, c.1681_1682delAC | p.L560X | ||||
| c.869G>A | p.R290H | ||||
| c.989T.>C | p.L330P | ||||
| deletions | 4 | ||||
| c.1579_1580+4delAGGTAAinsTCAT | 3 | 18 | Mol Hum Reprod 2016; 22: 35–45 | ||
| c.892C>T | p.R298C | ||||
| deletions | 11 | ||||
| not tested | not tested | not tested | 20 | Am J Hum Genet 2011; 88: 351–61 | |
| deletions | 15 | ||||
| not tested | not tested | not tested | 8 | Am J Hum Genet 2011; 88: 344–50 | |
| deletions | 7 | ||||
| c.2126+5G>A | - | 4 | 9 | The current study | |
| c.1720C>T | p.R574* | ||||
| c.1182_1184delATC | p.L394_S395delinsF | ||||
| c.368A>G | p.H123R | ||||
| c.1182_1186delATCTT | p.L394FfsX19 | ||||
| c.1553_1554delAT | p.Y518CfsX14 | ||||
| deletions | 5 | ||||
| SAPAT16 | c.848G>A | p.R283Q | 3 | 3 | Am J Hum Genet 2007; 81: 813–20 |
| deletion of 22.6 Kb | - | 2 | 19 | J Assist Reprod Genet 2016; 33: 815–20 | |
| PICK1 | c.G198A | p.G393R | 1 | 3 | Asian J Androl 2010; 12: 556–60 |
Treating the infertility of DPY19L2 variant-associated globozoospermic patients by ICSI with AOA
Finally, we wanted to evaluate the treatment of these DPY19L2 variant-associated globozoospermic patients. Because ICSI of globozoospermic spermatozoa may not be sufficient to overcome fertilization failure owing to a deficiency in oocyte activation capacity,29 we coupled ICSI with assisted oocyte activation (AOA) via calcium ionophore. ICSI with AOA was applied to five patients (P4–P8) at The Reproductive Medicine Center in The First Affiliated Hospital of Anhui Medical University, resulting in at least eight fertilized oocytes for each of the couples. Successful pregnancies and live birth were obtained for all the five couples (Table 3).
Table 3.
Results of intracytoplasmic sperm injection with assisted oocyte activation in patients with globozoospermia
| P4 | P5 | P6 | P7 | P8 | |
|---|---|---|---|---|---|
| Male age (year) | 33 | 34 | 29 | 25 | 26 |
| Female age (year) | 28 | 24 | 27 | 25 | 27 |
| Oocytes retrieved (n) | 20 | 10 | 15 | 14 | 22 |
| Mature oocytes (MII) (n) | 16 | 9 | 13 | 13 | 19 |
| Fertilized oocytes (n) | 10 | 8 | 9 | 9 | 14 |
| D3 embryos (n) | 10 | 8 | 9 | 9 | 14 |
| D5 embryos, n (embyo grade) | 2 (4BBX2,3BBX3) | 2 (4BB,3BB) | 4 (4AB,4BBX3,4BC,3BB) | 4 (4BC,3AB,3BA,3BC) | 2 (4BBX3,3BB) |
| D6 embryos, n (embyo grade) | 0 | 3 (4BB,4BB,4BC) | 0 | 0 | 0 |
| Fresh embryos transferred, day of embryo (embryo grade) | - | D5(4BB,3BB) | - | D5(3BB,3BA) | - |
| Frozen embryos transferred in the first cycle, day of embryo (embryo grade) | D5(4BB,3BB) | D6(4BB,4BB) | D5(4AB,4BB) | D5(4BC,3BC) | D5(4BB,4BB) |
| Livebirth, n (gender) | 1 (female) | 2 (male) | 2 (male) | 1 (female) | 1 (male) |
DISCUSSION
In the current study, we investigated a cohort of Chinese globozoospermic patients, who revealed high frequencies of DPY19L2 mutations, either deletion or point mutations. Our results revealed partial deletion of the DPY19L2 gene and identified six new variants, which might lead to the loss of function of DPY19L2.
Among the five new variants, two resulted in a premature stop codon, two led to amino acid substitutions, and one might lead to splicing defects of DPY19L2 mRNA. P3 carries c.2126+5G>A; this nucleotide substitution may lead to the alteration of the donor site in exon 21, most probably affecting splicing as predicted by Human Splicing Finder,30 and the mutant consensus value (CV) is 74.12, comparing to the WT 86.28. P3 contained a heterozygous variant of DPY19L2, which might lead to a dominant inactive form of DPY19L2. P6 carried a homozygous variant c.1720C>T, leading to premature termination codon (PTC) in DPY19L2 mRNA. The truncated DPY19L2 resulted in a large loss of protein after Ser574, which might disrupt its function in anchoring acrosome to the nuclear envelope. However, due to the lack of parental sequences for P6, it is possible that P6 contains one allele with c.1720C>T, while the other allele is deleted. Regardless of the cause, both possibilities are harmful to DPY19L2 function. Because blood samples of P9's parents are unavailable, it is difficult to conclude whether the above two deletions are compound heterozygous mutations or not. By considering the essential function of DPY19L2 in globozoospermia and the typical results of biallelic DPY19L2 variants, the two variants in P9 are probably compound heterozygous variants.
By analyzing all the point mutations, we found that these variants are likely enriched in certain regions, such as the 5th and 6th transmembrane domains. The 5th transmembrane domain of the DPY19L2 protein contains three point mutations, including two nonsense mutations and one missense mutation. This suggests the importance of this transmembrane domain and also indicates that this region might be more fragile. The other variant-enriched region is the 6th transmembrane domain, more specifically the 1182–1186 position on the cDNA sequence. Nucleotides in this region are frequently deleted in globozoospermic patients, which tend to cause frameshift (1183delT, 1182-1184delATC, and 1182-1186delATCTT). Although the number of the patients is limited, the distribution of these variants suggests the essential functions of these regions.
Numerous genes have been reported to be involved in sperm head shaping and related to globozoospermic phenotype, and DPY19L2 is the most frequently mutated gene in globozoospermic patients.18 We noticed that in the reported cases, the frequency of DPY19L2 variants in patients from Western countries was lower than that in Chinese patients. This discrepancy might arise from variability in data collection and analyses because some early studies only reported deletions, did not include point mutations, and did not perform WES for the globozoospermic patients without DPY19L2 deletion. Considering the lacking DPY19L2 point mutation data and the size of the cohort, the DPY19L2 mutation frequency in Western countries should be comparable to that in Chinese globozoospermic patients, making the DPY19L2 variant one of the major causes of globozoospermia globally.
If a spermatozoon lacks an acrosome, it cannot fertilize the oocyte, and the main technology to treat this syndrome is ICSI.31 Reports have suggested that the infertility of DPY19L2 mutation-associated globozoospermia could be overcome by ICSI,26 but the overall fertilization rates remain low.32,33,34 The exact pathogenic mechanism for the deficiency in oocyte activation capacity is still unknown, but the mechanism is speculated to be associated with the sperm-associated oocyte-activating factor (SOAF), phospholipase C zeta (PLCζ), which may play a role in Ca2+ flux during fertilization.35 Some studies have shown that reduced amounts or abnormal forms of PLCζ may contribute to fertilization failure by ICSI with some infertile men, including those with globozoospermia.35,36,37 In addition, some studies have shown that PLCζ is absent or significantly decreased in globozoospermia.35 By using ICSI followed by AOA with calcium ionophore, higher fertilization rates were achieved which might be a result of the restored Ca2+ flux required for globozoospermic sperm–oocyte activation.38 Hence our results, together with previous studies, demonstrate that ICSI together with AOA is a very efficient way to treat the infertility of those globozoospermic patients with DPY19L2 variants.
Many reported globozoospermic patients contain biallelic DPY19L2 variants, indicating that DPY19L2 variant-related globozoospermia is autosome recessively inherited.15,16,17,18,19 Hence, we suggest that the wife of the globozoospermic patient should be screened for possible DPY19L2 variants to avoid the globozoospermic risk of the male babies after ICSI treatment. Moreover, the couple should be informed of the globozoospermic risk of their baby in the genetic counseling.
AUTHOR CONTRIBUTIONS
YLS and FXZ collected the materials and clinic information, performed the phenotypic analysis of the samples, and wrote the manuscript. JY, LC, and WHT collected some of the samples and analyzed the phenotypes. SX performed the DPY19L2 deletion screening, sequence alignment, and statistical analysis. WKM conducted the whole-exome sequencing and subsequent analyses. ZGZ and XJH performed the ICSI experiments and JQ designed some of the experiments and coordinated the project. YXC and WL designed the project, supervised the whole research, and wrote the paper. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
ACKNOWLEDGMENTS
We thank all the patients and control individuals for their participation in the study. This work was supported by the National Key Technologies R&D Program of China (grant no. 2016YFA0500901), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDA16020701), National Nature Science of China (grant no. 31471277 and 91649202) to WL, and The Key Program in the Youth Elite Support Plan in Universities of Anhui Province (grant no. gxyqZD2016050) to FXZ.
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Dam AH, Feenstra I, Westphal JR, Ramos L, van Golde RJ, et al. Globozoospermia revisited. Hum Reprod Update. 2007;13:63–75. doi: 10.1093/humupd/dml047. [DOI] [PubMed] [Google Scholar]
- 2.Abou-Haila A, Tulsiani DR. Mammalian sperm acrosome: formation, contents, and function. Arch Biochem Biophys. 2000;379:173–82. doi: 10.1006/abbi.2000.1880. [DOI] [PubMed] [Google Scholar]
- 3.Hess RA, Renato de Franca L. Spermatogenesis and cycle of the seminiferous epithelium. Adv Exp Med Biol. 2008;636:1–15. doi: 10.1007/978-0-387-09597-4_1. [DOI] [PubMed] [Google Scholar]
- 4.Xiao X, Mruk DD, Wong CK, Cheng CY. Germ cell transport across the seminiferous epithelium during spermatogenesis. Physiology (Bethesda) 2014;29:286–98. doi: 10.1152/physiol.00001.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015;21:455–85. doi: 10.1093/humupd/dmv020. [DOI] [PubMed] [Google Scholar]
- 6.Ray PF, Toure A, Metzler-Guillemain C, Mitchell MJ, Arnoult C, et al. Genetic abnormalities leading to qualitative defects of sperm morphology or function. Clin Genet. 2017;91:217–32. doi: 10.1111/cge.12905. [DOI] [PubMed] [Google Scholar]
- 7.Dam AH, Koscinski I, Kremer JA, Moutou C, Jaeger AS, et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet. 2007;81:813–20. doi: 10.1086/521314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu G, Shi QW, Lu GX. A newly discovered mutation in PICK1 in a human with globozoospermia. Asian J Androl. 2010;12:556–60. doi: 10.1038/aja.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yatsenko AN, O’Neil DS, Roy A, Arias-Mendoza PA, Chen R, et al. Association of mutations in the zona pellucida binding protein 1 (ZPBP1) gene with abnormal sperm head morphology in infertile men. Mol Hum Reprod. 2012;18:14–21. doi: 10.1093/molehr/gar057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol. 2009;306:24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yao R, Ito C, Natsume Y, Sugitani Y, Yamanaka H, et al. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc Natl Acad Sci U S A. 2002;99:11211–6. doi: 10.1073/pnas.162027899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Wan H, Li X, Liu W, Chen Q, et al. Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Res. 2014;24:852–69. doi: 10.1038/cr.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbuz R, Zouari R, Pierre V, Ben Khelifa M, Kharouf M, et al. A recurrent deletion of DPY19L2 causes infertility in man by blocking sperm head elongation and acrosome formation. Am J Hum Genet. 2011;88:351–61. doi: 10.1016/j.ajhg.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pierre V, Martinez G, Coutton C, Delaroche J, Yassine S, et al. Absence of Dpy19l2, a new inner nuclear membrane protein, causes globozoospermia in mice by preventing the anchoring of the acrosome to the nucleus. Development. 2012;139:2955–65. doi: 10.1242/dev.077982. [DOI] [PubMed] [Google Scholar]
- 15.Koscinski I, Elinati E, Fossard C, Redin C, Muller J, et al. DPY19L2 deletion as a major cause of globozoospermia. Am J Hum Genet. 2011;88:344–50. doi: 10.1016/j.ajhg.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coutton C, Zouari R, Abada F, Ben Khelifa M, Merdassi G, et al. MLPA and sequence analysis of DPY19L2 reveals point mutations causing globozoospermia. Hum Reprod. 2012;27:2549–58. doi: 10.1093/humrep/des160. [DOI] [PubMed] [Google Scholar]
- 17.Elinati E, Kuentz P, Redin C, Jaber S, Vanden Meerschaut F, et al. Globozoospermia is mainly due to DPY19L2 deletion via non-allelic homologous recombination involving two recombination hotspots. Hum Mol Genet. 2012;21:3695–702. doi: 10.1093/hmg/dds200. [DOI] [PubMed] [Google Scholar]
- 18.Zhu F, Gong F, Lin G, Lu G. DPY19L2 gene mutations are a major cause of globozoospermia: identification of three novel point mutations. Mol Hum Reprod. 2013;19:395–404. doi: 10.1093/molehr/gat018. [DOI] [PubMed] [Google Scholar]
- 19.Ghedir H, Ibala-Romdhane S, Okutman O, Viot G, Saad A, et al. Identification of a new DPY19L2 mutation and a better definition of DPY19L2 deletion breakpoints leading to globozoospermia. Mol Hum Reprod. 2016;22:35–45. doi: 10.1093/molehr/gav061. [DOI] [PubMed] [Google Scholar]
- 20.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010;38:e131. doi: 10.1093/nar/gkq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Wang K. InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am J Hum Genet. 2017;100:267–80. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuentz P, Vanden Meerschaut F, Elinati E, Nasr-Esfahani MH, Gurgan T, et al. Assisted oocyte activation overcomes fertilization failure in globozoospermic patients regardless of the DPY19L2 status. Hum Reprod. 2013;28:1054–61. doi: 10.1093/humrep/det005. [DOI] [PubMed] [Google Scholar]
- 27.Scott L, Finn A, O’Leary T, McLellan S, Hill J. Morphologic parameters of early cleavage-stage embryos that correlate with fetal development and delivery: prospective and applied data for increased pregnancy rates. Hum Reprod. 2007;22:230–40. doi: 10.1093/humrep/del358. [DOI] [PubMed] [Google Scholar]
- 28.Alpha Scientists in Reproductive Medicine and ESHRE Special Interest Group of Embryology. The Istanbul consensus workshop on embryo assessment: proceedings of an expert meeting. Hum Reprod. 2011;26:1270–83. doi: 10.1093/humrep/der037. [DOI] [PubMed] [Google Scholar]
- 29.Rybouchkin AV, Van der Straeten F, Quatacker J, De Sutter P, Dhont M. Fertilization and pregnancy after assisted oocyte activation and intracytoplasmic sperm injection in a case of round-headed sperm associated with deficient oocyte activation capacity. Fertil Steril. 1997;68:1144–7. doi: 10.1016/s0015-0282(97)00378-6. [DOI] [PubMed] [Google Scholar]
- 30.Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, et al. Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrin A, Coat C, Nguyen MH, Talagas M, Morel F, et al. Molecular cytogenetic and genetic aspects of globozoospermia: a review. Andrologia. 2013;45:1–9. doi: 10.1111/j.1439-0272.2012.01308.x. [DOI] [PubMed] [Google Scholar]
- 32.Lundin K, Sjogren A, Nilsson L, Hamberger L. Fertilization and pregnancy after intracytoplasmic microinjection of acrosomeless spermatozoa. Fertil Steril. 1994;62:1266–7. [PubMed] [Google Scholar]
- 33.Liu J, Nagy Z, Joris H, Tournaye H, Devroey P, et al. Successful fertilization and establishment of pregnancies after intracytoplasmic sperm injection in patients with globozoospermia. Hum Reprod. 1995;10:626–9. doi: 10.1093/oxfordjournals.humrep.a136000. [DOI] [PubMed] [Google Scholar]
- 34.Sahu B, Ozturk O, Serhal P. Successful pregnancy in globozoospermia with severe oligoasthenospermia after ICSI. J Obstet Gynaecol. 2010;30:869–70. doi: 10.3109/01443615.2010.515321. [DOI] [PubMed] [Google Scholar]
- 35.Yoon SY, Jellerette T, Salicioni AM, Lee HC, Yoo MS, et al. Human sperm devoid of PLC, zeta 1 fail to induce Ca(2+) release and are unable to initiate the first step of embryo development. J Clin Invest. 2008;118:3671–81. doi: 10.1172/JCI36942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashir J, Jones C, Lee HC, Rietdorf K, Nikiforaki D, et al. Loss of activity mutations in phospholipase C zeta (PLCζ) abolishes calcium oscillatory ability of human recombinant protein in mouse oocytes. Hum Reprod. 2011;26:3372–87. doi: 10.1093/humrep/der336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor SL, Yoon SY, Morshedi MS, Lacey DR, Jellerette T, et al. Complete globozoospermia associated with PLCzeta deficiency treated with calcium ionophore and ICSI results in pregnancy. Reprod Biomed Online. 2010;20:559–64. doi: 10.1016/j.rbmo.2009.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rybouchkin A, Dozortsev D, Pelinck MJ, De Sutter P, Dhont M. Analysis of the oocyte activating capacity and chromosomal complement of round-headed human spermatozoa by their injection into mouse oocytes. Hum Reprod. 1996;11:2170–5. doi: 10.1093/oxfordjournals.humrep.a019071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DPY19L2 deletion screening of the globozoospermic patients. (a) The distribution of a-g loci on and around DPY19L2. (b) Genomic DNA was extracted from the blood of the patients and the reported seven loci of DPY19L2 were amplified. P1, P2, P4, P5, and P7 show complete or partial deletion of the DPY19L2 gene, while P3, P6, P8, and P9 show no deletion in the DPY19L2 gene.